Significance
Opportunities to study microevolution in wild populations are rare and challenging. Annual monitoring allowed us to capture both the prelude to and aftermath of one of the largest marine mass mortality events on record in a keystone marine species. Median mortality of 81% across populations was recorded along with significant allele frequency shifts at multiple loci in the adult population. Shifts were consistent across locations and also occurred in new recruits, with few exceptions. These results indicate a long-term species-wide change in allele frequencies will persist through future generations. Population genomic monitoring, at a time when marine diseases and mass mortalities are on the rise, will be essential for documenting rapid genetic shifts in response to chronic and extreme events.
Keywords: disease, marine, mass mortality, natural selection, rapid evolution
Abstract
Standing genetic variation enables or restricts a population’s capacity to respond to changing conditions, including the extreme disturbances expected to increase in frequency and intensity with continuing anthropogenic climate change. However, we know little about how populations might respond to extreme events with rapid genetic shifts, or how population dynamics may influence and be influenced by population genomic change. We use a range-wide epizootic, sea star wasting disease, that onset in mid-2013 and caused mass mortality in Pisaster ochraceus to explore how a keystone marine species responded to an extreme perturbation. We integrated field surveys with restriction site-associated DNA sequencing data to (i) describe the population dynamics of mortality and recovery, and (ii) compare allele frequencies in mature P. ochraceus before the disease outbreak with allele frequencies in adults and new juveniles after the outbreak, to identify whether selection may have occurred. We found P. ochraceus suffered 81% mortality in the study region between 2012 and 2015, and experienced a concurrent 74-fold increase in recruitment beginning in late 2013. Comparison of pre- and postoutbreak adults revealed significant allele frequency changes at three loci, which showed consistent changes across the large majority of locations. Allele frequency shifts in juvenile P. ochraceus (spawned from premortality adults) were consistent with those seen in adult survivors. Such parallel shifts suggest detectable signals of selection and highlight the potential for persistence of this change in subsequent generations, which may influence the resilience of this keystone species to future outbreaks.
As extreme disturbances potentially increase in both frequency and intensity (1, 2), a population’s capacity to respond with standing genetic variation will become increasingly important for rapid adaptation (3–8). Impacts will be mediated via physiological tolerances and life-history traits that may reflect underlying genetic differences (9), and these genetic differences—among and within populations—may amplify or mute a species’ sensitivity to change (4, 10). However, we know little about what determines which organisms are resilient or susceptible, whether or how impacted populations may recover, and if recovery increases resilience to future perturbations.
Standing genetic variation is shaped primarily by effective population size, selection, and life history of the species (11). Detecting changes in standing genetic variation due to selection in wild populations is challenging due to demographic processes in contemporary populations—such as range expansions, growth, and asymmetric migration resulting in complex spatial genetic patterns (12)—leading to potentially false conclusions about selective sweeps (13). A strategy to sidestep this problem involves identifying adaptive alleles in an ancestral population that have increased frequency in descendent populations; however, it is not always possible to know if a beneficial allele was secondarily introduced to the ancestral population (13). Additionally, selection can manifest in different ways in the genome: For example, a selected trait can be controlled by a few genes of large effect or many genes of small effect; in the latter case, the intensity of selection is diluted over many genes, resulting in only small changes in allele frequency (14). Marine taxa can pose particular challenges due to a suite of common traits—high fecundity, large population size, and high dispersal potential—acting to homogenize the gene pool and consequently restricting signals of selection to very small genomic regions (15); however, multiple studies have detected evidence of selection in such “typical” species (16).
A recent epizootic provided an opportunity to overcome many of these challenges. In 2013, a range-wide sea star wasting disease (SSWD) outbreak leading to mass mortality across the range of Pisaster ochraceus (17) created a rare opportunity to explore the genetic landscape in which selection acts, and to identify alleles that responded directly to the event through changes in frequency. A study of P. ochraceus immediately preceding the outbreak (18) was available to be coupled with samples following the disease outbreak, at multiple locations, along with quantitative surveys of mortality, to capture both the initial standing genetic variation and the aftermath of the largest documented noncommercial marine pandemic (19), which putatively was caused by a virus (17). Thus, individuals with different functional states (symptomatic–asymptomatic), and “parent” and “daughter” (sub)populations with differential responses (i.e., mortality rates), could be identified easily. Additionally, the epizootic offered the opportunity to explore how newly recruited P. ochraceus, which were spawned by preepizootic adults (SI Appendix, Fig. S1), did or did not show similar genetic changes as the adult population. Thus, we were able to measure the actual change from the original standing genetic stock and identify candidate loci under selection during this disease outbreak.
The goals of this study are threefold: (i) to quantify the magnitude of mortality and recruitment in north central California, (ii) ascertain whether selection may have shaped the surviving population of adult P. ochraceus, and (iii) test whether juvenile P. ochraceus recruiting during the mortality event are most genetically similar to the pre-SSWD population (i.e., no detectable selection in recruits) or the surviving population (i.e., selection in parallel to adults). In doing so, we aim to shed light on how this species, which in many ways is a typical marine species—large population size, broad range size, high gene flow, high fecundity, and high dispersal potential (20)—responds to an extreme event and what signature this may leave on the subsequent generation.
Results
Mortality and Recruitment in P. ochraceus.
