Significance
Internal timekeepers, called circadian clocks, are prevalent in all domains of life. Variation in circadian rhythms allows fine-tuning of an organism to its specific environment. Here we show that a mutation in LNK2, in addition to the already described mutation in EID1, was responsible for the deceleration of circadian rhythms in cultivated tomatoes. We show that the mutant alleles of both genes arose in the earliest cultivated types and were selected during the domestication process. Notably, both mutant alleles specifically affect light input to the clock, leading to a light-conditional clock deceleration. Such light-conditionality may be a widespread means to enhance resonance with changed day–night cycles at higher latitudes, despite the fixed 24-h period of the Earth.
Keywords: tomato, domestication, circadian rhythms, light signaling, phytochrome
Abstract
Circadian period and phase of cultivated tomato (Solanum lycopersicum) were changed during domestication, likely adapting the species to its new agricultural environments. Whereas the delayed circadian phase is mainly caused by allelic variation of EID1, the genetic basis of the long circadian period has remained elusive. Here we show that a partial deletion of the clock gene LNK2 is responsible for the period lengthening in cultivated tomatoes. We use resequencing data to phylogenetically classify hundreds of tomato accessions and investigate the evolution of the eid1 and lnk2 mutations along successive domestication steps. We reveal signatures of selection across the genomic region of LNK2 and different patterns of fixation of the mutant alleles. Strikingly, LNK2 and EID1 are both involved in light input to the circadian clock, indicating that domestication specifically targeted this input pathway. In line with this, we show that the clock deceleration in the cultivated tomato is light-dependent and requires the phytochrome B1 photoreceptor. Such conditional variation in circadian rhythms may be key for latitudinal adaptation in a variety of species, including crop plants and livestock.
Circadian clocks are endogenous timekeepers crucial for an optimal synchronization of physiological processes with the external environment (1, 2). Although circadian resonance, a clock period closely matched to the 24-h period of the Earth, enhances life span and fitness (3, 4), naturally occurring variation in circadian rhythms, manifesting as periods deviating from 24 h, appears to be important for local adaptation. This apparent paradox can be found in various organisms (5). For example, longer circadian periods seem advantageous at higher latitudes among natural accessions of Arabidopsis thaliana (6), Mimulus (7), or Drosophila (8, 9). Additionally, adaptive differentiation of circadian rhythms is driven by human selection. Domestication- and breeding-associated variation has been reported for various crop plants (7, 10, 11). In tomato, for example, two mutations led to a clock deceleration, likely adapting the cultivated species to the long summer days it encountered as it was carried away from its equatorial origin. One of these mutations, a single amino acid deletion in EID1, mainly delays the circadian phase, while the other one, not yet identified, primarily lengthens the circadian period (10). Here we identify a near-complete deletion of LNK2 as the period-lengthening mutation. We provide evidence that this mutation was selected during tomato domestication and reveal its coevolution with eid1. We further demonstrate that the pronounced alteration of circadian rhythms in cultivated tomatoes is light-dependent and requires the photoreceptor PHYB1.
Results
A Near-Complete Deletion of LNK2 Is Responsible for the Long Circadian Period in Cultivated Tomatoes.
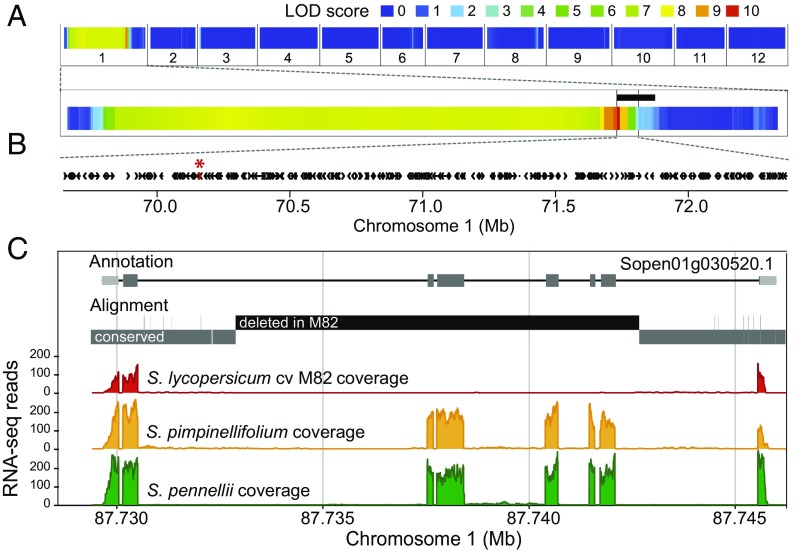
We previously identified a single genomic region on chromosome 1 responsible for the circadian period lengthening in cultivated tomato (10). This region is delimited on the right by the quantitative trait locus (QTL) detected in a cultivated tomato (cv. Moneymaker) x Solanum pimpinellifolium (wild tomato ancestor) recombinant inbred line (RIL) population and on the left by an introgression present in BIL497, a backcross inbred line from a cultivated tomato (cv. M82) x Solanum pennellii (wild tomato relative) population (10, 12, 13) (Fig. 1A and SI Appendix, Fig. S1). Analysis of expression profiles, polymorphisms, and functional descriptions of the 245 annotated genes within the region (Solyc01g068160 - Solyc01g080620) did not reveal any obvious candidate gene, and we considered the possibility that the causative gene is present only in the wild tomato species and has been lost during domestication. We therefore devised a method to identify such cases using the reference genome of the wild tomato relative S. pennellii (14) and transcriptome data from cultivated tomato, its wild ancestor S. pimpinellifolium, and S. pennellii (SI Appendix, SI Methods). We identified 11 genes that are at least partially deleted in cultivated tomato but present and expressed in both wild species (SI Appendix, Table S1). One of these genes, Solyc01g068560/Sopen01g030520, is located within the QTL region and shows a deletion of its middle five exons in cultivated tomato that leaves a severely truncated ORF annotated as unknown protein (Fig. 1 B and C). Strikingly, the full-length S. pennellii coding sequence unequivocally identified this gene as the homolog of the Arabidopsis circadian clock gene NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED 2 (LNK2, AT3G54500, SI Appendix, Table S2). The Arabidopsis lnk2 mutant exhibits a long circadian period (15), suggesting that the deletion of LNK2 in cultivated tomato may be responsible for the clock deceleration.
