Significance
Renal oncocytomas are benign kidney tumors with numerous mitochondria. Here, we analyze the mitochondrial (mtDNA) and nuclear genomes of these tumors. Our analysis finds mtDNA mutations in complex I (the first step in mitochondrial respiration) to be early genetic events that likely contribute to tumor formation. Since mtDNA mutations can lead to severe degenerative disorders, the cellular responses allowing renal oncocytoma cells to grow are important to consider. To properly understand authentic gene expression changes in tumors, we found it important to consider the gene expression pattern of the tumor’s cell of origin, the distal nephron. By doing so, we uncover alterations in glutathione synthesis and turnover that likely represent an adaptive metabolic response in renal oncocytoma.
Keywords: oncocytoma, complex I, mtDNA, glutathione, γ-glutamyl cycle
Abstract
Renal oncocytomas are benign tumors characterized by a marked accumulation of mitochondria. We report a combined exome, transcriptome, and metabolome analysis of these tumors. Joint analysis of the nuclear and mitochondrial (mtDNA) genomes reveals loss-of-function mtDNA mutations occurring at high variant allele fractions, consistent with positive selection, in genes encoding complex I as the most frequent genetic events. A subset of these tumors also exhibits chromosome 1 loss and/or cyclin D1 overexpression, suggesting they follow complex I loss. Transcriptome data revealed that many pathways previously reported to be altered in renal oncocytoma were simply differentially expressed in the tumor’s cell of origin, the distal nephron, compared with other nephron segments. Using a heuristic approach to account for cell-of-origin bias we uncovered strong expression alterations in the gamma-glutamyl cycle, including glutathione synthesis (increased GCLC) and glutathione degradation. Moreover, the most striking changes in metabolite profiling were elevations in oxidized and reduced glutathione as well as γ-glutamyl-cysteine and cysteinyl-glycine, dipeptide intermediates in glutathione biosynthesis, and recycling, respectively. Biosynthesis of glutathione appears adaptive as blockade of GCLC impairs viability in cells cultured with a complex I inhibitor. Our data suggest that loss-of-function mutations in complex I are a candidate driver event in renal oncocytoma that is followed by frequent loss of chromosome 1, cyclin D1 overexpression, and adaptive up-regulation of glutathione biosynthesis.
Renal oncocytoma (RO) is a rare and benign tumor, accounting for ∼5% of kidney tumors (1). RO is distinct from renal cell carcinomas (RCCs), which fall into three main histologic subtypes: clear cell (ccRCC), papillary (pRCC), and chromophobe (chRCC) (2–4). Molecular and histopathological observations suggest that ccRCC and pRCC arise from the proximal tubules of the nephron while chRCC and RO are distal in origin (5, 6). Recognition of RO is important as it can be cured surgically.
ROs are composed of large polygonal cells with granular eosinophilic cytoplasm, prominent nucleoli, and occasional nuclear atypia (7). The tumor cells are filled with abundant mitochondria and are termed “oncocytic.” Oncocytic neoplasms occur throughout the body but are most common in the thyroid and kidney (8). In electron micrographs, the numerous mitochondria of these tumors show morphological abnormalities, including increased size and centrally stacked cristae (8). In the kidney, mitochondrial accumulation is not only seen in RO but also in the eosinophilic variant of chRCC (9).
Genetic lesions both in the nuclear and mitochondrial genomes have previously been reported in RO. It has long been appreciated that oncocytic tumors, including RO, bear disruptive mutations in mtDNA genes encoding complex I subunits of the electron transport chain (10, 11). Sequencing of nuclear DNA in RO has revealed a low somatic mutation rate without recurrent nuclear gene mutations, although recurrent chromosome 1 loss and cyclin D1 rearrangement have been reported (12, 13).
In this study, we report the results of profiling DNA, RNA, and metabolites in RO to investigate the molecular and biochemical basis of these tumors. We performed whole exome sequencing (WES) on a cohort (termed ROMGH) of 19 histopathologically proven RO samples with matched normal kidney tissue. We then validated our findings by analyzing off-target mtDNA reads from a WES study by Durinck et al. (12) on a cohort of RO patients (termed ROGEN) in which mtDNA sequence analysis was not performed. We reanalyzed RNA-seq from this latter study taking cell of origin into consideration to spotlight tumor-associated gene expression changes. Finally, we performed mass spectrometry-based metabolomics on 10 additional fresh frozen RO samples from our cancer center to identify differentially abundant metabolites. Our integrative analysis reveals what appear to be the earliest genetic and biochemical changes in this benign tumor.
Nuclear Genomes in ROMGH
To search for somatic alterations in nuclear and mitochondrial genomes, we performed WES on 19 high purity formalin-fixed paraffin-embedded (FFPE) RO samples and adjacent normal kidney (ROMGH). Mean nuclear DNA coverage was 171× in normals and 212× in tumors with a relatively low nonsynonymous mutation rate of ∼15 mutations per sample (Dataset S1). MutSig analysis did not reveal any significantly mutated nuclear genes.
We then evaluated tumors for karyotypic alterations in the nuclear DNA. ROs were largely diploid with hemizygous loss of chromosome 1 or 1p in 12/19 patients (Fig. 1A and SI Appendix, Fig. S1A) and no recurrent focal copy number events. As previous studies detected cyclin D1 rearrangements leading to cyclin D1 overexpression (13), we assessed cyclin D1 via immunohistochemistry (IHC) (14), finding strong expression in 3/19, moderate expression in 4/19, and absent expression in 12/19 samples (SI Appendix, Fig. S1B and Dataset S1). Chromosome 1 was lost in 3/7 samples showing cyclin D1 overexpression by IHC (Fig. 1A). Together, 63% of RO tumors show chromosome 1 loss and 37% show cyclin D1 overexpression.