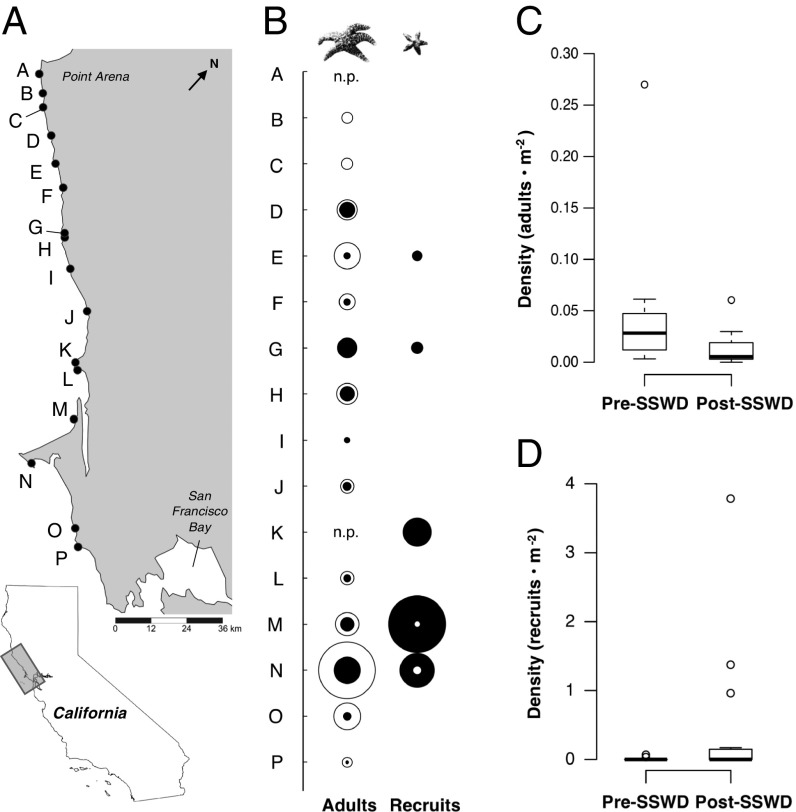
Following the outbreak of sea star wasting disease in 2013, median density of post-SSWD P. ochraceus adults (0.005·m−2; quartile Q1 0.003·m−2, Q3 0.018·m−2) was 81% lower than pre-SSWD densities (0.028·m−2; Q1 0.013·m−2, Q3 0.045·m−2; Wilcoxon signed-rank test, V = 105, df 1, P = 6.104 × 10−5; Fig. 1). For recruits (1.0 to 15.3 mm measured from center to arm tip; mean 5.9 mm; median 5.0 mm), there was a 74-fold increase in mean density (0.006·m−2 pre-SSWD; 0.401·m−2 post-SSWD) driven by a subset of sites (Fig. 1), though the change was not statistically significant due to high heterogeneity and an abundance of zeros.
Fig. 1.
Spatial and temporal variation in the abundance of P. ochraceus before and after the 2013 SSWD outbreak. (A) Map of study sites. (B) Densities of adults and recruits from before (white) and after (black) the outbreak shown by concentric bubbles; the area of the bubbles is proportional to the density (adults: maximum density 0.27·m−2; recruits: maximum density 3.78·m−2). Adult densities had a median decrease of 81% (range 9 to 100%). (C and D) Boxplots of mean density per square meter across surveyed sites for (C) adults and (D) <1-y recruits. n.p., no paired data. The map was created with SimpleMappr (www.simplemappr.net).
Sampling of Genetic Variation.
After filtering 63 individuals with low coverage, 374 samples were available for genetic analyses: 142 pre-SSWD adults, 126 post-SSWD adults, and 106 recruits (SI Appendix, Table S1). dDocent (21) identified a total of 3,546,608 variable sites. After filtering, we resolved 6,790 haplotypes (based on 7,303 SNPs) for 1,225 restriction site-associated DNA (RAD) loci for which direct allele frequency shifts were measured. No significant population genetic structure was found in pre-SSWD adults (FST = 0.0001) or post-SSWD adults (FST = −0.0037), and no clonality was detected across all samples (i.e., zero duplicated multilocus genotypes). Genetic diversity was consistent among samples: pre-SSWD (Hs = 0.434), post-SSWD adults (Hs = 0.432), and recruits (Hs = 0.433) (SI Appendix, Fig. S2).
Tests for Selection in Adult P. ochraceus.
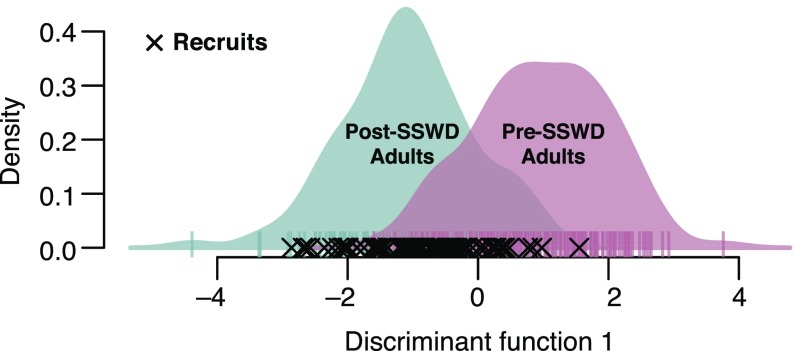
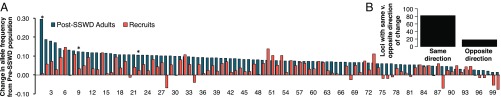
Discriminant analysis of principal components (DAPC) revealed genetic differences between pre- and post-SSWD adult P. ochraceus (Fig. 2). The top 100 discriminatory haplotypes identified in the DAPC analysis had a mean change in frequency of ±0.083 (SD 0.040) discriminating pre- from postepizootic P. ochraceus samples (Fig. 3); the mean frequency shift across all haplotypes was ±0.018 (SD 0.020). The BayeScan test for FST outliers yielded three outlier loci at a false discovery rate (FDR) of 0.10 (Fig. 3). These outlier loci were also detected within the top 100 discriminatory haplotypes from the DAPC analysis. Overall change in heterozygosity was negligible (Ho_PRE = 0.421; Ho_POST = 0.423), though heterozygosity for outlier loci did change in the surviving population, increasing at one locus and decreasing in the other two (SI Appendix, Fig. S3).
Fig. 2.
Discriminant analysis of principal components of 6,790 haplotypes from 1,225 ddRAD-generated loci comparing pre- (n = 142) and post-SSWD (n = 126) populations of adult P. ochraceus and assignment of <1-y-old recruits (indicated by x) (n = 106). The distribution of recruits is left-shifted relative to pre-SSWD adults. Cumulative variance explained by the number of retained principal components was 47.7%; one discriminant function was used (n groups − 1) in the DAPC (F statistic 276.4).
Fig. 3.
