Significance
Foraging and sleep are two conflicting behaviors in starved animals; however, it remains elusive how metabolic status governs sleep drive. In this study, we show that a biosynthetic pathway for the amino acid serine is transcriptionally up-regulated by starvation in adult fly brains. The behavioral response to genetic manipulation of key enzymes involved in serine metabolism supports the sleep-suppressing effect of serine in response to starvation. In a society where daily diet is becoming increasingly important to the sleep quality of individuals, our study defines an amino acid metabolic pathway that underlies adaptive sleep behaviors upon dietary stress.
Keywords: starvation, sleep regulation, serine
Abstract
Sleep and metabolism are physiologically and behaviorally intertwined; however, the molecular basis for their interaction remains poorly understood. Here, we identified a serine metabolic pathway as a key mediator for starvation-induced sleep suppression. Transcriptome analyses revealed that enzymes involved in serine biosynthesis were induced upon starvation in Drosophila melanogaster brains. Genetic mutants of astray (aay), a fly homolog of the rate-limiting phosphoserine phosphatase in serine biosynthesis, displayed reduced starvation-induced sleep suppression. In contrast, a hypomorphic mutation in a serine/threonine-metabolizing enzyme, serine/threonine dehydratase (stdh), exaggerated starvation-induced sleep suppression. Analyses of double mutants indicated that aay and stdh act on the same genetic pathway to titrate serine levels in the head as well as to adjust starvation-induced sleep behaviors. RNA interference-mediated depletion of aay expression in neurons, using cholinergic Gal4 drivers, phenocopied aay mutants, while a nicotinic acetylcholine receptor antagonist selectively rescued the exaggerated starvation-induced sleep suppression in stdh mutants. Taken together, these data demonstrate that neural serine metabolism controls sleep during starvation, possibly via cholinergic signaling. We propose that animals have evolved a sleep-regulatory mechanism that reprograms amino acid metabolism for adaptive sleep behaviors in response to metabolic needs.
Sleep and metabolism are interconnected processes that modulate each other, which is best exemplified during sleep deprivation conditions. During acute sleep deprivation, large amounts of lysolipids are found in the mouse cortex, indicating that membrane phospholipids are degraded during sleep loss (1). Metabolites such as tryptophan, serotonin, and taurine are increased in human plasma samples during acute sleep deprivation, and these increases may be associated with its antidepressive effects (2). While information from such metabolic studies of acute sleep deprivation is important, prolonged partial sleep loss is more prevalent in modern society. In rats and humans, prolonged sleep deprivation decreases two specific metabolites, diacylglycerol 36:3 and oxalic acid, which are restored to baseline levels after recovery sleep (3). Other studies further show that chronic sleep deprivation reduces blood leptin levels, induces hunger, and alters food choice in humans with a preference for sweet, salty, and high-carbohydrate foods (4, 5). These observations suggest that quantity of sleep affects metabolic processes and behaviors for reasons that are as yet unclear.
Conversely, changes in dietary composition influence the quantity and quality of sleep. Relative amounts of dietary sugar modulate arousal threshold and control sleep architecture in Drosophila (6–8). Gustatory perception of lower sugar concentration increases the number of sleep episodes whereas metabolic sensing of higher nutritional values in dietary sugar suppresses sleep partitioning (6). In addition, meal size and nutritional content contribute to postprandial sleep. Flies that eat more generally sleep more, and protein-rich foods promote sleep after meal consumption via leucokinin receptor neurons (9). The aforementioned studies highlight the important notion that various aspects of sleep can be regulated by diverse metabolic pathways.
The starvation state is an excellent model that illustrates the interaction between metabolism and sleep. As a behavioral response to starvation, animals suppress sleep and enhance locomotion to forage for new food sources (10, 11). Since the discovery that fruit flies suppress sleep during starvation (12), new genes and neural circuits are being identified that can modulate sleep during starvation. For example, circadian clock genes, Clock and cycle, limit starvation-induced sleep suppression in cryptochrome-expressing cells (12). Also, translin, a highly conserved RNA/DNA-binding protein, promotes wakefulness in leucokinin neurons during starvation (13). On the other hand, transgenic activation of gustatory neurons that sense sweetness is sufficient to restore sleep in starved flies (7, 8). However, despite recent advances, the molecular cues linking starvation and sleep remain largely unknown.
To identify genes that regulate starvation-induced sleep suppression, we profiled differentially expressed genes (DEG) in starved fly brains. DEG analysis identified genes involved in serine biosynthesis to be up-regulated in the brain poststarvation. Functional studies using genetic manipulation of key enzymes in the serine biosynthesis pathway revealed an essential role for serine in modulating sleep during starvation.
Results
Transcriptome Analyses of Starved Drosophila Brains.
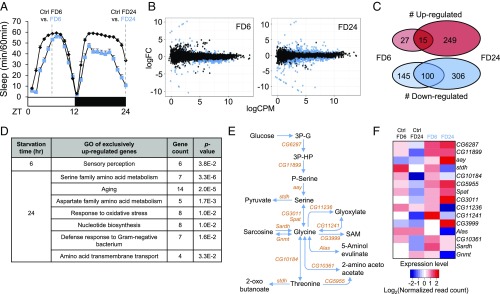
Physiological responses to starvation include differential gene expression (14, 15). We reasoned that some of those starvation-sensitive genes may play a specific role in sleep regulation under starved conditions. To identify sleep-regulatory genes that respond to starvation in the central nervous system but not in peripheral tissues, we decided to perform RNA sequencing (RNA-seq) (16) on the brains of Drosophila melanogaster. Accordingly, we searched for DEGs in fly brains starved for 6 [food deprivation 6 h (FD6)] and 24 h [FD 24 h (FD24)] (Fig. 1A and Dataset S1).