Fig. 1.
LNK2 colocalizes with the circadian period QTL and is partially deleted in cultivated tomato. (A) QTL analysis for circadian period in a S. pimpinellifolium x S. lycopersicum cv. MM RIL population identified a single significant locus on chromosome 1 (10). Logarithm of the odds (LOD) scores are given for the 12 tomato chromosomes; the genome-wide 5% significance threshold is 2.9. An introgression line with a precisely defined genomic fragment from the wild tomato relative S. pennellii (indicated by a black bar) exhibited the same short-period phenotype (SI Appendix, Fig. S1) and refined the left border of the QTL. (B) The 245 genes annotated in the tomato genome reference v2.4 present in the QTL region are depicted by black arrows; genes deleted in cultivated tomato but present in S. pennellii are shown in red and marked by an asterisk. (C) From top to bottom: gene model for LNK2 in the wild tomato species S. pennellii; graphical output of BLAST results comparing the genomic sequence of S. pennellii and cultivated tomato; and coverage plots of RNA-seq reads from cultivated tomato (red), S. pimpinellifolium (orange), and S. pennellii (green), aligned to the S. pennellii reference genome.
Expression of the truncated lnk2 transcript in tomato shows a robust diurnal oscillation with a peak at about 4 h after dawn, as described for the full-length homologs in Arabidopsis, poplar and rice (16) (SI Appendix, Fig. S2). This oscillation continues under circadian conditions, although with strong dampening in cultivated tomato, as reported for other clock-regulated genes (10) (SI Appendix, Fig. S2). A similar temporal expression pattern can be observed for the paralog LNK1 (Solyc01g105120), suggesting that the regulatory elements important for temporal gene expression are maintained in the truncated version of LNK2 (SI Appendix, Fig. S2).
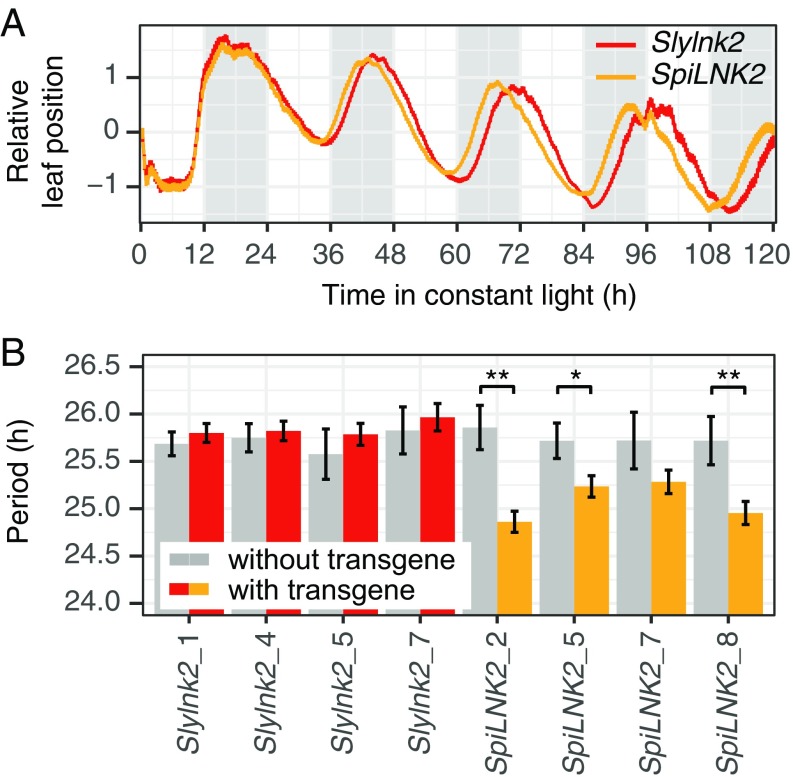
To test whether the LNK2 deletion is indeed underlying the period QTL, we transformed the cultivated tomato variety Moneymaker (MM) with either the complete LNK2 coding sequence from its wild ancestor S. pimpinellifolium or the truncated version present in cultivated tomato, driven by the native LNK2 promoter from MM. Circadian leaf movement analysis showed a significant period shortening of independent transgenic T2 families containing the wild species allele (P = 1.95e-06, one-way ANOVA with all families together), whereas lines with the truncated allele did not exhibit any differences (P = 0.634, one-way ANOVA with all families together; P = 9.93e-05 for the interaction between transgene presence and allele in a two-way ANOVA) (Fig. 2). The effect of the wild LNK2 transgene on period was lower than what was observed in the backcrossed inbred line BIL497 (Fig. 2 and SI Appendix, Fig. S1). This can be explained by variation in the expression levels of the transgene in the T2 lines, which negatively correlated with period length (SI Appendix, Fig. S3). In conclusion, the transgenic lines confirmed that the LNK2 deletion is underlying the QTL and thus is responsible for the major part of the period differentiation between wild and cultivated tomatoes.