(A) Difference in allele frequency for post-SSWD adults and recruits relative to the pre-SSWD adults for the top 100 discriminatory haplotypes (after filtering to retain a single haplotype per locus) identified in the DAPC analysis. The y = 0 line represents no change from pre-SSWD allele frequencies. Absolute value of difference is shown for post-SSWD adults. Asterisks indicate outlier loci from BayeScan (FDR 0.10)—from left to right: Loc1048_Hap1, Loc0379_Hap3, and Loc1166_Hap2. (B) Total haplotypes showing the same vs. opposite direction of frequency change in post-SSWD adults and recruits from pre-SSWD adult frequencies. Haplotype names are in Dataset S2.
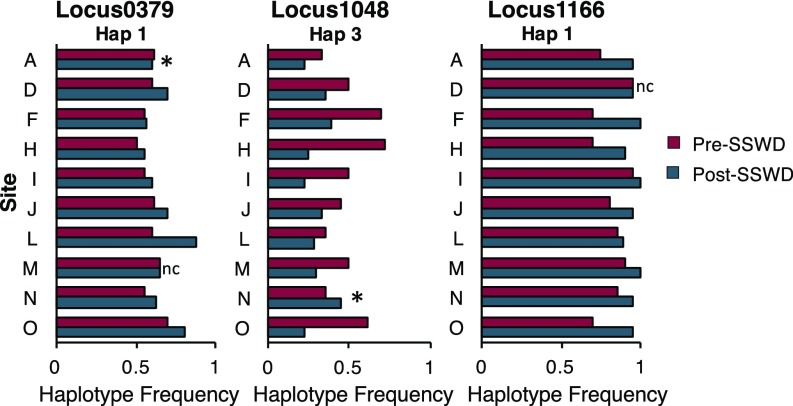
Allele frequency changes in the three outlier loci (Fig. 4) were significant after a sequential Bonferroni adjustment for three tests (i.e., three loci) (Locus0379: t = −2.93, df 9, P = 0.01664; Locus1048: t = 3.78, df 9, P = 0.00434; Locus1166: t = −4.49, df 9, P = 0.00151) and largely consistent across geographic locations, with the most common haplotype in each outlier locus showing consistent directional changes in frequencies at 8/10 locations (Locus0379) and 9/10 locations (Locus1048 and Locus1166) (Fig. 4 and SI Appendix, Fig. S4). However, despite the geographic consistency, these results were nonsignificant at α = 0.05 for Locus1048 and Locus1166—which each changed similarly at 9/10 locations (Fisher’s exact test: n = 10, df 1, P = 0.0704, power 0.72)—and for Locus0379, which changed similarly across 8/10 locations (Fisher’s exact test: n = 10, df 1, P = 0.1749, power 0.48).
Fig. 4.
Consistent change in allele frequency from pre- to post-SSWD adults across geographic locations for the most common haplotypes in the putatively selected outlier loci identified in BayeScan (FDR 0.10). Asterisks indicate the direction of allele frequency change differs from other geographic locations for a particular locus. Only sites with P. ochraceus samples of n ≥9 for both pre- and post-SSWD are included (SI Appendix, Table S1). Letters represent geographic sites; detailed locations are in SI Appendix, Table S1. nc, no change.
In a BLASTn query of the top 100 discriminating RAD loci, 85 loci mapped to echinoderm transcripts in EchinoDB (echinodb.uncc.edu) (22) (e < 1.0E-6) and 75 loci mapped to the Pycnopodia helianthoides transcriptome (23) (Dataset S1).
Estimating Genetic Affinity of New Recruits.
DAPC results show 84.0% of the new recruits collected following the outbreak of SSWD assign to postoutbreak adult P. ochraceus (Fig. 2 and SI Appendix, Fig. S5) (n = 106, α = 0.01, χ2 = 48.906, critical value = 6.635, df 1, power 1.00). Comparisons of the frequencies of the top 100 discriminant haplotypes also reveal a greater magnitude of difference between pre- and post-SSWD adults than pre-SSWD adults and recruits (Fig. 3). However, though the magnitude differs, the direction of change is largely consistent; we find 82% of haplotypes (n = 100, α = 0.01, χ2 = 40.960, critical value = 6.635, df 1, power 1.00) share the same direction of change in frequencies in post-SSWD adults and recruits (relative to pre-SSWD adults) (Fig. 3B). Additionally, of those 82% with a similar direction of change, 80% of recruit allele frequencies represent an intermediate frequency between pre- and post-SSWD adults (Fig. 3).
Discussion
P. ochraceus suffered elevated mortality, and experienced a coincident pulse of elevated recruitment, during the sea star wasting epizootic (Fig. 1). Direct comparisons of RADseq (RAD sequencing) data among pre- and post-SSWD adults and recruits revealed potential signals of selection as detected in parallel shifts in allele frequencies (Figs. 3 and 4). This was further corroborated by the BayeScan analysis at ∼0.2% of loci (Fig. 3) and the largely consistent change in allele frequency across geographic locations (Fig. 4). While newly recruited P. ochraceus were the progeny of pre-SSWD adults (SI Appendix, Fig. S1), shifts in allele frequencies in top candidate loci occurred in the same direction in the recruits as in the post-SSWD adult sample, relative to the pre-SSWD sample (Fig. 3); heterozygosity in recruits was intermediate between pre- and post-SSWD adults (SI Appendix, Fig. S3). These parallel shifts in allele frequencies in independent samples of adults and recruits suggest a significant selection event concurrent with the sea star wasting disease epizootic.
Putative Causes of Allele Frequency Changes in P. ochraceus.
Our empirical measurements of allele frequency changes in outlier and the 100 top discriminating loci during the decimation of P. ochraceus by an epizootic disease are consistent with theory, which would predict that selection rather than genetic drift or gene flow would drive differentiation in highly abundant and fecund species (14). We also were able to empirically rule out gene flow as a cause of allele frequency changes, because we sampled the surviving adults—likely ≥5 y old given their size (24)—from the same pool of adults sampled before the SSWD outbreak; adult P. ochraceus do not disperse. Likewise, while larval dispersal may have contributed immigrant alleles, these were unlikely to influence conclusions based on region-wide comparison of pre-SSWD adults and recruits, because P. ochraceus is well-mixed over large spatial scales (20) and the SSWD outbreak was range-wide. Moreover, while it can be difficult to distinguish between selection and drift using changes in allele frequencies between time-series samples (25), empirically it is improbable that drift leads to random allele frequency shifts in the same direction in post-SSWD adults and new recruits (Fig. 3). Consistency in the direction of allele frequency shifts in outlier loci across independently sampled geographic locations provides additional corroborating evidence consistent with selection (Fig. 4).
