Summary
CD6 is a type I T‐cell surface receptor that modulates antigen receptor signalling. Its activity is regulated by binding of its membrane proximal domain (domain 3) to a cell surface ligand, CD166. CD6 monoclonal antibodies (mAbs) specific for the membrane distal domain (domain 1) perturb CD6 function including itolizumab (Alzumab™), which has reached the clinic for treatment of autoimmune disease. We characterized molecular and functional properties of several CD6 mAbs including itolizumab to define potential mechanisms of action. Epitope mapping using the crystal structure of CD6 to design mutants identified two distinct binding sites on different faces of domain 1, one containing residue R77, crucial for MT605 and T12.1 binding and the other, E63, which is crucial for itolizumab and MEM98. Analysis of binding kinetics revealed that itolizumab has a lower affinity compared with other CD6 domain 1 mAbs. We compared potential agonistic (triggering) and antagonistic (blocking) properties of CD6 mAbs in assays where the mechanism of action was well defined. CD6 domain 1 and 3 mAbs were equally effective in triggering interleukin‐2 production by a cell line expressing a chimeric antigen receptor containing the extracellular region of CD6. CD6 domain 1 mAbs hindered binding of multivalent immobilized CD166 but were inferior compared with blocking by soluble CD166 or a CD6 domain 3 mAb. Characterization of CD6 mAbs provides an insight into how their functional effects in vivo may be interpreted and their therapeutic use optimized.
Keywords: CD166, CD6, immune therapies, immunomodulation, monoclonal antibody, T cell
Abbreviations
- CD4d3+4
CD4 domains 3 and 4
- IL‐2
interleukin‐2
- mAbs
monoclonal antibodies
- SPR
surface plasmon resonance
Introduction
A lack of characterization of the mode of action of biological therapeutic reagents including monoclonal antibodies (mAbs) hinders interpretation of effects and consequently further rational development. The leucocyte surface receptor CD6 primarily associated with expression on T cells, has been targeted using mAbs. The development of CD6 mAbs for therapeutic use has not been systematic. In the first clinical use of CD6 mAbs, the major mode of action was generally attributed to elimination of cells.1 More recently, it has been proposed that a therapeutic CD6 mAb is efficacious by perturbing CD6 function and that depletion is not the major mode of action.2, 3, 4 Among the leucocyte receptors, CD6 has the longest cytoplasmic region without catalytic activity so there is potential for significant modulation of signalling. Its activity is regulated by engagement of the T‐cell antigen receptor and its specific cell surface ligand, CD166. CD166, otherwise known as ALCAM (activated leucocyte cell adhesion molecule) has a broad distribution including antigen‐presenting cells.
Perturbation of CD6 function with mAbs or by genetic manipulation has revealed that CD6 has a role in both restraining and promoting the activation of immune cells.5, 6, 7, 8, 9 From a therapeutic standpoint, a key observation has been that CD6 mAbs generally suppress a strong immune response.6, 9, 10, 11 Inhibitory effects of CD6 mAbs have been interpreted as being the consequence of antagonistic effects caused by blocking co‐stimulatory interactions with CD1666, 9, 10, 11 and agonist activating effects as triggering signalling.5, 9, 12, 13 The mode of action by CD6 mAbs has not always been rigorously defined, leading to more than one interpretation of the effects of a CD6 mAb in a particular assay.6, 7 For the development of therapeutic modulation of CD6 activity it is imperative to understand how the pleiotropic effects of CD6 are regulated. A CD6 mAb, itolizumab, is now licensed for use to treat autoimmune disease and there is evidence that it is less toxic but also less effective than other reagents.3, 14 It is important to understand how CD6 mAbs perturb function if their use in immunotherapy is to be rationally optimized.
