Significance
The endoplasmic reticulum (ER) undergoes autophagy in response to starvation in a process utilizing Atg40, a cortical ER receptor that serves to recruit the machinery needed to assemble an autophagosome. Little is known about the components that work in conjunction with ER cargo receptors. Here we show that autophagy of the ER requires Lnp1, a protein that helps to maintain the structure of the ER by stabilizing rearrangements of the ER network. In the absence of Lnp1 or upon depolymerization of actin, Atg40 puncta fail to distribute to the cell interior, where the autophagy machinery resides, thus blocking incorporation of ER into autophagosomes. These studies show that the localization of Atg40 is critical for its function as a cargo receptor.
Keywords: selective autophagy, ER-shaping proteins, ER organization
Abstract
The endoplasmic reticulum (ER) forms a contiguous network of tubules and sheets that is predominantly associated with the cell cortex in yeast. Upon treatment with rapamycin, the ER undergoes degradation by selective autophagy. This process, termed ER-phagy, requires Atg40, a selective autophagy receptor that localizes to the cortical ER. Here we report that ER-phagy also requires Lnp1, an ER membrane protein that normally resides at the three-way junctions of the ER network, where it serves to stabilize the network as it is continually remodeled. Rapamycin treatment increases the expression of Atg40, driving ER domains marked by Atg40 puncta to associate with Atg11, a scaffold protein needed to form autophagosomes. Although Atg40 largely localizes to the cortical ER, the autophagy machinery resides in the cell interior. The localization of Atg40 to sites of autophagosome formation is blocked in an lnp1Δ mutant or upon treatment of wild-type cells with the actin-depolymerizing drug Latrunculin A. This prevents the association of Atg40 with Atg11 and the packaging of the ER into autophagosomes. We propose that Lnp1 is needed to stabilize the actin-dependent remodeling of the ER that is essential for ER-phagy.
The endoplasmic reticulum (ER) performs numerous essential cellular functions, such as protein folding, lipid synthesis, and calcium homeostasis (1, 2). The ER forms a single contiguous yet highly dynamic network of interconnected sheets and tubules that extends from the nuclear envelope, through the cytosol, to the cell periphery (3). The reticulons (Rtn1 and Rtn2) and DP1/Yop1 are evolutionarily conserved protein families that contain two transmembrane hairpin segments that insert as wedges into the lipid bilayer to induce the cylindrical curvature needed to form ER tubules and the edges of ER sheets (4, 5). The sheets and tubules remain interconnected by three-way junctions that are generated by ER tubule fusion. Tubule fusion is mediated by a GTPase called Atlastin in metazoans and Sey1 in yeast (6, 7). Our previous studies showed that the Lunapark protein Lnp1 resides at three-way junctions, where it serves to stabilize newly formed ER tubule rearrangements (8, 9).
ER stress induces the selective autophagy of the ER, a process termed ER-phagy (10). Selective autophagy pathways target damaged or superfluous cellular components for degradation via incorporation into an autophagosome, a double-membrane structure whose assembly requires the assembly of a hierarchy of ATG (autophagy-related) gene products (11). Each selective autophagy pathway employs a unique receptor that connects a targeted cargo to an autophagosome before it delivers its contents to a degradative compartment, the vacuole in yeast or lysosome in mammalian cells (12).
Recent studies suggest that a key role of the cargo receptor is to link cargo to Atg11, a scaffold protein that recruits the core Atg machinery that is needed to form an autophagosome (13, 14). ER-phagy requires the cargo receptor Atg40, a conserved reticulon family member that largely colocalizes with Rtn1 to ER tubules and the edges of ER sheets. A small population of Atg40 also resides on the nuclear ER. Consistent with its localization, Atg40 degrades cortical ER and inefficiently degrades nuclear ER. Nuclear ER degradation is primarily facilitated by the nucleophagy cargo receptor Atg39.
Mammalian ER-phagy cargo receptors FAM134B and RTN3 also localize to different ER subcompartments. FAM134B largely resides on ER sheets, while RTN3 is mainly found on ER tubules. Interestingly, Atg40 resides on ER tubules like RTN3 but has a similar domain structure to FAM134B. Atg40 and FAM134B contain a reticulon-like transmembrane hairpin domain and an AIM/LIR (Atg8-interacting motif/LC3-interacting region) motif that is essential for recruiting Atg8 (LC3 in mammals) to ER membranes targeted for degradation (15, 16). Atg8, which is conjugated to phosphatidylethanolamine before membrane recruitment, is one of two ubiquitin-like conjugating systems needed for autophagosome expansion (11). RTN3 contains six LIR motifs in its long amino-terminal domain that facilitates its interaction with LC3 (17). Not all cells, however, use FAM134B and RTN3 as cargo receptors. ER cargo receptors can also act in a tissue-specific manner. For example, the exocrine pancreas uses CCPG1 to degrade ER subdomains (18). Additionally, mammalian cells utilize a subunit of the translocon complex, Sec62, as a receptor during the recovery of cell stress (19).
Although Atg40 largely resides on the cortical ER, here we report that the localization of Atg40 puncta to the cell interior is critical for ER-phagy. Furthermore, we show that the distribution of Atg40 puncta to the cell interior is dependent on the actin cytoskeleton and Lnp1. Consistent with the proposal that ER-phagy requires Lnp1-dependent ER rearrangements, we find that ER-phagy and the appearance of ER-containing autophagosomes are diminished in the lnp1Δ mutant.
Results
Lnp1 Is Required for the Autophagy of the ER but Not Macroautophagy.