Fig. 2.
The partial deletion of LNK2 is responsible for the long circadian period of cultivated tomato. (A) Mean relative position of cotyledon tip under constant light of transgenic tomato T2 families carrying an ectopic LNK2 copy of S. pimpinellifolium (SpiLNK2, orange, n = 83) or S. lycopersicum (Slylnk2, red, n = 53). Line width shows SEM; shaded areas in the background indicate subjective nights. Data are from three independent experiments. (B) Mean circadian period estimates ± SEM (n = 3–27) of the same plants shown in A together with sister T2 lines without the transgene (colored in gray). Asterisks indicate significant differences for each T2 family (one-way ANOVA, *P < 0.05 **P < 0.01).
LNK2 Exhibits Signatures of Positive Selection Associated with Tomato Domestication.
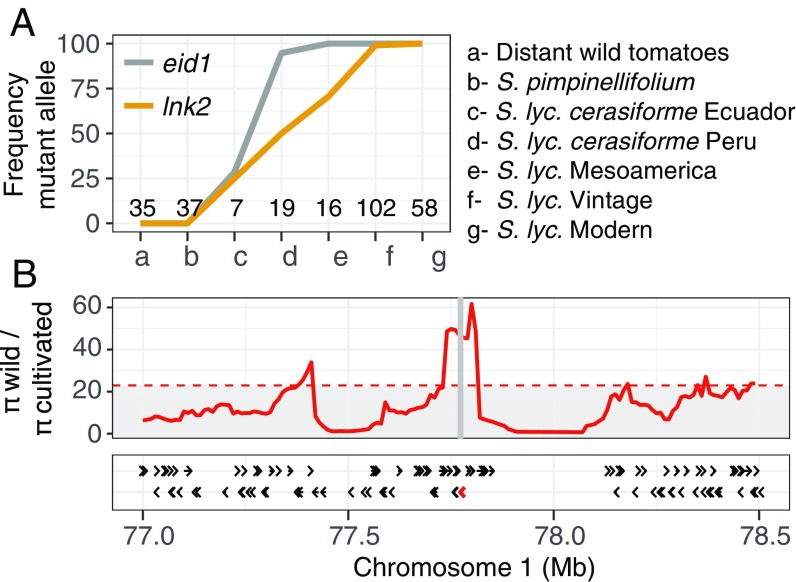
LNK2 sequence analysis in 419 tomato accessions, including members from 14 different species, showed that the LNK2 deletion occurs only within Solanum lycopersicum (Dataset S1). Analysis of a subset of 274 tomato accessions representing sequential domestication steps revealed an interesting evolutionary trajectory of the eid1 and lnk2 mutations during domestication and breeding (Fig. 3A and Datasets S2 and S3). Both mutations were absent in the wild tomato species, including the wild ancestor S. pimpinellifolium. They first appeared at low frequencies in the most ancient domesticated tomatoes, the Ecuadorian S. lycopersicum var. cerasiforme accessions (17). However, while the eid1 mutation rose to near fixation already in the next step of domestication, represented by the Peruvian S. lycopersicum var. cerasiforme accessions, the lnk2 mutation stayed at intermediate frequencies. It increased linearly and rose to fixation only in the vintage and modern tomato populations, which originated after the tomato was brought to Europe (Fig. 3A). This is consistent with previous results, suggesting that the phase shift occurred earlier in the domestication process than the period lengthening and that a delayed clock could be beneficial in northern latitudes (10). Together, these results show that the mutant lnk2 allele arose early during tomato domestication and increased in frequency to near fixation in the course of tomato breeding, suggesting that it may have been under positive selection.
Fig. 3.
The LNK2 deletion rose in frequency during tomato domestication and is located in a genomic region showing signatures of a selective sweep. (A) Frequencies of the lnk2 (orange) and eid1 (gray) mutant alleles in phylogenetic groups representing sequential domestication steps as defined in Dataset S3. Numbers of accessions in each group are shown along the x axis. (B) Pi ratio between wild and cultivated tomato in the region of LNK2 for 144 S. lycopersicum and 32 S. pimpinellifolium accessions. Ratios were calculated in windows of 100 kb and steps of 10 kb. Horizontal dashed line marks the chromosome-wide top 5% windows. The vertical gray bar represents the position of LNK2 in the reference genome. The Bottom graph shows annotated genes in the tomato reference genome v2.5 (black arrows), with LNK2 colored in red.
