Significance
Carbon fixation is arguably one of the most important metabolic processes on Earth. Stand-alone CP12 proteins are major players in the regulation of this pathway in all oxygenic photosynthetic organisms, yet their intrinsic disorder has so far hampered the capturing of a principal part of their structure. Here we provide structural insights into CP12 by investigating an uncharacterized CP12 fusion protein, CBS–CP12, which is widespread among cyanobacteria, and reveal a unique hexameric structure. Our data further extend the existing knowledge of the regulation of photosynthesis and carbon fixation by the CP12 protein family, suggesting a more versatile role of this protein family in global redox regulation, predominantly in bloom-forming cyanobacteria that pose major threats in lakes and reservoirs.
Keywords: crystal structure, hexamer, redox, Microcystis aeruginosa
Abstract
Cyanobacteria are important photosynthetic organisms inhabiting a range of dynamic environments. This phylum is distinctive among photosynthetic organisms in containing genes encoding uncharacterized cystathionine β-synthase (CBS)–chloroplast protein (CP12) fusion proteins. These consist of two domains, each recognized as stand-alone photosynthetic regulators with different functions described in cyanobacteria (CP12) and plants (CP12 and CBSX). Here we show that CBS–CP12 fusion proteins are encoded in distinct gene neighborhoods, several unrelated to photosynthesis. Most frequently, CBS–CP12 genes are in a gene cluster with thioredoxin A (TrxA), which is prevalent in bloom-forming, marine symbiotic, and benthic mat cyanobacteria. Focusing on a CBS–CP12 from Microcystis aeruginosa PCC 7806 encoded in a gene cluster with TrxA, we reveal that the domain fusion led to the formation of a hexameric protein. We show that the CP12 domain is essential for hexamerization and contains an ordered, previously structurally uncharacterized N-terminal region. We provide evidence that CBS–CP12, while combining properties of both regulatory domains, behaves different from CP12 and plant CBSX. It does not form a ternary complex with phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase. Instead, CBS–CP12 decreases the activity of PRK in an AMP-dependent manner. We propose that the novel domain architecture and oligomeric state of CBS–CP12 expand its regulatory function beyond those of CP12 in cyanobacteria.
Cystathionine β-synthase (CBS) domains are widespread structural domains, conserved either as single proteins (1, 2) or as domains in a wide range of functionally different cytosolic (3–5) and membrane proteins (6–8). The importance of CBS domains is reflected by the range of different hereditary diseases in humans caused by mutation in their sequence, including homocystinuria and retinitis pigmentosa (9, 10). Each domain folds as an antiparallel β-sheet flanked by helices on one side and contains the secondary structural elements α0-β1-α1-β2-β3-α2 (11). They always occur in pairs, forming a so-called CBS pair or Bateman module (11, 12). All structurally characterized CBS pairs, with the exception of a single archaeal representative, assemble in homodimers containing four CBS domains, oriented in a head-to-head or head-to-tail manner (11), such as CBSX1 and CBSX2, stand-alone CBS domain proteins of plants (1, 2). CBS domains are important sensors of cellular energy status, often binding adenine nucleotides as regulatory ligands (11). Plant CBSX proteins, for instance, are important partners in the thioredoxin (Trx)-mediated redox regulation of protein activity (1). Targets of Trx-regulated processes in photosynthetic organisms include proteins related to photosynthesis and oxidative stress response (13–15). By increasing the activity of all chloroplastic Trx, plant CBSX1 enhances carbon dioxide (CO2) fixation and carbohydrate synthesis, thus affecting plant development and stabilizing cellular redox homeostasis (1).
A bioinformatic analysis of 126 cyanobacterial genomes revealed the existence of CBS domain-containing proteins C-terminally fused to the small protein CP12 (chloroplast protein) (16). While these CBS–CP12 fusion proteins are to date only found in cyanobacteria, stand-alone CP12 proteins are conserved in all oxygenic photosynthetic organisms, namely plants, algae, and cyanobacteria (17–21). CP12 is an intrinsically disordered, regulatory protein of about 80 residues. It is characterized by the AWD_VEEL core sequence (22) and two N-terminal and C-terminal redox-sensitive cysteine pairs. Under oxidizing conditions, the cysteine pairs form disulfide bridges structuring two polypeptide loops required for the inactivation of two enzymes of the CO2 fixation pathway (Calvin cycle), phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), by forming a ternary GAPDH–CP12–PRK complex (17–19). The activation of both enzymes, and consequently CO2 fixation, is mediated by reduction via Trx (13), causing the dissociation of the ternary GAPDH–CP12–PRK complex. Structural information on CP12 is scarce due to its intrinsic disorder. Only about 20 residues of the C-terminal part of CP12 have been structurally characterized in a complex with GAPDH, indicating the disordered state of the N-terminal region even in the presence of its binding partner under oxidizing conditions (23, 24).
In contrast to CP12, the function of cyanobacterial CBS–CP12 fusion proteins is unknown. They typically occur in addition to a stand-alone CP12 and are divided into three variants according to their number of cysteine pairs in the CP12 domain (two, one, or none) (16). The CBS pairs present in these variants share a high degree of sequence similarity (16). It was hypothesized that the fusion of CP12 and CBS domains in cyanobacteria functionally connects them, implying another layer of complexity in the regulatory roles of both CP12 and CBS domains (16).