Fig. 1.
Transcriptome analyses of starved Drosophila brains identify up-regulation of serine biosynthesis pathway. (A) Schematic diagram depicting RNA-seq experimental design. Wild-type flies were fed 5% sucrose/1% agar (Ctrl) or deprived of sucrose for 6 and 24 h (FD6 and FD24). (B) Scatter plots demonstrate log counts per million (cpm) vs. log fold-change (FC) in expression of brains starved for 6 and 24 h. Blue dots represent DEGs (FDR < 0.05). (C) Venn diagram showing the number of genes that are regulated during short-term (FD6) and long-term (FD24) starvation. (D) Gene ontology analysis of genes that are up-regulated exclusively in FD6 and FD24. (E) Schematic diagram of the serine metabolic pathway. (F) Heat-map of expression level of genes involved in the serine metabolic pathway in fed (Ctrl FD6 and Ctrl FD24) and starved conditions (FD6 and FD24). Colors indicate the log2 values of normalized read counts.
Comparison of RNA expression profiles between starved and nonstarved brains revealed 287 and 670 DEGs [false discovery rate (FDR) < 0.05 threshold] after 6 and 24 h of starvation, respectively (Fig. 1B). Among the DEGs, 42 genes were up-regulated and 245 genes were down-regulated in FD6, while 264 genes were up-regulated and 406 genes were down-regulated in FD24 (Fig. 1C). Gene ontology (GO) analysis revealed that genes up-regulated exclusively during short-term starvation (FD6) were mainly relevant to sensory perception, while genes up-regulated exclusively during long-term starvation (FD24) were involved in biological processes such as amino acid metabolism, nucleotide metabolism, and amino acid transmembrane transport (Fig. 1D). In contrast, the GO classifications of down-regulated genes in either FD6 or FD24 samples were relatively diverse (Dataset S2). Consistent with the GO analysis, the starvation-induced genes in FD6 samples showed higher enrichment in brains (∼65%) than those in FD24 samples (∼20%) when their relative expression was compared between our control brain samples and previously published head samples (SI Appendix, Supplementary Materials and Methods) that contain peripheral tissues including fat body, sensory organs, and the compound eye, besides the brain (SI Appendix, Fig. S1 and Dataset S3). Finally, the regulation of 115 genes overlapped between FD6 and FD24 (SI Appendix, Fig. S2), and GO analysis of these genes revealed various biological processes to be enriched including mating behavior, axon development/neuron projection morphogenesis, and regulation of cell communication.
Intriguingly, astray (aay), a fly homolog of the rate-limiting phosphoserine phosphatase involved in serine biosynthesis (17, 18), was in the top 10 genes induced by long-term starvation. Therefore, we further examined the expression of genes involved in the serine metabolic pathway (Fig. 1E). Heat-map analysis revealed that genes involved in serine synthesis (CG6287, CG11899, and aay) were all up-regulated after 6 and 24 h of starvation (Fig. 1F).
aay-Dependent Serine Synthesis Is Required for Starvation-Induced Sleep Suppression.
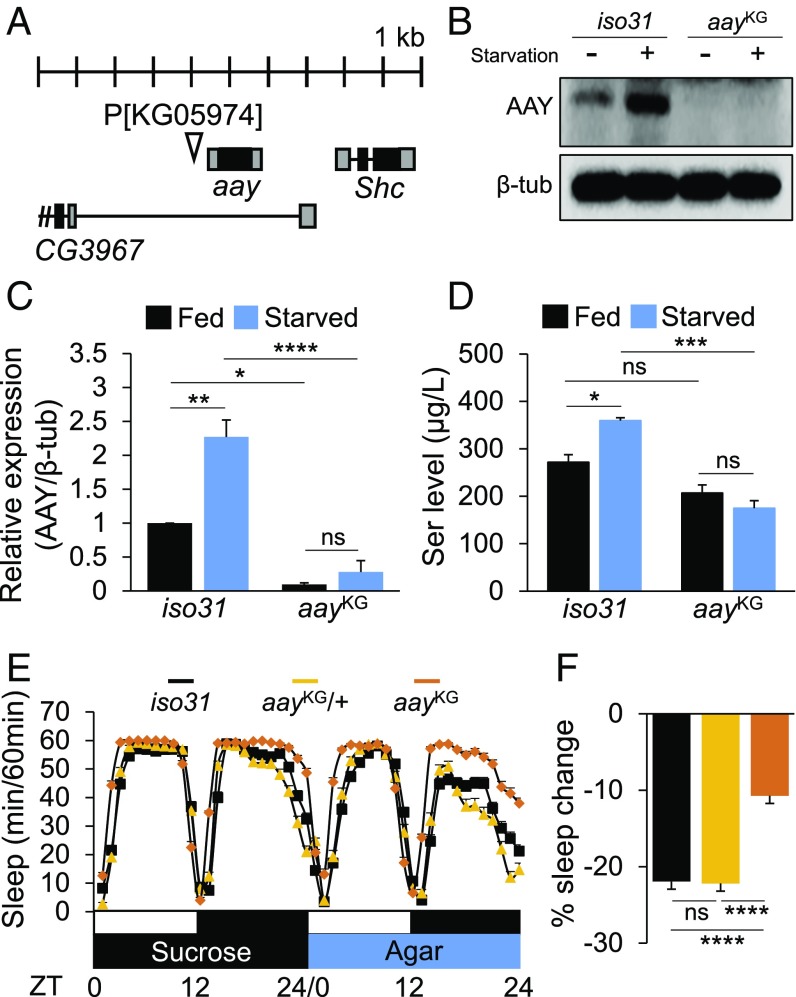
To determine genetically whether serine biosynthesis is important for starvation-relevant sleep regulation, we examined sleep behaviors in flies harboring a transgenic P-element insertion (KG05974, referred to as aayKG from hereafter) upstream of the 5′UTR of aay that reduced aay mRNA levels to ∼30% of iso31 control flies (Fig. 2A and SI Appendix, Fig. S3). Western blot analysis confirmed that AAY protein levels were significantly elevated in iso31 wild types after 24 h of starvation, while aayKG suppressed AAY expression (Fig. 2 B and C), thereby validating aayKG as a hypomorphic allele of aay. Consistent with AAY induction, starvation significantly elevated free serine levels in an aay-dependent manner (P = 0.0052, by two-way ANOVA) (Fig. 2D).