To further explore this possibility, we scanned the genomic region surrounding LNK2 for signatures of selective sweeps by comparing the genetic diversity (π) of cultivated and ancestral wild tomato (S. pimpinellifolium) using resequencing data (18). Remarkably, a genomic region of about 100 kb, including LNK2 and 19 neighboring genes (from Solyc01g068440 to Solyc01g068560), showed very low levels of genetic diversity, specifically in cultivated tomatoes (SI Appendix, Fig. S4). This leads to a high ratio of diversity between the ancestral wild and the cultivated species, which is indicative of a selective sweep (Fig. 3B). Although we cannot rule out random demography-associated effects being responsible for this pattern, the rise in allele frequency together with the genomic signatures of selection strongly suggest that the deletion in LNK2 and the resulting long circadian period have been targeted by selection during tomato domestication or breeding.
LNK2 and EID1 Require the Photoreceptor PHYB1 for the Deceleration of the Tomato Circadian Clock.
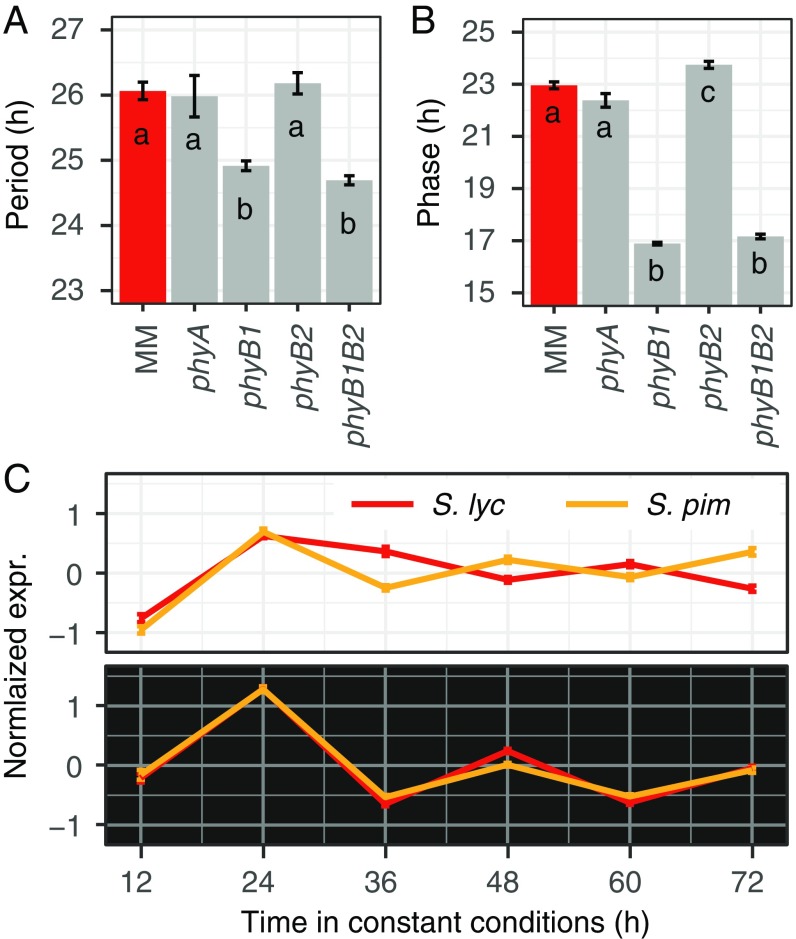
In Arabidopsis, the LNK1 and LNK2 genes have been implicated in phytochrome-mediated light input to the circadian clock (15, 19). Intriguingly, EID1, the gene underlying the other major QTL for the adaptive modulation of circadian rhythms in tomato, has also been described to play an important role in phytochrome signaling (20, 21). This suggests that selection may have specifically targeted the phytochrome-mediated light input pathway to the clock rather than the core of the clock itself. To explore this possibility we analyzed circadian leaf movements in mutants of the main phytochromes in tomato: phyA, phyB1, and phyB2 (22, 23). This analysis revealed a striking effect of PHYB1, but not PHYB2 or PHYA, on circadian rhythms. The circadian phase of the phyB1 single and the phyB1phyB2 double mutants was advanced by more than 5 h, and the period was shortened by more than 1 h (Fig. 4 A and B). Circadian rhythms of these lines thus resembled those of the wild ancestor S. pimpinellifolium, suggesting that the deceleration of circadian rhythms in cultivated tomato, caused by the mutations in EID1 and LNK2, may be PHYB1-dependent. Indeed, combining the phyB1 mutation with the different allelic states of EID1 and LNK2 demonstrated that the absence of PHYB1 completely abolished the effects of EID1 and LNK2 on circadian rhythms (SI Appendix, Fig. S5).
Fig. 4.