Here we report the structural and functional characteristics of a CBS–CP12 variant from the bloom-forming cyanobacterium Microcystis aeruginosa PCC 7806 (hereafter M. aeruginosa). We show that CBS–CP12 fusion proteins are widespread in cyanobacteria and particularly associated with a thioredoxin gene in bloom-forming species. The crystal structure of CBS–CP12 includes a complete structure of the N-terminal part of CP12 and reveals that CBS–CP12 forms a hexamer, unlike any other characterized CBS domain protein. We further show coregulated light-induced gene expression of CBS–CP12 with TrxA and an AMP-dependent inhibition of PRK activity by CBS–CP12 that counteracts the regulation by TrxA.
Results
The CBS–CP12 Protein Family Is Widespread in Cyanobacteria.
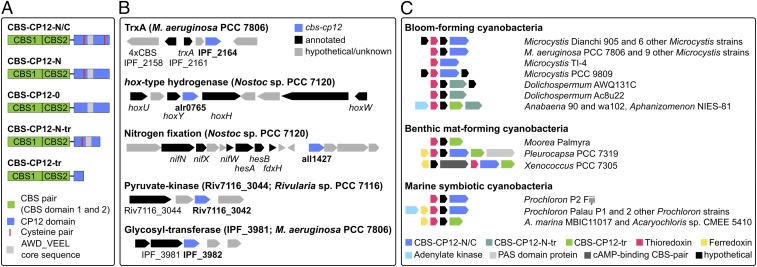
A reassessment of the occurrence of CBS–CP12 fusion proteins in 333 cyanobacterial genomes revealed the presence of five CBS–CP12 variants, the three previously described (16) and two variants with a shorter CP12 domain, CBS–CP12-N-tr and CBS–CP12-tr (Fig. 1A). The phylogenetic survey detected 232 single CBS–CP12 proteins and demonstrated their widespread distribution in all five morphological subsections of the cyanobacterial phylum, with the exception of the entire phylogenetic subclade C1 and the model strain Synechocystis sp. PCC 6803 (SI Appendix, Fig. S1). Truncated CBS–CP12 variants occur in the majority of strains along with the full-length variants. As previously shown (16), all CBS–CP12 variants coexist with one or more CP12 genes.
Fig. 1.
CBS–CP12 variants and gene neighborhood conservation. (A) Domain organization of the five CBS–CP12 variants. CBS–CP12-N/C contains both cysteine pairs in the CP12 domain, CBS–CP12-N only the N-terminal pair, and CBS–CP12-0 lacks both cysteine pairs. CBS–CP12-N-tr contains a truncated CP12 domain with the N-terminal cysteine pair, and CBS–CP12-tr possesses only a short CP12 domain without cysteine pairs. The distribution of the five CBS–CP12 variants within the cyanobacterial phylum is displayed in SI Appendix, Fig. S1. (B) Selected list of gene neighborhoods of CBS–CP12 genes in different cyanobacteria. Note that a CBS–CP12 gene is also copresent in the hydrogenase gene cluster of Synechococcus sp. PCC 7002. (C) Conservation of 12 gene cluster types featuring a CBS–CP12 and a TrxA gene. Note that one gene cluster type occurs in bloom-forming and marine symbiotic cyanobacteria (M. aeruginosa PCC 7806 and Prochloron P2 Fiji).
A gene neighborhood survey revealed an unexpected variety of gene clusters encoding a CBS–CP12 variant, including the hox-type hydrogenase and a nitrogen fixation (nif) gene cluster in Nostoc sp. PCC 7120 and a pyruvate kinase in Rivularia sp. PCC 7116 (Fig. 1B). Most commonly, CBS–CP12 genes were observed in the immediate vicinity of thioredoxin A-encoding genes (Fig. 1C). These joint CBS–CP12/TrxA gene clusters were predominantly detected in genomes of bloom-forming strains, but also in benthic mat-forming and marine symbiotic cyanobacteria. Some of these gene clusters additionally encode the electron transfer protein ferredoxin and adenylate kinase, an enzyme important for cellular energy homeostasis. In all cases, a CBS protein with four CBS domains is encoded upstream of the respective gene cluster (Fig. 1B). The different CBS–CP12/TrxA gene cluster types are distributed over four cyanobacterial subclades from unicellular strains (e.g., Microcystis and Acaryochloris strains) to multicellular strains capable of forming heterocysts for nitrogen fixation (e.g., Anabaena and Aphanizomenon strains).
CBS–CP12 Forms a Hexamer.
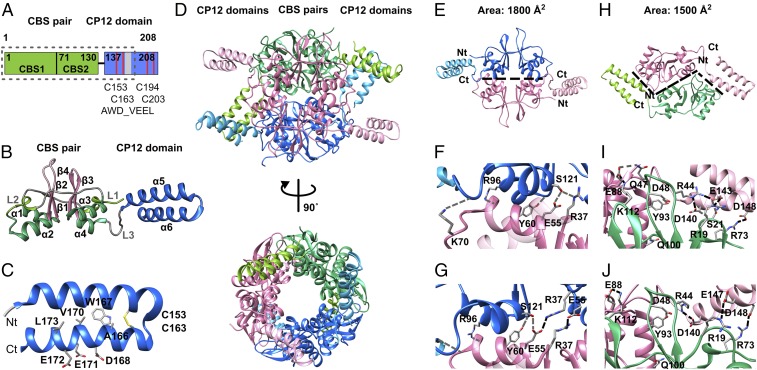
The CBS–CP12 protein (IPF_2164) of the CBS–CP12/TrxA gene cluster of M. aeruginosa (Fig. 1) was selected as representative of the unique fusion protein family for structural analysis. It consists of 205 residues, of which about 130 N-terminal residues form a CBS pair and about 70 C-terminal residues belong to the CP12 domain, as deduced from the sequence (Fig. 2A). CBS–CP12 was expressed in Escherichia coli as a Trx fusion protein (SI Appendix, Table S1). Three X-ray diffraction datasets were collected: a native and an ytterbium derivative in an orthorhombic space group with three monomers in the asymmetric unit to a resolution of 2.1 and 2.8 Å, respectively, and a native in a hexagonal space group with one monomer in the asymmetric unit to 2.5 Å (SI Appendix, Table S2).