The weaker signal of selection in juveniles, relative to adults, can be explained by the life history of P. ochraceus. The answer lies in reconstructing the timeline of reproduction, disease onset, and recruitment (SI Appendix, Fig. S1). P. ochraceus spawn from late March to May in central California (26, 27)—slightly later to the north, for example, late spring on San Juan Island (28) and May to early June in Oregon (29)—and have a pelagic duration of 6 to 8 wk (30), though this species is apparently capable of much longer larval periods (up to 32 wk) in the laboratory setting (31). This timeline would lead to intertidal settlement in approximately June through August. Even if we consider the earliest spawning (late March) (27) and shortest pelagic duration (6 wk) (26) published for P. ochraceus, the earliest settlement of recruits would have occurred in May 2014 (if spawned by post-SSWD adults). Although we did collect recruits during this month (SI Appendix, Fig. S1 and Table S2), we found they were the largest of all recruits sampled from December 2013 to May 2014 (SI Appendix, Fig. S6), suggesting these recruits had been settled for some time. Recruits for this study therefore would have been spawned in the previous year, but likely not after major mortality from SSWD was seen in adults. Observations of sea star wasting disease were first documented in this region in late summer 2013 (seastarwasting.org). Given the lability of life-history traits, one should also consider whether spawning could have occurred after adult mortality from SSWD, that is, fall 2013; however, the size distribution of recruits observed from December 2013 to May 2014 (SI Appendix, Fig. S6) indicates an earlier settlement, that is, during summer 2013 (SI Appendix, Fig. S1). The timeline indicates the recruits in this study settled during summer and then were exposed to SSWD for ∼4 to 12 mo in the intertidal zone before collection. The postoutbreak adults were sampled approximately 1 y after recruits were sampled, meaning they would have experienced additional exposure relative to recruits before collection (SI Appendix, Table S1). Thus, although juvenile P. ochraceus would have been spawned by pre-SSWD adults, they became genetically more similar to post-SSWD adults (Fig. 2 and SI Appendix, Fig. S5), as shown by the top 100 discriminating haplotypes in the DAPC analysis, because conditions were consistent with those seen in post-SSWD adults (Fig. 3). We consider sweepstakes reproductive success (32) and chaotic genetic patchiness (33) unlikely explanations of the observed patterns because both depend on chance mechanisms, which do not predict broad geographic consistency, and the same subset of adults being reproductively successful and surviving a disease outbreak. A multiyear recruitment study of another large-population high-fecundity spawning echinoderm, Strongylocentrotus purpuratus, revealed no evidence for the sweepstakes reproductive success hypothesis (34). The more probable explanation for all statistical outlier loci behaving consistently across the large majority of sites, and juveniles shifting heterozygosity consistent with the shift seen in post-SSWD adults, is that juveniles underwent similar, but less strong, selective pressure as adults consistent with SSWD.
The limitations of reduced representation genomic techniques—for example, the ability to detect signals of selection in polygenic traits (35) unless sweeps have been hard and recent, particularly when linkage disequilibrium is low (36–38)—and the challenges of studying the relative importance of selection in wild populations (39) were mitigated by the nature of our dataset. These challenges can result in a lack of power to detect selection, not an increase in false positives. Paired temporal samples allowed direct measurement of allele frequency changes between the pre- and post-SSWD samples and helped to maximize power.
Of the 75 RAD loci that mapped to the P. helianthoides transcriptome, 19 mapped to transcripts that were differentially expressed between asymptomatic and symptomatic P. helianthoides (23) (Dataset S1). While these results are consistent with a subset of putatively selected functional loci (Fig. 3), gene modeling and differential expression analyses are needed to describe the genomic context for the genetic shifts seen in P. ochraceus. Despite similar symptoms observed among asteroid species, there is potential for species-specific responses to SSWD. Responses to viral challenge experiments were not consistent among species, suggesting more complex and/or multiple etiologies are associated with SSWD (40).
The Future of P. ochraceus.
P. ochraceus suffered major mortality between 2013 and 2014 associated with the SSWD epizootic (Fig. 1) (41), but also experienced record recruitment rates up to 300 times previous rates (41). Sea star wasting disease is still present in P. ochraceus populations in 2018 (42) and likely still exerting selection on susceptible individuals in otherwise seemingly somewhat resilient populations. Despite recently elevated recruitment rates, these new recruits will not contribute to reproduction for some time; P. ochraceus do not reach maturity until ∼5 y of age (24), although maturation time could be shorter if P. ochraceus feed to satiation (43). This lag time makes predictions about recovery difficult, given continued persistence of the disease in P. ochraceus.
After a period of very low recruitment in P. ochraceus, ending in 2012 (Fig. 1) (41), elevated pulses of recruitment have continued through at least 2017 in this region. All recruits after 2013 would have been spawned by the post-SSWD population, suggesting the shift in allele frequency should be maintained in future generations. For subsequent generations, given long-term consequences of genetic drift are diminished in populations that bottleneck for only a small number of generations and subsequently expand rapidly (44, 45), it seems unlikely genetic drift will be important, given the large pulse of concurrent recruitment.
However, a number of factors could influence the trajectory of change in future populations. For example, elevated recruitment could introduce nonresistant alleles from other locations given heterogeneity in mortality and dilute the effects of selection for disease resistance (46). Additionally, sweepstakes reproduction is thought to be common in taxa with high fecundity and high larval mortality, whereby only a subset of adults contribute to successful cohorts of juveniles (32), potentially narrowing the gene pool on which selection can act in new recruits; however, we see no evidence of reduced genetic diversity in the recruits sampled, even within this single cohort (SI Appendix, Fig. S2). Genotyping of individuals from subsequent pulses of recruitment is needed to determine genetic relationships between adults and juveniles, and to help us better understand the range of possible futures for P. ochraceus in the aftermath of this outbreak of sea star wasting disease.