Itolizumab (Alzumab™) has been developed as a therapeutic drug for the treatment of psoriasis. It has been exploited for clinical use based on its immunosuppressive effects.2, 3 In a study of 26 psoriasis patients receiving multiple injections of the humanized version of ior t1, itolizumab, over the course of a year, the mAb was immunosuppressive; the reduction of the proliferation and production of pro‐inflammatory cytokines being statistically significant.15 Analysis of a similar number of rheumatoid arthritis patients also showed a suppressive effect of itolizumab, enhancing methotrexate treatment.16 Unlike lymphocyte numbers, which were transiently reduced, the reduction of inflammatory cytokine production was sustained, suggesting a mechanism other than depletion.16
CD6, a type I membrane protein, contains scavenger receptor cysteine‐rich domains in its extracellular region.17 The membrane proximal domain (domain 3) binds CD166 and the interaction has been well characterized.17, 18 CD6 mAbs that have been raised against cell surface CD6 are specific for the membrane distal domain (domain 1).17, 18 Immunization with soluble recombinant CD6 led to the production of CD6 domain 3 mAbs.6 We characterize the properties of CD6 mAbs including mapping the epitope for itolizumab on the crystal structure of CD6 and comparing their capacity for triggering through the extracellular region of CD6 and blocking ligand binding to CD166.
Materials and methods
Monoclonal antibodies
Monoclonal antibodies used were specific for: CD6 domain 1, itolizumab (human IgG1; Alzumab™; BioCon Ltd, Bangalore, India), a gift from Enrique Montero and BioCon), MEM98 (mouse IgG1; Bio‐Rad antibodies, Oxford, UK), UMCD6 (mouse IgG1; Santa Cruz Biotechnology, Dallas, TX) and MT605 (mouse IgG1; BD Pharmingen, Franklin Lakes, NJ); T12.1 (mouse IgG2a: ATCC, Manassas, VA): CD6 domain 3, OX126 (mouse IgG2a)6 and CD3, UCHT1 (Absolute Antibody). Control mAbs were CAMPATH‐1H (human IgG1 anti‐CD52, a gift from Herman Waldman) and OX21 (mIgG1).
Mutagenesis
Mutants of CD6 domain 1 were designed using the crystal structure of CD6 (17PDB: 5A2E) and pyMOL software. Mutagenesis of CD6 domain 1 residues to alanine was conducted using a pEFBOS vector encoding a chimeric protein containing the extracellular region of human CD6 (domains 1–3 and stalk; GenBank: U34623; Uniprot:P30203) and rat CD4 domains 3 and 4 (CD4d3+4) and the Q5 site directed mutagenesis SDM Kit (New England Biolabs, Hitchin, Herts) and verified by Sanger sequencing (SourceBioscience).6 Chimeric proteins, CD6–CD4d3+4 and mutants and as a control, CD4d3+4 were transiently expressed in 293T cells and used as tissue‐culture supernatant, which for kinetic analysis was concentrated and chimeric proteins biotinylated with BirA (Avidity, Aurora, CO).6
Surface plasmon resonance analysis
Surface plasmon resonance (SPR) analysis was carried out using a BIAcore 3000 using HBS‐EP buffer (GE Healthcare Biosciences AB, Uppsala, Sweden) at 10 μl/min. CD6–CD4d3+4 (~700 response units; RU) and rat CD4d3+4 (300 RU) were immobilized at approximately equivalent molar levels by injecting tissue culture supernatants over a rat CD4d3+4 mAb (OX68) amine‐coupled to a CM5 chip.6, 17 Antibodies in solution (10 μg/ml) were then injected at 25° for 60–180 seconds and allowed to dissociate for 120–180 seconds. For kinetic analysis at 37°, biotinylated chimeric proteins were immobilized via streptavidin.6 Monoclonal antibodies at 100, 33 or 10 nm were injected for 180 seconds at 50 μl/min and allowed to dissociate for ≥ 1200 seconds. The response over a control flow cell (CDd3+4) was subtracted and analysed with simultaneous global fitting of the association phases for different concentrations using a 1 : 1 Langmuir binding model in graphpad prism (Graphpad, San Diego, CA). The dissociation phases were fitted with two (itolizumab) and one (MEM98, MT605 and UMCD6) phase exponential decay models.
