Significance
Bacterial persisters are able to survive high concentrations of antibiotics that kill their genetically identical kin. Their tolerances are thought to arise from decreased activity of cellular processes, which limits damage from antibiotics. However, persistence to fluoroquinolones in growth-inhibited populations is not as cut-and-dried, with survivors of treatment exhibiting similar DNA damage as cells that die. In this article, we use a model system of persistence to reveal that the timing of events, such as DNA repair, following fluoroquinolone treatment is critical to survival and show that the same is true for WT populations. These data highlight the importance of processes following antibiotic treatments to persister phenotypes and establish that timing matters for genetically susceptible bacteria struggling to survive fluoroquinolone treatments.
Keywords: persistence, SOS response, DNA damage, MazF, starvation
Abstract
Bacterial persisters are subpopulations of phenotypic variants in isogenic cultures that can survive lethal doses of antibiotics. Their tolerances are often attributed to reduced activities of antibiotic targets, which limit corruption and damage in persisters compared with bacteria that die from treatment. However, that model does not hold for nongrowing populations treated with ofloxacin, a fluoroquinolone, where antibiotic-induced damage is comparable between cells that live and those that die. To understand how those persisters achieve this feat, we employed a genetic system that uses orthogonal control of MazF and MazE, a toxin and its cognate antitoxin, to generate model persisters that are uniformly tolerant to ofloxacin. Despite this complete tolerance, MazF model persisters required the same DNA repair machinery (RecA, RecB, and SOS induction) to survive ofloxacin treatment as their nongrowing, WT counterparts and exhibited similar indicators of DNA damage from treatment. Further investigation revealed that, following treatment, the timing of DNA repair was critical to MazF persister survival because, when repair was delayed until after growth and DNA synthesis resumed, survival was compromised. In addition, we found that, with nongrowing, WT planktonic and biofilm populations, stalling the resumption of growth and DNA synthesis after the conclusion of fluoroquinolone treatment with a prevalent type of stress at infection sites (nutrient limitation) led to near complete survival. These findings illustrate that the timing of events, such as DNA repair, following fluoroquinolone treatment is important to persister survival and provide further evidence that knowledge of the postantibiotic recovery period is critical to understanding persistence phenotypes.
Within clonal bacterial cultures, a subpopulation of cells known as persisters can transiently survive doses of antibiotics that kill their isogenic kin (1–3). Unlike resistant mutants, persisters are phenotypic variants, and their tolerances are not attributed to heritable genetic changes that allow them to grow in the presence of antibiotics. After the antibiotic is removed, persisters resume growth and replication and give rise to populations with similar susceptibility to the antibiotic used for treatment as cells in the original population (4). Given their potential to repopulate infections, persisters are considered to be important contributors to chronic and relapsing infections (5–7). To devise treatments against this threat, better understanding of persister physiology will be enabling (8–10).
Persisters are generally thought to originate from growth-arrested cells because such cells harbor less active cellular processes than their growing counterparts and thus suffer limited damage from antibiotic treatment. However, this paradigm is not generalizable to all types of persisters (2, 11). For example, Wakamoto et al. (12) demonstrated that persistence to isoniazid in Mycobacterium smegmatis depends on a dynamic balance between cell growth and stochastic expression of the prodrug activating enzyme, KatG. In addition, some classes of antibiotics, such as fluoroquinolones (FQs), which target topoisomerases and are on the World Health Organization’s essential medicine list (13), are capable of killing both growing and nongrowing bacteria (14). It was recently reported that, in stationary-phase cultures, persisters to ofloxacin (OFL) (a FQ) and cells that died from treatment exhibited indistinguishable DNA damage responses both during and after treatment (15). Further, those persisters required DNA repair machinery for survival, which suggested that they suffered OFL-induced DNA damage (15). These and other studies have established that growth arrest and persistence are not equivalent (16–19), which suggests that additional aspects of persister physiology must be considered to understand their impressive abilities to survive antibiotic exposure.
In a recent study, we used orthogonal expression of the MazF-MazE toxin–antitoxin (TA) system to generate model persisters for analysis (20). This model was constructed based on the importance of TA systems to persistence (21–24), and, notably, MazF accumulation has been employed by various groups to study antibiotic tolerance (25, 26). An important feature of our MazF persisters was that they were uniformly tolerant to OFL treatments (20) whereas ∼90% of stationary-phase populations were killed when treated with the same OFL dose (15). Furthermore, our model system recapitulated features of WT persisters, such as sensitivity toward aminoglycosides despite being tolerant to FQs and β-lactams (8, 10, 20, 27, 28). Based on these results, we hypothesized that MazF persisters could be used to understand how growth-arrested cells could tolerate OFL treatments and give rise to persisters.
In this study, we examined how MazF model persisters were able to achieve complete tolerance to OFL whereas growth-inhibited WT populations could not. We confirmed that MazF persisters required a similar repertoire of DNA repair enzymes as WT OFL persisters for survival and that, following treatment, they can experience DNA fragmentation and metabolic perturbations reflective of DNA damage, such as accumulation of dNTPs (29). We further provide evidence that resumption of growth and DNA synthesis was slower in MazF model persisters compared with a growth-arrested control that had lower persister levels, and we demonstrate that delaying DNA repair compromised MazF persister survival. Collectively, these observations suggested that the timing of events, such as DNA repair and reinitiation of growth, was important for survival from FQ damage in nongrowing cells. To examine the relevance of these findings to WT persister levels, we delayed growth resumption and DNA synthesis with nutrient deprivation, which can occur at infection sites (30–32), following FQ treatment, and discovered that such scenarios can increase survival of WT populations to near 100%. These results demonstrate that the timing of molecular events following FQ treatment is critical to the persistence phenotype and suggest that strategies to reduce FQ persistence in growth-arrested populations, which are notoriously difficult to treat, would be to stimulate DNA synthesis and/or slow DNA repair.
Results
MazF Persisters Require DNA Repair Systems to Survive OFL Treatment.
