Significance
Metastasis is the most common cause of cancer-related death. The function of reactive oxygen species (ROS) in metastasis is conflicting, and it has been observed to promote or inhibit metastasis. Thus, a more in-depth analysis is needed to assess the impact of ROS on metastasis. In this study, we find that local down-regulation of SIRT3 at the leading edge of migrating cells results in elevated ROS levels and activation of Src/focal adhesion kinase signaling. Further, collective cell migration and metastases formation are inhibited by SIRT3, while patient data sets point to an inverse correlation between SIRT3 and breast cancer metastasis.
Keywords: mitochondria, SIRT3, cancer, metabolism, cell migration
Abstract
Metastasis remains the leading cause of cancer mortality, and reactive oxygen species (ROS) signaling promotes the metastatic cascade. However, the molecular pathways that control ROS signaling relevant to metastasis are little studied. Here, we identify SIRT3, a mitochondrial deacetylase, as a regulator of cell migration via its control of ROS signaling. We find that, although mitochondria are present at the leading edge of migrating cells, SIRT3 expression is down-regulated during migration, resulting in elevated ROS levels. This SIRT3-mediated control of ROS represses Src oxidation and attenuates focal adhesion kinase (FAK) activation. SIRT3 overexpression inhibits migration and metastasis in breast cancer cells. Finally, in human breast cancers, SIRT3 expression is inversely correlated with metastatic outcome and Src/FAK signaling. Our results reveal a role for SIRT3 in cell migration, with important implications for breast cancer progression.
Metastasis is a complex multistep process, and local invasion of the primary tumor is important in the initiation of metastasis. Analysis of clinical samples of solid tumors demonstrates a role for collective cell migration during invasion into the surrounding tissue (1). Collective cell migration relies on the coordination of forces generated by the actomyosin cytoskeleton at integrin-based cell−extracellular matrix (ECM) and cadherin-based cell−cell adhesions (1). Src and focal adhesion kinase (FAK) signaling downstream of cell−ECM interactions enhance the strength of cell−cell adhesions and promote collective cell migration (2, 3).
Tumor cell−ECM interactions generate intracellular reactive oxygen species (ROS), leading to Src oxidation and activation (4). For instance, increased mitochondrial ROS activates Src kinase signaling to promote metastasis of cervical cancer and melanoma (5). ROS are a normal byproduct of mitochondrial metabolism and influence many aspects of cellular function (6, 7). While hyperelevated ROS levels can be toxic, ROS also promotes intracellular signaling pathways that enable cell proliferation and migration (8, 9). For instance, control of ROS levels by oncogene-induced expression of the transcriptional NRF2 antioxidant program drives the growth and survival of some tumors (10). In melanoma, high levels of ROS inhibit distant metastasis (11). However, other studies have found that ROS promote metastasis, underlining the conflicting function of ROS in metastasis (12). Given the important role of ROS in the metastatic cascade, a molecular understanding of how ROS is controlled in migrating tumor cells is of critical importance.
Cells have evolved mechanisms to balance ROS levels. SIRT3, an NAD+-dependent mitochondrial deacetylase that promotes efficient oxidative metabolism, is a key regulator of mitochondrial ROS production and detoxification. We and others have previously shown that SIRT3 functions as a tumor suppressor by decreasing ROS levels, reducing stress-induced genomic instability, and repressing cancer-related metabolic reprogramming (13–16). Although many studies have demonstrated the tumor-suppressive role of SIRT3, its function in cell migration and metastasis is conflicting and requires further research (17–19). Given the role of SIRT3 in controlling ROS homeostasis, and the evidence that SIRT3 loss promotes breast tumor cell proliferation, we probed whether SIRT3 regulates cell migration and breast cancer metastasis. Our studies demonstrate that SIRT3 regulates ROS levels and is important in repressing Src oxidation and Src/FAK signaling to inhibit cell migration and metastasis.
Results
SIRT3 Expression Is Repressed During Collective Cell Migration.