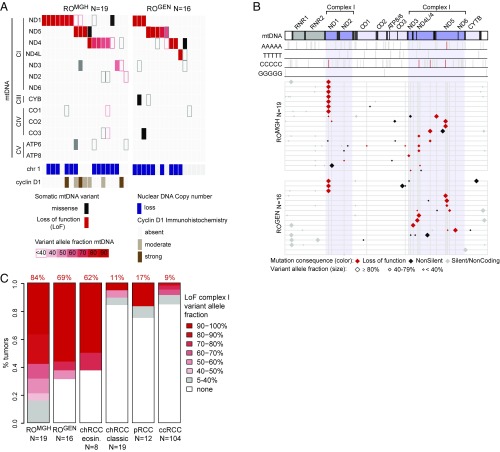
Fig. 1.
Analysis of mitochondrial and nuclear genomes in RO. (A) Somatic mutations in mtDNA for RO samples (columns) across two cohorts (ROMGH and ROGEN). Somatic protein-altering mutations in the 13 mtDNA genes are shown with color (red, LoF; black, missense) and shade to indicate VAF (see legend). OXPHOS complexes are indicated (CI/III/IV/V: complex I/III/IV/V, respectively). Chromosome 1 and cyclin D1 status are shown (see legend). (B) Position of mutations on an mtDNA molecule from position 0 (Left) to 16,569 (Right). Top rows show positions of 5 base-pair homopolymeric tracts (gray bars); red bars highlight sites of recurrent mutation (≥2 samples). (Bottom) Shows all somatic mutations as diamonds (see legend) in each RO sample (horizontal lines) from both cohorts (ROMGH and ROGEN) by position. (C) Stacked barplots show the percent of kidney tumors with complex I LoF mutations at ≥40% VAF. LoF, loss-of-function.
Mitochondrial Genomes in ROMGH
We next analyzed off-target mtDNA reads from WES to identify somatic mtDNA variants in ROMGH (Methods). We obtained 61× mean coverage of the mtDNA in normals and 114× coverage in tumors (Dataset S1). Consistent with a prior pan-cancer analysis of mtDNA across 31 different types of cancer, we observed a G > A and T > C bias on the light strand, suggestive of a replicative mutational process (Dataset S2) (15).
We find that 16/19 tumors harbor protein-truncating mtDNA mutations with variant allele fraction (VAF) ≥ 0.4 (Fig. 1A). Strikingly, these all occur in complex I genes (MT-ND1, MT-ND5, MT-ND4, MT-ND3), with no loss-of-function (LoF) mutations in complexes III, IV, or V at this VAF (Fig. 1A). Importantly, these LoF mutations had high VAF, a surrogate for heteroplasmy (fraction of mutated mtDNA molecules). These LoF mutations were typically short indels occurring at specific mtDNA sites (Fig. 1B). Of the 16 LoF mutations with VAF ≥0.4, one was a nonsense mutation (m.10126; MT-ND3:pTrp23) and 15 were short indels (1–2 base pairs) typically within 4–6 base-pair homopolymeric DNA tracts often resulting in early frameshift mutations (Dataset S3). Many mutations were recurrent, including seven mutations at m.3565; MT-ND1:p.Thr87, three mutations at m.10946; MT-ND4:p.Thr63, and two mutations at m.11866; MT-ND4:p.Thr370 (Fig. 1B and SI Appendix, Fig. S2). We note that homopolymeric tracts occur in all mtDNA genes (Fig. 1B), so the recurrent mutations are not simply due to increased prevalence of such tracts within complex I subunits.
The remaining 3/19 samples also likely harbor deleterious complex I mutations (Dataset S3). One tumor had a complex I LoF event at 0.13 VAF in a sample with only 20% tumor purity by pathology estimate, consistent with likely high heteroplasmy in the tumor (SI Appendix, Fig. S3). The remaining 2/19 samples harbor missense mutations in complex I subunits: MT-ND4.pE123K at 0.36 VAF and MT-ND1.pW179M at 0.89 VAF. Both missense mutations were at highly conserved residues (identical in at least 34/44 aligned vertebrates) and based on structural modeling of bacterial complex I (16) are predicted to impair complex I proton transport (MT-ND4.pE123K) or disrupt an accessory subunit interaction site (MT-ND1.pW179M) (17).
Mitochondrial Genomes in ROGEN
The above analysis of ROMGH revealed that 19/19 samples had evidence for selection of disabling complex I mutations, hence making complex I loss a universal feature of these tumors. A large subset, but not all, of these samples also exhibited loss of chromosome 1 and cyclin D1 overexpression. We validated our finding of complex I mtDNA mutations in a previously published RO cohort (ROGEN) in which the mtDNA was not evaluated (12). Analysis of 16 RO samples with sufficient mtDNA coverage revealed 69% of samples showed recurrent, high VAF LoF events in MT-ND1, MT-ND4, and MT-ND5 (Fig. 1 and Dataset S3). In three instances, frameshifts even occurred at the same mtDNA position in ROMGH and ROGEN (Fig. 1B and SI Appendix, Fig. S2). Importantly, in ROGEN, chromosome 1 loss occurred in 9/16 samples, all of which had mutations in complex I (Fig. 1A). Together, these two cohorts suggest that complex I mtDNA mutations may be inciting driver events in RO, with secondary recurrent chromosome 1 loss.