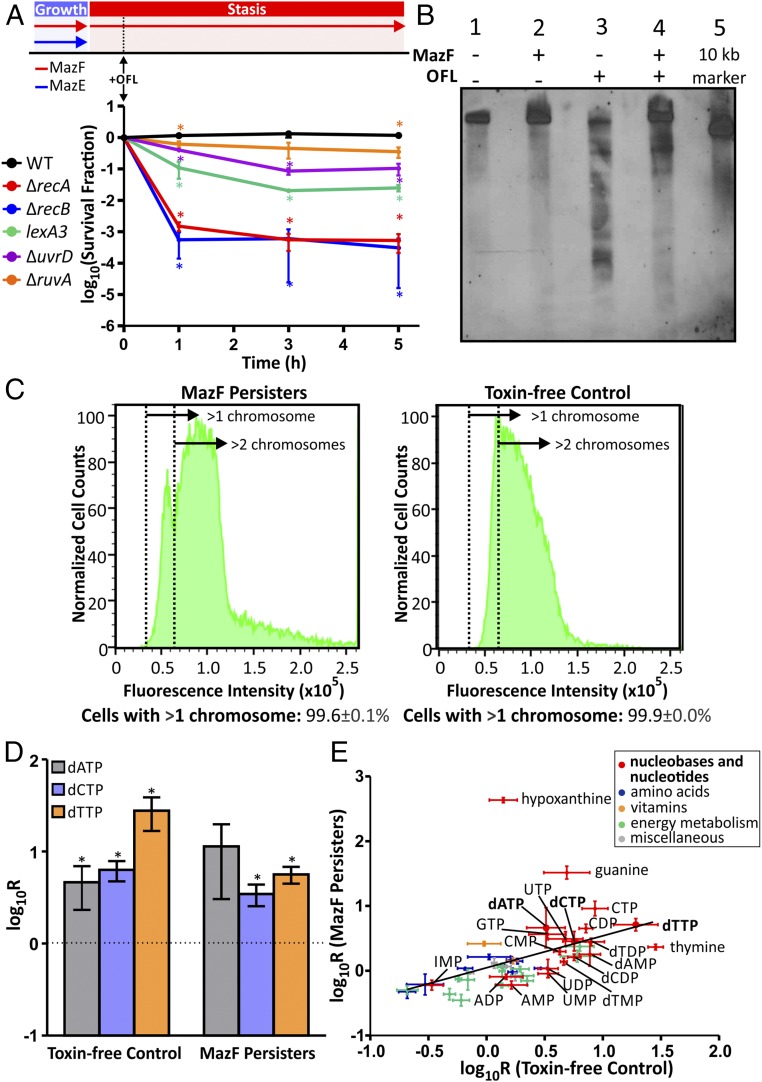
We previously generated an Escherichia coli strain that would allow us to produce MazF model persisters (20); in this strain, the endogenous chromosomal copy of mazEF was deleted, and mazE and mazF were returned to the chromosome under orthogonal control from two independent, inducible promoters [arabinose-inducible PBAD for mazE and anhydrotetracycline (aTc)-inducible PLtetO1 for mazF]. We observed that MazF accumulation resulted in reversible growth inhibition and that MazF-arrested cells were completely tolerant to two classes of antibiotics: β-lactams (e.g., ampicillin) and fluoroquinolones (e.g., OFL). By comparison, the survival of a toxin-free control expressing MazE alone but induced with the same concentration of aTc as MazF-arrested cells and cells coexpressing MazE and MazF decreased by four orders of magnitude following OFL treatment (SI Appendix, Fig. S1A). These findings suggested that the MazF persisters could be a suitable model system to study persistence to FQs in nongrowing populations. To further assess this capability, we assayed whether similar DNA repair functions were needed by MazF and WT persisters to survive OFL treatment (Fig. 1A). We deleted or mutated a number of key genes whose products were involved in homologous recombination and the SOS response that we had previously examined for involvement in OFL persistence within stationary-phase WT cultures (ΔrecA, lexA3, ΔrecB, ΔrecF, ΔrecN, and ΔruvA) (15). In addition, to cover a wider breadth of DNA repair pathways, we also examined the effects of deleting uvrD, which encodes a helicase involved in methyl-directed mismatch repair, nucleotide excision repair, and implicated in replication fork restart (33); sulA, which inhibits cell division during the SOS response (34); and mutM, nfo, and ung, which are involved in base excision repair (35).
Fig. 1.
MazF persisters recapitulate aspects of WT OFL persister physiology. (A) MazF persisters depend on DNA repair to survive OFL treatment. Strain WM03 and its derivatives, which harbor mutations in DNA repair genes, were cultured to exponential phase while expressing MazE (induced with arabinose) and MazF (induced with anhydrotetracycline), and then arabinose was removed to allow accumulation of MazF and growth stasis. During stasis, OFL was added, and, at designated time points, OFL and aTc were removed, and cells were recovered on media supplemented with arabinose for MazE induction. Mutations or deletion of genes involved in the SOS response (recA and lexA), repair of DNA DSBs (recB and ruvA), and nucleotide excision repair (uvrD) significantly reduced the survival following OFL treatment, despite MazF-induced stasis. (B) The integrity of an 11-kb region encompassing dnaA-dnaN-recF-gyrB (dnaA locus) was examined before and 30 min after OFL treatment by Southern blotting. Before OFL administration, the dnaA locus was intact in the toxin-free control (strain WM02 grown in the presence of aTc; lane 1) and in MazF persisters (lane 2). After OFL treatment, fragmentation was observed in the antibiotic susceptible toxin-free control (lane 3) and in MazF persisters (lane 4). A 10-kb DNA marker was included (lane 5) for size comparisons. The Southern blot shown is representative of at least three biological replicates. Another replicate of the Southern blot is shown in SI Appendix, Fig. S1F. (C) MazF persisters and toxin-free controls have more than one chromosome at the time of OFL treatment, and most have more than two chromosomes. Controls used to determine the fluorescence intensities corresponding to different chromosomal units are shown in SI Appendix, Fig. S1G. (D) Abundances of metabolites extracted from MazF persisters and toxin-free control before and 1 h after OFL treatment were normalized by culture density (OD600) at the time of extraction. Ratios of changes in metabolite abundances (R) were calculated by dividing the OD600-normalized abundance of each metabolite at 1 h following treatment by the normalized abundance before treatment. Both MazF persisters and toxin-free control accumulated deoxyribonucleotides (dNTPs) posttreatment, which is an indicator of DNA damage. (E) The metabolites that are depicted exhibited statistically significant (P ≤ 0.05) increases or decreases in relative abundance at 1 h post-OFL treatment compared with their relative abundance pretreatment for at least one of the strains (MazF persisters or toxin-free control). The regression line shown has the equation y = 0.4929x + 0.047 (r = 0.4573, P = 0.0010). These data depict three or more biological replicates, and error bars represent SEM.