The clock modulation of cultivated tomato is PHYB1-dependent and exhibits light-conditionality. (A and B) Mean circadian period (A) and phase (B) estimates ± SEM (n = 10–47) of the cultivated tomato variety Moneymaker (MM, red) and the phyA and phyB mutants. Different letters in each bar indicate significant differences (P < 0.05, one-way ANOVA and Tukey’s post hoc HSD test). Each genotype was analyzed in two independent experiments. (C) Mean normalized and scaled expression (±SEM) of 109 genes whose expression levels are controlled by the circadian clock and peak at dawn (ZT0). RNA-seq was performed on biological duplicates from S. lycopersicum and S. pimpinellifolium seedlings collected every 12 h for 3 d in constant light (Top) or dark (Bottom). Dark or white backgrounds represent constant dark or light conditions.
To further characterize the role of phytochrome B in the tomato circadian clock, we analyzed the transcriptional responses of the phyB1 and phyB2 single and the phyB1phyB2 double mutants by RNA-seq. Both single mutants exhibited a relatively small number of differentially expressed genes compared with the double mutant, demonstrating partial redundancy of the two phytochrome B photoreceptors (SI Appendix, Fig. S6A). Interestingly, the expression of several tomato clock genes, including PRR5, ELF4, FKF1, and all four LNK genes, was significantly reduced (adjusted P < 0.05) in the phyB1 but not in the phyB2 mutant (SI Appendix, Fig. S6B). While these results are in line with phyB1 and not phyB2 affecting tomato circadian rhythms (Fig. 4 A and B), the rather minor reduction of LNK2 expression in the phyB1 mutant (SI Appendix, Fig. S6C) is probably not sufficient to explain the epistasis of phyB1 over LNK2.
The complete epistasis of the phyB1 mutant over EID1 (SI Appendix, Fig. S5) highlights a different light-signaling mechanism in tomato compared with A. thaliana, where EID1 is associated with the far-red light photoreceptor PHYA (24). A lengthening of the circadian period of PHYB1-overexpressing tomato lines (25), on the other hand, is consistent with the negative role of EID1 on phytochrome signaling described in A. thaliana (24) (SI Appendix, Fig. S7). Considering the effect of EID1 on PHYB1 signaling, allelic variation of EID1 might be expected to impact LNK2 function. However, the absence of a genetic interaction between the EID1 and LNK2 loci argues against this possibility (SI Appendix, Fig. S8). In summary, mutations in EID1 and LNK2 appear to impact phytochrome-mediated light input to the clock and decelerate circadian rhythms of the cultivated tomato in a phytochrome B1-dependent manner.
The involvement of EID1 and LNK2 in light signaling raises the question of whether the mutant eid1 and lnk2 alleles may have been selected to change light signaling rather than circadian rhythms. To address this question, we examined hypocotyl growth, which represents one of the best-studied light-mediated developmental traits in plants. Lines with the mutant eid1 allele exhibited significantly shorter hypocotyls than lines with the ancestral allele (t test, P < 0.001), which is in concordance with enhanced PHYB1 signaling caused by the mutation in EID1 (SI Appendix, Fig. S9). However, the mutant lnk2 allele led to a hypocotyl lengthening (t test, P < 0.001, SI Appendix, Fig. S9), as described for A. thaliana (15). The two QTL thus have opposing effects with respect to photomorphogenesis but act synergistically with respect to circadian rhythms, suggesting that selection of the eid1 and lnk2 variants targeted circadian rhythms and not light signaling.
Modulation of the Circadian Clock in Cultivated Tomato Is Light-Conditional.
Phytochrome molecules are activated by light but revert to an inactive conformational state under darkness (26). Given the phytochrome-dependency of the domestication-associated variation in circadian rhythms, we wondered whether the circadian rhythm differentiation caused by EID1 and LNK2 requires light and would be lost under darkness. Since tomato does not exhibit circadian leaf movements in the absence of photosynthetic active radiation (SI Appendix, Fig. S10), we used RNA-seq to assess circadian rhythms in the dark. For this, we took advantage of a high-resolution transcriptome time-course performed in constant light (10). These data allowed us to group tomato genes that show rhythmic gene expression under diurnal and circadian conditions according to their time of maximal expression during the 24-h daily cycle (SI Appendix, SI Methods and Dataset S4). By only considering these a priori defined rhythmic genes of known phase, we were able to monitor the molecular circadian clock, despite sampling at low temporal resolution. Specifically, we analyzed expression of the defined genes in RNA-seq samples obtained from cultivated tomato and the wild ancestor S. pimpinellifolium grown in constant light or dark conditions at 12-h intervals. As expected, we found marked differences in the expression of circadian-regulated genes between cultivated tomato and its wild ancestor when plants were grown under constant light (Fig. 4C and SI Appendix, Fig. S11). These differences are consistent with the clock deceleration observed in S. lycopersicum under constant light conditions (10). In contrast, expression of circadian-regulated genes in plants grown under constant darkness appeared basically identical between the two species, strongly suggesting light-conditionality of the domestication-driven clock modulation (Fig. 4C and SI Appendix, Fig. S11). A higher-resolution qRT-PCR expression time-course of the core clock gene LHY further supported these observations (SI Appendix, Fig. S12). While we cannot rule out subtle differences in circadian period or phase between wild and cultivated tomato under constant darkness, we can conclude that the striking alteration of the circadian clock in cultivated tomato depends on light and the photoreceptor PHYB1.