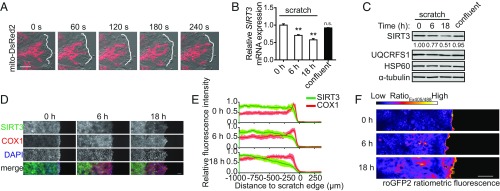
To examine the role of SIRT3 in cell migration, we investigated the localization of mitochondria and SIRT3 expression at the leading edge of migrating cells. We first visualized mitochondria during cell migration in MCF10A breast epithelial cells by using a mitochondrial-tagged DsRed2 fluorophore for live-cell imaging. Consistent with previous studies (20, 21), a subset of mitochondria localized toward the leading edge of migrating cells (Fig. 1A and SI Appendix, Fig. S1A and Movie S1). To assess whether the levels of SIRT3 changed upon initiation of migration, we applied multiple scratches in a confluent MCF10A monolayer. Unexpectedly, SIRT3 mRNA and protein levels were decreased at 6 and 18 h postscratch (Fig. 1 B and C). By contrast, levels of the complex III UQCRFS1 subunit and the mitochondrial heat shock protein 60 (HSP60) remained unaltered (Fig. 1C), indicating that total mitochondrial content did not decrease during migration. To determine whether SIRT3 expression is decreased specifically in cells at the leading edge, we performed immunofluorescence (IF) during MCF10A scratch assays. SIRT3 protein expression was lower in migrating cells at the edge of the scratch, compared with cells distal to the scratch (Fig. 1 D and E). Consistent with our previous observation that reduced SIRT3 expression occurred independently of changes in total mitochondrial content, the intensity of complex IV subunit (COX1) of the electron transport chain did not decrease during migration (Fig. 1 D and E). Together, these results show that migratory cells decrease expression of SIRT3.
Fig. 1.
SIRT3 expression is down-regulated during collective cell migration. (A) Mitochondria in MCF10A cells were labeled with mito-DsRed2 and visualized during scratch assays using live-cell confocal microscopy at the indicated time points. White outlines trace cell boundary at the scratch edge. (Scale bar, 10 μm.) (B) SIRT3 mRNA levels were quantified using qRT-PCR at indicated time points after scratches were made in the MCF10A cell monolayer (n = 3). (C) Protein expression during MCF10A scratch assays was measured by Western blot at indicated times. The relative SIRT3 band intensity normalized to α-tubulin was calculated using ImageJ. (D) Levels of SIRT3 and COX1 protein during MCF10A scratch assays at the indicated time points were measured using IF and confocal microscopy. Nuclei were stained with DAPI. (Scale bar, 100 μm.) (E) Fluorescence intensities from D were quantified across the x axis of the images using ImageJ (n = 12 to 16). (F) Cellular ROS levels in MCF10A were measured during scratch assays at the indicated time points using the ratiometric sensor roGFP2. (Scale bar, 100 μm.) For all panels, error bars ±SEM: **P < 0.01.
As SIRT3 regulates ROS levels, we examined whether reduced SIRT3 expression might correlate with enhanced ROS production in migrating cells. Utilizing a fluorescent ratiometric redox sensor roGFP2 (21, 22), we found that ROS levels were elevated at the scratch edge (Fig. 1F). To rule out the effect of proliferation on cell migration, we stained cells with the proliferation marker Ki-67 during scratch assays. We found that migratory and nonmigratory cells were equally proliferative (SI Appendix, Fig. S1B), suggesting that the increased ROS at the scratch edge was not correlated with higher proliferative potential.
SIRT3 Inhibits Collective Cell Migration by Decreasing ROS.
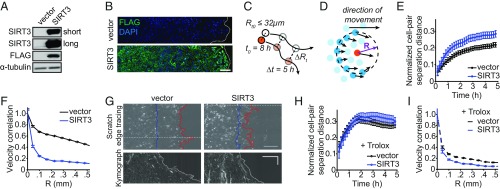
To test whether SIRT3 expression affects cell migration, we stably overexpressed SIRT3 in MCF10A cells (Fig. 2A) containing nuclei labeled with histone 2B (H2B)-mCherry and performed a scratch assay. Automatic tracking of labeled nuclei permitted analysis of cellular trajectories during cell migration (23). We first established that the overexpressed SIRT3 protein is not down-regulated during collective migration (Fig. 2B). Nuclear tracking revealed that the vast majority of cell trajectories of control MCF10A cells at the scratch edge were parallel to neighboring cells, whereas SIRT3 overexpression disrupted this coordinated movement (SI Appendix, Fig. S2A). Quantification of cell migration behavior showed that SIRT3 overexpression increased the normalized cell-pair separation distance (Fig. 2 C and E), thus indicating that cells with higher levels of SIRT3 were migrating away from each other over a 5-h period during the scratch assay. Furthermore, SIRT3 overexpression also decreased the velocity correlation between neighboring cells (Fig. 2 D and F), meaning that increased SIRT3 levels abrogated cell−cell migration coordination. Since coordinated cellular trajectory promotes directional movement of the collective epithelial cell layer, we also measured the total distance traveled by the MCF10A cell sheet by tracing of the scratch edge. Kymographs of the cell sheet during scratch assays revealed that control MCF10A cell sheet traveled farther than SIRT3-overexpressing cells (Fig. 2G). Quantification of this process showed that SIRT3 overexpression reduced the total distance covered by the cells (SI Appendix, Fig. S2B). To confirm that the changes in cell migration with SIRT3 overexpression were dependent on the catalytic activity of SIRT3, we compared the cell migratory parameters of those cells overexpressing wild-type (WT) SIRT3 to those overexpressing the catalytic mutant of SIRT3 (H248Y). We found that the overexpression of SIRT3 H248Y in MCF10A cells led to a lower normalized cell-pair separation distance (SI Appendix, Fig. S2C) and higher velocity correlation (SI Appendix, Fig. S2D) than the overexpression of SIRT3 WT. Knockdown of SIRT3 with shRNA did not alter the migratory parameters of MCF10A cells (SI Appendix, Fig. S2 E and F), suggesting that these cells are maximally coordinated with the endogenous down-regulation of SIRT3. Together, these data demonstrate that SIRT3 inhibits collective cell migration by disrupting intercellular coordination.