Deletion of genes encoding base excision repair enzymes did not affect OFL persister levels in MazF persisters, and neither did ΔrecF, ΔrecN, or ΔsulA mutations (SI Appendix, Fig. S1B). Deletion of ruvA and uvrD significantly (P < 0.05) reduced the survival of MazF persisters following OFL treatment, resulting in 3- and 11-fold decreases in the survival fraction, respectively (Fig. 1A). However, loss of LexA activation, RecA, or RecB had the most dramatic effect on MazF persister tolerance; survival of the lexA3 mutant was reduced by ∼50-fold, and survival of ΔrecA and ΔrecB mutants decreased by >1,000-fold (Fig. 1A). Complementation of recA (SI Appendix, Fig. S1C) and recB (SI Appendix, Fig. S1D) in these deletion mutants with plasmid-borne copies of these genes restored OFL persistence. These findings demonstrated that MazF model persisters depended on a similar repertoire of DNA repair enzymes as a MazE-MazF coexpression control (SI Appendix, Fig. S1E) and OFL persisters derived from WT stationary-phase cultures, where functional RecA, RecB, and cleavable LexA were the most important DNA damage and repair systems tested (15). These data provided strong genetic evidence for the use of this model system to interrogate the survival strategies of persisters to OFL in growth-inhibited WT populations.
MazF Persisters Experience DNA Breaks Following OFL Treatment.
Given that the whole population of MazF persisters was tolerant to OFL, we could use population-wide measurements to evaluate whether they suffered DNA damage as a result of treatment. To demonstrate that drug-stabilized DNA–gyrase complexes, which are the cause of lethal DNA double-stranded breaks (DSBs) (36), were formed in MazF model persisters following OFL treatment, we measured cleavage sites in an 11-kb stretch of genomic DNA in OFL-treated cells. We used Southern blotting to examine the integrity of the region encompassing dnaA-dnaN-recF-gyrB as this locus was previously shown to be fragmented as a result of treatment with another quinolone antibiotic, oxolinic acid (37). We observed DNA fragmentation in MazF persisters after exposure to OFL although we note that damage was reduced compared with the toxin-free, growing control, which bears only an inducible MazE and was readily killed (Fig. 1B and SI Appendix, Fig. S1F). Interestingly, these data and the dependence of MazF persister survival on RecB and RecA suggested that MazF persisters suffered DNA DSBs that were repaired via homologous recombination. Since homologous recombination requires an intact copy of a damaged locus to perform efficient repair (38, 39), we used PicoGreen staining to quantify the chromosomal content of MazF persisters at the time of treatment (Fig. 1C; see SI Appendix, Fig. S1G for controls). We observed that more than 99% of MazF persisters, as well as toxin-free controls, contained greater than one chromosome before OFL treatment, with the vast majority containing greater than two chromosomes, which confirms that the MazF persisters had sufficient genomic material for recombinatorial repair. Further, we observed a continuous distribution of fluorescence intensities from MazF persisters, as opposed to the discrete peaks that are indicative of integer abundances of chromosomes (SI Appendix, Fig. S1G), which suggested that many MazF persisters contained partially replicated chromosomes.
To complement the genetic and physiological data described above, we used liquid chromatography-mass spectrometry (LC-MS) to compare the metabolic responses of MazF persisters to OFL treatment to those of an identically treated, toxin-free control. A previous study suggested that the accumulation of dNTPs following exposure to genotoxic stress is a metabolic response of cells experiencing DNA damage and requiring translesion synthesis (29). After 1 h of OFL treatment, we observed that both treated populations accumulated a number of nucleobases and nucleotides in response to OFL treatment. Specifically, dATP, dCTP, and dTTP were found to accumulate (Fig. 1D) whereas dGTP could not be reliably measured with the separation method used here due to similarities between its retention time and those of its more abundant isomer ATP (40). On the more global scale, we found a positive correlation (correlation coefficient of 0.4573, P ≤ 0.05) between the changes in relative abundance of metabolites from the OFL-tolerant MazF persisters and the OFL-susceptible toxin-free control (Fig. 1E and SI Appendix, Fig. S2) although we note that the response in the MazF persisters was dampened compared with the control. Results from this metabolomic analysis are in agreement with observations from our genetic analysis and Southern blotting experiments, which demonstrated that MazF persisters experienced DNA damage from OFL treatment despite their extraordinary tolerance to the antibiotic. These hallmarks of DNA damage and the requirement for the same DNA repair systems as WT persisters for survival suggests that MazF persisters recapitulate essential features of their WT counterparts and thus constitute a good model system for the study of OFL persistence in nongrowing WT populations.
MazF Persisters Respond Slowly Following the Removal of OFL.
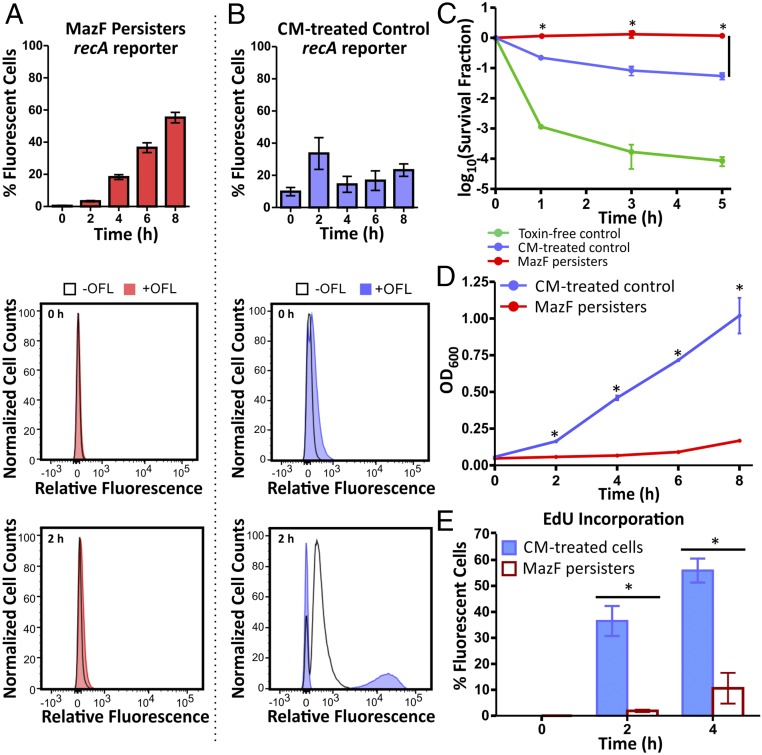
Our group previously demonstrated that the postantibiotic recovery period was critical for the survival of WT OFL persisters originating from a nongrowing population as DNA repair machineries and SOS induction were only necessary following the removal of OFL (15). Focusing in on this critical period for persisters, we monitored the expression of an SOS reporter following the conclusion of OFL treatment. We used an SOS reporter comprised of GFP under the regulation of a recA promoter (PrecA), which was validated previously to reflect SOS induction (15), and quantified its fluorescence every 2 h for 8 h during recovery from OFL in LB that included arabinose for MazE induction (Fig. 2 A and B and SI Appendix, Figs. S3 and S4). In the toxin-free control, which was replicative and translationally active when the antibiotic was added, cells were fluorescent during OFL treatment. Approximately 97% of this population remained highly fluorescent during the 8-h recovery period (SI Appendix, Figs. S3A and S4A). When SOS induction in MazF persisters was assayed, we observed a long delay without appreciable fluorescence before 4 h (Fig. 2A and SI Appendix, Figs. S3B and S4B). To confirm that this fluorescence in MazF persisters was attributable to SOS induction, the reporter was monitored in MazF persisters carrying a lexA3 mutation (SI Appendix, Figs. S3C and S4C). As anticipated, fluorescence was not appreciably induced in this lexA3 mutant throughout the course of the experiment. As MazF interrupts translation and causes growth inhibition, we used a chloramphenicol (CM)-treated, MazF-free system as a growth-inhibited control for comparison (Fig. 2B and SI Appendix, Figs. S3D and S4D). Interestingly, we observed that SOS induction in MazF persisters and CM-treated cells differed in both intensity and timing. Approximately 33.6 ± 10.0% (Fig. 2B and SI Appendix, Fig. S3D) of CM-treated cells were highly fluorescent 2 h following OFL removal. By comparison, 3.3 ± 0.4% (Fig. 2A and SI Appendix, Fig. S3B) of MazF persisters were fluorescent above background at this time. The fluorescent subpopulation of MazF persisters increased over time after 4 h of recovery (Fig. 2A and SI Appendix, Figs. S3B and S4B), but the magnitude of fluorescence was dampened compared with responsive CM-treated cells (Fig. 2B and SI Appendix, Figs. S3D and S4D).