Discussion
Here we show that a nearly complete deletion of LNK2 was selected during domestication to lengthen the circadian period of cultivated tomatoes. Similarly to the mutation in EID1 delaying circadian phase, the deletion is absent in any wild tomato species analyzed and first observed at low frequencies in the early domesticated types, the S. lycopersicum var. cerasiforme accessions from Ecuador. However, while the mutation in EID1 rose to near fixation already in the next step of domestication, represented by the S. lycopersicum var. cerasiforme accessions from Peru, the deletion of LNK2 was only fixed after the tomato was imported to Europe in the 16th century. This pattern of evolution, together with the participation of both mutations in light-signaling pathways dependent on PHYB1, suggests that perception and timing of light inputs played important roles during tomato domestication.
An involvement of phytochromes in the induction of LNK2 expression by light has been demonstrated for A. thaliana (15). In accordance with this, we observe reduced LNK2 expression in phyB1 mutants in tomato. However, this reduction appears too subtle for explaining the epistasis of the phyB1 mutant over LNK2, suggesting an additional role of PHYB1 for LNK2 activity beyond direct transcriptional regulation. Interestingly, further analysis of differential gene expression in tomato phytochrome mutants revealed that established molecular functions of LNK2, like the activation of the afternoon-phased clock genes ELF4, FKF1, and PRR5 (15, 19) are also impaired by the lack of PHYB1, highlighting conserved roles of LNK2 between tomato and A. thaliana. Another feature conserved between the two species and thus across more than 100 My of independent evolution is the effect of PHYB on circadian phase (27, 28) and period (29–31). A leading phase (27) and a short period (30) reported for A. thaliana phyB mutants grown under white light resemble our results obtained with the tomato phyB1 mutant lines.
Considering the adaptive advantage of a clock that resonates with the environment (4, 32), a conditional clock modulation may represent an ideal solution for adapting to changed day/night cycles at higher latitudes. A slow clock under normal light conditions may allow taking full advantage of long days, while a fast clock under darkness fits the short nights. In concordance, selection during tomato domestication or breeding specifically targeted mutations in two genes mediating light input to the circadian clock, EID1 and LNK2. This resulted in a pronounced deceleration of the tomato circadian rhythms that depends on light and the photoreceptor PHYB1.
In conclusion, positive selection of mutations in two genes mediating light input to the clock conditionally altered tomato circadian rhythms, potentially enhancing synchronization between internal and external daily cycles. Such conditional alterations of circadian rhythms may play a broad role in enabling latitudinal adaptation and could reconcile the apparent paradox of natural circadian variation, despite the fixed 24-h periodicity of the external environment.
Materials and Methods
Plant Material.
The phytochrome mutants (22, 23) and the cultivated variety Moneymaker were obtained from the University of California Davis C.M. Rick Tomato Genetics Resource Center (TGRC). The PHYB1-overexpressor was kindly provided by A. R. van der Krol, Wageningen University, Wageningen, The Netherlands (25), the S. pimpinellifolium recombinant inbred lines (RILs) and respective genotype data by A. W. van Heusden, Wageningen University, Wageningen, The Netherlands (33), and the S. pennellii backcross inbred lines (BILs) by Dani Zamir, Hebrew University of Jerusalem, Jerusalem (12, 13). The EID1 near isogenic line (NIL) has been described before (10).
Growth Conditions.
After an initial treatment with saturated trisodium phosphate (Na3PO4) for 15 min, tomato seeds were kept in water for 3 d in the dark. On the third day they were sown on standard soil and randomized. For more detail regarding the growth conditions, see SI Appendix, SI Methods.
Image Capture and Analysis.
This has been described in detail elsewhere (10, 34). Briefly, pictures were taken at an interval of 20 min for 5 d using Pentax Optio WG-1 digital cameras. Picture analysis was performed using the software ImageJ (35). Estimates for circadian variables were obtained via fast Fourier transform nonlinear least-squares analysis (36) using BioDare (37). We excluded the first 24 h from the analysis to remove potential noise caused by the transfer from the entrainment chamber to the imaging chamber. Only seedlings with relative amplitude error (RAE) values below 0.20 were used for statistical analyses.
Genome Comparison.
To identify expressed genes that have been completely or partially deleted during tomato domestication, we took advantage of the high-quality reference genome of the wild tomato species S. pennellii (14) and transcriptome data from the wild species S. pennellii (LA0716) and S. pimpinellifolium (LA1589) and the S. lycopersicum M82 cultivar (38). The final list includes only 11 genes, half of them annotated as hypothetical proteins (SI Appendix, Table S1). For details, see SI Appendix, SI Methods.
Cloning of LNK2.
We cloned the LNK2 cDNA of S. pimpinellifolium LA1589 and S. lycopersicum cv. MM, including 290 bp of its 3′ UTR, under control of the MM native promoter (∼2.4 kb upstream sequence from the LNK2 start codon) using the MultiSite Gateway Pro-2.0 Kit (Life Technologies), the destination vector pGWB1 (39), and Phusion High-Fidelity DNA polymerase (New England Biolabs). For more information, see SI Appendix, SI Methods.
qRT-PCR Experiments.