Fig. 2.
Starvation-induced aay expression elevates free serine levels in heads and supports sleep suppression during starvation. (A) Schematic diagram illustrating KG05974 insertion near the 5′UTR of aay locus. (B) Western blot of head extracts in iso31 and aayKG flies during fed and starved conditions. (C) Quantification of Western blots (n = 3) [F(1,8) = 12.92; P = 0.0070]. (D) Serine concentrations in head extracts of iso31 control and aayKG flies during fed and starved conditions (n = 3) [F(1,8) = 14.53; P = 0.0052]. (E) Average sleep traces of iso31 control (black; n = 88), aayKG/+ (light orange; n = 80), and aayKG (orange; n = 78) flies. (F) Percentage sleep change during starvation in iso31 and aayKG/+ controls vs. aayKG flies. ns, P > 0.05; *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. All error bars represent SEM.
Importantly, aayKG flies showed a modest increase in baseline sleep during fed condition (SI Appendix, Fig. S4), while starvation-induced sleep suppression was substantially compromised compared with isogenic iso31 wild-type or aayKG/+ controls (Fig. 2 E and F). Furthermore, aayKG flies exhibited shorter latency to night-time sleep onset in fed condition, as well as an inability to increase the sleep latency during starvation (SI Appendix, Fig. S5). Waking activity was modestly reduced in aayKG flies, compared with either iso31 or aayKG/+ controls (SI Appendix, Fig. S6A). However, negative geotaxis assay (SI Appendix, Supplementary Materials and Methods) revealed that aayKG flies showed climbing activity comparable to iso31 control flies (SI Appendix, Fig. S6B), arguing against the idea that sleep phenotypes observed in aayKG flies are due to their sickness or defects in general locomotion. In addition, we mechanically sleep-deprived wild-type and aayKG flies during the night-time (ZT12–ZT24) and analyzed their sleep rebound on the following day (ZT0–ZT12) (SI Appendix, Supplementary Materials and Methods). Under these conditions, iso31 control and aayKG flies exhibited similar sleep rebound (SI Appendix, Fig. S7), indicating that aay mutants have intact sleep homeostasis to modulate sleep behaviors upon sleep deprivation, but their sleep response to starvation was specifically disrupted.
We next contemplated the possibility that deeper sleep depth in aayKG flies might be responsible for their sleep phenotypes in either fed or starved conditions. When we assessed arousal threshold by a light pulse at night (ZT18) (SI Appendix, Supplementary Materials and Methods), aayKG flies displayed decreased arousability compared with iso31 control flies during fed condition (SI Appendix, Fig. S8A). Accordingly, the higher arousal threshold in aayKG flies is consistent with their longer baseline sleep and possibly higher sleep drive in fed condition. By contrast, aay mutants exhibited similar arousability to iso31 control flies during starvation (SI Appendix, Fig. S8A). It is thus unlikely that aay mutants exhibit reduced starvation-induced sleep suppression due to their deeper sleep. These results also indicate aay-dependent and aay-independent regulation of arousal threshold in fed and starved conditions, respectively. Nonetheless, we do not exclude the possibility that a smaller pool of sleeping flies during starvation influences the behavioral assessment of arousability in different genetic backgrounds (SI Appendix, Fig. S8B). Using an independent long-sleeping transgenic line, we further validated that higher baseline sleep in fed condition does not necessarily cause resistance to starvation-induced sleep suppression (SI Appendix, Fig. S9).
aay and stdh Function Along a Common Genetic and Metabolic Axis for Sleep Regulation.