Fig. 2.
SOS induction, growth, and DNA synthesis are delayed in MazF persisters following OFL treatment. Expression of an SOS reporter was quantified in (A) MazF persisters and (B) toxin-free cells treated with translation inhibitor chloramphenicol (CM) during recovery from OFL treatment in LB with arabinose. The bar graphs (Top) depict the percentage of cells with fluorescence intensities above a nonfluorescent negative control (lexA3 mutant without OFL treatment) measured at each time (determined from histograms in SI Appendix, Fig. S3 B and D). The histograms show the fluorescence distributions of cells from each population at 0 and 2 h. Compared with the CM-treated control, MazF persisters exhibited a delayed SOS response following OFL removal. Three biological replicates were performed, and the histograms shown here are representative of one of those replicates. (C) While CM treatment enhanced OFL persister levels compared with untreated cells, survival of CM-treated cells was 10-fold less than the survival of MazF persisters, which were completely tolerant to OFL. (D) Compared with MazF persisters, CM-treated cultures increased in density faster following OFL treatment. (E) Incorporation of EdU in newly synthesized DNA of CM-treated cells and MazF persisters recovering from OFL was determined by flow cytometry. The experiments shown in this figure were repeated at least three times, and the asterisks (*) denote a statistically significant (P ≤ 0.05) difference in survival fraction, OD600, or fluorescence between CM-treated controls and MazF persisters at each time point.
MazF-model persisters and CM-treated toxin-free controls were both translationally inhibited before and during OFL treatment, and both populations suffered less DNA fragmentation compared with growing and translationally active cells (Fig. 1B and SI Appendix, Fig. S5A). Additionally, both populations have sufficient chromosomal copies to undergo repair by homologous recombination (Fig. 1C and SI Appendix, Fig. S5B) yet the SOS response they executed following OFL removal differed considerably. Compared with MazF persisters, where the entire population survived OFL treatment, ∼90% of CM-treated cells were killed (Fig. 2C), resulting in survival that was comparable with other growth-inhibited WT populations (15). During recovery from OFL treatment, we found that CM-treated cells increased in culture density within 2 h of OFL and CM removal (Fig. 2D) whereas MazF persisters, which needed to accumulate sufficient MazE to overcome the inhibitory effect of MazF, experienced an extended lag. The faster increase in culture density and induction of a GFP SOS reporter in CM-treated cells compared with MazF persisters led us to hypothesize that DNA synthesis also resumes earlier in the CM-treated population. Under such a scenario, DNA synthesis might proceed before the repair of damaged DNA, which could lead to replication fork collapse and compromised survival as polymerases hit unrepaired lesions.
To test our hypothesis that resumption of DNA synthesis is delayed in MazF persisters compared with CM-treated controls, we utilized 5-ethynyl-2-deoxyuridine (EdU), a thymidine analog, to detect newly synthesized DNA (see SI Appendix, Fig. S6 A and B for controls) (41). EdU was introduced in the recovery media (LB with arabinose for MazE induction), and it can be taken up and incorporated into newly synthesized DNA in cells. The alkyne functional group on the nucleobase can form a covalent bond with an azide group conjugated on an Alexa fluorophore, allowing DNA synthesis to be detected by an increase in fluorescence using flow cytometry. Supporting our hypothesis, we detected DNA synthesis in 36.5 ± 5.8% of CM-treated cells within 2 h of OFL removal whereas 1.9 ± 0.5% of MazF persisters had resumed synthesis by this time (Fig. 2E and SI Appendix, Fig. S6C). After 4 h, 55.9 ± 4.6% of CM-treated cells and 10.6 ± 5.9% of MazF persisters were detected to have resumed synthesis. These results demonstrate that CM-treated cells resumed DNA synthesis significantly faster than MazF persisters following OFL treatment.
Timing of DNA Repair Following OFL Treatment Impacts Survival.