We set up 10-µL reactions in triplicate in Bio-Rad 384-well PCR plates (Bio-Rad Laboratories) using IQ SYBR Green Master Mix (Bio-Rad Laboratories) and 500 ng cDNA, which was synthesized from 1 µg of total RNA using SuperScript II Reverse Transcriptase (Life Technologies) following the manufacturer’s instructions using Oligo(dT)18 primers. qRT-PCR was performed using the Bio-Rad CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories). Expression was calculated using the standard curve method and the AP-2 complex subunit mu (Solyc08g006960, CAC) as an internal control to normalize transcript abundance (40). Primers are listed in SI Appendix, Table S3.
Genotyping the Deletions in LNK2 and EID1.
We genotyped the deletions present in LNK2 and EID1 using short reads available from 426 tomato accessions (18, 41). Reads were aligned to the reference genomes of S. lycopersicum v2.50 and S. pennellii v2 and the causative mutations determined by probabilistic indel calling (EID1) or by differences in coverage profiles (LNK2). For details, see SI Appendix, SI Methods.
Classification of Resequenced Tomato Accessions into Phylogenetic Groups.
More than 1,000 tomato accessions have been previously classified into phylogenetic groups using 8,700 genome-wide SNPs genotyped with the SolCAP Infinium Chip (17). These variants were integrated with polymorphisms extracted from resequencing data available for 426 tomato accessions (18, 41). We selected a final dataset of 1,956 genome-wide variants genotyped across 1,412 accessions to generate a neighbor-joining tree that allowed us to classify resequenced accessions in phylogenetic groups that are relevant to tomato domestication. Further information is given in SI Appendix, SI Methods.
Signatures of Selection.
To test for signatures of selection in the chromosomal region of LNK2, we measured the level of genetic diversity (π) of chromosome 1, employing resequencing data (18) of 144 S. lycopersicum (Processing, Fresh, and Vintage groups) and 32 S. pimpinellifolium accessions (Ecuador and Peru groups) classified in our phylogenetic analysis (Datasets S2 and S3). Short read alignments were used to call variants on chromosome 1 with GATK v3.5 with default settings. Nucleotide diversity was calculated using VCFtools (42) on biallelic SNPs with a minor allele frequency above 0.05 in 100-kb windows and 10-kb steps. Pi ratios between wild and cultivated groups, as well as the 5% chromosome-wide threshold, were calculated and plotted using custom R scripts.
Hypocotyl Measurements.
After pretreatment (see Growth Conditions), seeds were sown in a 48-cell growing tray, holding ∼4.3 L of standard soil in total. The tray was then placed into a controlled environment chamber (Percival Scientific) under constant red light provided by LED light panels. The day of germination was recorded for each seedling, and hypocotyl lengths were measured 10 d after germination.
Genetic Interaction Analyses Between the Circadian Rhythm QTL and PHYB1.
To test for genetic interactions between the circadian rhythm QTL EID1 and LNK2 and the PHYB1 gene, we crossed the near isogenic lines (NILs) harboring the wild-type allele of either of the two QTL (EID1 NIL = rec47, and LNK2 NIL = BIL497), both described in detail before (10), with a phyB1 mutant line containing the mutant eid1 and lnk2 alleles (23). The resulting F1 lines were self-pollinated to generate segregating F2 populations. The four allelic combinations were used for circadian leaf movement analysis. For further details, see SI Appendix, SI Methods.
RNA Sequencing.
Total RNA from leaf samples was extracted with the RNeasy Plant Mini Kit (QIAGEN). Libraries were prepared according to the Illumina TruSeq RNA protocol and sequenced on the Illumina HiSeq platform (Illumina, Inc.) at the Genome Center of the Max Planck Institute for Plant Breeding Research Cologne. For details about the phyB and circadian time-course RNA-seq experiments and RNA-seq data analysis, see SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank all members of the J.M.J.-G. and M.K. laboratories and A. de Montaigu for helpful comments and discussions. We specially thank U. Tartler, D. Hamacher, M. Pohe, and A. Lautscham for technical assistance and the Genome Center at the Max Planck Institute for Plant Breeding Research for their help with sequencing. We acknowledge funding to N.A.M., J.M.J.-G., and M.K. from a core grant from the Max Planck Society. L.Z. was funded by an International Max Planck Research School PhD fellowship. J.M.J.-G. received funding from the German Research Foundation under the German–Israeli Project Cooperation program (Deutsche Forschungsgemeinschaft, Deutsch-Israelische Projektkooperation, Project FE552/12-1) and the Agence Nationale de la Recherche (ANR) Tremplin-ERC 2017 program (Project Ritmo ANR-17-ERC2-0013-01). The Institut Jean-Pierre Bourgin benefits from the support of the LabEx Saclay Plant Sciences (Grant ANR-10-LABX-0040-SPS).
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-seq data have been deposited on the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra [accession nos. SRP133995 (phyB mutants) and SRP124605 (low-resolution light and dark time-courses)].
See Commentary on page 6888.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801862115/-/DCSupplemental.