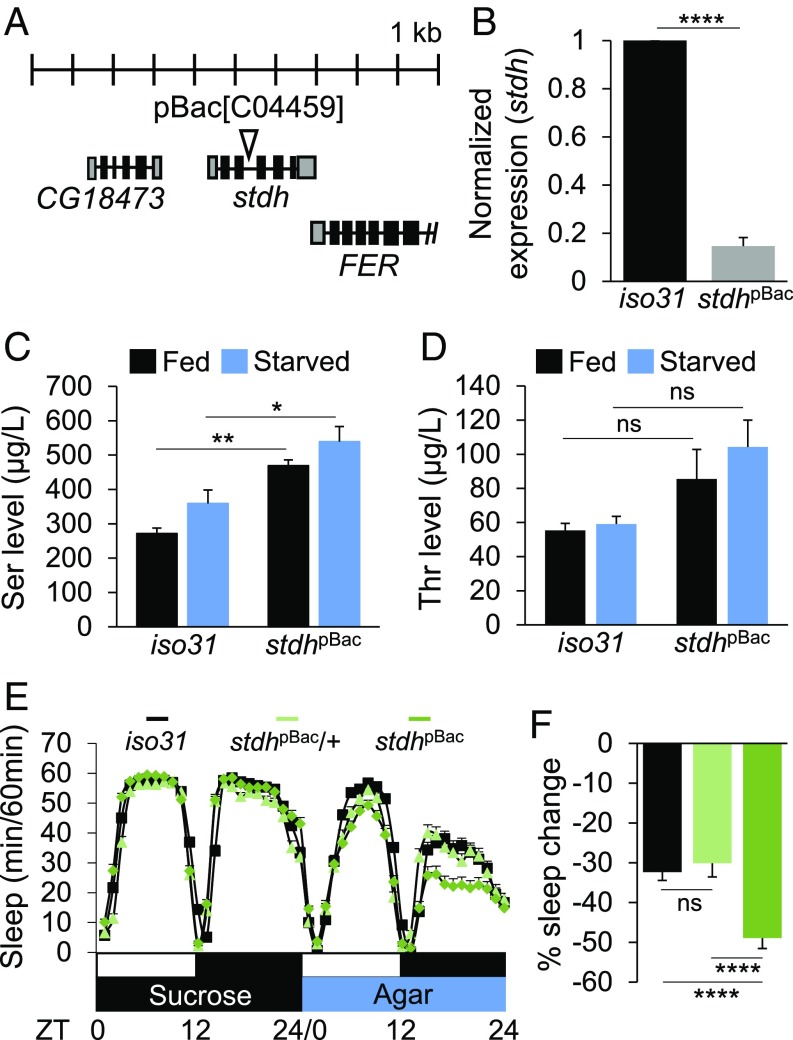
CG8129 likely encodes a fly homolog of serine/threonine dehydratase (referred to as stdh hereafter) that breaks down serine and threonine to pyruvate and 2-oxobutanoic acid, respectively (19–21). We isolated a transgenic fly with a piggyBac insertion in the intron of stdh (C04459, referred to as stdhpBac from hereafter), which strongly reduced stdh mRNA levels in homozygous mutants (Fig. 3 A and B). Hypomorphic mutant phenotypes were biochemically validated by higher serine levels in stdhpBac flies compared with iso31 control in either fed or starved conditions (Fig. 3C). We detected no significant increase in threonine levels in stdhpBac flies (Fig. 3D), although much lower concentrations of free threonine in fly heads might have been limiting for its detectability in our biochemical analyses. We found that stdhpBac flies showed baseline sleep comparable to iso31 wild-type or stdhpBac/+ flies in fed condition (SI Appendix, Fig. S10), while sleep suppression by starvation was exacerbated (Fig. 3 E and F). In addition, latency to sleep onset was increased in stdhpBac flies compared with controls during starvation, but not in fed condition (SI Appendix, Fig. S11). Collectively, these data thus suggest that stdh specifically limits sleep suppression during starvation, without affecting baseline sleep.
Fig. 3.
stdh metabolizes free serine and limits starvation-induced sleep suppression. (A) Schematic diagram illustrating C04459 insertion in the intron of the stdh locus. (B) stdh transcript levels are strongly repressed in stdhpBac flies (n = 3). (C) Serine and (D) threonine concentrations in head extracts of iso31 control and stdhpBac flies during fed and starved conditions (n = 3). (E) Average sleep traces of iso31 (black; n = 91), stdhpBac/+ (light green; n = 81), and stdhpBac flies (green; n = 83). (F) Percentage sleep change during starvation in iso31 and stdhpBac/+ controls vs. stdhpBac flies. ns, P > 0.05; *P < 0.05; **P < 0.005; ****P < 0.0001. All error bars represent SEM.