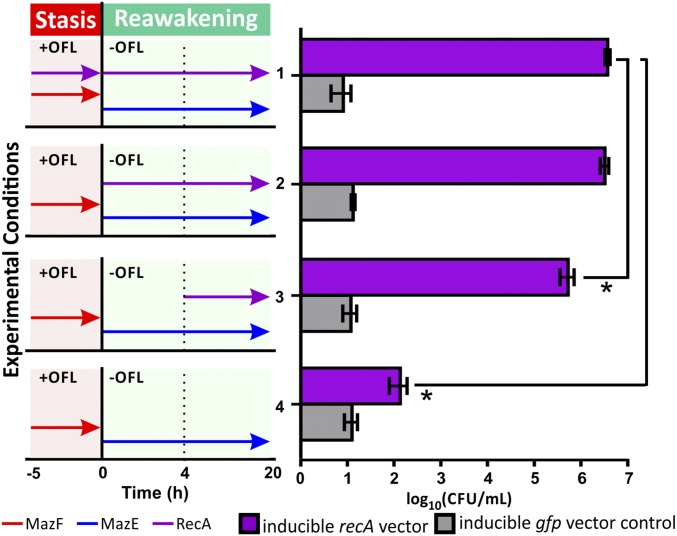
Given that MazF persisters were slow to reinitiate DNA synthesis and resume growth following OFL treatment compared with CM-inhibited controls, we hypothesized that the enhanced survival of MazF-arrested cells might be attributable to the timing of DNA repair and growth-related processes following OFL treatment. To investigate this, we used a ΔrecA mutant bearing an inducible, plasmid-borne copy of recA, which enabled us to control the timing of DNA repair with isopropyl-β-d-1-thiogalactopyranoside (IPTG). Using this strain, we induced RecA expression during OFL treatment and at different times during recovery in MazF persisters (Fig. 3). Consistent with our previous observations in WT stationary-phase persisters, we found that RecA is only necessary during recovery from OFL treatment (15) as the survival of MazF persisters that expressed RecA throughout the course of treatment and recovery (condition 1) is comparable with the survival of the population that expressed RecA only during recovery from OFL (condition 2). When RecA expression was delayed until 4 h after OFL removal (condition 3), which was after culture density had begun to increase in MazF persisters (Fig. 2D) and DNA synthesis had resumed (Fig. 2E), the survival of MazF persisters decreased sevenfold, compared with cells that expressed RecA throughout the course of recovery. With this reduction, the survival of MazF persisters approached that of the CM-inhibited and WT stationary phase populations. Similar to our findings in Fig. 1, lack of RecA expression throughout the course of treatment and recovery resulted in a 104- to 105-fold decrease in cfus (condition 4). These results show that DNA repair is only needed during recovery in MazF persisters and that, when it is delayed to the point at which growth-associated processes, such as DNA synthesis, would resume, survival of the population is compromised.
Fig. 3.
Delaying DNA repair impacts survival of MazF persisters following OFL treatment. RecA expression was induced at different times during OFL treatment and recovery in MazF persisters. MazF persisters that expressed RecA throughout the course of OFL treatment and during recovery (condition 1) and persisters that only expressed RecA during recovery from OFL treatment (condition 2) exhibited equivalent survival. Without RecA expression, MazF persisters were readily killed (condition 4). When RecA expression was delayed for 4 h after removal of OFL, we observed a sevenfold decrease in survival (condition 3). Three biological replicates were performed for this experiment, and the asterisks (*) denote statistically significant (P ≤ 0.05) differences in log-transformed cfu counts.
Timing Impacts Persistence to FQs in Nongrowing WT Populations.
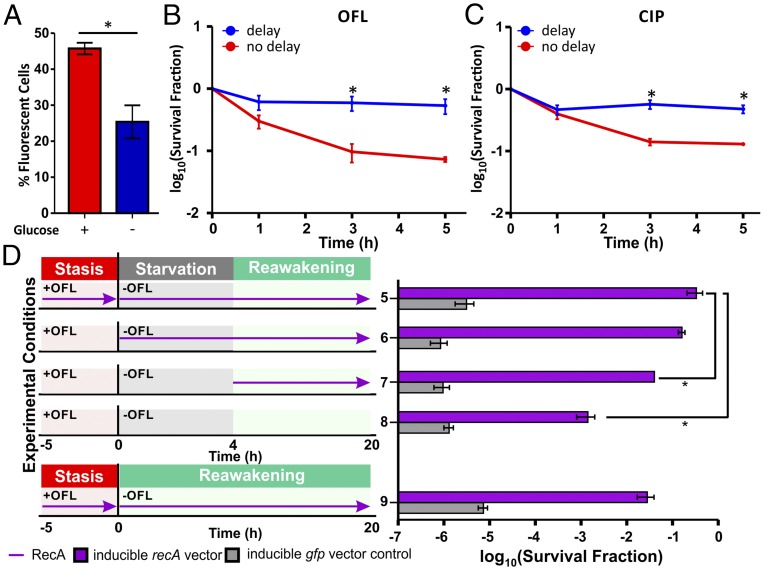
Based on our results with the MazF model persisters, we examined whether delaying DNA synthesis and growth following FQ treatment in WT populations would increase their survival by providing DNA repair machinery a longer time to repair damage. To do this, we treated WT E. coli MG1655 that had been deprived of glucose and were nongrowing with OFL (SI Appendix, Fig. S7 A and B). Following removal of the antibiotic, cells were either inoculated immediately on nutritive agar that supported growth or they were inoculated on nonnutritive agar that lacked carbon sources before being transferred onto nutritive agar. We found that 4 h posttreatment, 45.8 ± 1.6% of cells on nutritive agar resumed DNA synthesis whereas significantly fewer cells (25.4 ± 4.6%) resumed DNA synthesis on nonnutritive agar (Fig. 4A and SI Appendix, Fig. S7C). We observed that delaying growth resumption of WT cells from FQ-treated, nongrowing cultures by nutrient limitation significantly increased survival, compared with cells that were exposed to nutrients immediately following antibiotic removal. Such delay increased survival of OFL persisters by eightfold (Fig. 4B) and ciprofloxacin (CIP) persisters by fourfold (Fig. 4C). For populations exposed to FQ while growing exponentially, posttreatment starvation did not increase persistence (SI Appendix, Fig. S7D). Remarkably, delaying growth resumption of OFL-treated, nongrowing cultures resulted in near-complete tolerance in the population. To further examine the impact of the duration of posttreatment starvation on survival, we demonstrated that delaying growth resumption by starvation for 3 h was sufficient to significantly increase survival (SI Appendix, Fig. S7E).
Fig. 4.
Starvation following FQ treatment delays resumption of DNA synthesis and enhances persister levels in a RecA-dependent manner. (A) After treating growth-inhibited E. coli MG1655 with OFL, cells were inoculated on nutritive media containing glucose or on nonnutritive media (without glucose), and incorporation of EdU into newly synthesized DNA during this 4-h posttreatment recovery period was quantified. After treating growth-inhibited WT cells with (B) OFL or (C) CIP, cells were inoculated on nonnutritive media for 4 h after antibiotic removal before transfer onto growth-supporting nutritive media (these media did not contain EdU or thymidine). Compared with cells that were immediately inoculated onto nutritive media after treatment, inoculation onto nonnutritive media enhanced OFL persister levels by eightfold and CIP persister levels by fourfold. (D) Delaying DNA repair reduces persistence following OFL treatment. RecA expression was induced at different times before, during, and after OFL treatment in MG1655ΔrecA, which was starved of glucose and was growth-inhibited at the time of OFL administration. Cells that expressed RecA throughout the course of OFL treatment and during recovery (condition 5, purple bar) and cells that only expressed RecA during recovery from OFL treatment (condition 6, purple bar) exhibited comparable tolerance to OFL. When RecA expression was delayed for 4 h after removal of OFL, we observed an eightfold decrease in survival (condition 7, purple bar), similar to the survival of cells that were plated on nutritive media immediately after OFL removal (condition 9, purple bar). Without RecA expression, persisters decreased by three orders of magnitude (condition 8, purple bar). In the absence of RecA (gray bars), inoculation on nonnutritive media did not enhance persistence. These experiments were repeated at least three times, and asterisks (*) denote statistically significant (P ≤ 0.05) differences in the percentage of fluorescent cells or log-transformed survival fractions.