Cell lines
Mouse hybridoma cells expressing 2B4 TCR (Reay line) were transduced using a pFBneo retroviral vector encoding a chimeric receptor consisting of the extracellular region of human CD6 (Genbank: U34623) fused to the cytoplasmic region of mouse CD3 ζ‐chain (CD6‐ζ).6, 19 The join was CD6/mouse ζ: FILL/RAKFSR. The cells were cultured in 5–10% fetal calf serum/Dulbecco's modified Eagle's medium containing 0·5–1 mg/ml G418.
Enzyme‐linked immunosorbent assay
CD6 mAbs (50 μl) in phosphate‐buffered saline were immobilized on plastic round‐bottomed 96‐well plates at 4° for 18 hr, and unbound material flicked out. 2B4 hybridoma cells 105 in 200 μl were added, incubated at 37° for 18 hr and interleukin‐2 (IL‐2) production measured by a sandwich enzyme‐linked immunosorbent assay.
Flow cytometry
CD6 domain 3 mAb (OX126) and negative control (OX21) coupled to fluorescein isothiocyanate (FITC) in house and stored at −20° were used at 5 μg/ml. Cells were incubated, and pre‐incubated where applicable, with binding reagents for at least 30 min at 0° before fixation with 2% paraformaldehyde and analysis of mean fluorescence intensity on a FACSCalibur. CD6 mAbs and OX126‐FITC were added to cells simultaneously. Concentrated tissue‐culture supernatant containing soluble biotinylated hCD166 was produced in 293T cells with a pHL‐hCD166 avi‐His vector constructed with the extracellular region of human CD166 (CD166) or human CD166 lacking domain 1 (CD166 d2‐5) and BirA (Avidity, Aurora, CO).6, 20, 21, 22 The amount used to saturate avidin‐coated yellow fluorescent beads (Spherotech, Lake Forest, IL) was determined empirically. As CD166 is also homophilic, coated beads were sonicated to reduce aggregates and 10 μl of the mix was added to 5 × 105 cells in a flat‐bottomed well and the plate was centrifuged for 20 min at 4° to ensure contact, incubated for a further 40 min at 0° and then diluted with fixative.22 Optimization of binding by multivalent complexes of soluble CD166 was also determined empirically for each preparation of biotinylated protein by incubating 1 μl Quantum Red streptavidin (Sigma, St Louis, MO: 156 μm) with soluble CD166 or CD166 d2‐5 in a final volume of 50 μl/sample for at least 1 hr at 0–4° before adding to cells pre‐incubated with mAbs or soluble CD166 VVC 17 and a 3 immunoglobulin‐like domain control protein,signal regulatory protein α (courtesy of Deborah Hatherley) at a final concentration of 10 μm. After incubation, cells were spun and resuspended in fixative.
Results
The specificity of CD6 domain 1 mAbs is defined by single point mutants R77, E63 and R61
We have previously defined two well‐differentiated epitopes involving R77 and E63 respectively for CD6 domain 1 mAbs, MT605 and MEM98.17 Here we extend our analysis using five CD6 domain 1 mAbs and a panel of CD6 domain 1 mutants selected as surface residues on the crystal structure of CD6.17 Monoclonal antibodies were injected over immobilized CD6 (Fig. 1a–d). We show that itolizumab fails to bind E63A, indicating similar specificity to MEM98 and providing an explanation of why itolizumab is effective at blocking MEM98 binding in sequential injections over CD6 on the same sensor chip (see Supplementary material, Fig. S1). Itolizumab and MEM98 differed in recognition of R61A, itolizumab binding being affected but not MEM98, differentiating between these two mAbs.