Mitochondrial Genomes Across RCC
We next asked if mtDNA mutations were specific to RO or if they occurred in RCC variants. We analyzed WES off-target mtDNA reads from 8 eosinophilic chRCC, 19 classic chRCC, and 12 pRCC from Durinck et al. as well as 104 ccRCC from the TCGA (2) (Dataset S2). Specifically, we analyzed the seven complex I genes compared with the six other oxidative phosphorylation (OXPHOS) genes for LoF mutations. This mtDNA analysis revealed a large fraction of complex I LoF mutations in ROMGH (84%), ROGEN (69%), and eosinophilic chRCC (62%)—precisely the renal tumors known to exhibit mitochondrial accumulation (Fig. 1C and Dataset S2). These complex I LoF mutations are likely drivers of pathology because they (i) occur preferentially in complex I compared with other OXPHOS complexes (SI Appendix, Fig. S4A), and (ii) have significantly higher VAF compared with silent or noncoding variants (SI Appendix, Fig. S4B). This contrasts with a recent pan-cancer analysis across 31 different cancers in which LoF mtDNA mutations exhibited lower VAF compared with silent mutations (15). Thus, the analysis of five kidney tumors as well as 1,664 pan-cancer samples shows that RO and eosinophilic chRCC are unique for their high VAF, LoF complex I mtDNA mutations.
As functional alterations in mtDNA can also result from copy number variation, we used quantitative PCR to measure mtDNA copy number in our ROMGH cohort. We found a significant increase in mtDNA copy number in RO (mean ∼ 2,700 copies per cell) compared with normal kidney (mean ∼ 1,900 copies per cell; SI Appendix, Fig. S5A). It was notable the copy number increase was less than twofold considering the numerous mitochondria in these tumors. We found the fraction of WES reads aligning to mtDNA correlated strongly with mtDNA copy number estimated by quantitative PCR (SI Appendix, Fig. S5B). Thus, we extended our analysis using WES reads to other RCC variants, finding no significant differences between tumors and normals except in ccRCC where there was a striking decrease in mtDNA copies (SI Appendix, Fig. S5C). We note that such analyses across RCC subtypes will be influenced by differences in tumor purity, nuclear copy number, and importantly mtDNA copy number in the cell of origin across tumors.
Transcriptional Profiles in RO
We next sought to characterize gene expression changes in RO to relate them back to primary genetic lesions in mtDNA complex I genes. Previous groups have reported significant up-regulation of mitochondrial biogenesis genes, including PPARGC1α and OXPHOS subunits, as well as marked elevation in AMP-activated protein kinase (AMPK) subunits (PRKAB2, PRKAG2) and its targets (ACACA, ACACB) (12, 13). Since AMPK activation may result from complex I dysfunction, we measured the levels of AMPKα Thr172 phosphorylation, a modification required for AMPK activation (18), in tumors and normal kidney.
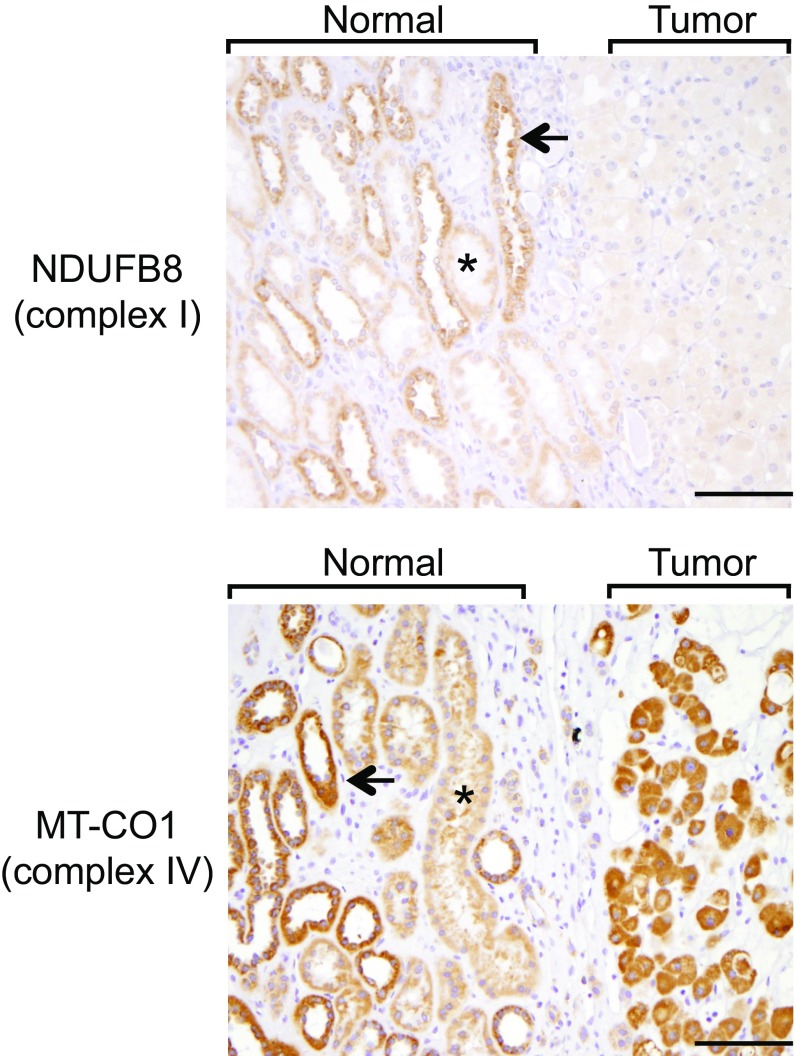
Although we saw strong AMPK activation in tumors as previously reported (SI Appendix, Fig. S6), on closer examination, the signal was increased compared with normal kidney proximal tubules but comparable to normal distal tubules (SI Appendix, Fig. S6 and Supplementary Materials and Methods for tubule definitions). Since RO derives from the distal nephron (6), we concluded the apparent AMPK phosphorylation enrichment was not due to a tumor versus normal distal tubule difference, but rather a difference in distal tubules compared with the other cortical nephron segments (estimated by volume in rat to be roughly 68% proximal tubules, 15% distal tubules, 8% glomeruli and capillaries, and 9% interstitial and extracellular space in the subcapsular cortex; ref. 19). The increased staining in distal tubules relative to proximal tubules was also evident for the OXPHOS subunits NDUFB8 (a subunit of complex I) and MT-CO1 (a subunit of complex IV), with subsequent attenuation of only the complex I signal in tumors consistent with their pattern of mtDNA mutation (Fig. 2). These observations raise the possibility that prior reports (12, 13) of differential gene expression in RO may have suffered from extensive cell-of-origin bias.