Fig. 2.
SIRT3 inhibits collective cell migration of MCF10A cells. (A) Immunoblot of MCF10A cells expressing control or a vector expressing SIRT3 fused to the molecular tag FLAG (SIRT3-FLAG). Long and short exposures are shown of the SIRT3 blot. (B) Levels of overexpressed SIRT3-FLAG protein during scratch assays in MCF10A at 18 h were measured using IF and confocal microscopy. Nuclei were stained with DAPI. (Scale bar, 100 μm.) (C) Schematic of normalized cell-pair separation distance: cell pairs that were less than 32 μm apart at 8 h after scratch formation were tracked for the next 5 h, and the change in their separation was measured. (D) Schematic of velocity correlation: Velocity trajectories of a central cell and all cells at a certain distance (R) from the central cell were compared over a 1-h period during scratch assay. (E) Normalized cell-pair separation distance from 8 h to 13 h after scratch formation in untreated control and SIRT3-overexpressing MCF10A cells. (F) Velocity correlation at 12 h in untreated control and SIRT3-overexpressing MCF10A cells. (G) Scratch edge tracing at 0 h (blue) and 17 h (red) after scratch assay (Top) and kymographs of the cell edge at indicated positions from 0 to 22 h (Bottom). (Horizontal scale bar, 100 μm; vertical scale bar, 5 h.) (H) Normalized cell-pair separation distance in control and SIRT3-overexpressing cells treated with 1 mM Trolox. (I) Velocity correlation in control and SIRT3-overexpressing cells treated with 1 mM Trolox.
Since SIRT3 expression decreased while mitochondria were recruited to the leading edge of cells and ROS levels were increased at the scratch edge, we hypothesized that SIRT3 inhibits collective cell migration by reducing ROS levels and that oxidant scavenging could recapitulate the effects of SIRT3 overexpression on cell migration. Treatment with antioxidants had little effect on SIRT3-overexpressing cells. However, treatment with Trolox, a water-soluble vitamin E analog, disrupted the linear and parallel movement of control cells (SI Appendix, Fig. S2A) and increased the normalized cell-pair separation distance (Fig. 2H) while decreasing the velocity correlation (Fig. 2I), similar to the effects of SIRT3 overexpression. In addition, treatment with N-acetylcysteine or the manganese superoxide dismutase mimetic, MnTBAP, also inhibited collective cell migration (SI Appendix, Fig. S2 G–J), similar to SIRT3 overexpression. Together, our data show that SIRT3 regulates collective cell migration through modulation of ROS levels.
SIRT3 Represses Src/FAK Signaling.
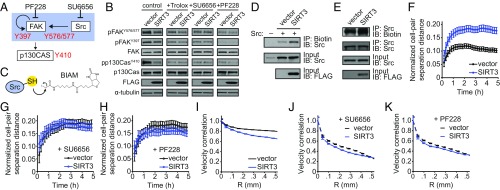
To probe the mechanism(s) by which SIRT3 attenuated cell migration in a ROS-dependent manner, we examined signaling pathways sensitive to ROS. Activation of Src kinase and FAK have been shown to promote collective cell migration (2, 24). FAK is activated by autophosphorylation of Tyr397, a site recognized by Src kinase, which then phosphorylates FAK on Tyr576/577 (Fig. 3A) (24). Indeed, phosphorylation of FAK on Tyr576/577 was decreased upon SIRT3 overexpression, while phosphorylation of FAK on Tyr397 remained unaltered (Fig. 3B). Consistent with reduced FAK activity, SIRT3 overexpression led to a decrease in the phosphorylation of p130Cas, a downstream target of the Src/FAK signaling complex (Fig. 3 A and B).