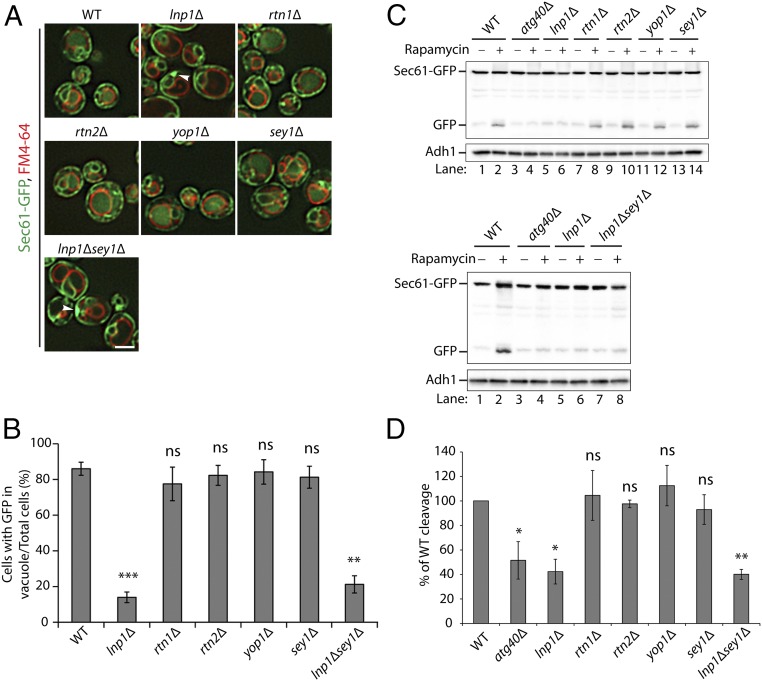
To ask if ER-shaping proteins play a role in ER-phagy, we examined the vacuolar delivery and degradation of Sec61-GFP in all known ER-shaping mutants 16 h after rapamycin treatment. We found that vacuolar GFP fluorescence failed to increase in the lnp1Δ mutant after rapamycin treatment (Fig. 1 A and B). Cytoplasmic Sec61-labeled ER patches were also seen in a fraction of the mutant cells (see arrowheads in Fig. 1A). These cytoplasmic ER patches are discussed in more detail below. Interestingly, the defect in ER-phagy was specific for the loss of Lnp1, as the rtn1Δ, rtn2Δ, yop1Δ, and sey1Δ mutants did not display a defect in the translocation of Sec61-GFP to the vacuole (Fig. 1 A and B). To confirm this result by a biochemical method, we assessed the cleavage of Sec61-GFP. Western blot analysis revealed that lnp1Δ, but none of the other ER-shaping mutants, was defective in the rapamycin-induced cleavage of Sec61-GFP to GFP (Fig. 1C, Top and Fig. 1D). In an earlier study, we reported that the ER morphology defect of lnpΔ was partially suppressed by deletion of SEY1; however, we observed no restoration in the delivery of Sec61-GFP to the vacuole (Fig. 1 A and B) or in the cleavage of Sec61-GFP (Fig. 1C, Bottom and Fig. 1D) in the lnp1Δ sey1Δ double mutant.
Fig. 1.
Loss of Lnp1, but not Rtn1, Rtn2, Yop1, or Sey1, disrupts the degradation of Sec61 in rapamycin-treated cells. (A) WT and mutant cells expressing Sec61-GFP were grown in SC media containing rapamycin (200 ng/mL) for 16 h at 30 °C, and the vacuoles were stained with FM4-64. Subsequently, cells were harvested and directly examined by fluorescence microscopy. Arrowheads point to a patch of Sec61-GFP at the cell periphery. (Scale bar, 2 μm.) (B) The percent of cells in A with GFP in the vacuole was quantified. Error bars represent SEM for three separate experiments. **P < 0.01, ***P < 0.001, Student’s t test; ns, nonsignificant. (C) Cells expressing Sec61-GFP were grown in SC medium containing rapamycin (200 ng/mL) for 0 or 12 h at 30 °C. The lysates were prepared as described in Materials and Methods, and the cleaved GFP product was analyzed by Western blot analysis. (D) Quantitation of the percent of cleaved GFP divided by total Sec61-GFP for the 12-h rapamycin-treated samples in C. The data were normalized to the WT control. Error bars represent SEM from three separate experiments. *P < 0.05, **P < 0.01, Student’s t test; ns, nonsignificant.
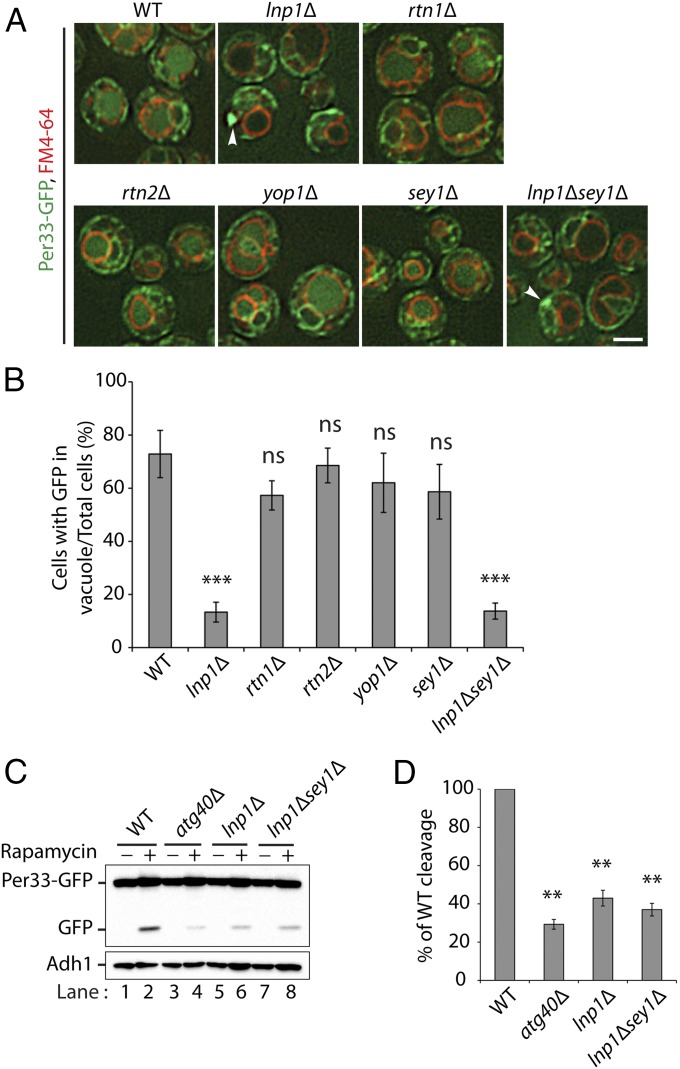
We then used a second marker protein, Per33, to confirm the ER-phagy defect in the lnp1Δ mutant. Per33, like Sec61, resides on the cortical and nuclear ER (20). We observed a defect in the translocation of Per33-GFP to the vacuole in the lnp1Δ mutant and the lnp1Δ sey1Δ double mutant but not in the rtn1Δ, rtn2Δ, yop1Δ, and sey1Δ single mutants (Fig. 2 A and B). The defect in lnp1Δ and the lnp1Δ sey1Δ double mutant was confirmed biochemically by monitoring the cleavage of Per33-GFP to GFP (Fig. 2 C and D). Therefore, while the loss of Sey1 partially restores the ER morphology defect in the lnp1Δ mutant, it cannot suppress the ER-phagy defect.