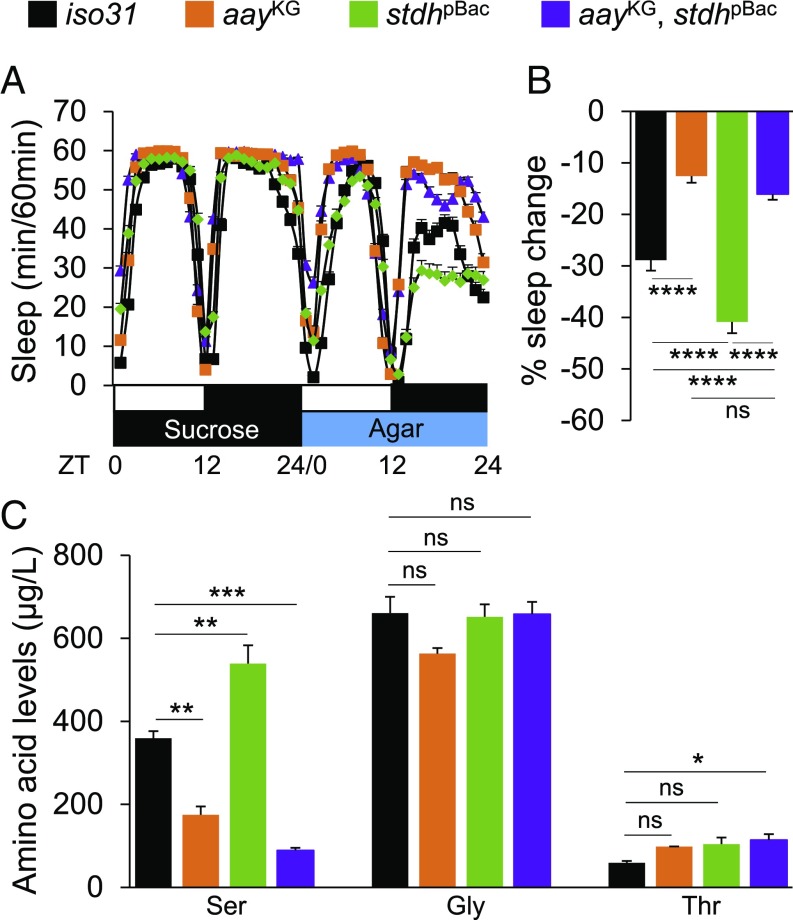
We next generated a double-mutant line harboring both aayKG and stdhpBac alleles to determine if aay and stdh control sleep during starvation via a common sleep-regulatory pathway. We found that the double mutants exhibited reduced starvation-induced sleep suppression, similar to aay single mutants (Fig. 4 A and B), indicating that aay mutation masks the starvation-dependent exaggeration of sleep suppression in stdhpBac flies (P = 0.0144, by two-way ANOVA). To further examine if starvation-induced sleep suppression was relevant to serine levels, we assessed the individual levels of all detectable amino acids in the heads of iso31 control and each mutant during fed and starved conditions (SI Appendix, Table S1). While significant changes in the amount of several amino acids were detectable in different genetic or nutrient conditions, serine levels were most consistent with the starvation-induced sleep suppression phenotypes of each single or double mutant. In fact, aay mutation masked the elevated serine levels in stdhpBac flies (P = 0.0009, by two-way ANOVA) (Fig. 4C). Possible explanations for other amino acids dysregulated in aay or stdh mutants (e.g., glutamate, glycine, and lysine) could include the partial overlap of their metabolic pathways with aay- or stdh-mediated biochemical reactions or physiological compensation for the genetic loss of aay- or stdh-dependent amino acid metabolism. Also, a relatively small number of biological replications (n = 3) do not exclude the possibility of experimental variations. Regardless, we clearly observed that the starvation-induced sleep suppression phenotypes in each mutant strongly correlated to serine levels. Collectively, these data demonstrate that aay and stdh function along a common genetic and metabolic axis to modulate sleep during starvation.
Fig. 4.
aay and stdh act in the same genetic pathway to titrate free serine levels and control sleep during starvation. (A) Average sleep traces of iso31 (black; n = 116), aayKG (orange; n = 81), stdhpBac (green; n = 104), and double mutants (purple; n = 160). (B) aayKG masks the exaggerated starvation-induced sleep suppression of stdhpBac flies [F(1,457) = 6.035; P = 0.0144]. (C) Comparison of serine, glycine, and threonine levels in head extracts of iso31 and mutants in starved condition (n = 3). ns, P > 0.05; *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. All error bars represent SEM.
Serine Metabolic Pathway Implicates Cholinergic Signaling via Nicotinic Acetylcholine Receptors in Sleep Suppression During Starvation.
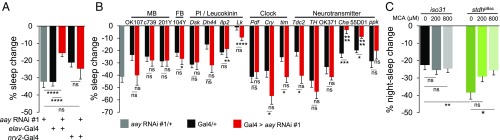
To understand the neural basis of how the serine metabolic pathway contributes to starvation-relevant sleep behaviors, we determined which aay-expressing cells are important for starvation-induced sleep suppression. We first confirmed that pan-neuronal expression of an RNA interference (RNAi) transgene against aay significantly decreased aay mRNA levels in fly head extracts (SI Appendix, Fig. S12). Consistent with a role of neuronal aay in sleep regulation, pan-neuronal, but not pan-glial depletion of aay, phenocopied aayKG flies during starvation (Fig. 5A and SI Appendix, Fig. S13). Similar results were obtained using two additional RNAi transgenes that target different regions of the aay transcript (SI Appendix, Fig. S14), strongly suggesting that the RNAi phenotypes are not due to off-target effects. In addition, pan-neuronal depletion of aay transcripts by all three RNAi transgenes caused longer sleep duration in fed condition (SI Appendix, Fig. S15), similar to aayKG flies.
Fig. 5.
Sleep regulation by the serine metabolic pathway implicates cholinergic signaling. (A) Percentage sleep change during starvation in flies with pan-neuronal and glial knockdown of aay transcripts (aay RNAi #1/+, n = 37; elav-Gal4/+, n = 46; elav-Gal4 > aay RNAi #1, n = 42; nrv2-Gal4/+, n = 29; nrv2-Gal4 > aay RNAi #1, n = 21). (B) Percentage sleep change during starvation when aay transcripts are depleted in mushroom body (MB), fan-shaped body (FB), pars intercerebralis (PI), leucokinin, clock, and various neurotransmitter-related regions in the brain (n = 7–33). (C) Dose-dependent effect of MCA administration on stdhpBac flies during starvation. The 200 μM MCA effect: [F(1,281) = 3.637; P = 0.0575] and 800 μM MCA effect: [F(1,284) = 6.071; P = 0.0143]. iso31 0 μM, n = 71; iso31 200 μM, n = 79; iso31 800 μM, n = 81; stdhpBac 0 μM, n = 66; stdhpBac 200 μM, n = 69; and stdhpBac 800 μM, n = 70. ns, P > 0.05; *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. All error bars represent SEM.