Fig. 3.
SIRT3 represses Src/FAK signaling by down-regulating ROS. (A) Model of Src-FAK signaling and the inhibitors used in this study. (B) Immunoblots of phospho-Tyr397, phospho-Tyr576/577 and total FAK, and phospho-Tyr410 and total p130Cas, as well as of FLAG tag and α-tubulin in control and SIRT3-FLAG overexpressing MCF10A cells treated with Trolox (1 mM), SU6656 (5 μM), or PF228 (1 μM). (C) Schematic of BIAM labeling of reduced Src protein. (D) Levels of reduced Src protein (overexpressed) in control and SIRT3-FLAG overexpressing MCF10A cells were measured by biotin labeling of thiol moieties using iodoacetamide. (E) Levels of reduced endogenous Src protein in control and SIRT3-FLAG overexpressing MCF10A cells were measured using biotin labeling. (F–H) Normalized cell-pair separation distance from 8 h to 13 h after scratch formation in (F) untreated control and SIRT3-overexpressing MCF10A cells and (G) cells treated with 5 μM SU6656 or (H) 1 μM PF228. (I–K) Velocity correlation at 12 h in untreated control and SIRT3-overexpressing MCF10A cells (I), as well as cells treated with 5 μM SU6656 (J) or 1 μM PF228 (K). For all panels, error bars represent 95% confidence interval, and nonoverlapping bars are statistically significant with P < 0.05 (n = 4 to 6 wells per group).
Since FAK phosphorylation was diminished only on sites regulated by Src, we hypothesized that SIRT3 inhibits Src activity, which is induced by protein oxidation in addition to phosphorylation (4, 25). To test whether SIRT3 alters the redox state of Src protein, we used biotinylated iodoacetamide (BIAM) to label reduced cysteine thiol side chains (Fig. 3C). SIRT3 overexpression lowered the level of oxidized Src as measured by increased BIAM labeling of both the ectopically expressed Src protein (Fig. 3D) and the endogenous Src protein (Fig. 3E). Furthermore, treatment with Trolox also decreased the levels of FAK phosphorylation on Tyr576/577, recapitulating the effects of SIRT3 overexpression (Fig. 3B). Collectively, these data suggest that SIRT3 attenuates Src/FAK signaling through modulation of Src oxidation.
To examine the involvement of Src/FAK signaling in the SIRT3-mediated control of cell migration, we tested the effect of Src and FAK inhibitors on SIRT3 phenotypes. Treatment with inhibitors of Src (SU6656) and FAK (PF228) reduced the levels of Tyr576/577 FAK and p130Cas phosphorylation in control cells to that of the SIRT3-overexpressing cells (Fig. 3 A and B). Furthermore, Src and FAK inhibitors did not alter collective migration parameters in cells overexpressing SIRT3 (Fig. 3 F–K). Treatment with dasatinib, a tyrosine kinase inhibitor that targets Src, although with less specificity, led to similar outcomes in terms of signaling and collective migration (SI Appendix, Fig. S3). These findings demonstrate that Src and FAK inhibitors phenocopy the effect of SIRT3 overexpression.
SIRT3 Levels Are Altered in Metastatic Breast Cancer Cells.
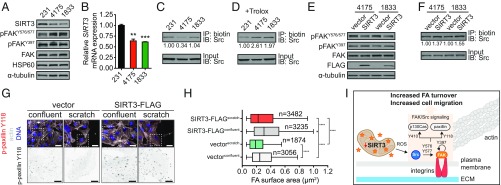
The observation that SIRT3 regulates collective cell migration via Src/FAK signaling led us to test whether cells with enhanced metastatic capabilities might display altered SIRT3 levels. We took advantage of the well-characterized lung (LM2-4175) and bone (BoM-1833) metastatic breast cancer cell lines derived from MDA-MB-231 cells (26, 27). Although MDA-MB-231 cells lack E-cadherin expression, and are therefore deficient for strong cell−cell adhesion and collective migration in vitro (28), Src/FAK signaling plays a prometastatic role in these cells. FAK signaling mediates the formation of filopodium-like protrusions that promote lung metastasis of breast cancer cells including MDA-MB-231 cells (29, 30). Furthermore, enhanced Src activity promotes the survival and outgrowth of MDA-MB-231 cells in the bone microenvironment (31). Consistent with their enhanced metastatic abilities, the lung and bone metastatic cells exhibited an increase in Src-specific FAK phosphorylation (Fig. 4A). Moreover, these metastatic clones had lower levels of SIRT3 mRNA and protein expression compared with parental MDA-MB-231 cells (Fig. 4 A and B). HSP60 levels were comparable between these cell lines, indicating that the decrease in SIRT3 expression was not due to lower mitochondrial content in the metastatic cells (Fig. 4A).