Fig. 2.
Per33-GFP degradation is defective in the lnp1∆ mutant. (A) WT and mutant cells expressing Per33-GFP were grown for 16 h at 30 °C in SC media containing rapamycin (200 ng/mL), and the vacuoles were stained with FM4-64. Subsequently, the cells were harvested and directly examined by fluorescence microscopy. Arrowheads point to a patch of Per33-GFP at the cell periphery. (Scale bar, 2 μm.) (B) The percent of cells in A with GFP in the vacuole was quantified. ***P < 0.001, Student’s t test; ns, nonsignificant. (C) Western blot analysis showing the cleavage of Per33-GFP in WT, atg40Δ, lnp1Δ, and lnp1Δ sey1Δ mutant cells after 0 or 12 h of rapamycin treatment at 30 °C. (D) Quantitation of the percent of cleaved GFP divided by total Per33-GFP for WT and mutant cells treated with rapamycin for 12 h. The data were normalized to the WT control. Error bars represent SEM from three separate experiments. **P < 0.01, Student’s t test.
Although the rtn1Δ, rtn2Δ, and yop1Δ single mutants do not disrupt ER-phagy (Figs. 1 and 2), ER tubules still form in these mutants. A near-complete loss of ER tubules is only achieved when RTN1, RTN2, and YOP1 are all disrupted in the same haploid strain (4, 9). When the rtn1Δ rtn2Δ yop1Δ triple mutant was examined, we found it exhibited only a modest decrease in Sec61-GFP (SI Appendix, Fig. S1 A and B) and Per33-GFP cleavage (SI Appendix, Fig. S1 C and D). Therefore, of all of the ER-shaping proteins, only the loss of Lnp1 has dramatic consequences for ER-phagy. The less severe consequences of disrupting ER tubule formation for ER-phagy are discussed in more detail below.
As rapamycin induces macroautophagy in addition to ER-phagy, we asked if Lnp1 is also needed for macroautophagy. Two different assays were performed to address this question. When macroautophagy is induced by starvation, cytosolic GFP-Atg8 is lipidated and incorporated into the autophagosomal membrane before it is delivered to the vacuole (21). Although translocation of GFP-Atg8 to the vacuole was completely disrupted in the atg1Δ mutant, no defect was observed in the lnp1Δ and atg40Δ mutants (SI Appendix, Fig. S2 A and B). To confirm this fluorescence localization result with a biochemical assay, we monitored the activity of a second macroautophagy marker, Pho8Δ60, a cytosolic form of vacuolar alkaline phosphatase that is activated in the vacuole when macroautophagy is induced (21). In contrast to the atg1Δ mutant, the lnp1Δ and atg40Δ mutants did not exhibit a significant decrease in Pho8Δ60 activity with respect to the wild-type (WT) (SI Appendix, Fig. S2C). Thus, using two different macroautophagy assays, the translocation of GFP-Atg8 to the vacuole and the activation of Pho8Δ60, we found that Lnp1 does not act in macroautophagy and therefore its role is specific to ER-phagy.
Although previous studies showed that Lnp1 binds to several proteins that contain reticulon domains (9), we could not coprecipitate Lnp1 with Atg40 (SI Appendix, Fig. S3A). Furthermore, we did not observe colocalization of Atg40 puncta and Lnp1 puncta during rapamycin treatment (SI Appendix, Fig. S3B). We also did not detect coprecipitation of Atg40 with Rtn1 during ER-phagy induction (SI Appendix, Fig. S3C). Thus, Lnp1 does not appear to exert its effects on ER-phagy via a direct interaction with Atg40. Consistent with the observation that Atg40 does not interact with Lnp1 or Rtn1, we found that the loss of Atg40 did not enhance the ER structural defects in the lnp1Δ and rtn1Δ mutants (SI Appendix, Fig. S4).
Lnp1 Regulates the Number of Sec61-Containing Autophagosomes That Are Formed During ER-Phagy.
To begin to address the role of Lnp1 in ER-phagy, we asked if the lnp1Δ mutant impacts the number of ER-containing autophagosomes that are formed. Autophagosome biogenesis starts with the assembly of a double-membrane structure called the phagophore. As the phagophore expands it engulfs proteins and membranes that are targeted for degradation and then seals these components from the cytosol to form an autophagosome. The contents of the autophagosome are then delivered to the lysosome or vacuole for degradation (11).