We next tested a number of Gal4 transgenes to deplete aay expression in specific subsets of neurons and map a neural locus important for aay-dependent sleep regulation. While most sleep-relevant Gal4 drivers did not show any significant sleep phenotypes during starvation, aay depletion by either ChAT 7.4-Gal4 (Cha) (22) or 55D01-Gal4, an enhancer transgene from a genetic locus of vesicular acetylcholine transporter, significantly reduced starvation-induced sleep suppression (Fig. 5B), without affecting baseline sleep in fed condition (SI Appendix, Fig. S16). We further employed a ChAT-Gal80 transgene (23) that blocks the expression of the aay RNAi transgene possibly in cholinergic neurons. Nonetheless, starvation-induced sleep suppression was significantly reduced by the pan-neuronal depletion of aay, even in the presence of the ChAT-Gal80 transgene (SI Appendix, Fig. S17). Although these Gal4 and Gal80 transgenes might not be expressed exclusively in cholinergic neurons, our observations suggest the hypothesis that aay-dependent serine biosynthesis in cholinergic neurons might be necessary, but not sufficient, for regulating sleep during starvation.
To examine the possible implication of cholinergic signaling in sleep regulation by the serine metabolic pathway, we tested if oral administration of mecamylamine (MCA) (SI Appendix, Supplementary Materials and Methods), a nicotinic acetylcholine receptor antagonist (24), could affect starvation-induced sleep suppression. Interestingly, MCA rescued the exaggerated sleep suppression in starved stdhpBac flies during night-time in a dose-dependent manner, while it had no significant effects on iso31 control flies (Fig. 5C and SI Appendix, Fig. S18). These data indicate that the exaggerated sleep-suppressing effect of stdh mutation during starvation requires nicotinic acetylcholine receptor function, thereby supporting a possible role for cholinergic signaling in serine metabolism-dependent sleep regulation during starvation.
Discussion
The genetic and cellular components that link sleep and metabolism have been unveiled by traditional genetic approaches in animal models including Drosophila (8, 25, 26); yet there are likely more uncharacterized genes and pathways that regulate this important process. We have demonstrated that starvation induces serine biosynthesis in the transcript, enzymes, and free amino acid levels. Genetic mutants of aay and stdh exhibited decreased and increased serine levels, respectively, and a positive correlation between serine levels and starvation-induced sleep suppression was evident. Collectively, our molecular and genetic evidences implicate endogenous serine in the brain as a wake-promoting signal during starvation.
It has been documented that the lack of food availability immediately suppresses Drosophila sleep (13), suggesting that sensory processes might be involved in sleep suppression at the initial stages of starvation. In fact, our GO enrichment analyses revealed that short-term starvation selectively induces the transcripts of several genes implicated in sensory perception in the brain. On the other hand, prolonged starvation elevates the brain expression of genes involved in various cellular processes such as amino acid metabolism and nucleotide biosynthesis, indicating that long-term starvation might not be simply a severer version of short-term starvation. We thus speculate that starvation-induced sleep suppression might be regulated by two distinctive processes: a fast-acting mechanism perceives the lack of food via a sensory input and temporally suppresses sleep, while slow metabolic processes sustain suppressed sleep states to ensure that food sources with nutritional value are obtained.
What will be the mechanistic basis by which endogenous serine suppresses sleep during starvation? Fat storage and expenditure have been associated with starvation resistance and locomotor activities during starvation (27), yet our biochemical analyses indicate that the sleep-modulatory effects of serine upon starvation are unlikely to involve alterations in lipid metabolism (SI Appendix, Figs. S19 and S20). Intriguingly, amino acid transporters in the brain are emerging as important players in the direct regulation of various neural activities and relevant physiological functions (28–30). Therefore, we reason that a dedicated transporter might facilitate the mobilization of serine to its site of action to suppress sleep during starvation. It is noteworthy that long-term starvation selectively induces the brain expression of genes implicated in amino acid transmembrane transport, which possibly sensitizes Drosophila sleep to the wake-promoting effects of serine. This hypothesis partially explains why stdh mutants, which have high basal levels of serine in heads, could exhibit their sleep suppression phenotypes specifically in starved condition.
Serine is a major precursor to d-serine (31), which in turn acts as a potent coagonist of the N-Methyl-d-aspartic acid (NMDA) receptor (32, 33). However, it is unlikely that serine promotes starvation-induced sleep suppression via NDMA receptors because NMDA receptors are known to promote sleep in flies (34). Besides acting as a precursor to d-serine synthesis, serine is also used for synthesizing bioactive lipids such as phosphatidylserine, which are enriched in the inner leaflet of neural plasma membranes (35). Interestingly, phosphatidylserine increases acetylcholine release from cortical slices of rats (36, 37). Given the potential implication of nicotinic acetylcholine receptors downstream of stdh-dependent sleep effects, we reason that serine might support cholinergic signaling to wake flies up during starvation.