Fig. 4.
SIRT3 attenuates Src/FAK signaling in breast cancer. (A) Immunoblots of SIRT3, phospho-Tyr397, phospho-Tyr576/577, total FAK, HSP60, and α-tubulin in parental MDA-MB-231 (231) cells, as well as lung metastatic LM2-4175 (4,175) and bone metastatic BoM-1833 (1,833) cells. (B) SIRT3 mRNA levels in the above-mentioned breast cancer cell lines were quantified using qRT-PCR (n = 3). Statistical significance was determined using the Student’s t test. (C and D) Levels of reduced endogenous Src protein in the above breast cancer cells, (C) untreated or (D) with 1 mM Trolox, were measured by biotin labeling of thiol moieties using iodoacetamide. Biotin labeling normalized to total Src protein was calculated using ImageJ. (E) Immunoblots of phospho-Tyr397, phospho-Tyr576/577, total FAK, FLAG tag, and α-tubulin in 4,175 and 1,833 cells with or without SIRT3-FLAG overexpression. (F) Levels of reduced endogenous Src protein in 4,175 and 1,833 cells with or without SIRT3-FLAG overexpression. Biotin labeling normalized to total Src protein was calculated using ImageJ. (G) Representative images of focal adhesions in SIRT3-overexpressing and control MDA-MB-231 cells in scratch assays (Top). The phospho-paxillin channel of the boxed area (Bottom). [Scale bars denote 25 μm (Top) and 10 μm (Bottom).] (H) Focal adhesion surface area assessed through phospho-paxillin staining in SIRT3-overexpressing and control MDA-MD-231 cells. Statistical significance was determined using ANOVA while correcting for multiple comparison. (I) Model of regulation of SCR-dependent focal adhesion signaling by SIRT3. For all panels, error bars ±SD: *P < 0.05; **P < 0.01; ***P < 0.001; P < 0.0001.
To test whether reduced SIRT3 expression corresponded to an increase in Src oxidation, we examined the redox state of Src. While Src was more oxidized in the lung metastatic cells but not in the bone metastatic cells (Fig. 4C), both lines were responsive to Trolox treatment, as Src oxidation decreased compared with parental MDA-MB-231 cells (Fig. 4D). Consequently, we tested whether restored SIRT3 expression could reverse the increased FAK activation in these metastatic cells. SIRT3 overexpression repressed FAK phosphorylation on Tyr576/577 in both cell lines but had no effect on Tyr397 phosphorylation, confirming that SIRT3 repressed Src activity downstream of FAK autophosphorylation (Fig. 4E). SIRT3 overexpression also decreased Src oxidation in both metastatic cell lines (Fig. 4F), suggesting that SIRT3 attenuates Src/FAK signaling in metastatic breast cancer cells by inhibiting Src oxidation. Interestingly, we did not observe a difference in MDA-MB-231 cell migration after SIRT3 knockdown, which may be explained by the already low levels of SIRT3 of basal-like breast cancer cells (SI Appendix, Fig. S4 A–C, G, and H).
Src regulates cellular migration through substrates other than FAK. For example, Src phosphorylates and activates paxillin to modulate focal adhesion assembly and migration (32). To examine the role of SIRT3 on Src/paxillin activation, we examined the size of focal adhesions. SIRT3 overexpression caused an increase in focal adhesion size compared with the control cells (Fig. 4 G and H). SIRT3-mediated increase in focal adhesion size is consistent with reduced FAK activation (Fig. 3B) that is required for focal adhesion turnover. Increased focal adhesion stability retards cell motility, which was also observed in this system with SIRT3 overexpression, without affecting proliferation in this time frame (SI Appendix, Fig. S4 D–F). Taken together, our studies suggest a mechanism whereby reduced SIRT3 expression, and subsequent increased ROS production, stimulates Src/FAK signaling at the focal adhesion to induce cell migration (Fig. 4I).