Fig. 2.
IHC analysis of OXPHOS in normal kidney and RO. Representative images of a tumor with adjacent normal kidney in the same field (demarcated by brackets) from ROMGH cohort (n = 19 normals and n = 19 tumors) with indicated primary antibodies for complex I and complex IV. A single representative proximal tubule (*) and distal tubule (arrow) are indicated in each image (not all proximal and distal tubules in each image are labeled). (Magnification: 200×.) (Scale bars: 50 μm).
To investigate if a pervasive cell-of-origin bias besets prior RO gene expression analysis, we developed a heuristic approach to identify transcripts genuinely altered in tumors. Our approach is conceptually simple and is conservative: ROs arise from the distal nephron, whereas the normal cortex consists of mostly proximal tubules (estimated to represent ∼60–90% of the cortex; refs. 19 and 20). To spotlight genes whose expression is genuinely elevated or reduced in tumors compared with their cell of origin, we aimed to identify genes with higher expression in tumors versus normal kidney, yet not elevated in distal tubules compared with proximal segments. Although it has been suggested that RO arises from the intercalated cells of the collecting duct, a recent study found high correlation of gene expression profiles between RO and numerous distal nephron segments (6, 13). Thus, we favored a more conservative comparison of gene expression between proximal and distal nephron segments to account for potential bias while remaining inclusive of distal nephron sites from which RO may arise.
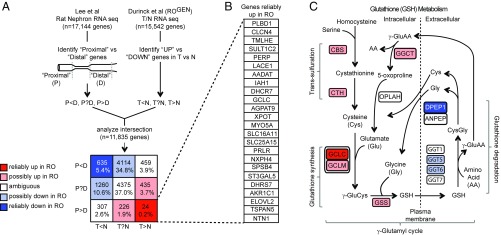
Using a previously published RNA-seq atlas of the rat nephron (21), we used a fivefold expression difference to score each gene as increased in proximal versus distal tubules (P>D), increased in distal versus proximal tubules (P<D), or ambiguous (P?D; Fig. 3A and Methods). We concurrently analyzed RNA-seq data from 12 tumor/normal pairs from ROGEN to identify transcripts that are differentially abundant between tumor and normal (twofold difference at P < 1e-5). In this way we could classify each transcript as increased in tumor versus normal (T>N), decreased in tumor versus normal (T<N), or ambiguous (T?N; Methods). Our strategy allowed us to parse 11,835 transcripts represented in both studies into one of nine disjoint categories (Fig. 3A). Shown in red are genes that we infer to be reliably up-regulated in RO in a way not obviously related to cell of origin; shown in blue are genes we infer are reliably reduced in RO and not obviously biased by cell of origin. We substantiated the validity of this heuristic method by revisiting known nephron markers as well as genes argued to be differentially expressed in RO (SI Appendix, Figs. S7 and S8 and Supplementary Text).
Fig. 3.
Analysis of RO transcriptional profiles with respect to cell of origin. (A) Heuristic method to spotlight genes reliably up or down in RO relative to normal distal tubules using two publicly available datasets. Transcripts from the rat nephron RNA-seq atlas were scored as distal (P<D), ambiguous (P?D), or proximal (P>D). Transcripts from RO versus normal kidney RNA-seq were scored as down in tumor (T<N), ambiguous (T?N), or up in tumor (T>N). Intersecting these datasets yields n = 11,835 transcripts that are common to both datasets and can be organized on a 3 × 3 grid. See legend for interpretation of each box by color. (B) List of 24 genes reliably up (T>N, P>D) in RO. (C) Schematic for glutathione metabolism with transsulfuration, glutathione synthesis, glutathione degradation, and the γ-glutamyl cycle. The γ-glutamyl cycle includes all steps except for the transsulfuration reactions. Each gene symbol is color coded by the scheme in A.
What then are genes whose expression is reliably altered in RO compared with normal distal tubules? We focused our attention on the 24 dark red (“reliably up”) and 635 dark blue (“reliably down”) genes whose expression is in the opposite direction to what would be predicted based on cell of origin (Fig. 3A and Dataset S4). We observed reliable up-regulation of the rate-limiting enzyme in glutathione biosynthesis, GCLC (Fig. 3B) (22). This was noteworthy because glutathione was the most up-regulated metabolite detected by metabolome profiling of RO, a finding confirmed by IHC (described below).