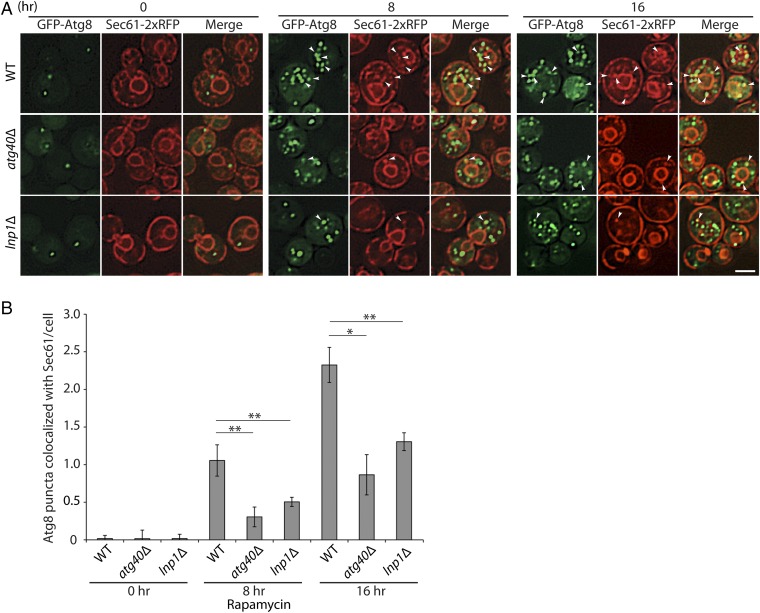
To measure autophagosomes that contain ER, we marked autophagosomes with GFP-Atg8 and ER with Sec61-2xRFP. Membrane-associated Atg8 localizes to the phagophore assembly site, phagophore, and mature autophagosomes (22). To ensure that we were quantitating fully formed autophagosomes and not autophagosomal precursors and to increase the number of autophagosomes we were analyzing, we disrupted YPT7 in all strains studied. Ypt7 is a Rab GTPase that is needed for the fusion of autophagosomes with the vacuole (22). The loss of Ypt7 therefore blocks autophagosome turnover and leads to an increase in the number of mature autophagosomes per cell. Consistent with the proposal that the GFP-Atg8–tagged puncta that colocalize with Sec61-2xRFP represent ER-containing autophagosomes, we only observed these structures in WT cells following treatment with rapamycin (Fig. 3). The number of GFP-Atg8 puncta that colocalized with Sec61-2xRFP increased after 8 and 16 h of rapamycin treatment (Fig. 3B). Previous studies demonstrated that the degradation of membrane proteins residing on both the cortical and nuclear ER is partially blocked in the atg40Δ mutant (16). Consistent with this observation, the atg40Δ mutant partially disrupted the formation of Sec61-containing autophagosomes at the 8- and 16-h time points (Fig. 3). A significant decrease in the number of GFP-Atg8 puncta that colocalized with Sec61-2xRFP was also observed at both time points in the lnp1Δ mutant (Fig. 3) relative to WT, implying that the loss of Lnp1 severely reduces the number of Sec61-containing autophagosomes that are formed during ER-phagy.
Fig. 3.
Loss of Lnp1 leads to a decrease in autophagosomes containing Sec61. (A) Autophagosomes, marked with GFP-Atg8, that also contain Sec61-2xRFP, were observed in WT, atg40Δ, and lnp1Δ mutant cells that were deleted for YPT7. The disruption of YPT7, which blocks autophagosome fusion, ensured that only autophagosomes were quantitated. Yeast cells expressing GFP-Atg8 and Sec61-2xRFP were grown for 0, 8, and 16 h at 30 °C in SC media containing rapamycin (200 ng/mL). The cells were harvested and examined directly by fluorescence microscopy. Arrowheads point to Sec61-2xRFP puncta that colocalize with GFP-Atg8 puncta. (Scale bar, 2 μm.) (B) The percent of GFP-Atg8–marked membranes that colocalized with Sec61-2xRFP in A was quantified. Error bars represent SEM from three separate experiments. *P < 0.05, **P < 0.01, Student’s t test.
Lnp1 and the Cytoskeleton Are Needed to Bring Atg40 Puncta into Proximity with the Autophagy Machinery.
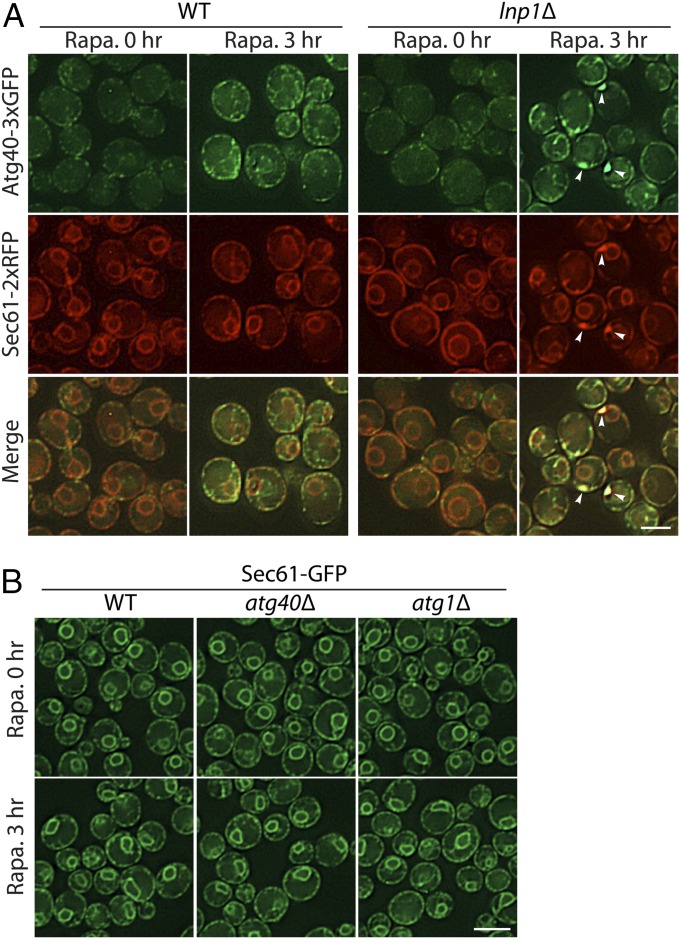
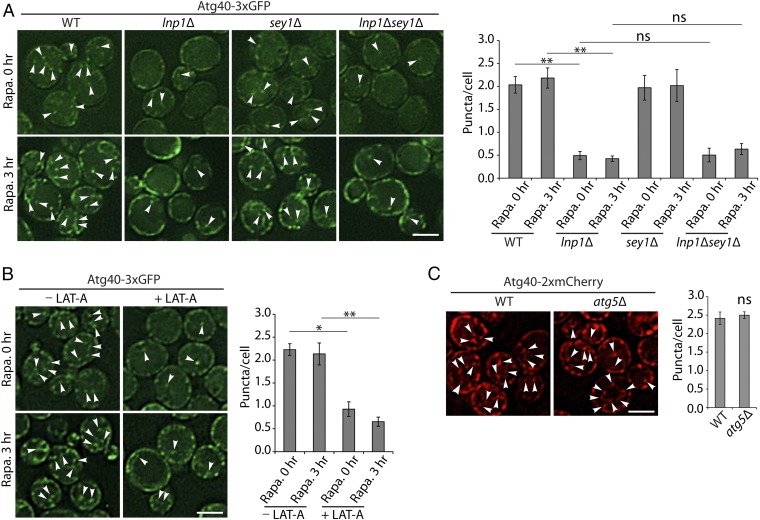
Next, we sought to understand how Lnp1 promotes the formation of Sec61-containing autophagosomes. Under normal growth conditions, Atg40 resides in punctate structures closely associated with the peripheral ER (16); however, as previously reported, the level of Atg40 undergoes a transient increase in expression during rapamycin treatment (SI Appendix, Fig. S5). As the level of Atg40 increases, Atg40 puncta within the cell interior become more apparent (Figs. 4A and 5A). We reasoned that this distribution may be important for Atg40 function, as the Atg machinery resides in the interior of the cell (23). Interestingly, we observed a reduction in