A major concern for species affected by environmental change is that adaptation is outpaced by environmental change (47), but currently our understanding of environmental stressors is incomplete. Although temperature has not been clearly linked to initial SSWD emergence it is perceived to play a role in exacerbating the disease (42), but whether the role is in elevated temperature (19, 48) or reduced temperature (41), or some combination, is unresolved. Additional environmental factors known to influence P. ochraceus include CO2 (49) and salinity (50), but how these might interact with SSWD is currently unknown. Clear genetic responses to SSWD have been documented in the sea star P. helianthoides, with increased expression of genes associated with immune pathways (23). It is conceivable, if observed shifts in allele frequencies in P. ochraceus are linked with increased survivorship, leading to reproductive success, that P. ochraceus may fare better in future outbreaks of SSWD if caused by the same pathogen. Insight into this issue might be gleaned by comparing the recent outbreak of SSWD with prior geographically restricted outbreaks (51–53) and ongoing monitoring. Nonetheless, given its large population size, extensive gene flow, and the high standing stock of genetic variation on which selection can act (54), P. ochraceus seems to have a propensity to persist and respond to perturbation.
Materials and Methods
Ecological Surveys.
Quantitative surveys of P. ochraceus were conducted at 16 sites (Fig. 1 and SI Appendix, Table S1) the year preceding (August 2012 to May 2013) and following (December 2013 to May 2014, January 2014, and December 2014 to May 2015) the 2013 SSWD outbreak. At each site, we sampled two rocky intertidal areas (usually one on either side of a beach or headland and separated by ∼100 m), using quadrats to estimate juvenile abundance and transects to target larger adult P. ochraceus. [N.B. Sites A and K, Arena Cove and Bodega Reserve (Fig. 1 and SI Appendix, Table S1), only had paired quadrat surveys.] All specimens were georeferenced using a Garmin C60x GPS (±3-m precision). Data were reported as the number of individuals per square meter per site (SI Appendix, Table S2).
Quadrats.
We exhaustively searched 32 to 40 1-m-square quadrats per site (i.e., 16 to 20 per each of two areas within a site), recording GPS waypoint, time, percent cover of major substrate and macrophytes, and abundances and sizes of P. ochraceus. Individuals were categorized as juvenile/recruit if their length from center to arm tip was ≤20 mm. Quadrat locations were selected by first finding one of the target habitat types—surf grass, low-zone red algae, coralline turf, cobble or boulder field, or urchin pools either empty or occupied—selecting a starting point haphazardly, and then using a random numbers table (range 1 to 10 m) to choose remaining quadrat locations.
Transects.
To quantify changes in mature P. ochraceus density, we conducted GPS-tracked 2- or 4-m-wide swath transects nested in each of two areas at each site. From a distance, an approximate starting point and orientation (with landmarks) for the starting transect were selected. Transects ran from the most shoreward to the most seaward possible suitable habitat at ∼10-m intervals along the shore, particularly targeting the low intertidal zone when the tide was maximally receded, with as many transects being done as permitted by the tide. The GPS autorecorded a trackpoint every 6 s. To reduce error in estimates of the length and position of swathes (commonly ±3 m), we smoothed tracks by averaging across windows of two consecutive trackpoints and removed outlying trackpoints that led to a Euclidean distance ≥8 m, since these were likely due to temporary dropouts in satellite signal. We calculated total transect search area by multiplying the adjusted transect length by swath width. To estimate the change in density from pre- to post-SSWD P. ochraceus on transects, we used individuals with at least a 40-mm radius (which was chosen based on the minimum size of adult P. ochraceus observed at each site in 2012 to 2013 surveys; SI Appendix, Fig. S7) to ensure calculations were as inclusive of juveniles and adults as possible but influenced minimally by recruits of the previous 2 y (43).
Statistical analyses.
Normality was tested in adult and recruit datasets using the Shapiro–Wilk normality test using stats v.3.3.2 in R (55). Given normality was rejected in adults and recruits (Shapiro–Wilk normality test; adults: W = 0.502, P = 1.195 × 10−8; recruits: W = 0.324, P = 4.992 × 10−11), the nonparametric Wilcoxon signed-rank test was used to test for a change in density between pre- and post-SSWD outbreak samples using stats v.3.3.2 in R (55).
Sampling of Genetic Variation.
We sampled tissue from mature P. ochraceus before the onset of sea star wasting disease at each of 16 locations from August 2012 to May 2013 (Fig. 1 and SI Appendix, Table S1). Complementary samples of mature adults (January 2014 and December 2014 to May 2015) and new recruits (<1 y; December 2013 to May 2014) were collected after the mid-2013 onset of SSWD. Tissue collection consisted of approximately a half-dozen tube feet or 2 to 3 mm of arm tip, targeting 10 adults and all recruits; samples were immediately preserved in 95% ethanol (SI Appendix, Table S1). DNA was extracted using a silica-based filter plate (5053; PALL) (56). DNA (50 to 100 ng) in 25 μL for each specimen was submitted to the Genomic Sequencing and Analysis Facility (GSAF) at the University of Texas at Austin for quantitation, normalization, double digestion with the EcoRI and MspI restriction enzymes (57), size selection for 300 ± 50 bp using custom bead preparation (GSAF), adaptor ligation, purification, and 2 × 150 paired-end sequencing on an Illumina HiSeq 4000.
Sequences were demultiplexed using process_radtags from Stacks v.1.35 (58), allowing a maximum of two mismatches in the barcode. Raw sequences were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) (accession no. SRP136569). dDocent v.2.2.13 (21) was used to trim and map reads and genotype SNPs using default parameters. Trimmed reads were directly mapped to the P. ochraceus draft genome (genome.ucsc.edu/), which was generated and assembled by Dovetail Genomics using Chicago, HiRise, and HiC technologies.
Genotyped SNPs underwent additional filtering, modified from Puritz et al. (59). Sequences were filtered using VCFtools v.0.1.15 (https://vcftools.github.io/index.html) (60) and custom scripts (https://github.com/jpuritz/dDocent/tree/master/scripts) (59). The final filtered vcf file had a 98% genotype call rate across all individuals (with maximum allowed missing loci per individual of 13%), minimum depth of 20, and minor allele frequency of at least 0.01. Haplotypes were then called and filtered for potential paralogs or if missing in more than 10% of individuals using rad_haplotyper v.1.1.8 (61). The output genepop file of haplotypes was converted to Genetix and BayeScan format using PGDSpider v.2.1.0.3 (62) for subsequent analyses. Haplotype frequencies were calculated using popgenreport in PopGenReport v.3.0.0 in R (63). Heterozygosity, gene diversity (Hs), and FST were calculated using basic.stats in Hierfstat v.0.04-22 in R (64). Clonality was tested using clonecorrect after removing loci with missing data using poppr v.2.5.0 in R (65).