Closer inspection revealed up-regulation of the entire transsulfuration and glutathione synthesis programs (CBS, CTH, GCLC, GCLM, GSS) even after cell of origin was considered (Fig. 3C). Moreover, there were distinct changes in the glutathione degradation phase of the γ-glutamyl cycle that would favor glutathione accumulation. The γ-glutamyl transferase genes involved in the first step of glutathione degradation showed preferential expression in the proximal tubules (GGT1), down-regulation in tumors (GGT5, GGT6), or ambiguous expression (GGT7) (Fig. 3C). The dipeptidase, DPEP1, involved in the second step of glutathione degradation was down-regulated in tumors despite being a distal tubule gene (23). Furthermore, there was up-regulation of glutathione exporters (ABCC4, CFTR) and the SLC7A11 subunit of the xCT cystine-glutamate antiporter (SI Appendix, Fig. S9). The coordinated up-regulation of intracellular glutathione synthesis alongside the down-regulation of extracellular glutathione degradation demarcates a change in the γ-glutamyl cycle expected to boost glutathione levels in tumors.
Metabolic Profiles in RO
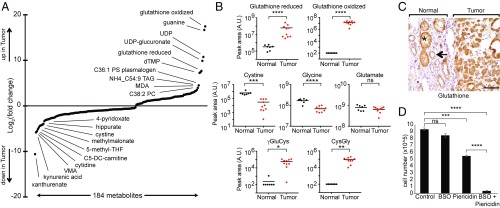
To complement the DNA and RNA analysis, we performed metabolic profiling on an additional cohort of fresh frozen RO (n = 10) and normal kidney (n = 7) samples (Dataset S5), recognizing the results will certainly be impacted by cell-of-origin effects. We used four liquid chromatography-tandem mass spectrometry (LC-MS) methods to interrogate a total of 592 metabolites of confirmed identity, of which 554 were detected in at least 20% of our samples (Dataset S5). Of these 554 metabolites, 184 were differentially abundant between tumors and normals at a nominal P value <0.05 (Dataset S5). These 184 metabolites (75 elevated and 109 reduced in tumors) are shown in Fig. 4A and listed in Dataset S5.
Fig. 4.
Metabolic profiles of RO. (A) Distribution of fold change of 184 metabolites with significant differential abundance (nominal P value <0.05) in our second RO cohort of fresh frozen tumors (n = 10) versus normal kidney (n = 7). The 10 most elevated and reduced metabolites in tumors are labeled. (B) Relative abundance of metabolites related to glutathione metabolism are shown for normals and tumors. y axis shows peak area in arbitrary units (A.U.) and horizontal bars indicate mean. (C) Representative glutathione IHC sample at the interface of tumor with normal kidney (indicated by brackets) from our ROMGH cohort (n = 19 normal kidney; n = 19 tumors). A single representative proximal tubule (*) and distal tubule (arrow) are indicated. (Magnification: 200×.) (Scale bar: 50 μm.) (D) Cell growth assay in HEK293T cells treated with a vehicle control, 100 μM BSO, 1 μM piericidin, or BSO plus piericidin. All conditions were counted in triplicate wells, and the assay was repeated in five independent experiments with a representative experiment shown. Mean cell number is shown with SEM. P values in B and D are *P < 0.01, **P < 0.001, ***P < 5e-4, ****P < 1e-5 by two-tailed Student’s t test.
From the distribution of fold change, it was clear that metabolites related to glutathione were among the most altered (Fig. 4A). Both oxidized and reduced glutathione were strongly elevated in tumors relative to normals (Fig. 4B). Reduced glutathione showed a striking 209-fold increase and oxidized glutathione, which was not detected in normals, was reproducibly detected at high levels in tumors. The glutathione result, which is consistent with our RNA analysis (Fig. 3C), is unlikely a cell-of-origin artifact since glutathione levels are actually higher in the proximal compared with distal tubules (24). Using IHC, we validated in our ROMGH cohort that glutathione levels are increased in human proximal relative to distal tubules and that tumors show increased glutathione staining compared with distal tubules (Fig. 4C). Hence, abundant glutathione levels are a genuinely acquired feature of RO.
This marked and reliable up-regulation of glutathione prompted us to examine other metabolites related to glutathione metabolism (Fig. 4B). Cystine, a precursor for cysteine, which is the rate-limiting substrate in glutathione synthesis (22), was one of the most decreased metabolites with an ∼18-fold reduction in tumors (Fig. 4 A and B). Taurine, a byproduct of cysteine degradation, was decreased in tumors, suggesting the drop in cystine was more likely due to utilization rather than degradation. Glycine, another metabolite involved in glutathione biosynthesis, was also significantly lower in tumors (Fig. 4B).
Based on the transcriptional changes in glutathione synthesis and degradation (increased GCLC and decreased DPEP1; Fig. 3C), we predicted an elevation in the dipeptides γ-glutamyl-cysteine (γGluCys, the product of GCLC) and cysteinyl-glycine (CysGly, a substrate of DPEP1). We interrogated our full scan mass spectra using retention time standards to evaluate levels of γGluCys and CysGly. As predicted by gene expression, we observed a marked increase in both dipeptides in tumors (Fig. 4B), consistent with our transcriptional profile and further highlighting the robust change in the γ-glutamyl cycle in tumors.
Finally, we sought to determine whether increased glutathione synthesis is related to loss of mitochondrial complex I activity. Given that there are no available RO cell lines, we used human embryonic kidney HEK293T cells as an experimental model system. We treated HEK293T cells either with buthionine sulfoximine (BSO; an inhibitor of GCLC) or piericidin (an inhibitor of complex I), or with the two drugs in combination. Cells treated with either drug alone experienced no or mildly impaired cell growth, respectively (Fig. 4D). However, cells treated with both BSO and piericidin showed a massive decrease in cell number (Fig. 4D). The synergism between BSO and piericidin argues that flux through the glutathione biosynthesis pathway is required in the face of complex I dysfunction and that the increased glutathione in RO represents a metabolic profile adaptive to loss of complex I.
Discussion
Collectively, combined DNA, RNA, and metabolite profiling of RO reveals recurrent complex I loss to be an early and likely driving event that is accompanied by frequent loss of chromosome 1, cyclin D1 overexpression, and increased glutathione.