the number of Atg40 puncta in the cell interior in both untreated and rapamycin-treated lnp1Δ mutant and lnp1Δ sey1Δ double mutant cells relative to WT (Figs. 4A and 5A). Furthermore, the patches of Sec61-2xRFP observed in lnp1Δ cells colocalized with Atg40-3xGFP and were consistently found at the cell cortex (Fig. 4A). No Sec61 patches were observed in atg40Δ or atg1Δ mutant cells, indicating that patch formation is specific to the loss of Lnp1 function in rapamycin-treated cells and not a general response to a defect in ER-phagy (Fig. 4B). Interestingly, while Atg40-GFP puncta are still present in the cell interior in the rtn1Δ rtn2Δ yop1Δ triple mutant (SI Appendix, Fig. S6A), the nuclear localization of Atg40, which is barely apparent in WT cells, became more prominent in the triple mutant (SI Appendix, Fig. S6A). When we quantitated the Atg40-GFP puncta in the cell interior in the rtn1Δ rtn2Δ yop1Δ triple mutant there was a modest decrease relative to WT (Fig. 6B), suggesting that the shift of Atg40-3xGFP to the nuclear envelope in the triple mutant (SI Appendix, Fig. S6) led to the small decrease in ER-phagy observed in this strain.
Fig. 4.
Patches of ER containing both Sec61 and Atg40 accumulate at the cortex in the lnp1Δ mutant. (A) WT and lnp1Δ mutant cells expressing Atg40-3xGFP and Sec61-2xRFP were incubated for 0 or 3 h at 30 °C in SC media containing rapamycin (200 ng/mL). Arrowheads mark the cytoplasmic Sec61-labeled ER patches observed in roughly 15% of the rapamycin-treated mutant cells. These patches colocalize with Atg40. (B) Sec61-labeled patches were not seen in atg40Δ or atg1Δ strains that were treated with rapamycin for 0 or 3 h. [Scale bars, 3 μm (A) and 5 μm (B).]
Fig. 5.
Fewer Atg40 puncta are observed in the cell interior in the lnp1Δ mutant and in LAT-A–treated cells. (A) Cells expressing Atg40-3xGFP were untreated (0 h) or treated for 3 h with rapamycin. Arrowheads mark Atg40 puncta located in the cell interior. (A, Right) Quantitation of Atg40-3xGFP puncta in the cell interior. Error bars represent SEM from three separate experiments. Puncta were quantitated from the following number of cells: WT at 0 h, n = 379; WT at 3 h, n = 454; lnp1∆ at 0 h, n = 511; lnp1∆ at 3 h, n = 325; sey1∆ at 0 h, n = 361; sey1∆ at 3 h, n = 288; lnp1∆ sey1∆ at 0 h, n = 306; lnp1∆ sey1∆ at 3 h, n = 413. **P < 0.01, Student’s t test; ns, nonsignificant. (B) Cells were treated with rapamycin with or without 100 µM LAT-A as described in Materials and Methods. Arrowheads mark Atg40 puncta located in the cell interior. (B, Right) Quantitation of Atg40-3xGFP puncta in the cell interior. Error bars represent SEM from three separate experiments. Puncta were quantitated from the following number of cells: −LAT-A at 0 h with rapamycin, n = 348; −LAT-A at 3 h with rapamycin, n = 292; +LAT-A at 0 h with rapamycin, n = 420; +LAT-A at 3 h with rapamycin, n = 314. *P < 0.05, **P < 0.01, Student’s t test. (C) WT and atg5Δ mutant cells were treated for 3 h with rapamycin. Arrowheads mark the Atg40 puncta located in the cell interior. (C, Right) Quantitation of Atg40-2xmCherry puncta in the cell interior. Error bars represent SEM from three separate experiments. Puncta were quantitated from the following number of cells: WT, n = 319; atg5∆, n = 280; ns, nonsignificant. (Scale bars, 3 μm.)
Fig. 6.