Low SIRT3 Levels Are Consistent with High Metastasis.
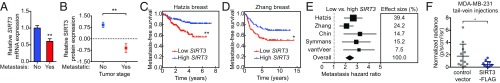
We next probed whether SIRT3 expression was altered in metastatic tumors from breast cancer patients. SIRT3 mRNA expression was reduced in tumor from breast cancer patients diagnosed with metastatic disease (26) (Fig. 5A). To assess the correlation between SIRT3 protein expression and metastatic outcome, we obtained a human breast tumor lysate array generated from primary breast cancer samples and adjacent normal mammary tissues. Breast cancers that had metastasized expressed lower levels of SIRT3 protein compared with those that were not metastatic (Fig. 5B). Furthermore, analysis of metastasis-free survival of breast cancer patients (33) revealed that tumors with low SIRT3 mRNA expression had worse metastasis-free survival, with a hazard ratio of 2.436 compared with those with high SIRT3 mRNA expression (95% confidence interval of 1.433 to 4.068; Fig. 5C and SI Appendix, Fig. S5A). Analyses of additional datasets comparing SIRT3 DNA copy number (34, 35) and mRNA expression (36, 37) also revealed that higher SIRT3 levels correlate with improved metastasis-free survival (Fig. 5 D and E and SI Appendix, Fig. S5 B–D), further supporting the correlation between low SIRT3 levels and increased metastatic potential in breast cancer.
Fig. 5.
SIRT3 is inversely correlated with metastasis in breast cancer. (A) SIRT3 protein fold change in breast cancer samples compared with normal tissue in metastatic (Stage IV) versus nonmetastatic (Stages I to III) breast cancers from the human breast cancer lysate array. (B) Relative SIRT3 mRNA expression in tumors from breast cancer patients with or without metastases at 5 y determined using Oncomine cancer microarray database. (C and D) Kaplan−Meier curves depicting metastasis-free survival in patients with primary breast cancers of high (top quartile SIRT3 expression, blue) versus low (bottom quartile SIRT3 expression, red). Significance was calculated using log-rank test. (E) Forest plot summarizing metastasis hazard ratio ±95% confidence interval comparing breast cancers of low versus high SIRT3 levels. Effect size was determined by the number of samples in each dataset. (F) Control or SIRT3-overexpressing MDA-MB-231 cells were injected i.v. through the tail vein. Luciferase-dependent bioluminescence imaging quantified to detect metastatic take in the lungs. Statistical significance was determined using the Student’s t test at 5 wk postinjection. For all panels, error bars ±SD: *P < 0.05; **P < 0.01.
Because breast cancer patient data indicate that low SIRT3 expression correlates with greater metastasis, we examined whether SIRT3 overexpression in MDA-MB-231 cells can suppress their ability to colonize the lungs in an in vivo tail vein injection model. Indeed, colonization of MDA-MB-231 cells in the lung was suppressed by SIRT3 overexpression compared with cells expressing the vector control, as measured by bioluminescence imaging of luciferase activity (Fig. 5F). Together, our findings demonstrate that SIRT3 regulates breast cancer metastases formation in vivo.
Discussion
The majority of cancer-related deaths in solid tumors arise from metastatic relapse and clinical complications of disseminated disease, yet our understanding of the metastatic process remains incomplete. Increased mitochondrial ROS has been shown to promote metastasis via activation of Src (5), and, therefore, finding regulators of ROS production may be critical to battle metastatic disease. Here, we provide evidence that SIRT3 inhibits cell migration and metastases formation. We demonstrate that down-regulation of SIRT3, but not total mitochondrial content, during epithelial cell migration increases local ROS levels and activates Src/FAK signaling. Furthermore, we show that SIRT3 expression is down-regulated in metastatic breast cancer cell lines, which corresponds with increased Src/FAK signaling. SIRT3 overexpression in these metastatic cells reverses this hyperactive signaling cascade by reducing Src oxidation. Finally, we find that overexpression of SIRT3 can inhibit metastases formation in vivo. These findings are in line with our observations that reduced SIRT3 levels correlate with decreased metastasis-free survival in clinical datasets. Together, these findings elucidate a pathway linking SIRT3 to cancer cell migration.