Our work points to complex I loss being the earliest detected genetic alteration that subsequently undergoes positive selection, a feature of a driver event. Supporting this claim are the high prevalence across two independent cohorts of recurrent, high VAF complex I LoF mutations. The high VAF contrasts a pan-cancer analysis showing selection against deleterious mtDNA mutations (15) and argues for positive selection in RO similar to the tissue-specific enrichment of protein-altering mtDNA variants in a more recent pan-cancer analysis (25). The high VAF mutations are unlikely to occur by simple drift, as we do not see such mtDNA mutations in genes encoding subunits of complexes III–V. However, we cannot exclude that disruptive mutations in these downstream complexes may be incompatible with cell viability, while complex I loss may be tolerated as it sits at a branch point for entry into the electron transport chain. Finally, although the disruptive complex I mutations described here have features of candidate driver events, the completely benign nature of RO suggests that these complex I events correlate with a notable lack of aggressive tumor behavior.
The recurrent chromosome 1 loss and cyclin D1 overexpression appear to be later genetic events that may complement complex I loss. Our assertion is supported by the fact that while all 19/19 ROMGH samples harbor complex I mutations, only a subset harbor chromosome 1 and cyclin D1 changes. Moreover, in ROGEN, all samples with chromosome 1 loss occur in complex I mutant samples. In Drosophila, complex I dysfunction leads to a G1-S cell-cycle checkpoint as part of a retrograde response triggered by increased reactive oxygen species (26). Forced expression of cyclin D1 could provide a mechanism to overcome this mitochondrial cell-cycle checkpoint. Loss of chromosome 1 is a frequent event in cancer that is thought to indicate the presence of an important tumor suppressor in this region (27). Therefore, we speculate that alterations in either chromosome 1 or cyclin D1 help to further enable tumor formation following complex I loss.
Collectively, our work suggests that complex I—whose classical role is in energy metabolism—can act like a tumor suppressor (10, 28). Consistent with this notion, cellular models with impaired complex I show potentiation of tumor growth (29) and diminished apoptosis (30). A recent study identified the mitochondrial protein LACTB as a tumor suppressor (31), which is notable since phylogenetic profiling and biochemical studies from our laboratory showed that LACTB is required for complex I activity (32). Although the presence of recurrent inactivating mutations at the same mtDNA position is unusual for a tumor suppressor, such homopolymeric hotspots for indels have been reported in cancer (33). Moreover, homopolymeric frameshifts allow for rapid gene inactivation and adaptation in bacteria, raising the possibility that a similar process occurs in the mtDNA of RO (34). Paradoxically, while the inactivating LoF complex I mutations reported here are enriched in RO, inherited mtDNA missense mutations in complex I can lead to rare, degenerative respiratory chain disorders (35). Why germ-line complex I loss leads to encephalomyopathy while its tissue specific somatic loss is observed in cancer remains an important and unsolved problem.
The current study, employing RNA-seq and metabolomics, demonstrates marked differential expression for genes and metabolites involved in the γ-glutamyl cycle. By accounting for cell-of-origin effects, we uncovered marked up-regulation of transcripts involved in glutathione synthesis, including the rate-limiting enzyme GCLC, and down-regulation, or maintained absence of expression, of genes involved in glutathione degradation. These gene expression alterations create an imbalance in the γ-glutamyl cycle that would favor glutathione production. This assertion is supported by our metabolomics revealing a striking increase in glutathione, a finding corroborated by an independent report showing a similar rise in glutathione in RO that was published since the submission of this manuscript (36). We utilized IHC to validate that the increased glutathione is not due to a cell-of-origin effect, hence confirming that glutathione metabolism is authentically rewired in RO.
It is notable that the mammalian retrograde response to damaged mitochondria is characterized by increased transsulfuration, redirecting homocysteine toward glutathione synthesis (37). The altered flux to glutathione as part of this retrograde response fits with our finding that inhibition of glutathione biosynthesis causes a pronounced growth defect when complex I is disrupted and suggests that this response is likely adaptive. Why increased biosynthesis of glutathione is adaptive in the face of complex I loss is not known. One possibility is that flux through the pathway is critical, while another possibility is that the biosynthetic products aid in defending against oxidative stress (22, 26). Emerging evidence points toward oncogenic pathways, such as activated PI(3)K/AKT, up-regulating glutathione to cope with oxidative stress and confer a growth advantage (38). The increased γGluCys and CysGly highlight a prominent change in the γ-glutamyl cycle—a cycle coordinating the intracellular and extracellular recycling of glutathione and its constituent amino acids (39). While the elevated CysGly levels suggest impaired glutathione degradation, this dipeptide may also provide a source of cysteine, the rate-limiting substrate for glutathione, in tumors (40). Thus, loss of complex I may create a metabolic response, punctuated by a dramatic increase in glutathione, that supports cell growth analogous to TCA cycle mutant tumors (41).
Our study underscores the importance of accounting for cell of origin in molecular analyses of renal tumors. Our current heuristic approach is simple and conservative; however, there are important limitations such as the inclusive grouping of all distal nephron sites together when RO may derive from a particular distal segment. Although new computational methods are now available for deconvoluting gene expression profiles from cellular mixtures, these methods make assumptions about the linearity of gene expression and rely on the fidelity of reference expression profiles (42). To our knowledge, an RNA-seq atlas of human microdissected kidney and matched whole kidney is currently not available. In the future, as atlases of single human cells become richer and the true cell of origin of RO is experimentally verified, it may be possible to perform more principled gene expression analysis to expose additional molecular changes authentic to these unusual tumors.