Fig. 2.
Structure and interfaces of hexameric CBS–CP12. (A) Domain organization of CBS–CP12 (IPF_2164) of M. aeruginosa. The dotted box indicates the boundaries of the protein visible in the native crystal structure. The numbering refers to the heterologously expressed protein (SI Appendix, Table S1). (B) Cartoon representation of the monomeric subunit, as obtained from the native orthorhombic crystal structure. The helices in the CBS domain are colored yellow-green (linkers L1 and L2) and green, and the sheets are colored pink. The helices in the CP12 domain are colored blue. The secondary structural elements are sequentially labeled. (C) Cartoon representation of the CP12 domain highlighting the two cysteine residues C153 and C163 and the AWD_VEEL core sequence, as obtained from the native orthorhombic crystal structure. The N and C termini, as visible in the structure, are labeled Nt and Ct, respectively. (D) Cartoon representation of hexameric CBS–CP12 in two different orientations related by a 90° rotation around a vertical axis. The canonical dimers are colored pink, blue, and green, respectively. The respective CP12 domains are colored light pink, light blue, and yellow-green, respectively. (E) Canonical CBS:CBS interface formed by residues in the CBS pairs only. (F and G) Close-up view of the CBS:CBS interface in (F) the native orthorhombic structure showing five hydrogen bonds (gray dashed lines) and (G) the native hexagonal structure showing four hydrogen bonds and two salt bridges (black dashed lines). (H) CBS–CP12:CBS–CP12 interface formed between the CBS pairs and adjacent CP12 domain. (I and J) Close-up view of the CBS–CP12:CBS–CP12 interface in (I) the native orthorhombic structure showing five salt bridges and five hydrogen bonds, formed between the first α-helix of the CP12 domain (α5) and the opposite CBS pair and (J) the native hexagonal structure showing four hydrogen bonds and two salt bridges. Hydrogen bonds (gray dashed lines), salt bridges (black dashed lines), and area are stated as calculated by PISA (25). Numbering of residues refers to the heterologously expressed protein (SI Appendix, Table S1).
The CBS pair consists of two CBS domains, each showing a typical secondary structural arrangement (11, 12), excluding the first β-strand and preceded by a short α-helix in the linker region (linker-α1-β1-β2-α2) (Fig. 2B). The β-strands form an antiparallel β-sheet, and the two α-helices are located on one side of the sheet. The CBS pair is connected to the CP12 domain with a third linker of about 10 residues. Notably, the N-terminal part of CP12 is ordered (Fig. 2 and SI Appendix, Fig. S2) and forms two α-helices (α5-α6), of which the latter contains the AWD_VEEL core sequence (22). Both helices are connected by an intramolecular disulfide bridge between residues Cys153 and Cys163 (Fig. 2C). About 30 C-terminal residues are not visible in the electron density.
Remarkably, the biological assembly of CBS–CP12 is a hexamer, composed of the monomers of the asymmetric unit and the symmetry equivalent (Fig. 2D and SI Appendix, Fig. S2). The six subunits form a cylinder-shaped protein. The central ring is composed of the CBS pairs, while the CP12 domains protrude from the perimeter of the ring (Fig. 2D). There are differences in the N-terminal regions of the CP12 domain in the three structures, the α-helices being tilted at about 40 to 50° toward the CBS pair (SI Appendix, Fig. S2), reflecting the intrinsic plasticity of the CP12 domain and also stabilizing effects by lattice interaction in the different crystal forms.
The hexameric assembly of CBS–CP12 is based on the establishment of two different interaction interfaces (Fig. 2 E–J). One interface is similar to the known CBS:CBS interface and based exclusively on interactions between residues of the CBS pairs of two opposite subunits, leading to a head-to-tail assembly (Fig. 2E). As calculated by PISA (25), it is formed by the four α-helices and comprised mainly of hydrogen bonds (Fig. 2 F and G), similar to plant CBSX1 and CBSX2 (1, 2). In contrast, the second interface is formed between the CBS pairs and the CP12 domains of adjacent subunits, showing a prominent shape complementarity (Fig. 2H). The area of this interface is smaller compared with the CBS:CBS interface but involves several salt bridges, predominantly formed between the first α-helix (α5) of the CP12 domain and the opposite CBS pair (Fig. 2 I and J). Despite the different tilt of the CP12 domain toward the CBS pair between both native structures (SI Appendix, Fig. S2), an almost identical set of salt bridges is established (Fig. 2 I and J). Even though the three CBS–CP12 variants featured in the various CBS–CP12/TrxA gene cluster types vary in length (Fig. 1), 11 of the 16 identified residues involved in the CBS–CP12:CBS–CP12 interface are conserved (SI Appendix, Fig. S3).