Through our study, we propose a mechanism by which down-regulation of SIRT3 levels promotes cell migration by increasing ROS levels and activating prometastatic Src signaling. These observations support the findings from our previous study, in which cell communication, extracellular matrix receptor interaction, and integrin-mediated cell adhesion were top hits in the GSEA (Gene Set Enrichment Analysis) comparing tissues from WT and SIRT3-null mice (15). It would be interesting for future studies to uncover how SIRT3 is down-regulated during cell migration and in metastatic breast cancer cells. Interestingly, while SIRT3 levels are reduced in metastatic cells, SIRT3 is not completely lost, suggesting that cells may benefit from maintaining SIRT3 expression during the metastatic cascade. One explanation is that metastasizing cells in the bloodstream and visceral organs experience high levels of oxidative stress that can inhibit the metastatic process (15), and therefore rely on the residual SIRT3 expression to balance toxic ROS levels and maintain cell viability.
Previous studies in fibroblasts demonstrated the importance of mitochondria in energy production that supports cytoskeletal remodeling required for cellular migration (38). Our study adds a surprising layer to this model, as SIRT3, which promotes energy homeostasis, appears down-regulated at the leading edge of migrating cells. Thus, it appears that there are system-dependent differences for how mitochondria mediate cell migration by intricately balancing ATP and ROS levels.
In summary, we have found that SIRT3 inhibits collective cell migration, down-regulates prometastatic Src/FAK signaling, reduces in vivo metastases formation, and is inversely correlated with metastatic outcome in human breast cancer. Through this study, we posit that breast cancers with lower SIRT3 expression are more aggressive and that inhibition of the relevant pathways, including Src/FAK signaling, could improve clinical outcome. In this regard, SIRT3 could prove to be a useful biomarker for disease prognosis as well as predicting response to targeted therapy.
Materials and Methods
Live-Cell Imaging.
Fluorescent time-lapse images for scratch assay analysis were collected on a Ti-E inverted motorized microscope with integrated Perfect Focus System (Nikon) equipped with a 20× Plan Apo 0.75 NA objective lens, a linear-encoded motorized stage (Nikon), SOLA fluorescence light source (Lumencor), fast excitation and emission filter wheels (excitation 560/40 nm and emission 630/75 nm for RFP (Red Fluorescent Protein)-mCherry; Chroma Technologies), fast transmitted and epifluorescence light path shutters (SmartShutter; Sutter Instrument), ORCA-AG cooled CCD (charged-coupled device) camera (Hamamatsu Photonics), and a custom-built 37 °C microscope incubator enclosure with 5% CO2 delivery. Detailed information on data acquisition is provided in SI Appendix, Supplemental Experimental Procedures.
ROS Measurement.
MCF10A cells with the ROS sensor roGFP2 were generated by retroviral infection with pBabe containing roGFP2 cDNA and propagated as single-cell clones to ensure uniform overexpression of the sensor between all cells. Cells were imaged, as described in Live-Cell Imaging, with dual excitation at quad 402-525 (402/15 nm) and YFP (500/20 nm). Relative ROS levels were analyzed by taking the ratiometric images using ImageJ.
Src Oxidation Assay.
MCF10A cells were transfected with pSG5 vector containing chicken c-Src (kindly provided by P. Chiarugi, University of Florence, Florence, Italy) using Lipofectamine LTX reagent (Life Technologies) according to manufacturer’s instructions, and experiments were performed 48 h after transfection. Cells were lysed with deoxygenated lysis buffer containing 50 mM Tris⋅HCl (pH 7.5), 0.5% Triton X-100, and 150 mM NaCl supplemented with protease inhibitor and 100 μM EZ-link Iodoacetyl-LC-Biotin (Pierce). To remove excess iodoacetyl biotin, proteins were precipitated by adding five volumes of −20 °C methanol and incubating on ice for 20 min. The samples were centrifuged at max speed for 10 min at 4 °C, supernatants were discarded, and pellets were air-dried and then resuspended in lysis buffer. Protein concentrations were measured and normalized. Samples were incubated with either NeutrAvidin agarose beads (Pierce) or Src antibody (Cell Signaling) followed by EZview Red Protein G Affinity Gel (Sigma). The beads were washed four to five times and resuspended in loading buffer for Western blot.
Image Analyses and Statistics.
Analyses of cell tracking were performed as previously described (33). All error bars indicate errors of the mean at 95% confidence level. Nonoverlapping error bars in these plots therefore indicate statistical significance with P < 0.05. For all other scratch assays, n = 6 and n = 800 to 1,500 were analyzed per experiment, where n represents the total number of cells measured from N experiments.
Statistics.