Materials and Methods
We provide a brief description of methods below. Full details are available in SI Appendix, Supplementary Materials and Methods.
Sample Acquisition.
Specimens were obtained with Partners Human Research Committee institutional review board (IRB) approval (protocol no. 2011P000023) of RO and matched normal kidney from patients undergoing surgical resection. DNA for sequencing was isolated from n = 19 FFPE RO samples with matched normal kidney (ROMGH). A second cohort of fresh frozen specimens of RO with matched normal kidney whenever possible was also obtained under the same IRB protocol. All samples from both cohorts have been deidentified.
Whole Exome Sequencing and Analysis.
Exomes were sequenced at the Broad Institute using Illumina SureSelect with paired 75 base-pair reads. A BAM file was produced using the Picard pipeline (broadinstitute.github.io/picard), which aligns tumor and normal sequences to the hg19 human genome build from raw Illumina reads using the BWA aligner. BAM files were uploaded into the Firehose pipeline (archive.broadinstitute.org/cancer/cga/Firehose), which manages input and output files to be executed by GenePattern. Artifacts introduced by DNA oxidation during sequencing or from FFPE were computationally removed using a filter-based method (43). MuTect (version 1.1.6) was applied to identify somatic single-nucleotide variants (44). Arm-level somatic copy number alterations were analyzed by Allelic CapSeg, an updated iteration of HapSeg.
Analysis of mtDNA from Exomes.
All WES reads mapping to GRCh37 chromosome MT or 1,049 nuclear sequences of mitochondrial origin (NUMTs) (45) were realigned to mtDNA revised Cambridge Reference Sequence (NC_012920 equivalent to GRCh37 chromosome MT) using GATK version 3.3 (46), BWA version 0.7.10 (47), and Picard Tools version 1.119 (broadinstitute.github.io/picard/). Heteroplasmic variants were detected via the GATK HaplotypeCaller assuming a mixture of 100 chromosomes, which estimated VAF, equivalent to heteroplasmy in normal samples and in 100% pure tumor samples.
mtDNA Copy Number Analysis.
Tumor and normal kidney DNA were analyzed by multiplex TaqMan real-time quantitative PCR per protocol (37). WES data were also used to calculate mtDNA copy number by estimating the ratio of reads aligning to the mtDNA and to the entire genome using samtools idxstats (48).
Tumor/Normal RNA-Seq Analysis.
Paired-end RNA-seq fastq files for RO from a previously published study were obtained with permission from the European Genome-phenome Archive (12). Only the 12 pairs of matched tumor/normal samples for which the mtDNA sequence was also analyzed were studied.
Cell-of-Origin Bias Correction.
Genes were defined as being up (T>N) or down (T<N) in tumors if there was at least a twofold difference in expression with a Benjamini–Hochberg adjusted P value <1.0 × 10−5. All other genes were classified as having an unclear difference between tumors and normals (T?N). To account for cell-of-origin effects, an atlas of microdissected rat nephron segments from Lee et al. (21) was analyzed. Proximal tubules were defined as S1 proximal tubule (S1), S2 proximal tubule (S2), and S3 proximal tubule (S3) segments while distal tubules were defined as cortical thick ascending limb, distal convoluted tubule, connecting tubule, and cortical collecting duct segments. A median proximal and distal value per gene was then calculated using the median RPKM values for the above segments. Genes were defined as being proximal (P>D) or distal (P<D) if there was at least a fivefold difference in median between the defined proximal and distal segments. All other genes were designated as having no, or unclear, difference between proximal and distal regions (P?D). The intersection of genes included in both datasets (Durinck et al. and Lee et al.), mapped by gene symbol, were then analyzed to look for reliable gene expression changes that were not due to a cell-of-origin bias.
Metabolite Profiling.
Tissue pieces (17–35 mg) from fresh frozen tumor and normal kidney samples were homogenized in 4 μL of water per milligram of sample using a bead mill (TissueLyser II; Qiagen). Metabolite profiles were measured using a combination of four liquid chromatography-tandem mass spectrometry (LC-MS) methods. We applied two polar metabolite profiling methods and a lipid method similar to those described in Mascanfroni et al. (49).
Immunohistochemistry.
Primary antibodies were as follows: P-AMPK (Cell Signaling 2535), HNF4α (Cell Signaling 3113), MTCO1 (Abcam ab14705), NDUFB8 (Abcam ab110246), CD117 (Cell marque catalog no. 117R-18), and cyclin D1 (Cell marque catalog no. 241R-18). For cell-of-origin interpretation, proximal tubules (including both the convoluted and straight segments, i.e., S1, S2, and S3 segments) were identified morphologically as tubules lined by a simple columnar epithelium with abundant apical cytoplasm, prominent brush borders, and often frothy tubular lumens. Distal tubules were identified by the presence of simple cuboidal epithelium that was variably attenuated.
Cell Growth Assay.
HEK293T cells were grown in DMEM (11995; Thermo Fisher) supplemented with 10% FBS (Sigma), penicillin-streptomycin, and 50 μg/mL uridine at 37 °C and 5% CO2. HEK293T cells were plated into six well plates and grown for 48 h in the presence of vehicle control (DMSO), BSO (100 μM; Sigma), piericidin A (1 μM; Santa Cruz), or a combination of BSO plus piericidin and cell number was then determined using a Coulter Counter.
Statistical Analysis.
Data are reported as mean unless otherwise specified. For two-group comparisons, the two-sample Student’s t test was used unless otherwise indicated in the figure.
Data Availability.
Somatic variants from exome sequencing, RNA-seq analysis with cell-of-origin interpretation, and metabolite profiling are made fully available in Datasets S1–S5.