References
- 1.Hsu PY, Harmer SL. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014;19:240–249. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyse CA, Coogan AN, Selman C, Hazlerigg DG, Speakman JR. Association between mammalian lifespan and circadian free-running period: The circadian resonance hypothesis revisited. Biol Lett. 2010;6:696–698. doi: 10.1098/rsbl.2010.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hut RA, Paolucci S, Dor R, Kyriacou CP, Daan S. Latitudinal clines: An evolutionary view on biological rhythms. Proc Biol Sci. 2013;280:20130433. doi: 10.1098/rspb.2013.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michael TP, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 7.Greenham K, et al. Geographic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms. 2017;32:26–34. doi: 10.1177/0748730416679307. [DOI] [PubMed] [Google Scholar]
- 8.Joshi DS. Latitudinal variation in locomotor activity rhythm in adult Drosophila ananassae. Can J Zool. 1999;77:865–870. [Google Scholar]
- 9.Pittendrigh CS, Takamura T. Latitudinal clines in the properties of a circadian pacemaker. J Biol Rhythms. 1989;4:217–235. [PubMed] [Google Scholar]
- 10.Müller NA, et al. Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet. 2016;48:89–93. doi: 10.1038/ng.3447. [DOI] [PubMed] [Google Scholar]
- 11.Yarkhunova Y, et al. Selection during crop diversification involves correlated evolution of the circadian clock and ecophysiological traits in Brassica rapa. New Phytol. 2016;210:133–144. doi: 10.1111/nph.13758. [DOI] [PubMed] [Google Scholar]
- 12.Ofner I, Lashbrooke J, Pleban T, Aharoni A, Zamir D. Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. Plant J. 2016;87:151–160. doi: 10.1111/tpj.13194. [DOI] [PubMed] [Google Scholar]
- 13.Fulop D, et al. A new advanced backcross tomato population enables high resolution leaf QTL mapping and gene identification. G3 (Bethesda) 2016;6:3169–3184. doi: 10.1534/g3.116.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger A, et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet. 2014;46:1034–1038. doi: 10.1038/ng.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rugnone ML, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA. 2013;110:12120–12125. doi: 10.1073/pnas.1302170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 17.Blanca J, et al. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics. 2015;16:257. doi: 10.1186/s12864-015-1444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014;46:1220–1226. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 19.Xie Q, et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell. 2014;26:2843–2857. doi: 10.1105/tpc.114.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 2001;15:939–944. doi: 10.1101/gad.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenden B, et al. Light inputs shape the Arabidopsis circadian system. Plant J. 2011;66:480–491. doi: 10.1111/j.1365-313X.2011.04505.x. [DOI] [PubMed] [Google Scholar]
- 22.van Tuinen A, Kerckhoffs LH, Nagatani A, Kendrick RE, Koornneef M. Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol Gen Genet. 1995;246:133–141. doi: 10.1007/BF00294675. [DOI] [PubMed] [Google Scholar]
- 23.Weller JL, Schreuder ME, Smith H, Koornneef M, Kendrick RE. Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant J. 2000;24:345–356. doi: 10.1046/j.1365-313x.2000.00879.x. [DOI] [PubMed] [Google Scholar]
- 24.Büche C, Poppe C, Schäfer E, Kretsch T. eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- 25.Husaineid SS, et al. Overexpression of homologous phytochrome genes in tomato: Exploring the limits in photoperception. J Exp Bot. 2007;58:615–626. doi: 10.1093/jxb/erl253. [DOI] [PubMed] [Google Scholar]
- 26.Quail PH. Phytochrome: A light-activated molecular switch that regulates plant gene expression. Annu Rev Genet. 1991;25:389–409. doi: 10.1146/annurev.ge.25.120191.002133. [DOI] [PubMed] [Google Scholar]
- 27.Salomé PA, et al. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 2002;129:1674–1685. doi: 10.1104/pp.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Montaigu A, et al. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc Natl Acad Sci USA. 2015;112:905–910. doi: 10.1073/pnas.1422242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 30.Palágyi A, et al. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol. 2010;153:1834–1845. doi: 10.1104/pp.110.153031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 33.Kazmi RH, et al. Complex genetics controls natural variation among seed quality phenotypes in a recombinant inbred population of an interspecific cross between Solanum lycopersicum × Solanum pimpinellifolium. Plant Cell Environ. 2012;35:929–951. doi: 10.1111/j.1365-3040.2011.02463.x. [DOI] [PubMed] [Google Scholar]
- 34.Müller NA, Jiménez-Gómez JM. Analysis of circadian leaf movements. Methods Mol Biol. 2016;1398:71–79. doi: 10.1007/978-1-4939-3356-3_7. [DOI] [PubMed] [Google Scholar]
- 35.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 37.Moore A, Zielinski T, Millar AJ. Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. Methods Mol Biol. 2014;1158:13–44. doi: 10.1007/978-1-4939-0700-7_2. [DOI] [PubMed] [Google Scholar]
- 38.Koenig D, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA. 2013;110:E2655–E2662. doi: 10.1073/pnas.1309606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 40.Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aflitos S, et al. 100 Tomato Genome Sequencing Consortium Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014;80:136–148. doi: 10.1111/tpj.12616. [DOI] [PubMed] [Google Scholar]
- 42.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.