Tests for Selection in Adult P. ochraceus.
Four main approaches were used to identify changes between pre- and post-SSWD populations and test for signals of selection: (i) a discriminant analysis of principal components (66), (ii) an FST-based outlier method implemented in BayeScan (67), (iii) changes in heterozygosity, and (iv) consistency in allele frequency change across separate geographic locations. Pre- and post-SSWD adult P. ochraceus populations were defined a priori.
The DAPC was performed on adult P. ochraceus (SI Appendix, Table S1) using dapc in adegenet v.2.0.2 in R (66) to identify loci distinguishing pre- and post-SSWD populations. The DAPC evaluates shifts at all haplotypes at a locus, making this analysis potentially more sensitive to detecting selection than FST-based outlier analyses, which do not permit haplotype-specific comparisons. Pre- and post-SSWD groups were defined a priori. After running the a-score spline interpolation, which identified the optimal number of principal components to retain to be 124, we retained 89 to follow the <N/3 rule to avoid overfitting (68). The proportion of conserved variance was 47.7%, and one discriminant function (n groups − 1) was used. Haplotypes were sorted by loading from the DAPC, and haplotypes with the highest loading per locus were retained. The top 100 haplotypes in this list were used to calculate changes in allele frequency in pre- and post-SSWD adults and recruits. As a preliminary exploration of the potential identity of these putatively selected loci, we used BLASTn (e < 1.0E-6) against EchinoDB (the transcript database for echinoderms) [echinodb.uncc.edu (22)] and the P. helianthoides transcriptome (23).
BayeScan v.2.1 (67, 69) was used to test for candidate loci under selection by using the difference in allele frequencies between pre- and post-SSWD adult samples. Default parameters were employed: a thinning interval of 10 and 20 pilot runs for 5,000 iterations, with an additional burn-in of 50,000; prior odds for the neutral model was 10. The q value was calculated for each locus and a false discovery rate of 0.10, an analog of the P value, was used to determine significance of outlier loci. To test whether geographic signals of selection might be present, we conducted two additional BayeScan analyses (default parameters) defining populations by geographic location (SI Appendix, Table S1) within each time point (i.e., pre- and post-SSWD populations separately). No significant changes in any loci were found at FDR 0.10, though sample sizes on a geographic location basis are small.
Heterozygosity was calculated across all loci and then separately for each outlier locus to identify whether heterozygosity was changing with SSWD.
Consistency in adult allele frequency change in the most common haplotype across geographic locations was assessed in any outlier loci identified by BayeScan using two approaches: a two-tailed paired t test—comparing pre- and post-SSWD adults at each location—with a sequential Bonferroni adjustment for the number of loci (70) and a Fisher’s exact test (FET). Our criteria for including a geographic location in these tests was that it had ≥9 pre- and ≥9 post-SSWD adults (which yielded n = 10 locations; SI Appendix, Table S1). For the FET, the change in allele frequency from pre- to post-SSWD for each location (n = 10) was classified as either “changing similarly” (i.e., meaning it shared a consistent direction of change with the overall change in frequency detected in the t test), or “not changing similarly” (i.e., no change or a change in the opposite direction). We then compared these with the null expectation of random change, that is, the equal probability of similar or dissimilar allele frequency change—for example, five locations with a similar shift and five with a dissimilar or no shift. We then used these values to conduct the FET. Recruits were excluded from this analysis because a portion of allele frequency changes from pre-SSWD adults could be immigrant.
Estimating Genetic Affinity of New Recruits.
To assess whether recruits were most genetically similar to pre- or post-SSWD adults, recruits were assigned by transformation using the centering and scaling of the adult data from the two-group DAPC using the same discriminant coefficients as the adults (68). Recruit assignment was evaluated using posterior membership probabilities with the predict.dapc function in adegenet v.2.0.2. We used separate χ2 tests against a null hypothesis of (i) equal probability the recruits will assign to the pre- and post-SSWD adult samples and (ii) equal probability recruit allele frequencies will be in the opposite or same direction of change as the post-SSWD adult sample, relative to the pre-SSWD adult population. Achieved power was calculated using G*Power 3 (71). Specific changes in allele frequencies for the top 100 discriminatory haplotypes (using only one haplotype per locus) identified in the DAPC analysis were calculated and compared in pre- and post-SSWD adults and recruits. For loci identified as outliers in adults by BayeScan, we also compared heterozygosity in recruits with pre- and post-SSWD adults.
Supplementary Material
Acknowledgments
We thank the volunteers who helped in the field or lab for annual surveys: S. Abboud, A. Ahuja, F. Armstrong, J. Bayardo-Guzman, C. Berg, A. Calderon, L. Chavez, B. Cornwell, J. Doornenbal, E. Ernst, M. Friedrich, A. Fryjoff-Hung, B. Gaylord, L. Gómez-Daglio, R. Gonzàlez-Gil, E. Green, R. Grosberg, A. Guzman, B. Jellison, C. Johnson, K. Jones, C. Jorgensen, L. Jurgens, C. Karuppiah, S. Knapp, S. Masclin, K. McClintock, H. Mondo, S. Mylavarapu, M. Parekh, M. Park, E. Ramirez, A. Rosso, S. Rutsaert, S. Sanchez, B. Scholz, M. Rocha de Souza, C. Stilphen, H. Swift, E. Tucker, S. Ul-Hasan, G. Verhaegen, D. Wallace, G. Wallace, J. Wallace, K. Weston, J. Wilson, and L. Wynkoop. Feedback from two anonymous reviewers helped improve this manuscript. This work was funded by NSF Awards OCE-1243970 and OCE-1737381, California Sea Grant College Program Award R/ENV-223PD, the UC Merced Graduate & Research Council, and Environmental Systems Summer fellowships that helped facilitate this work. The California Department of Fish and Wildlife, California State Parks, and National Park Service provided permits and access to field sites. Computation time was provided by the MERCED cluster at UC Merced, funded by NSF Award ACI-1429783.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information short read archive (accession no. SRP136569). Commented bioinformatics code, raw SNP calls, filtered SNPs, and final haplotype calls are deposited at University of California, Merced Dash (https://doi.org/10.6071/M3WW84).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800285115/-/DCSupplemental.