Significance between groups was evaluated using Student’s unpaired t test unless otherwise noted.
Animal Experiment Approval.
All procedures involving animals were performed in accordance with the regulations of Boston Children's Hospital Institutional Animal Care and Use Committee (protocol 12-11-2308R) and Harvard Medical School Institutional Animal Care (protocol IS00000668).
Supplementary Material
Acknowledgments
We thank Joan Massagué (Howard Hughes Medical Institute, Memorial Sloan-Kettering Cancer Center) and Paola Chiarugi (University of Florence) for providing reagents. We also thank members of the M.C.H., G.D., Brugge, and S.S.M. laboratories for helpful discussions and technical assistance, especially F. Kyle Satterstrom for analysis of SIRT3 expression. We thank the Nikon Imaging Center at Harvard Medical School for help with microscopy. Analyses of cell migration assays were performed on the Orchestra cluster supported by the Harvard Medical School Research Information Technology Group. J.J.L. was supported as a Howard Hughes Medical Institute Medical Research Fellow. E.Z. was supported by the American Heart Association (Award 15POST25560077). M.C.H. is supported by National Institutes of Health Grant R01DK103295 from the National Institute of Diabetes and Digestive and Kidney Diseases, Grant R01CA213062 from the National Cancer Institute, the Ludwig Center at Harvard, and the Glenn Foundation for Medical Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800440115/-/DCSupplemental.
References
- 1.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Rico C, Pincet F, Thiery JP, Dufour S. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. J Cell Sci. 2010;123:712–722. doi: 10.1242/jcs.047878. [DOI] [PubMed] [Google Scholar]
- 3.Canel M, et al. Quantitative in vivo imaging of the effects of inhibiting integrin signaling via Src and FAK on cancer cell movement: Effects on E-cadherin dynamics. Cancer Res. 2010;70:9413–9422. doi: 10.1158/0008-5472.CAN-10-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giannoni E, Buricchi F, Raugei G, Ramponi G, Chiarugi P. Intracellular reactive oxygen species activate Src tyrosine kinase during cell adhesion and anchorage-dependent cell growth. Mol Cell Biol. 2005;25:6391–6403. doi: 10.1128/MCB.25.15.6391-6403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 6.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vyas S, Zaganjor E, Haigis MC. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu J, Dawes IW. Redox control of cell proliferation. Trends Cell Biol. 2012;22:592–601. doi: 10.1016/j.tcb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Tochhawng L, Deng S, Pervaiz S, Yap CT. Redox regulation of cancer cell migration and invasion. Mitochondrion. 2013;13:246–253. doi: 10.1016/j.mito.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 10.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 13.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong SM, et al. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene. 2015;34:2115–2124. doi: 10.1038/onc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang GC, et al. The expression and related clinical significance of SIRT3 in non-small-cell lung cancer. Dis Markers. 2017;2017:8241953. doi: 10.1155/2017/8241953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, et al. The SIRT 3 expression profile is associated with pathological and clinical outcomes in human breast cancer patients. Cell Physiol Biochem. 2014;34:2061–2069. doi: 10.1159/000366401. [DOI] [PubMed] [Google Scholar]
- 19.Ren T, et al. MCU-dependent mitochondrial Ca2+ inhibits NAD+/SIRT3/SOD2 pathway to promote ROS production and metastasis of HCC cells. Oncogene. 2017;36:5897–5909. doi: 10.1038/onc.2017.167. [DOI] [PubMed] [Google Scholar]
- 20.Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson GT, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 23.Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol. 2012;199:545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: In command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 25.Lim SK, Choi YW, Lim IK, Park TJ. BTG2 suppresses cancer cell migration through inhibition of Src-FAK signaling by downregulation of reactive oxygen species generation in mitochondria. Clin Exp Metastasis. 2012;29:901–913. doi: 10.1007/s10585-012-9479-z. [DOI] [PubMed] [Google Scholar]
- 26.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 28.Volakis LI, et al. Loss of myoferlin redirects breast cancer cell motility towards collective migration. PLoS One. 2014;9:e86110. doi: 10.1371/journal.pone.0086110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2012;2:706–721. doi: 10.1158/2159-8290.CD-11-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibue T, Brooks MW, Weinberg RA. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell. 2013;24:481–498. doi: 10.1016/j.ccr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XH, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 33.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–3801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 35.Chin SF, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 37.Symmans WF, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuler MH, et al. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol Biol Cell. 2017;28:2159–2169. doi: 10.1091/mbc.E16-10-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.