To study the role of the CP12 domain in the oligomerization of CBS–CP12 in more detail, we constructed two proteins truncating either the CBS pair (CP12del, residues 134 to 208) or the CP12 domain (CBSdel, residues 1 to 133) (SI Appendix, Table S1) and used analytical size-exclusion chromatography (SEC) to determine their oligomeric status. As expected, CBS–CP12 eluted as a hexameric protein, while CP12del and CBSdel did not form oligomers larger than dimers (SI Appendix, Fig. S4 A and B). Additional verification of the oligomerization of CBS–CP12 was obtained by native mass spectrometry (MS). CBS–CP12 appeared with an average mass of 138.9 kDa, corresponding to a hexamer, and no other oligomeric forms were detected (SI Appendix, Fig. S4F).
CBS–CP12 Has Two Distinct Binding Sites for AMP.
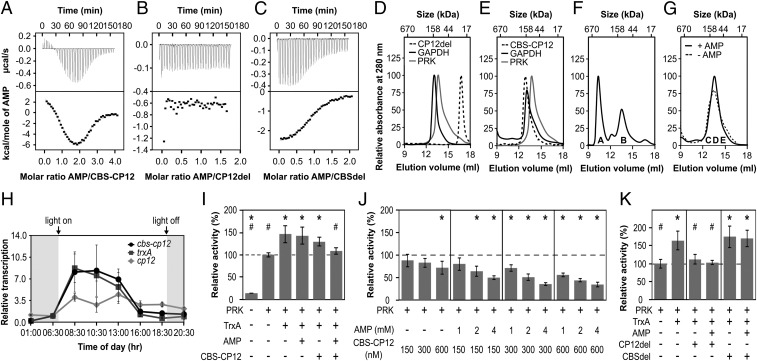
CBS domains are known to bind a wide range of ligands (11). Using isothermal titration calorimetry (ITC), we tested the interaction of CBS–CP12 with several small ligands and found that it exhibits a high specificity for AMP and does not bind ADP, ATP, or cAMP (Fig. 3A and SI Appendix, Fig. S4 H–J). Each CBS–CP12 monomer has two nonidentical binding sites for AMP with different thermodynamic parameters (Fig. 3A and SI Appendix, Table S3). The best fit corresponds to a two-independent site-binding model. The binding event at site 1 is endothermic with a binding affinity of 0.41 µM, while the binding event at site 2 is exothermic and shows a four times lower binding affinity for AMP. In both cases, a stoichiometry of one AMP per binding site was observed, amounting to two molecules of AMP bound per CBS–CP12 monomer and hence 12 AMP molecules bound per CBS–CP12 hexamer. Analytical SEC indicated that AMP does not affect the oligomerization of CBS–CP12 (SI Appendix, Fig. S4E). Native MS revealed that in the presence of AMP the vast majority of CBS–CP12 appeared as a hexamer and only a small amount as a dimer, with a mass indicating the saturated binding of four ligands (SI Appendix, Fig. S4G).
Fig. 3.
Biochemical properties and gene expression of CBS–CP12. (A–C) ITC titration of (A) 400 µM AMP into 20 µM CBS–CP12, (B) 800 µM AMP into 100 µM CP12del, and (C) 700 µM AMP into 70 µM CBSdel. The black lines (Bottom) are the best fit to a two-site model (A) or one-site model (C). Thermodynamic data of three technical replicates are provided in SI Appendix, Table S3. (D and E) Individual S200 SEC elution profiles of (D) CP12del, GAPDH, and PRK and (E) CBS–CP12, PRK, and GAPDH. CP12del elutes at 22.5 ± 0.5 kDa, CBS–CP12 as a hexamer at 143.5 ± 13.6 kDa, PRK at 99.2 ± 7.5 kDa (theoretical monomer size 38 kDa), and GAPDH at 139.2 ± 3.7 kDa (theoretical monomer size 37 kDa) (mean ± SD, n ≥ 2). (F) SEC elution profile of an equimolar mixture (30 µM, subunit base) of GAPDH, PRK, and CP12del with 25 mM DTTox and 0.5 mM NAD. The ternary GAPDH–CP12–PRK complex elutes at ∼569 kDa (peak A). Peak B contains GAPDH and PRK not bound by CP12 (n = 1). (G) SEC elution profile of an equimolar mixture (30 µM, subunit base) of GAPDH, PRK, and CBS–CP12 with 25 mM DTTox, 0.5 mM NAD, ±0.5 mM AMP. GAPDH, PRK, and CBS-CP12 coelute in fraction C-E. (n = 1). (F and G) The denaturing SDS-PAGE of peak A and B and fractions C-E is provided in SI Appendix, Fig. S4K. (H) Diurnal rhythm of the relative transcription of cbs–cp12, trxA, and cp12 in M. aeruginosa (mean ± SD, n = 3). The experiment was repeated twice with consistent results (SI Appendix, Table S4). (I) Relative activity of 300 nM PRK upon addition of 600 nM TrxA, 2 mM AMP, and 300 nM CBS–CP12. (J) Relative activity of 300 nM PRK upon addition of different concentrations of CBS–CP12 and AMP. (K) Relative activity of 300 nM PRK upon addition of 600 nM TrxA, 2 mM AMP, 300 nM CP12del, and 300 nM CBSdel. Data in I–K are mean ± SD, n ≥ 5; *P ≤ 0.01 compared with 300 nM PRK, #P ≤ 0.01 compared with 300 nM PRK + 600 nM TrxA, Student’s t test.