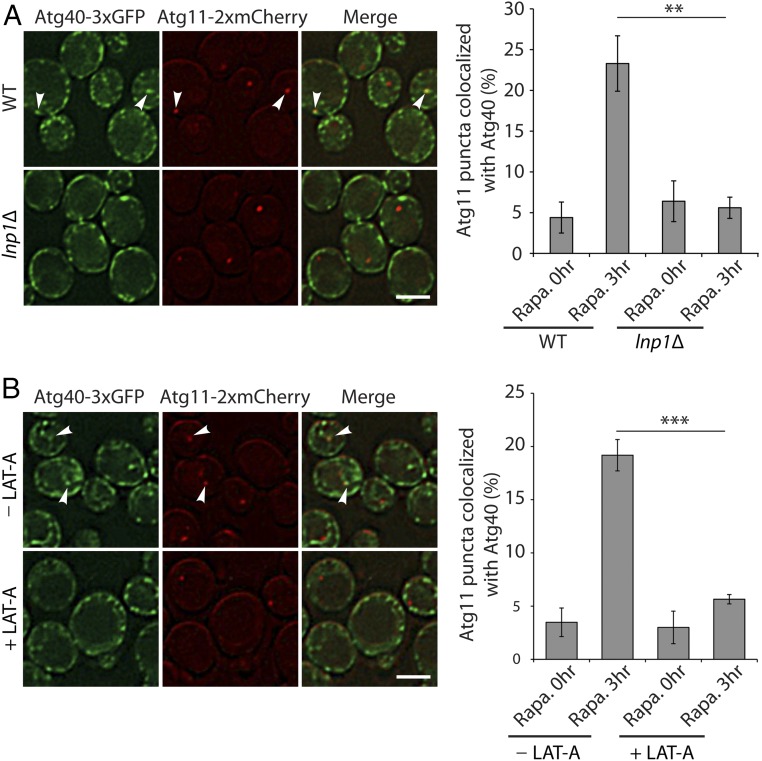
Atg40 and Atg11 fail to colocalize in the lnp1∆ mutant and in LAT-A–treated cells during the induction of ER-phagy with rapamycin. (A) WT and lnp1Δ cells expressing Atg40-3xGFP and Atg11-2xmCherry were incubated for 3 h at 30 °C in SC media containing rapamycin (200 ng/mL) before they were directly examined by fluorescence microscopy. Arrowheads point to Atg11 puncta colocalized with Atg40 puncta. (A, Right) Quantitation of the percent of Atg11-2xmCherry puncta colocalized with Atg40-3xGFP puncta after 0 or 3 h of rapamycin treatment. Error bars represent SEM from three separate experiments. **P < 0.01, Student’s t test. (B) Same as A, only WT cells were treated for 3 h with rapamycin in the absence or presence of 100 µM LAT-A. Arrowheads point to Atg11 puncta colocalized with Atg40 puncta. (B, Right) Quantitation of the percent of Atg11-2xmCherry puncta colocalized with Atg40-3xGFP puncta after 0 or 3 h of rapamycin treatment. Error bars represent SEM from three separate experiments. ***P < 0.001, Student’s t test. (Scale bars, 2 μm.)
Since previous studies showed that Latrunculin A (LAT-A), a drug that inhibits actin polymerization, blocks ER-phagy and ER dynamics (10, 24), we asked if the actin cytoskeleton is also required for the localization of Atg40 puncta within the cell interior. Consistent with the hypothesis that actin is required for the distribution of Atg40 puncta, we found that Atg40 puncta remain at the cell periphery in rapamycin-treated cells incubated with LAT-A (Fig. 5B). Impeding general autophagy in the atg5Δ mutant, however, did not alter the distribution of Atg40 puncta (Fig. 5C). Together, these findings suggest that both Lnp1 and filamentous actin are required for the subcellular distribution of Atg40 puncta.
Cargo receptors are known to link cargo to the autophagy machinery via their ability to interact with Atg11 (14), the scaffold protein for assembly of the core autophagy machinery (13). If the receptor fails to interact with Atg11, autophagosome formation is blocked (13). A possible explanation for the reduction in Sec61-containing autophagosome formation in the lnp1Δ mutant is that loss of Lnp1 impedes the association of Atg40 with Atg11. To address this possibility, we examined the colocalization of Atg40 with Atg11 during ER-phagy induction in WT, the lnp1Δ mutant, and LAT-A–treated cells. Greater than 20% of the Atg11-2xmCherry puncta were found to colocalize with Atg40-3xGFP puncta 3 h after WT cells were treated with rapamycin (Fig. 6). In contrast, Atg11-2xmCherry and Atg40-3xGFP failed to colocalize in the lnp1Δ mutant (Fig. 6A). Blocking actin polymerization with LAT-A also disrupted the colocalization of Atg11-2xmCherry with Atg40-3xGFP puncta (Fig. 6B). Together, these observations imply that Atg40 puncta require both Lnp1 and the actin cytoskeleton to come into proximity with Atg11. Failure to do so is predicted to lead to a defect in the assembly of core autophagy machinery adjacent to the ER. Collectively, these findings imply that the decrease in Sec61-containing autophagosomes observed in the lnp1Δ mutant may be a consequence of the impaired localization of Atg40 puncta to sites in the cell interior where Atg11 resides.
Discussion
Although autophagy was initially thought to be a nonselective process, recent studies have revealed it plays an important role in selectively degrading damaged organelles and protein aggregates that are too large to be degraded by the proteasome. Selective autophagy also modulates organelle number in response to environmental cues. Here we address the role of ER-shaping proteins in ER-phagy, the selective degradation of the ER.
We have found that the loss of Lnp1 leads to a dramatic reduction in ER-phagy. Lnp1 is a member of the Lunapark family (25). Members of this protein family have a zinc finger at their carboxy termini that is needed for homodimerization and two hairpin transmembrane domains at their amino termini. (9, 26). Earlier studies have shown that Lnp1 localizes to the three-way junctions of the ER network in both lower and higher eukaryotic cells and, in its absence, the cortical ER collapses into a compact, sheet-like meshwork (8, 9). Like members of the DP1/Yop1 family, the loss of Lnp1 has been reported to cause neurological defects (27).
In 2015, we reported that Lnp1 affects ER network dynamics (8). Because the small size of yeast made it challenging to image sheets and tubules, we tagged mammalian Lunapark with GFP in COS-7 cells to learn more about the role of Lnp1 in ER dynamics. In these studies, the ER was followed as it formed new polygons through tubule–tubule fusion and underwent polygon loss through ring closure. We found that, although Lnp is predominantly at three-way junctions, not all junctions contain Lnp (8). Furthermore, we reported that Lnp puncta move along tubules from one junction to another. By focusing specifically on newly formed three-way junctions, a striking finding was noted: Nascent junctions that acquired Lnp were found to be more stable than those junctions that failed to acquire Lnp. Junctions that failed to acquire Lnp were more mobile and much more likely to undergo loss through ring closure. Thus, Lnp stabilizes tubule rearrangements and, in its absence, junctions and tubules are less abundant (8, 9, 28). Together, these findings suggest that Lnp plays an important role in remodeling the ER via its ability to stabilize nascent three-way junctions. The observation that Lnp1 is required for ER-phagy is interesting in light of its proposed role in ER remodeling.