References
- 1.Kharin VV, Zwiers FW, Zhang X, Wehner M. Changes in temperature and precipitation extremes in the CMIP5 ensemble. Clim Change. 2013;119:345–357. [Google Scholar]
- 2.Jentsch A, Kreyling J, Beierkuhnlein C. A new generation of climate‐change experiments: Events, not trends. Front Ecol Environ. 2007;5:365–374. [Google Scholar]
- 3.Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst. 2006;37:637–669. [Google Scholar]
- 4.Kelly MW, Sanford E, Grosberg RK. Limited potential for adaptation to climate change in a broadly distributed marine crustacean. Proc Biol Sci. 2012;279:349–356. doi: 10.1098/rspb.2011.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 6.Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell-Staton SC, et al. Winter storms drive rapid phenotypic, regulatory, and genomic shifts in the green anole lizard. Science. 2017;357:495–498. doi: 10.1126/science.aam5512. [DOI] [PubMed] [Google Scholar]
- 9.Munday PL, Warner RR, Monro K, Pandolfi JM, Marshall DJ. Predicting evolutionary responses to climate change in the sea. Ecol Lett. 2013;16:1488–1500. doi: 10.1111/ele.12185. [DOI] [PubMed] [Google Scholar]
- 10.Foo SA, Dworjanyn SA, Poore AG, Byrne M. Adaptive capacity of the habitat modifying sea urchin Centrostephanus rodgersii to ocean warming and ocean acidification: Performance of early embryos. PLoS One. 2012;7:e42497. doi: 10.1371/journal.pone.0042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellegren H, Galtier N. Determinants of genetic diversity. Nat Rev Genet. 2016;17:422–433. doi: 10.1038/nrg.2016.58. [DOI] [PubMed] [Google Scholar]
- 12.Riginos C, Crandall ED, Liggins L, Bongaerts P, Treml EA. Navigating the currents of seascape genomics: How spatial analyses can augment population genomic studies. Curr Zool. 2016;62:581–601. doi: 10.1093/cz/zow067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Gagnaire PA, Gaggiotti OE. Detecting polygenic selection in marine populations by combining population genomics and quantitative genetics approaches. Curr Zool. 2016;62:603–616. doi: 10.1093/cz/zow088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagnaire PA, et al. Using neutral, selected, and hitchhiker loci to assess connectivity of marine populations in the genomic era. Evol Appl. 2015;8:769–786. doi: 10.1111/eva.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanford E, Kelly MW. Local adaptation in marine invertebrates. Annu Rev Mar Sci. 2011;3:509–535. doi: 10.1146/annurev-marine-120709-142756. [DOI] [PubMed] [Google Scholar]
- 17.Hewson I, et al. Densovirus associated with sea-star wasting disease and mass mortality. Proc Natl Acad Sci USA. 2014;111:17278–17283. doi: 10.1073/pnas.1416625111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurgens LJ, et al. Patterns of mass mortality among rocky shore invertebrates across 100 km of northeastern Pacific coastline. PLoS One. 2015;10:e0126280. doi: 10.1371/journal.pone.0126280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenlord ME, et al. Ochre star mortality during the 2014 wasting disease epizootic: Role of population size structure and temperature. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150212. doi: 10.1098/rstb.2015.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harley CD, Pankey MS, Wares JP, Grosberg RK, Wonham MJ. Color polymorphism and genetic structure in the sea star Pisaster ochraceus. Biol Bull. 2006;211:248–262. doi: 10.2307/4134547. [DOI] [PubMed] [Google Scholar]
- 21.Puritz JB, Hollenbeck CM, Gold JR. dDocent: A RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ. 2014;2:e431. doi: 10.7717/peerj.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janies DA, et al. EchinoDB, an application for comparative transcriptomics of deeply-sampled clades of echinoderms. BMC Bioinformatics. 2016;17:48. doi: 10.1186/s12859-016-0883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuess LE, et al. Up in arms: Immune and nervous system response to sea star wasting disease. PLoS One. 2015;10:e0133053. doi: 10.1371/journal.pone.0133053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menge BA. Brood or broadcast? The adaptive significance of different reproductive strategies in the two intertidal sea stars Leptasterias hexactis and Pisaster ochraceus. Mar Biol. 1975;31:87–100. [Google Scholar]
- 25.Feder AF, Kryazhimskiy S, Plotkin JB. Identifying signatures of selection in genetic time series. Genetics. 2014;196:509–522. doi: 10.1534/genetics.113.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris RH, Abbott DP, Haderlie EC. Intertidal Invertebrates of California. Stanford Univ Press; Stanford, CA: 1980. [Google Scholar]
- 27.Farmanfarmaian A, Giese AC, Boolootian RA, Bennett J. Annual reproductive cycles in four species of West Coast starfishes. J Exp Zool. 1958;138:355–367. [Google Scholar]
- 28.Mauzey KP. Feeding behavior and reproductive cycles in Pisaster ochraceus. Biol Bull. 1966;131:127–144. [Google Scholar]
- 29.Sanford E, Menge BA. Reproductive output and consistency of source populations in the sea star Pisaster ochraceus. Mar Ecol Prog Ser. 2007;349:1–12. [Google Scholar]
- 30.Strathmann MF. Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast: Data and Methods for the Study of Eggs, Embryos, and Larvae. Univ of Washington Press; Seattle: 2017. [Google Scholar]
- 31.Strathmann R. Length of pelagic period in echinoderms with feeding larvae from the northeast Pacific. J Exp Mar Biol Ecol. 1978;34:23–27. [Google Scholar]
- 32.Hedgecock D. Does variance in reproductive success limit effective population size of marine organisms? In: Beaumont A, editor. Genetics and Evolution of Aquatic Organisms. Chapman & Hall; London: 1994. pp. 122–134. [Google Scholar]
- 33.Johnson MS, Black R. Chaotic genetic patchiness in an intertidal limpet, Siphonaria sp. Mar Biol. 1982;70:157–164. [Google Scholar]
- 34.Flowers JM, Schroeter SC, Burton RS. The recruitment sweepstakes has many winners: Genetic evidence from the sea urchin Strongylocentrotus purpuratus. Evolution. 2002;56:1445–1453. doi: 10.1111/j.0014-3820.2002.tb01456.x. [DOI] [PubMed] [Google Scholar]