The CP12 domain CP12del did not show any interaction with AMP, while CBSdel was clearly able to bind AMP but had only one binding site (Fig. 3 B and C). CBSdel binds AMP in an exothermic reaction, similar to the reaction observed at site 2 of CBS–CP12. The binding affinity of CBSdel for AMP is about seven times lower compared with site 2, and 32 times lower for site 1 of CBS–CP12 (SI Appendix, Table S3). Contrary to CBS–CP12 only one AMP is bound per CBSdel monomer, indicating that two CBS domains form one AMP-binding site, as shown for CBSX2 (26).
CBS–CP12 Does Not Replace Canonical CP12.
Using analytical SEC and CP12del as a surrogate for canonical CP12, we investigated the ability of CBS–CP12 to bind recombinant GAPDH (IPF_4508) and PRK (IPF_5236) of M. aeruginosa (SI Appendix, Table S1). The elution peaks of both enzymes overlap with CBS–CP12 but are separated from CP12del (Fig. 3 D and E). Under oxidizing conditions and the presence of NAD, the incubation of CP12del with GAPDH and PRK led to the emergence of a peak with an estimated mass of 569 kDa (Fig. 3F). Denaturing SDS/PAGE confirmed that the majority of GAPDH and PRK formed a ternary complex with CP12del (SI Appendix, Fig. S4K). Under the same conditions, and also with AMP, CBS–CP12 coeluted with GAPDH and PRK but did not form a ternary complex (Fig. 3G and SI Appendix, Fig. S4K). There was also no evidence from pull-down experiments for complex formation of CBS–CP12 with any other protein of M. aeruginosa protein extract (SI Appendix, Fig. S5 A–C) and TrxA under different conditions (SI Appendix, Fig. S5 D–I).
Cbs–cp12 and trxA Are Coexpressed and CBS–CP12 Regulates PRK Activity in Tandem with AMP.
Using RT-PCR experiments, the expression of CBS–CP12 and TrxA genes in comparison with the canonical CP12 gene (IPF_0141) was analyzed in a 12/12-h day/night cycle (Fig. 3H and SI Appendix, Table S4). In agreement with previous data (27), canonical CP12 does not show strong variations in its gene expression. In contrast, the gene expression of CBS–CP12 and TrxA follows the same circadian rhythm and shows a clear light-induced expression, reaching their maximum early in the morning (Fig. 3H). Their gene expression is furthermore strongly induced under high-light stress and slightly repressed under iron-limiting conditions (SI Appendix, Fig. S4L).
It has been demonstrated that plant CBSX1 regulates the activity of a Calvin cycle enzyme through Trx (1). We used the Calvin cycle enzyme and Trx target PRK to examine the effect of CBS–CP12 and AMP on its activity as well as its regulation through TrxA (Fig. 3 I–K). TrxA increased PRK activity by 46% (Fig. 3I), as described previously (28). While AMP and CBS–CP12 alone did not affect this TrxA stimulation of PRK, when combined they almost completely abolished the stimulation of PRK activity by TrxA (Fig. 3I). This negative effect is also observed without TrxA, and depends strongly on the concentration of CBS–CP12 and AMP. Alone, AMP had only negligible effects on PRK (SI Appendix, Fig. S4M), while higher CBS–CP12 concentrations slightly reduced PRK activity (Fig. 3J). Together, however, increasing concentrations of CBS–CP12 and AMP gradually and significantly diminished PRK activity up to 65% (Fig. 3J). This AMP-dependent behavior of CBS–CP12 is not evident for either individual domain; CP12del completely inhibits the stimulation of PRK activity by TrxA, while CBSdel did not have an effect, with or without AMP present (Fig. 3K).
Discussion
The discovery of CBS–CP12 fusion proteins in cyanobacteria (16) has led to speculations about their function in the regulation of metabolism, in particular in relation to the photosynthetic regulator CP12 (29). The present study not only unveils a structurally unique CBS domain protein and the previously elusive structure of the N-terminal part of a CP12 protein but also suggests a more versatile role of CP12 proteins in cyanobacteria beyond its established role in ternary complex formation with the Calvin cycle enzymes GAPDH and PRK.
Multifaceted roles of CP12 proteins in cyanobacteria are supported by the fact that CBS–CP12 genes occur not only in addition to CP12 (16) but also in a variety of gene neighborhoods, often unrelated to photosynthesis (Fig. 1B). In the model strain Nostoc sp. PCC 7120, two CBS–CP12 genes (Fig. 1B) are coexpressed with genes of the hox-type hydrogenase (30) and nif gene cluster (31), respectively. This suggests an involvement of these proteins in the respective metabolic pathway rather than regulation of PRK and GAPDH.
The most prevalent gene neighborhood observed for CBS–CP12 proteins connects the fusion protein with thioredoxin (Fig. 1C) and indicates a close alliance of both proteins. As stand-alone CP12 and CBS proteins are antagonists in the thioredoxin-dependent regulation of GAPDH and PRK in plants, we were intrigued by the question of whether CBS–CP12 would rather act like CP12 or plant CBSX. Our results clearly show that, contrary to CP12 (17–21) and CP12del, CBS–CP12 does not form a ternary complex with GAPDH and PRK (Fig. 3 D–G). However, both CBS–CP12 and CP12del negatively affect the stimulation of PRK by TrxA (Fig. 3 I and K). While this is solely redox-dependent for CP12del, it strongly depends on the concentration of AMP for CBS–CP12 (Fig. 3 J and K). This is in agreement with the observed binding of AMP by CBS–CP12 but not CP12del (Fig. 3 A and B).