To demonstrate that the increase in survival observed in cells subjected to posttreatment starvation was dependent on DNA repair, we showed that delaying growth resumption in mutants lacking RecA failed to enhance survival (Fig. 4D, gray bars). Moreover, when we induced RecA expression using an IPTG-inducible, plasmid-borne copy of recA in this ΔrecA mutant, we observed that delaying RecA expression until after starvation concluded (Fig. 4D, condition 7, purple bar) decreased survival by eightfold, compared with cells that expressed RecA throughout the course of OFL treatment and posttreatment recovery (Fig. 4D, condition 5, purple bar). Furthermore, without RecA during the starvation period (Fig. 4D, condition 7, purple bar), posttreatment starvation failed to increase survival over that observed in controls that were not starved and expressed RecA (Fig. 4D and SI Appendix, Fig. S7F, conditions 9 and 10, purple bars). These data support our hypothesis that the timing of DNA repair and growth-related processes following antibiotic exposure is a feature of importance for persistence to FQs in WT populations.
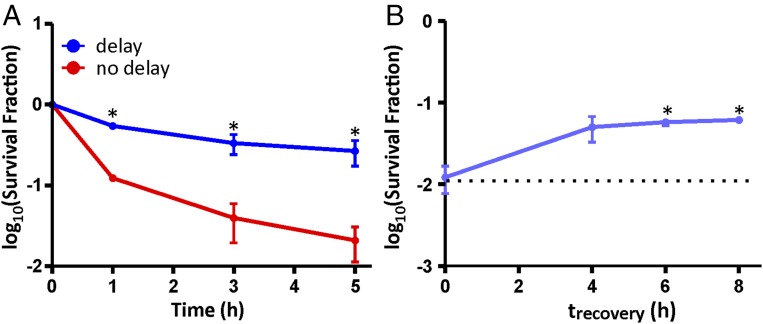
In addition to examining the impact of delaying growth resumption on FQ persistence in low-density nongrowing cultures that were starved of glucose at the time of treatment, we assessed the effects of stalling growth-related processes in persisters originating from stationary-phase planktonic cultures (Fig. 5A). Similar to our observations for the low-density populations, we found that delaying growth resumption of OFL persisters from stationary-phase cultures increased survival by 13-fold. As persisters have been implicated in the recalcitrance of infections involving biofilms (7, 42), we investigated whether the timing of posttreatment molecular events impact survival of cells occupying this more physiologically and clinically relevant niche. We demonstrated that, in glucose-starved colony biofilms treated with OFL, stalling growth resumption by starvation for at least 6 h after treatment concluded increased persistence fivefold, compared with cells from biofilms that were immediately plated on nutritive media after antibiotic removal (Fig. 5B and SI Appendix, Fig. S8). Collectively, these results show that the timing of molecular events following the conclusion of FQ treatments can have a significant impact on the survival of bacteria originating from growth-inhibited planktonic cultures and colony biofilms.
Fig. 5.
Stalling growth resumption by starvation following OFL treatment enhances persisters originating from planktonic stationary phase cultures and colony biofilms. (A) When OFL-treated WT cells from stationary phase cultures were inoculated on nonnutritive media for 4 h after antibiotic removal before being transferred onto nutritive media, survival increased by 13-fold compared with populations that were immediately inoculated onto nutritive media after treatment. (B) When OFL-treated cells from colony biofilms were starved of carbon sources for at least 6 h following treatment, persister levels significantly increased by fivefold. Three biological replicates were performed for each experiment, and asterisks (*) denote statistically significant (P ≤ 0.05) differences in persister levels between “no delay” (indicated by dashed line) and “delay” samples.
Discussion
For some classes of antibiotics, such as FQs, growth arrest and reduced target activity alone cannot explain persistence (15). FQs target topoisomerases, such as DNA-bound DNA gyrase, which is involved in introducing negative supercoiling in DNA during replication and transcription (43). As nongrowing cells often retain transcriptional activity, FQs have the capacity to kill both growing and nongrowing cells (14, 15, 44). Our results confirm that persisters are not spared DNA damage during FQ treatment and that posttreatment DNA repair is necessary for survival, which is consistent with previous findings from our group (15). Delving deeper into this phenomenon here, we found that the survival of nongrowing cells treated with FQs hinges on the timing of DNA repair and resumption of growth-related processes, such as DNA synthesis. Delaying growth resumption through carbon source starvation enhanced survival following FQ treatment to almost 100%, and that enhancement depended on when RecA was available to function relative to resumption of growth. These findings suggest that the coordination and relative timing of molecular events during the postantibiotic period differentiate FQ persisters from cells that die. Single-cell technologies, such as time-lapse microscopy and fluorescence-activated cell sorting, will be central to efforts to understand how persisters orchestrate survival from these near-death experiences, just as they have been critical for delineating important features of persistence to other antibiotics within different environments (16, 18, 45, 46). To complement these technologies, model systems, such as the hipA7 and hipQ mutants, have proven to be quite valuable (45, 47), and all indications suggest that MazF persisters could be equally useful.
Here, we observed that MazF persisters exhibited reduced and delayed expression of recA compared with CM-treated cultures following OFL treatment. Conceivably, the need to accumulate MazE to overcome MazF inhibition provided the time delay in MazF persisters whereas the CM-treated control could resume translation and growth more rapidly upon removal of the chemical inhibitor. We suspect that the delay gave repair enzymes sufficient time to mend DNA damage before reawakening events rendered unrepaired lesions lethal. Accordingly, we postulate that this might be a common feature of FQ persisters formed by type II TA modules where a proteinaceous antitoxin must be synthesized to resume growth, which would effectively add a time delay to many growth-associated processes. Indeed, TA modules have been implicated in the formation of many types of persisters, and many bacterial species have been found to harbor genes encoding TA systems that target diverse cellular processes (22–24, 48–50). Comparing the schedules of repair and reawakening in FQ persisters generated with different toxins could shed light on features of post-FQ recovery that enable survival and the extent of their generality.