Supplementary Material
Acknowledgments
We thank Zenon Grabarek for complex I crystal structure analysis, Andrew Markhard for figure assistance, and Ruslan Sadreyev for his sequencing technical advice. Results in Fig. 1 are in part based upon data generated by the The Cancer Genome Atlas Research Network (https://cancergenome.nih.gov/), specifically the Clear cell kidney carcinoma (KIRC) dataset. We thank members of the V.K.M. laboratory for valuable feedback. This work was supported by an Ellison Foundation Award. R.K.G. was supported by an American Society of Clinical Oncology Young Investigator Award, a Claire and John Bertucci Fellowship, and NIH Grant P50 CA101942-13. V.K.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711888115/-/DCSupplemental.
References
- 1.Kuroda N, Toi M, Hiroi M, Shuin T, Enzan H. Review of renal oncocytoma with focus on clinical and pathobiological aspects. Histol Histopathol. 2003;18:935–942. doi: 10.14670/HH-18.935. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linehan WM, et al. Cancer Genome Atlas Research Network Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis CF, et al. The Cancer Genome Atlas Research Network The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Reports. 2016;14:2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Störkel S, et al. Intercalated cells as a probable source for the development of renal oncocytoma. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;56:185–189. doi: 10.1007/BF02890016. [DOI] [PubMed] [Google Scholar]
- 7.Kryvenko ON, Jorda M, Argani P, Epstein JI. Diagnostic approach to eosinophilic renal neoplasms. Arch Pathol Lab Med. 2014;138:1531–1541. doi: 10.5858/arpa.2013-0653-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–642. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Latham B, Dickersin GR, Oliva E. Subtypes of chromophobe cell renal carcinoma: An ultrastructural and histochemical study of 13 cases. Am J Surg Pathol. 1999;23:530–535. doi: 10.1097/00000478-199905000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Gasparre G, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 11.Simonnet H, et al. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis. 2003;24:1461–1466. doi: 10.1093/carcin/bgg109. [DOI] [PubMed] [Google Scholar]
- 12.Durinck S, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi S, et al. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Reports. 2015;13:1895–1908. doi: 10.1016/j.celrep.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: A publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju YS, et al. ICGC Breast Cancer Group; ICGC Chronic Myeloid Disorders Group; ICGC Prostate Cancer Group Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3:e02935. doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley SA, et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller W, Rittinger M. Quantitative morphology of the rat kidney. Int J Biochem. 1980;12:17–22. doi: 10.1016/0020-711x(80)90035-x. [DOI] [PubMed] [Google Scholar]
- 20.Guder WG, Wagner S, Wirthensohn G. Metabolic fuels along the nephron: Pathways and intracellular mechanisms of interaction. Kidney Int. 1986;29:41–45. doi: 10.1038/ki.1986.6. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol. 2015;26:2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenach PA, et al. Dipeptidase 1 (DPEP1) is a marker for the transition from low-grade to high-grade intraepithelial neoplasia and an adverse prognostic factor in colorectal cancer. Br J Cancer. 2013;109:694–703. doi: 10.1038/bjc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall AM, Crawford C, Unwin RJ, Duchen MR, Peppiatt-Wildman CM. Multiphoton imaging of the functioning kidney. J Am Soc Nephrol. 2011;22:1297–1304. doi: 10.1681/ASN.2010101054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandhi S, et al. Heteroplasmic shifts in tumor mitochondrial genomes reveal tissue-specific signals of relaxed and positive selection. Hum Mol Genet. 2017;26:2912–2922. doi: 10.1093/hmg/ddx172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 27.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalakonda S, et al. Monoallelic loss of tumor suppressor GRIM-19 promotes tumorigenesis in mice. Proc Natl Acad Sci USA. 2013;110:E4213–E4222. doi: 10.1073/pnas.1303760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JS, et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Compton S, et al. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function. J Biol Chem. 2011;286:20297–20312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keckesova Z, et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017;543:681–686. doi: 10.1038/nature21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannakis M, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsi RH, Bowen BM, Wiedmann M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics. 2010;11:102. doi: 10.1186/1471-2164-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 36.Kürschner G, et al. Renal oncocytoma characterized by the defective complex I of the respiratory chain boosts the synthesis of the ROS scavenger glutathione. Oncotarget. 2017;8:105882–105904. doi: 10.18632/oncotarget.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao XR, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016;5:e10575. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lien EC, et al. Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat Cell Biol. 2016;18:572–578. doi: 10.1038/ncb3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meister A. Glutathione, metabolism and function via the gamma-glutamyl cycle. Life Sci. 1974;15:177–190. doi: 10.1016/0024-3205(74)90206-9. [DOI] [PubMed] [Google Scholar]
- 40.Dringen R, Pfeiffer B, Hamprecht B. Synthesis of the antioxidant glutathione in neurons: Supply by astrocytes of CysGly as precursor for neuronal glutathione. J Neurosci. 1999;19:562–569. doi: 10.1523/JNEUROSCI.19-02-00562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erez A, DeBerardinis RJ. Metabolic dysregulation in monogenic disorders and cancer–Finding method in madness. Nat Rev Cancer. 2015;15:440–448. doi: 10.1038/nrc3949. [DOI] [PubMed] [Google Scholar]
- 42.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, Schroeder R, Ko A, Stoneking M. Fidelity of capture-enrichment for mtDNA genome sequencing: Influence of NUMTs. Nucleic Acids Res. 2012;40:e137. doi: 10.1093/nar/gks499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mascanfroni ID, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat Med. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Somatic variants from exome sequencing, RNA-seq analysis with cell-of-origin interpretation, and metabolite profiling are made fully available in Datasets S1–S5.