The expression of the cortical ER transmembrane protein Atg40 is induced by rapamycin treatment. How Atg40 recognizes the autophagy machinery, which resides in the cell interior, is unknown. Based on the findings we present here, we propose that the ER undergoes obligatory remodeling to facilitate ER-phagy. This event may involve the rearrangement of an ER domain, marked by Atg40, from the cortex to the cell interior. We show that both the actin cytoskeleton and Lnp1 are needed for the appearance of Atg40 puncta within the cell interior. Specifically, we found that disruption of the actin cytoskeleton with LAT-A or loss of Lnp1 function alters the steady-state distribution of Atg40 puncta in the cell. These findings are consistent with the proposed role of Lnp1 in stabilizing ER tubule rearrangements (8) and previous studies showing that LAT-A blocks ER-phagy (10). In the absence of Lnp1, the ER rearrangements necessary for ER-phagy might initiate but cannot be stabilized, which may lead to the formation of cortical patches containing both Sec61 and Atg40 in some cells. A modest reduction in ER-phagy in the rtn1Δ rtn2Δ yop1Δ triple mutant was also observed during the course of our studies. We speculate that the loss of highly curved membranes in this mutant results in the redistribution of Atg40 to the nuclear membrane. This in turn leads to a small decrease in the number of Atg40 puncta in the cell interior and ER-phagy.
We propose that the localization of Atg40 puncta to the cell interior is needed to enable Atg40 to interact with the Atg machinery. Consistent with this proposal, we find that Atg40 puncta fail to colocalize with Atg11 in the lnp1Δ mutant or in LAT-A–treated cells. Interestingly, it was recently reported that the induction of mitophagy leads to the appearance of some punctate mitochondria near the cell center that colocalize with the autophagy machinery. The punctate mitochondria near the center of the cell were not observed when mitophagy was disrupted (29).
The role of cargo receptors is to link cargo to Atg11, the scaffold for the core Atg machinery (13, 14). If the receptor fails to interact with Atg11, cargo cannot be packaged into autophagosomal membranes. The failure of Atg40 to colocalize with Atg11 in the lnp1Δ mutant could explain why we observe fewer autophagosomes that contain Sec61 in the lnp1Δ mutant. As Lnp1 is not required for macroautophagy or for autophagosome formation per se, an inability to package Sec61 into autophagosomes is the most likely explanation for the observed decrease in the number of Sec61-containing autophagosomes in the lnp1Δ mutant.
In summary, here we report that the presence of Atg40-containing ER structures within the cell interior is dependent on the actin cytoskeleton and Lnp1. Our findings support the proposal that the internal cellular localization of Atg40 is critical for ER-phagy. Currently, the precise role of Lnp1 in ER-phagy is unknown; however, an attractive possibility is that Lnp1 plays a role in stabilizing these structures in a manner that is analogous to the way that it stabilizes newly formed three-way junctions.
Materials and Methods
Strains and Growth Conditions.
All strains used in this study are listed in SI Appendix, Table S1. To induce ER-phagy, yeast cells were grown to OD600 0.1 to 0.2 in synthetic complete media (SC) and treated with 200 ng/mL rapamycin for 12 to 24 h at 30 °C. For the macroautophagy assays, cells were grown to OD600 0.5 and then shifted to synthetic media containing 2% glucose without ammonium sulfate and amino acids (SD-N media) for 1 to 2 h.
Fluorescence Microscopy.
Approximately 1.0 OD600 unit of cells was pelleted at 800 × g, resuspended in 50 to 200 μL of growth medium, and examined on an Axio Imager Z1 fluorescence microscope (Zeiss) using a 100× oil-immersion objective. Images were captured with an AxioCam MRm digital camera and AxioVision software (Zeiss), and deconvolved using OpenLab software (Improvision). For the Latrunculin A experiments, the drug (10 mM stock in DMSO) was added to a final concentration of 100 µM and incubated for 30 min before rapamycin was added.
For the fluorescence ER-phagy assays, cells were imaged as described above, and the percent of cells with GFP in the vacuole/total number of cells was calculated for each sample.
FM4-64 Staining.
To visualize vacuoles, yeast cells were harvested and resuspended in YPD media to an optical density of OD600 1.0. FM4-64 was then added to a final concentration of 1.6 μM. After incubation with FM4-64 at 30 °C for 30 to 60 min, cells were washed twice with YPD media and viewed by fluorescence microscopy.
Cleavage Assays.
To observe the cleavage of Sec61-GFP or Per33-GFP during the induction of ER-phagy, cells were grown to OD600 0.1 to 0.2 in SC media and treated with 200 ng/mL rapamycin for 0 or 12 h at 30 °C. Subsequently, the cells (5 OD600 units) were pelleted, resuspended in 0.1 M NaOH, and incubated for 5 min at room temperature. For the Sec61-GFP cleavage assays, samples were heated at 100 °C in sample buffer (62.5 mM Tris⋅HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol, pH 6.8) for 10 min. For the Per33-GFP cleavage assays, samples were solubilized in cracking buffer (50 mM Tris⋅HCl, pH 6.8, 8 M urea, 5% SDS, 5% 2-mercaptoethanol) and subsequently heated in sample buffer at 55 °C for 10 min. All samples were subjected to SDS/PAGE and the cleaved GFP product was probed by immunoblotting with anti-GFP mouse monoclonal antibody (1:3,000 dilution; clones 7.1 and 13.1; Roche). As a loading control, Adh1 was analyzed using anti-Adh1 rabbit polyclonal antibody (1:10,000 dilution; AB1202; EMD Millipore).
Vacuolar Alkaline Phosphatase Activity.
Alkaline phosphatase assays (Pho8Δ60) were performed as described previously (21). Briefly, yeast cells were grown to OD600 0.5 to 1.0, washed twice with water, resuspended in SD-N media, and incubated at 30 °C for 2 h. Cells (5.0 OD600 units) were harvested and resuspended in 300 μL of ice-cold lysis buffer (20 mM Pipes, pH 7.2, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 μM ZnSO4, 1 mM PMSF). Glass beads, equal to the volume of the sample, were added and the samples were vortexed for 1 min, followed by another 1-min incubation at 4 °C. This was repeated four more times before the lysates were centrifuged at 15,700 × g for 5 min at 4 °C. The supernatant (100 μL) was transferred to a tube containing 400 μL of assay buffer (1.25 mM p-nitrophenyl phosphate, 250 mM Tris⋅HCl, pH 8.5, 0.4% Triton X-100, 10 mM MgSO4, 10 μM ZnSO4) and incubated at 37 °C for 20 min. At the end of the incubation, the reaction was terminated with 500 μL of stop buffer (1 M glycine/KOH, pH 11.0) before OD400 was measured. Activity was normalized to equal protein concentration.