The inhibition of PRK via AMP has previously been reported (32). In contrast, PRK of M. aeruginosa is not directly regulated by AMP (SI Appendix, Fig. S4M) but through a CBS–CP12–AMP complex (Fig. 3J) that also operates in combination with TrxA (Fig. 3I). This provides an additional layer in the regulation of photosynthesis, possibly sensing and conveying the cellular energy status through AMP and counteracting the redox regulation of photosynthesis by TrxA when the energy level of the cell is low (SI Appendix, Fig. S6). Notably, CBS–CP12 acts as an AMP-dependent inhibitor, while plant CBSX1 acts as an AMP-dependent activator (1).
CBS–CP12 and TrxA exhibit a light-dependent coexpression in M. aeruginosa (Fig. 3H and SI Appendix, Fig. S4L), unlike canonical CP12, the second CBS–CP12 variant (IPF_3982), and a CBS domain protein (IPF_2158) upstream of CBS–CP12 and TrxA (Fig. 1B) but similar to several photosynthetic Trx target proteins (13–15, 27, 33). The concerted gene expression of CBS–CP12 and TrxA is also evident in the symbiotic strain Acaryochloris marina (34), indicating they form part of an operon. In this context it is interesting to note that CBS–CP12/TrxA gene clusters are predominant in scum-forming strains that are exposed to extreme high-light conditions at the surface of lakes and in benthic mat-forming and marine symbiotic cyanobacteria. Although they inhabit entirely different niches, their unifying feature is the formation of densely populated communities with gradients of light, oxygen, and inorganic nutrients fluctuating in a daily cycle (35–37).
Conservation of the CBS–CP12/TrxA gene cluster, gene expression pattern, and regulation of PRK activity points to an antagonistic interaction of CBS–CP12 and TrxA. It furthermore indicates that CBS–CP12 acts as a redox-dependent, regulatory sensor for intracellular AMP levels, similar to other CBS domains (11). A redox-regulatory role of CBS–CP12 in these cyanobacteria is supported by a recent proteomic study of M. aeruginosa showing an increased accumulation of CBS–CP12 in the redox-sensitive, microcystin-deficient mutant ∆mcyB (38).
The hexamerization of CBS–CP12 is based on genetic fusion of a CBS pair with a CP12 protein (Fig. 1), a process described earlier to lead to homooligomeric proteins (39). As shown for several disordered proteins and domains (40, 41), the CP12 domain is essential for hexamerization (Fig. 2). Individually, but also when incubated together, CP12del and CBSdel do not form oligomers larger than dimers (SI Appendix, Fig. S4 A–C). It should be noted that the CBS–CP12 hexamer is otherwise stable under all conditions tested, that is, native, reduced, with AMP, GAPDH, and PRK, or TrxA (Fig. 3 E and G and SI Appendix, Fig. S4 D, E, and G). The disordered nature of CP12 is reflected by the ∼30 C-terminal residues not visible in either crystal structure (SI Appendix, Fig. S2), while the involvement of the CP12 domain in the CBS–CP12:CBS–CP12 interface (Fig. 2) has stabilized the N-terminal part and enabled its structural characterization. This region of stand-alone CP12 proteins is highly flexible and has eluded all previous attempts to solve its structure using X-ray crystallography (23, 24). Interestingly, a recent study showed that the N-terminal region of CP12 of Chlamydomonas reinhardtii oscillates between a disordered state and a folded, helical structure in its free, oxidized form (42). When the C-terminal region of the algal CP12 is bound to GAPDH, the N-terminal region assumes an extended conformation (42), contrary to plant CP12, where the binding of GAPDH leads to a compaction of the N-terminal region (43). All of these examples illustrate the conformational flexibility within the extended family of intrinsically disordered CP12 proteins of different organisms, either in their free form or caused by the interaction with their respective inter- or intramolecular interaction partner.
We show that the N-terminal part of CP12 forms two long α-helices, as proposed (44), which are connected by a disulfide bridge (Fig. 2C). The AWD_VEEL core sequence, located in α-helix 6, is not involved in the CBS–CP12:CBS–CP12 interface (Fig. 2 H–J). The residues of the CP12 domain responsible for hexamer formation are located in α-helix 5, and are, together with their counterparts in the CBS pair, conserved in some CBS–CP12 variants of strains harboring a CBS–CP12/TrxA gene cluster (SI Appendix, Fig. S3), suggesting that oligomerization may be a common feature of these proteins. Further experiments are required to reveal if CBS–CP12 proteins present in other gene clusters (Fig. 1B) also form high-order oligomers.