Cholinergic neurons comprise a large portion of the fly brain, and it has been shown that each cholinergic subpopulation mediates different aspects of sleep regulation. For example, cholinergic mushroom body α/β core neurons promote baseline sleep, whereas neighboring cholinergic α/β surface/posterior and γ neurons suppress baseline sleep (38). On the other hand, a cholinergic subpopulation that expresses the gene pickpocket promotes homeostatic rebound sleep (39). Interestingly, our neural mapping similarly revealed locus-specific effects of aay on sleep behaviors. While pan-neuronal aay contributes to sleep behaviors in either fed or starved conditions, aay depletion in a narrower population of cells using cholinergic Gal4 drivers reduced sleep suppression during starvation, but did not affect baseline sleep in fed condition. These data suggest the possibility that aay might differentially modulate sleep, depending on the neuronal loci and availability of food.
In summary, we have identified that starvation-induced elevation of serine biosynthesis in neurons plays a crucial role in sleep regulation during starvation, possibly via signaling through nicotinic acetylcholine receptors. Our study stands out from previous studies in that we have focused purely on metabolic pathways in neurons that can regulate sleep behaviors under metabolic stress conditions. Further investigation of serine-mediated sleep suppression and its downstream neurological effectors will provide a platform on which molecular and neural links between sleep and metabolism can be elucidated.
Methods
Behavioral Analysis.
Sleep behavior was measured using the Drosophila Activity Monitor (DAM) system (Trikinetics). Five- to seven-day-old male flies were individually loaded into 65 × 5 mm glass tubes plugged with 5% sucrose/1% agar in PCR tubes for 4 d. On the fifth day, PCR tubes with 1% agar replaced previous food at ZT0 (lights on), and flies were starved for 24 h within the DAM to measure sleep during starvation. Unless mentioned otherwise, percentage sleep change was calculated as the change in sleep during 24 h of starvation compared with the previous fed day. Data were analyzed using a custom written Excel macro (40).
RNA-Seq and Differential Gene Expression Analysis.
Two biological replicates for each experimental condition (Ctrl FD6, Ctrl FD24, FD6, and FD24) were used to perform RNA-seq analysis. Eighty brains per experimental condition were manually dissected in PBS, and brains were kept in ice-cold RNA-Later ICE (Invitrogen) during the duration (2 h per group) of brain collection. Control and starved brain samples at each time point were collected on different days, to exclude possible circadian time-dependent effects on gene expression. Total RNA was extracted using TRIzol (Invitrogen). Library construction was carried out using a TruSeq RNA Prep kit (Illumina). RNA-seq was performed on a Hiseq2500 (Illumina) at DNA Link, resulting in 20–60 million 101-bp reads per sample. TopHat 2.0.9 (41) was used to align RNA-seq reads to the BDGP5.77 reference genome. Cufflinks 2.1.1 (42) was used to estimate read count for the BDGP5.77 transcriptome model.
Western Blot Analysis.
For Western blot analysis, 12 h:12 h light:dark entrained male flies were frozen at −80 °C, and 20 heads were separated and lysed in lysis buffer (25 mM Tris-Cl pH 7.5, 300 mM NaCl, 1 mM PMSF, 1 mM DTT, 0.5% Nonidet P-40, and protease/phosphatase inhibitor mixture) and resolved by SDS/PAGE. For probing of Western blots, rabbit anti-AAY (SI Appendix, Supplementary Materials and Methods) and mouse anti-tubulin (Cell Signaling) were both used at 1:2,000. HRP-conjugated secondary antibodies (Cell Signaling) were used at 1:20,000 and visualized with ECL Plus (Thermo). Western blots were quantified using Image Studio Lite.
Amino Acid Analysis.
Individual 5–7-d-old male flies, entrained in 12 h:12 h light:dark cycles, were collected. Twenty heads were separated and homogenized in a total of 600 µL of HPLC-grade water. Samples were centrifuged at 4 °C for 10 min at 12,000 × g, and 400 µL of supernatant was retrieved. Subsequently, 1:1 trichloroacetic acid was added to deproteinize samples. n-hexane was added to remove lipids and nonpolar substances, and the bottom phase was passed through a 0.2-µM syringe filter to remove trace impurities. Ion exchange resin #2622 column (Hitachi) was attached to the Hitachi l-8900 Amino Acid Analyzer (Hitachi), and ninhydrin-derivatized amino acids were detected with a UV detector.
Supplementary Material
Acknowledgments
We thank Eun Young Suh at the Center for Research Facilities, Chungnam National University for amino acid analysis. This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Ministry of Science and Information & Communication Technology (MSIT) of Republic of Korea [2016R1A2B4011111 (to J.C.); 2016R1E1A2914795 (to C.L.); 2017R1E1A2A02066965 (to C.L.); and 2016R1A6A3A11932215 (to J.L.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719033115/-/DCSupplemental.