Immunoprecipitation.
Yeast cells were grown to early log phase at 30 °C. To induce ER-phagy, cells were grown to OD600 0.1 to 0.2 and treated with 200 ng/mL rapamycin at 30 °C. Cells were pelleted at 2,300 × g, washed once with ice-cold water, and resuspended in spheroplasting buffer (1.4 M sorbitol, 50 mM NaPi, pH 7.4, 50 mM 2-mercaptoethanol, 10 μg/OD600 Zymolyase-100T). The resuspended cells were incubated at 37 °C for 30 min with gentle shaking and pelleted through a chilled sorbitol cushion (1.7 M sorbitol, 20 mM NaPi, pH 7.4), and the pellet was resuspended in lysis buffer (25 mM NaPi, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor, 1% Triton X-100, pH 7.4) using a dounce homogenizer (40 strokes). The lysates were centrifuged at 37,000 × g for 20 min at 4 °C to remove cell debris. For the dithiobis(succinimidyl propionate) (DSP) experiments, lysates were incubated on ice with 200 μg/mL DSP for 60 min. Tris⋅HCl (pH 7.6; 100 mM) was added and the samples were incubated for 15 min on ice to quench excess cross-linker.
Lysates were incubated overnight at 4 °C with anti-FLAG antibody (clone M2; F1804; Sigma) before protein G agarose (Thermo) resin was added to the lysates and incubated at room temperature for 2 h. The protein G agarose was pelleted at 2,300 × g for 30 s and washed three times with ice-cold lysis buffer that contained 0.5% Triton X-100. The resin was heated to 100 °C in sample buffer (62.5 mM Tris⋅HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol, pH 6.8) for 5 min. The eluted protein was subjected to SDS/PAGE and immunoblotted with anti-HA (1:2,000 dilution; clone HA.11, MMS-101R; Covance), anti-Rtn1 (1:1,000 dilution; laboratory-generated antibody), and anti-FLAG (1:2,000 dilution; clone M2, F1804; Sigma) antibodies. The secondary antibodies used were goat anti-mouse IgG-HRP (1:10,000 dilution; W402B; Promega) or donkey anti-rabbit IgG-HRP (1:10,000 dilution; NA9340V; GE Healthcare).
Analysis of Atg40 Expression Levels During Rapamycin Treatment.
Cells expressing Atg40-3xFLAG were grown in SC media to OD600 0.1 to 0.2 at 30 °C before rapamycin (200 to 400 ng/mL) was added. Samples (50 OD600 units) were harvested at the indicated time points and washed twice with 0.5 mL of ice-cold water. The cell pellets were resuspended in 200 µL of lysis buffer (25 mM Hepes, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor, 0.5% Triton X-100, pH 7.4) and vortexed five times in the presence of glass beads (1 min each time). The cell debris and beads were removed by centrifugation at 15,700 × g, and the protein concentration of the supernatant was measured using the Bradford assay. Samples were heated to 100 °C in sample buffer (62.5 mM Tris⋅HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol, pH 6.8) for 5 min, subjected to SDS/PAGE, and immunoblotted with anti-HA (1:2,000 dilution; clone HA.11, MMS-101R; Covance), anti-GFP (1:2,000 dilution; clones 7.1 and 13.1; Roche), and anti-Adh1 (1:10,000 dilution; AB1202; EMD Millipore) antibodies. The secondary antibodies used were goat anti-mouse IgG-HRP (1:10,000 dilution; W402B; Promega) or donkey anti-rabbit IgG-HRP (1:10,000 dilution; NA9340V; GE Healthcare).
Statistical Analysis.
P values were calculated using the Student’s t test. The statistical significance reported is from three or more independent experiments and is presented as mean values. The error bars in the figures represent SEM, as specified in the legends.
Supplementary Material
Acknowledgments
We thank Serena Cervantes for the initial characterization of the ER-shaping mutants for defects in ER-phagy, and Randy Hampton for helpful discussions. Salary support for S.F.-N., S.C., Y.C., and S.P. was provided by the National Institute of General Medical Sciences (NIGMS) under Awards 1R01GM115422 and 1R01GM115422. Salary support for P.J.N. was provided by NIGMS under Awards 1R01GM35370 and 1R01GM082861.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805032115/-/DCSupplemental.
References
- 1.Du Y, Ferro-Novick S, Novick P. Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci. 2004;117:2871–2878. doi: 10.1242/jcs.01286. [DOI] [PubMed] [Google Scholar]
- 2.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25:428–433. doi: 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 5.De Craene JO, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, et al. Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2015;112:418–423. doi: 10.1073/pnas.1423026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol. 2012;14:707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamasaki M, Noda T, Baba M, Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 12.Anding AL, Baehrecke EH. Cleaning house: Selective autophagy of organelles. Dev Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torggler R, et al. Two independent pathways within selective autophagy converge to activate Atg1 kinase at the vacuole. Mol Cell. 2016;64:221–235. doi: 10.1016/j.molcel.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 16.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 17.Grumati P, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017;6:e25555. doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MD, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018;44:217–232.e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fumagalli F, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 20.Chadrin A, et al. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 22.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 24.Prinz WA, et al. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 26.Casey AK, Chen S, Novick P, Ferro-Novick S, Wente SR. Nuclear pore complex integrity requires Lnp1, a regulator of cortical endoplasmic reticulum. Mol Biol Cell. 2015;26:2833–2844. doi: 10.1091/mbc.E15-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghila L, Gomez M. The evolutionarily conserved gene LNP-1 is required for synaptic vesicle trafficking and synaptic transmission. Eur J Neurosci. 2008;27:621–630. doi: 10.1111/j.1460-9568.2008.06049.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Tukachinsky H, Romano FB, Rapoport TA. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife. 2016;5:e18605. doi: 10.7554/eLife.18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita SI, et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J Cell Biol. 2016;215:649–665. doi: 10.1083/jcb.201605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.