As with most high-order assemblies, the CBS–CP12 hexamer probably derived from dimeric proteins, as indicated by the smaller buried surface area and binding energy of the unique CBS–CP12:CBS–CP12 interface compared with the conserved CBS:CBS interface (Fig. 2 E and H) (45, 46). The fusion of CBS with CP12 and hexamerization of CBS–CP12 may be explained by the need to create a protein with a new function (39, 47) while retaining the properties of the individual domains: (i) CBS–CP12 does not substitute CP12 in the formation of the ternary complex with GAPDH and PRK (Fig. 3 D–G). (ii) The CP12 domain is as essential for hexamerization of CBS–CP12 (Fig. 2) as CP12 is for ternary complex formation. (iii) CBS–CP12 binds AMP, similar to CBS domain proteins (1, 11), but has an additional binding site. The deletion of the CP12 domain affects the number of binding sites in CBSdel and also the binding affinity for AMP (Fig. 3 A–C and SI Appendix, Table S3). This suggests that the CP12 domain is involved in AMP binding. (iv) CBS–CP12 inhibits PRK activity in an AMP-dependent manner, unlike CP12del (Fig. 3 I–K). The CP12 domain is, however, essential for the transfer of the AMP signal to the target protein, as CBSdel alone does not have any effect on PRK activity (Fig. 3K). The functional separation of CBS–CP12 and canonical CP12 is likely further enhanced by differential gene expression (Fig. 3H), preventing CP12 from redox-regulating PRK through TrxA in vivo, as observed for CP12del (Fig. 3K).
Based on our results, we propose that cyanobacterial CP12 proteins are not exclusively dedicated to the interaction with GAPDH and PRK, and that CBS–CP12 fusion proteins perform additional functions. Given the widespread distribution among cyanobacteria (16), clarifying the role of CBS–CP12 proteins encoded in other gene clusters and the involvement of the other proteins encoded in the various CBS–CP12/TrxA gene cluster types (Fig. 1) will be of interest. Understanding the principles of the regulation of metabolic processes can aid in the management of harmful blooms, as caused by M. aeruginosa, and provide insights for metabolic engineering of photosynthesis as well as hydrogen production and nitrogen fixation in cyanobacteria.
Materials and Methods
Cloning, expression in E. coli, and purification of proteins using affinity and size-exclusion chromatography, crystallization and structure determination of CBS–CP12, ITC, analytical SEC, native MS, quantitative RT-PCR, PRK and pull-down assays, and bioinformatic studies are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
C.H. thanks H. C. P. Matthijs, D. Passon, and V. Pogenberg for their helpful discussions, input, and advice. The help of S. Staak and A. Weiz is highly appreciated. C.H. also thanks the EMBL for financial and infrastructure support. C.A.K. and F.C. are supported by the US Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021 and the National Science Foundation (IOS 1557324). E.D. is supported by German Research Foundation (DFG) Contract Di910/10-1. C.E. acknowledges the support of the Erasmus scholarship and the EMBL. The FT-ICR MS facility is supported by Biocenter Finland/Biocenter Kuopio and the European Union Regional Fund (Grant A70135). J.J. acknowledges support from the European Union’s Horizon 2020 research and innovation program (Grant Agreement 731077). The crystallographic data were collected at synchrotron beamline P13 operated by the EMBL at the PETRA III storage ring (DESY).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The experimental structure factor amplitudes and atomic coordinates of the derived crystal structural models reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5NMU, 5NPL, and 5NVD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806668115/-/DCSupplemental.
References
- 1.Yoo KS, et al. Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell. 2011;23:3577–3594. doi: 10.1105/tpc.111.089847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeong BC, Park SH, Yoo KS, Shin JS, Song HK. Crystal structure of the single cystathionine β-synthase domain-containing protein CBSX1 from Arabidopsis thaliana. Biochem Biophys Res Commun. 2013;430:265–271. doi: 10.1016/j.bbrc.2012.10.139. [DOI] [PubMed] [Google Scholar]
- 3.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 4.Xiao B, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sintchak MD, et al. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85:921–930. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 6.Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- 7.Meyer S, Savaresi S, Forster IC, Dutzler R. Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5. Nat Struct Mol Biol. 2007;14:60–67. doi: 10.1038/nsmb1188. [DOI] [PubMed] [Google Scholar]
- 8.Biemans-Oldehinkel E, Mahmood NABN, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci USA. 2006;103:10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraus JP, et al. Cystathionine β-synthase mutations in homocystinuria. Hum Mutat. 1999;13:362–375. doi: 10.1002/(SICI)1098-1004(1999)13:5<362::AID-HUMU4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 10.Bowne SJ, et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:559–568. doi: 10.1093/hmg/11.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ereño-Orbea J, Oyenarte I, Martínez-Cruz LA. CBS domains: Ligand binding sites and conformational variability. Arch Biochem Biophys. 2013;540:70–81. doi: 10.1016/j.abb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 13.Marri L, et al. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- 14.Florencio FJ, Pérez-Pérez ME, López-Maury L, Mata-Cabana A, Lindahl M. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth Res. 2006;89:157–171. doi: 10.1007/s11120-006-9093-5. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Pérez ME, Mata-Cabana A, Sánchez-Riego AM, Lindahl M, Florencio FJ. A comprehensive analysis of the peroxiredoxin reduction system in the cyanobacterium Synechocystis sp. strain PCC 6803 reveals that all five peroxiredoxins are thioredoxin dependent. J Bacteriol. 