References
- 1.Hinard V, et al. Key electrophysiological, molecular, and metabolic signatures of sleep and wakefulness revealed in primary cortical cultures. J Neurosci. 2012;32:12506–12517. doi: 10.1523/JNEUROSCI.2306-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies SK, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci USA. 2014;111:10761–10766. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weljie AM, et al. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci USA. 2015;112:2569–2574. doi: 10.1073/pnas.1417432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: Sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–850. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- 5.Nedeltcheva AV, et al. Sleep curtailment is accompanied by increased intake of calories from snacks. Am J Clin Nutr. 2009;89:126–133. doi: 10.3945/ajcn.2008.26574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linford NJ, Chan TP, Pletcher SD. Re-patterning sleep architecture in Drosophila through gustatory perception and nutritional quality. PLoS Genet. 2012;8:e1002668. doi: 10.1371/journal.pgen.1002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasegawa T, et al. Sweetness induces sleep through gustatory signalling independent of nutritional value in a starved fruit fly. Sci Rep. 2017;7:14355. doi: 10.1038/s41598-017-14608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linford NJ, Ro J, Chung BY, Pletcher SD. Gustatory and metabolic perception of nutrient stress in Drosophila. Proc Natl Acad Sci USA. 2015;112:2587–2592. doi: 10.1073/pnas.1401501112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy KR, et al. Postprandial sleep mechanics in Drosophila. eLife. 2016;5:e19334. doi: 10.7554/eLife.19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, et al. Octopamine mediates starvation-induced hyperactivity in adult Drosophila. Proc Natl Acad Sci USA. 2015;112:5219–5224. doi: 10.1073/pnas.1417838112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene AC, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami K, et al. Translin is required for metabolic regulation of sleep. Curr Biol. 2016;26:972–980. doi: 10.1016/j.cub.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinke I, Schütz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thimgan MS, Seugnet L, Turk J, Shaw PJ. Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep. 2015;38:801–814. doi: 10.5665/sleep.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borkenhagen LF, Kennedy EP. The enzymatic exchange of L-serine with O-phospho-L-serine catalyzed by a specific phosphatase. J Biol Chem. 1959;234:849–853. [PubMed] [Google Scholar]
- 18.Tabatabaie L, Klomp LW, Berger R, de Koning TJ. L-serine synthesis in the central nervous system: A review on serine deficiency disorders. Mol Genet Metab. 2010;99:256–262. doi: 10.1016/j.ymgme.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa H, et al. Human liver serine dehydratase. cDNA cloning and sequence homology with hydroxyamino acid dehydratases from other sources. J Biol Chem. 1989;264:15818–15823. [PubMed] [Google Scholar]
- 20.Akopov MA, Kagan ZC, Berezov TT, Filiptsev PIa. [Kinetic and allosteric properties of L-threonine-L-serine dehydratase from human liver] Biokhimiia. 1979;44:282–292. [PubMed] [Google Scholar]
- 21.Datta P, Goss TJ, Omnaas JR, Patil RV. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: Homology with other dehydratases. Proc Natl Acad Sci USA. 1987;84:393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- 23.Diao F, et al. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bacher I, Wu B, Shytle DR, George TP. Mecamylamine–A nicotinic acetylcholine receptor antagonist with potential for the treatment of neuropsychiatric disorders. Expert Opin Pharmacother. 2009;10:2709–2721. doi: 10.1517/14656560903329102. [DOI] [PubMed] [Google Scholar]
- 25.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010;8:e1000466. doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metaxakis A, et al. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014;12:e1001824. doi: 10.1371/journal.pbio.1001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Octopamine controls starvation resistance, life span and metabolic traits in Drosophila. Sci Rep. 2016;6:35359. doi: 10.1038/srep35359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosjean Y, Grillet M, Augustin H, Ferveur JF, Featherstone DE. A glial amino-acid transporter controls synapse strength and courtship in Drosophila. Nat Neurosci. 2008;11:54–61. doi: 10.1038/nn2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manière G, Ziegler AB, Geillon F, Featherstone DE, Grosjean Y. Direct sensing of nutrients via a LAT1-like transporter in Drosophila insulin-producing cells. Cell Rep. 2016;17:137–148. doi: 10.1016/j.celrep.2016.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tărlungeanu DC, et al. Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell. 2016;167:1481–1494.e18. doi: 10.1016/j.cell.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolosker H, Blackshaw S, Snyder SH. Serine racemase: A glial enzyme synthesizing D-serine to regulate glutamate-N-methyl-D-aspartate neurotransmission. Proc Natl Acad Sci USA. 1999;96:13409–13414. doi: 10.1073/pnas.96.23.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mothet JP, et al. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc Natl Acad Sci USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panatier A, et al. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 34.Tomita J, Ueno T, Mitsuyoshi M, Kume S, Kume K. The NMDA receptor promotes sleep in the fruit fly, Drosophila melanogaster. PLoS One. 2015;10:e0128101. doi: 10.1371/journal.pone.0128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. 2013;1831:543–554. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Pedata F, Giovannelli L, Spignoli G, Giovannini MG, Pepeu G. Phosphatidylserine increases acetylcholine release from cortical slices in aged rats. Neurobiol Aging. 1985;6:337–339. doi: 10.1016/0197-4580(85)90013-2. [DOI] [PubMed] [Google Scholar]
- 37.Vannucchi MG, Pepeu G. Effect of phosphatidylserine on acetylcholine release and content in cortical slices from aging rats. Neurobiol Aging. 1987;8:403–407. doi: 10.1016/0197-4580(87)90034-0. [DOI] [PubMed] [Google Scholar]
- 38.Yi W, et al. A subset of cholinergic mushroom body neurons requires Go signaling to regulate sleep in Drosophila. Sleep. 2013;36:1809–1821. doi: 10.5665/sleep.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidner G, et al. Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol. 2015;25:2928–2938. doi: 10.1016/j.cub.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Processing sleep data created with the Drosophila activity monitoring (DAM) system. Cold Spring Harb Protoc. 2010;2010:pdb.prot5520. doi: 10.1101/pdb.prot5520. [DOI] [PubMed] [Google Scholar]
- 41.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.