Using the knowledge gained from the MazF system, we hypothesized that delaying growth and DNA synthesis following OFL treatment would have an impact on the survival of nongrowing WT populations. Recognizing that nutrient limitation is a stress that can be encountered by pathogens during an infection and could suspend growth and DNA synthesis (51), we assayed whether carbon source deprivation following FQ treatment impacted survival. For populations that were nutrient-deprived and thus growth-inhibited before and during treatment, the impact was significant. These observations were true for OFL- and CIP-treated populations and point to the importance of a timing component with respect to FQ-induced DNA repair and the processes of replication. However, it is important to note that this phenomenon only pertained to populations that were nongrowing before and during FQ treatment as subjecting populations that were growing exponentially before FQ treatment to posttreatment starvation did not increase persistence (SI Appendix, Fig. S7D). Interestingly, this suggests that FQ persisters in nutrient-replete and nutrient-depleted environments are different, which agrees with previous observations of the role of TisB in persister formation (24).
In addition to experiments with planktonic cultures, we showed that starvation boosted FQ persistence in cells originating from carbon-starved colony biofilms. As biofilms account for 65 to 80% of hospital-treated infections and are a source of relapsing infections (52, 53), these findings suggest that the coordination of DNA repair and growth-related processes following FQ treatment will also impact survival of bacteria occupying this clinically relevant lifestyle. One interesting question raised by these and the planktonic data concerns whether additional types of nutrient deprivation, besides carbon source limitation, provide similar enhancements in survival in FQ-treated populations. Such knowledge could prove useful for the development of strategies that modulate nutrient availability as a means to improve antibiotic lethality (8, 10, 54).
Collectively, the study presented here emphasizes the importance of understanding how persisters repair damaged cellular components, reactivate metabolic processes, and resume replication after antibiotic treatment as the metabolic and repair processes that occur during this time period can dictate whether a bacterium survives or dies. Further, the data provide additional justification for impairing DNA repair pathways and the SOS response as a means to potentiate FQ killing and eradicate FQ persisters (55, 56).
Materials and Methods
Bacterial Strains.
Bacterial strains used in this study were derivatives of E. coli MG1655. Strains, plasmids, and primers used for strain construction in this work are detailed in SI Appendix, Table S1. Procedures for strain construction and culture conditions are described in SI Appendix, SI Materials and Methods.
Planktonic Persistence Assays.
Planktonic persistence assays were carried out with cultures grown in Gutnick media (57) with 10 mM glucose as the sole carbon source (with 300 ng/mL aTc for MazF induction or 300 ng/mL aTc and 100 mM arabinose for MazF-MazE coinduction in strain WM03) or growth-inhibited cultures grown in Gutnick media with 0.5 mM glucose. MazF persisters and glucose-starved E. coli MG1655 were treated ∼2 h after growth inhibition whereas CM-inhibited cells were treated 2 h after inhibitor administration. Exponentially growing toxin-free controls were cultured to an OD600 ∼ 0.2 to 0.4 at the time of treatment as this density matched those of MazF persisters and CM-inhibited cells at the time of growth arrest. Stationary phase cultures were grown for 16 h before treatment. All cultures were treated with 5 μg/mL OFL or 1 μg/mL CIP for 5 h. The cfus were enumerated from samples collected before FQ was added and periodically during treatment as detailed in SI Appendix, SI Materials and Methods.
Colony Biofilm Persistence Assays.
Colony biofilms were cultured following a protocol similar to the one described in Amato and Brynildsen (28). E. coli MG1655 was cultured in Gutnick media with 10 mM glucose for 16 h. Following overnight growth, cells were diluted to an OD600 of 0.01 in Gutnick media with 2.5 mM glucose as the sole carbon source. Then, 100 μL of this culture was inoculated onto sterile 25-mm Supor hydrophilic polyethersulfone filter discs with 0.2-μm pores (VWR International) overlaid on Gutnick agar pads. Biofilms were cultured at 37 °C for 16 h, and biofilms were treated with 200 μL of a 10 μg/mL OFL solution that was applied directly onto the filters. Before OFL treatment and at designated times during treatment and posttreatment recovery, cells were dislodged from filters for cfu enumeration as detailed in the SI Appendix, SI Materials and Methods.
SOS Reporter Assays.
To quantify SOS induction using recA promoter activity as a readout during recovery from OFL treatment, planktonic persistence assays were performed as described above, and OFL was removed after 5 h of treatment. Cells were then inoculated into LB with 100 mM arabinose (for MazE induction) and kanamycin (for plasmid retention). At designated time points, cells were fixed with paraformaldehyde, and fluorescence from PrecA-gfp was quantified using an LSRII flow cytometer. Details of culture and treatment conditions, cell fixation procedure, and flow cytometer settings and controls are provided in SI Appendix, SI Materials and Methods.
Liquid Chromatography-Mass Spectrometry Metabolomic Analysis.
Before OFL administration and 1 h following OFL treatment, cultures were collected for metabolite extraction using ice-cold 40:40:20 methanol:acetonitrile:H2O (all HPLC-grade). Metabolites were dried under nitrogen gas, resuspended in HPLC-grade H2O, and analyzed by reversed-phase ion-pairing liquid chromatography coupled to a stand-alone orbitrap mass spectrometer (Exactive; Thermo Scientific) by negative-ion mode electrospray ionization (58). Metabolite peaks that matched both exact mass-to-charge ratios and retention times of authenticated standards were quantified and extracted using MAVEN software (59). Details of sample preparation and analysis are provided in SI Appendix, SI Materials and Methods.
Southern Blotting.
The integrity of the dnaA-gyrB locus before and after 30 min of treatment with 5 μg/mL OFL was examined by Southern blotting (37). Following genomic DNA extraction, digestion with HindIII (New England Biolabs), and gel electrophoresis, DNA was transferred onto a Nytran membrane using the GE TurboBlotter system (GE Healthcare Life Sciences). Probe hybridization and immunoblotting were performed using the Roche DIG High Prime DNA Labeling and Detection Kit (Sigma-Aldrich). Chemiluminescence was detected after exposing the membrane to a Hyblot CL autoradiography film (Denville Scientific Inc.). Sample and blot preparation are detailed in SI Appendix, SI Materials and Methods.
PicoGreen Staining.