2009;191:7477–7489. doi: 10.1128/JB.00831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley DN, Raines CA, Kerfeld CA. Comparative analysis of 126 cyanobacterial genomes reveals evidence of functional diversity among homologs of the redox-regulated CP12 protein. Plant Physiol. 2013;161:824–835. doi: 10.1104/pp.112.210542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marri L, Trost P, Pupillo P, Sparla F. Reconstitution and properties of the recombinant glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase supramolecular complex of Arabidopsis. Plant Physiol. 2005;139:1433–1443. doi: 10.1104/pp.105.068445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wedel N, Soll J. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc Natl Acad Sci USA. 1998;95:9699–9704. doi: 10.1073/pnas.95.16.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oesterhelt C, et al. Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant Cell Physiol. 2007;48:1359–1373. doi: 10.1093/pcp/pcm108. [DOI] [PubMed] [Google Scholar]
- 21.Graciet E, et al. The small protein CP12: A protein linker for supramolecular complex assembly. Biochemistry. 2003;42:8163–8170. doi: 10.1021/bi034474x. [DOI] [PubMed] [Google Scholar]
- 22.Groben R, et al. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynth Res. 2010;103:183–194. doi: 10.1007/s11120-010-9542-z. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura H, et al. Structure basis for the regulation of glyceraldehyde-3-phosphate dehydrogenase activity via the intrinsically disordered protein CP12. Structure. 2011;19:1846–1854. doi: 10.1016/j.str.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Fermani S, et al. Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J Biol Chem. 2012;287:21372–21383. doi: 10.1074/jbc.M112.350355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Jeong BC, Park SH, Yoo KS, Shin JS, Song HK. Change in single cystathionine β-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding. J Struct Biol. 2013;183:40–46. doi: 10.1016/j.jsb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Straub C, Quillardet P, Vergalli J, de Marsac NT, Humbert JF. A day in the life of Microcystis aeruginosa strain PCC 7806 as revealed by a transcriptomic analysis. PLoS One. 2011;6:e16208. doi: 10.1371/journal.pone.0016208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rault M, Gontero B, Ricard J. Thioredoxin activation of phosphoribulokinase in a chloroplast multi-enzyme complex. Eur J Biochem. 1991;197:791–797. doi: 10.1111/j.1432-1033.1991.tb15973.x. [DOI] [PubMed] [Google Scholar]
- 29.López-Calcagno PE, Howard TP, Raines CA. The CP12 protein family: A thioredoxin-mediated metabolic switch? Front Plant Sci. 2014;5:9. doi: 10.3389/fpls.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sjöholm J, Oliveira P, Lindblad P. Transcription and regulation of the bidirectional hydrogenase in the cyanobacterium Nostoc sp. strain PCC 7120. Appl Environ Microbiol. 2007;73:5435–5446. doi: 10.1128/AEM.00756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehira S, Ohmori M, Sato N. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2003;10:97–113. doi: 10.1093/dnares/10.3.97. [DOI] [PubMed] [Google Scholar]
- 32.Marsden WJN, Codd GA. Purification and molecular and catalytic properties of phosphoribulokinase from the cyanobacterium Chlorogloeopsis fritschii. Microbiology. 1984;130:999–1006. [Google Scholar]
- 33.Muramatsu M, Hihara Y. Acclimation to high-light conditions in cyanobacteria: From gene expression to physiological responses. J Plant Res. 2012;125:11–39. doi: 10.1007/s10265-011-0454-6. [DOI] [PubMed] [Google Scholar]
- 34.Hernández-Prieto MA, Lin Y, Chen M. The complex transcriptional response of Acaryochloris marina to different oxygen levels. G3. 2017;7:517–532. doi: 10.1534/g3.116.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibelings BW, Maberly SC. Photoinhibition and the availability of inorganic carbon restrict photosynthesis by surface blooms of cyanobacteria. Limnol Oceanogr. 1998;43:408–419. [Google Scholar]
- 36.Kuehl M, et al. Microenvironmental ecology of the chlorophyll b-containing symbiotic cyanobacterium Prochloron in the didemnid ascidian Lissoclinum patella. Front Microbiol. 2012;3:1–18. doi: 10.3389/fmicb.2012.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jørgensen BB, Revsbech NP, Blackburn TH, Cohen Y. Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl Environ Microbiol. 1979;38:46–58. doi: 10.1128/aem.38.1.46-58.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zilliges Y, et al. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One. 2011;6:e17615. doi: 10.1371/journal.pone.0017615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 40.Faust O, Bigman L, Friedler A. A role of disordered domains in regulating protein oligomerization and stability. Chem Commun (Camb) 2014;50:10797–10800. doi: 10.1039/c4cc03863k. [DOI] [PubMed] [Google Scholar]
- 41.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Launay H, et al. Cryptic disorder out of disorder: Encounter between conditionally disordered CP12 and glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 2018;430:1218–1234. doi: 10.1016/j.jmb.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Del Giudice A, et al. Unravelling the shape and structural assembly of the photosynthetic GAPDH-CP12-PRK complex from Arabidopsis thaliana by small-angle X-ray scattering analysis. Acta Crystallogr D Biol Crystallogr. 2015;71:2372–2385. doi: 10.1107/S1399004715018520. [DOI] [PubMed] [Google Scholar]
- 44.Gardebien F, Thangudu RR, Gontero B, Offmann B. Construction of a 3D model of CP12, a protein linker. J Mol Graph Model. 2006;25:186–195. doi: 10.1016/j.jmgm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch M. The evolution of multimeric protein assemblages. Mol Biol Evol. 2012;29:1353–1366. doi: 10.1093/molbev/msr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishi H, Hashimoto K, Madej T, Panchenko AR. Evolutionary, physicochemical, and functional mechanisms of protein homooligomerization. Prog Mol Biol Transl Sci. 2013;117:3–24. doi: 10.1016/B978-0-12-386931-9.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.