- 35.de Villemereuil P, Frichot É, Bazin É, François O, Gaggiotti OE. Genome scan methods against more complex models: When and how much should we trust them? Mol Ecol. 2014;23:2006–2019. doi: 10.1111/mec.12705. [DOI] [PubMed] [Google Scholar]
- 36.Pritchard JK, Di Rienzo A. Adaptation—Not by sweeps alone. Nat Rev Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowry DB, et al. Breaking RAD: An evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol Ecol Resour. 2017;17:142–152. doi: 10.1111/1755-0998.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowry DB, et al. Responsible RAD: Striving for best practices in population genomic studies of adaptation. Mol Ecol Resour. 2017;17:366–369. doi: 10.1111/1755-0998.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth B, Charlesworth D. Population genetics from 1966 to 2016. Heredity (Edinb) 2017;118:2–9. doi: 10.1038/hdy.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hewson I, et al. Investigating the complex association between viral ecology, environment, and northeast Pacific sea star wasting. Front Mar Sci. 2018;5:77. [Google Scholar]
- 41.Menge BA, et al. Sea star wasting disease in the keystone predator Pisaster ochraceus in Oregon: Insights into differential population impacts, recovery, predation rate, and temperature effects from long-term research. PLoS One. 2016;11:e0153994. doi: 10.1371/journal.pone.0153994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miner CM, et al. Large-scale impacts of sea star wasting disease (SSWD) on intertidal sea stars and implications for recovery. PLoS One. 2018;13:e0192870. doi: 10.1371/journal.pone.0192870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monaco CJ, Wethey DS, Helmuth B. A dynamic energy budget (DEB) model for the keystone predator Pisaster ochraceus. PLoS One. 2014;9:e104658. doi: 10.1371/journal.pone.0104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 45.Zenger KR, Richardson BJ, Vachot-Griffin AM. A rapid population expansion retains genetic diversity within European rabbits in Australia. Mol Ecol. 2003;12:789–794. doi: 10.1046/j.1365-294x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 46.Lenormand T. Gene flow and the limits to natural selection. Trends Ecol Evol. 2002;17:183–189. [Google Scholar]
- 47.van Oppen MJ, Oliver JK, Putnam HM, Gates RD. Building coral reef resilience through assisted evolution. Proc Natl Acad Sci USA. 2015;112:2307–2313. doi: 10.1073/pnas.1422301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohl WT, McClure TI, Miner BG. Decreased temperature facilitates short-term sea star wasting disease survival in the keystone intertidal sea star Pisaster ochraceus. PLoS One. 2016;11:e0153670. doi: 10.1371/journal.pone.0153670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gooding RA, Harley CDG, Tang E. Elevated water temperature and carbon dioxide concentration increase the growth of a keystone echinoderm. Proc Natl Acad Sci USA. 2009;106:9316–9321. doi: 10.1073/pnas.0811143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Held MB, Harley CD. Responses to low salinity by the sea star Pisaster ochraceus from high‐and low‐salinity populations. Invertebr Biol. 2009;128:381–390. [Google Scholar]
- 51.Eckert GL, Engle JM, Kushner DJ. Proceedings of the Fifth California Islands Symposium. US Dept of the Interior, Minerals Management Service, Pacific Outer Continental Shelf Region; Camarillo, CA: 2000. Sea star disease and population declines at the Channel Islands; pp. 390–393. [Google Scholar]
- 52.Becker BJ. Status and Trends of Ecological Health and Human Use of the Cabrillo National Monument Rocky Intertidal Zone (1900–2005) Natl Park Serv; Seattle: 2006. [Google Scholar]
- 53.Bates AE, Hilton BJ, Harley CD. Effects of temperature, season and locality on wasting disease in the keystone predatory sea star Pisaster ochraceus. Dis Aquat Organ. 2009;86:245–251. doi: 10.3354/dao02125. [DOI] [PubMed] [Google Scholar]
- 54.Lynch M. Scaling expectations for the time to establishment of complex adaptations. Proc Natl Acad Sci USA. 2010;107:16577–16582. doi: 10.1073/pnas.1010836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team . R: A Language and Environment for Statistical Computing. R Found Stat Comput; Vienna: 2016. [Google Scholar]
- 56.Ivanova NV, Dewaard JR, Hebert PD. An inexpensive, automation‐friendly protocol for recovering high‐quality DNA. Mol Ecol Resour. 2006;6:998–1002. [Google Scholar]
- 57.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puritz JB, Gold JR, Portnoy DS. Fine-scale partitioning of genomic variation among recruits in an exploited fishery: Causes and consequences. Sci Rep. 2016;6:36095. doi: 10.1038/srep36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willis SC, Hollenbeck CM, Puritz JB, Gold JR, Portnoy DS. Haplotyping RAD loci: An efficient method to filter paralogs and account for physical linkage. Mol Ecol Resour. 2017;17:955–965. doi: 10.1111/1755-0998.12647. [DOI] [PubMed] [Google Scholar]
- 62.Lischer HE, Excoffier L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics. 2012;28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- 63.Adamack AT, Gruber B. PopGenReport: Simplifying basic population genetic analyses in R. Methods Ecol Evol. 2014;5:384–387. [Google Scholar]
- 64.Goudet J. Hierfstat, a package for R to compute and test hierarchical F‐statistics. Mol Ecol Resour. 2005;5:184–186. [Google Scholar]
- 65.Kamvar ZN, Brooks JC, Grünwald NJ. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet. 2015;6:208. doi: 10.3389/fgene.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jombart T, Ahmed I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jombart T, Collins C. A Tutorial for Discriminant Analysis of Principal Components (DAPC) Using Adegenet 2.0.0. Imperial College London–MRC Centre for Outbreak Analysis and Modelling; London: 2015. [Google Scholar]
- 69.Foll M. BayeScan v2.1 user manual. Ecology. 2012;20:1450–1462. [Google Scholar]
- 70.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 71.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.