Cells were cultured following the conditions described for persistence assays to the time of OFL treatment, and a fraction of each culture was fixed with 70% ethanol for 3 h. After removing the ethanol and drying the cell pellets, cells were resuspended in PBS and adjusted to OD600 ∼ 0.4. PicoGreen staining was performed following a previously published protocol (41). Cells were stained with 100 μL of a PicoGreen solution (Thermo-Fisher Scientific) that was diluted in 25% dimethyl sulfoxide (prepared in MilliQ water) for 3 h (in the dark). Cells were further diluted with 1 mL of a 1,000-fold diluted PicoGreen solution, and the fluorescence of each sample was analyzed by flow cytometry. See SI Appendix, SI Materials and Methods for details.
Click-IT EdU Assay.
Cells were cultured and subjected to persistence assays as described above. For these experiments, 50 μg/mL thymidine was added to culture media during the growth period to increase EdU incorporation (SI Appendix, Fig. S6 A and B), and 60 μg/mL EdU was used for incorporation (SI Appendix, Fig. S6C). After 5 h of treatment with 5 μg/mL OFL, antibiotics and thymidine were removed, and persisters were inoculated into LB with 100 mM arabinose and 60 μg/mL EdU while WT populations were inoculated onto filters on top of Gutnick media agar pads with 60 μg/mL EdU (or 60 μg/mL thymidine in controls) in the presence or absence of glucose. Click labeling with Alexa Fluor 647 was carried out following a protocol modified from Ferullo et al. (41). At designated time points, cells were collected and fixed in 90% methanol following the manufacturer’s recommendation. See SI Appendix, SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Junyoung Park and Joshua Rabinowitz for assistance and advice on mass spectrometry, Christina DeCoste (Princeton Flow Cytometry Resource Center) for assistance with flow cytometry, and Gary Laevsky (Princeton Confocal Imaging Facility) for assistance with microscopy. We also thank the National BioResource Project (National Institute of Genetics, Japan) for distribution of the Keio collection and Theresa C. Barrett for providing pPrecA-recA. This work was supported by Army Research Office Grant W911NF-15-1-0173 (to M.P.B.), a Charles H. Revson Foundation Fellowship in Biomedical Science (to W.W.K.M.), and Princeton University start-up funds (to M.P.B.). This content is solely the responsibility of the authors and does not necessarily represent the views of the funding agencies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804218115/-/DCSupplemental.
References
- 1.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 2.Van den Bergh B, Fauvart M, Michiels J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev. 2017;41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 4.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 5.Van den Bergh B, et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol. 2016;1:16020. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 6.Michiels JE, Van den Bergh B, Verstraeten N, Fauvart M, Michiels J. In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob Agents Chemother. 2016;60:4630–4637. doi: 10.1128/AAC.00757-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: Clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 8.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon BP, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orman MA, Brynildsen MP. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob Agents Chemother. 2013;57:4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison KR, Brynildsen MP, Collins JJ. Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol. 2011;14:593–598. doi: 10.1016/j.mib.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakamoto Y, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 13.Sharland M, et al. 21st WHO Expert Committee on Selection and Use of Essential Medicines Classifying antibiotics in the WHO essential medicines list for optimal use-be AWaRe. Lancet Infect Dis. 2018;18:18–20. doi: 10.1016/S1473-3099(17)30724-7. [DOI] [PubMed] [Google Scholar]
- 14.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother. 1991;35:1824–1828. doi: 10.1128/aac.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Völzing KG, Brynildsen MP. Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. MBio. 2015;6:e00731-15. doi: 10.1128/mBio.00731-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orman MA, Brynildsen MP. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry TC, Brynildsen MP. Development of persister-FACSeq: A method to massively parallelize quantification of persister physiology and its heterogeneity. Sci Rep. 2016;6:25100. doi: 10.1038/srep25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pu Y, et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell. 2016;62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roostalu J, Jõers A, Luidalepp H, Kaldalu N, Tenson T. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 2008;8:68. doi: 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mok WW, Park JO, Rabinowitz JD, Brynildsen MP. RNA futile cycling in model persisters derived from MazF accumulation. MBio. 2015;6:e01588-15. doi: 10.1128/mBio.01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 22.Rotem E, et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA. 2010;107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berghoff BA, Hoekzema M, Aulbach L, Wagner EG. Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol Microbiol. 2017;103:1020–1033. doi: 10.1111/mmi.13607. [DOI] [PubMed] [Google Scholar]
- 24.Dörr T, Vulić M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi A, Dewan PC, Siddique SA, Varadarajan R. MazF-induced growth inhibition and persister generation in Escherichia coli. J Biol Chem. 2014;289:4191–4205. doi: 10.1074/jbc.M113.510511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luidalepp H, Jõers A, Kaldalu N, Tenson T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol. 2011;193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amato SM, Brynildsen MP. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One. 2014;9:e93110. doi: 10.1371/journal.pone.0093110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderl JN, Zahller J, Roe F, Stewart PS. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47:1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernier SP, et al. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013;9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinney JD, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 33.Carter AS, Tahmaseb K, Compton SA, Matson SW. Resolving Holliday junctions with Escherichia coli UvrD helicase. J Biol Chem. 2012;287:8126–8134. doi: 10.1074/jbc.M111.314047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huisman O, D’Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 35.Sung JS, Mosbaugh DW. Escherichia coli uracil- and ethenocytosine-initiated base excision DNA repair: Rate-limiting step and patch size distribution. Biochemistry. 2003;42:4613–4625. doi: 10.1021/bi027115v. [DOI] [PubMed] [Google Scholar]
- 36.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franco RJ, Drlica K. DNA gyrase on the bacterial chromosome. Oxolinic acid-induced DNA cleavage in the dnaA-gyrB region. J Mol Biol. 1988;201:229–233. doi: 10.1016/0022-2836(88)90449-4. [DOI] [PubMed] [Google Scholar]
- 38.Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P, et al. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharm Res. 2009;26:1504–1515. doi: 10.1007/s11095-009-9863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 43.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gefen O, Fridman O, Ronin I, Balaban NQ. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc Natl Acad Sci USA. 2014;111:556–561. doi: 10.1073/pnas.1314114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 46.Orman MA, Brynildsen MP. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat Commun. 2015;6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korch SB, Hill TM. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: Effects on macromolecular synthesis and persister formation. J Bacteriol. 2006;188:3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheverton AM, et al. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leplae R, et al. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 53.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 54.Gutierrez A, et al. Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol Cell. 2017;68:1147–1154.e3. doi: 10.1016/j.molcel.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 55.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci USA. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wigle TJ, et al. Inhibitors of RecA activity discovered by high-throughput screening: Cell-permeable small molecules attenuate the SOS response in Escherichia coli. J Biomol Screen. 2009;14:1092–1101. doi: 10.1177/1087057109342126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gutnick D, Calvo JM, Klopotowski T, Ames BN. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.