Figure 1.
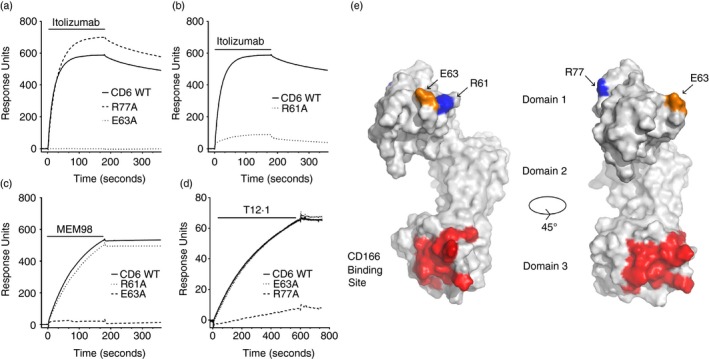
CD6 monoclonal antibodies (mAbs) bind epitopes on different faces of CD6 domain 1. (a–d) Surface plasmon resonance sensograms show binding traces for itolizumab, MEM98 and T12.1 injection (bars) over wild‐type or mutated CD6–CD4d3+4 chimeric proteins immobilized via a CD4d3+4 mAb at 25°. The background signal over CD4d3+4 (control surface) is subtracted from each trace. Conformation of CD6 was confirmed by OX126 binding or reactivity with the other domain 1 mAbs (data not shown). Mutants and their effects on antibody binding are summarized in Table 1. (e) Residues that affect MEM98 (E63, in orange), MT605 (R77, in blue) or itolizumab (E63 and R61, in blue) binding are highlighted on the structure of CD6 shown as a solvent‐excluded surface, as is the CD166 binding site (in red) in the third domain. A second orientation rotated through 45° with respect to the first is shown for clarity.
The specificity of the CD6 domain 1 mAb, T12.1 was dependent on R77 and not E63, in a similar way to MT605. Binding of mAbs specific for the defined epitopes was relatively independent of the other, neither itolizumab nor MEM98 had a marked effect on MT605 (see Supplementary material, Fig. S1A) and vice versa (see Supplementary material, Fig. S1B). Mapping the mutations that altered mAb binding onto the structure of CD6 shows that R77 and E63/R61 are well separated lying on different faces (Fig. 1e).17
None of the CD6 domain 1 mutants compromised UMCD6 binding (Table 1). We confirmed the specificity of UMCD6 for domain 1 by showing that it did not bind to chimeric CD6 containing rat CD6 domain 1 (data not shown and see Supplementary material, Fig. S1C).5, 23, 24 In sequential injections over the same sensor chip, UMCD6 binding was hindered by preinjection of CD6 domain 1 mAbs specific for both the two defined epitopes and vice versa (see Supplementary material, Fig. S1).23, 24 It was not definitive from these data with which of the two epitopes the specificity of UMCD6 overlapped. To clarify the specificity of UMCD6, we tested for competitive binding by saturating levels of UMCD6 on cells expressing CD6. Pre‐incubation of cells with UMCD6 or MEM98 blocked binding of itolizumab (see Supplementary material, Fig. S1C). Neither T12.1 not MT605 blocked binding of itolizumab (see Supplementary material, Fig. S1C).
Table 1.
The specificity of CD6 domain 1 monoclonal antibodies is defined by single point mutants R77, E63 and R61
| Residue | Itolizumab | MEM98 | MT605 | T12.1 | UMCD6 |
|---|---|---|---|---|---|
| R46 | Not expressed/not conformational | ||||
| N49 | + | + | + | + | + |
| S53 | + | + | + | + | + |
| R61 | − | + | + | + | + |
| E63 | − | − | + | + | + |
| S76 | + | + | + | + | + |
| R77 | + | + | − | − | + |
| R84 | + | + | + | + | + |
| E103 | + | + | + | + | + |
| N112 | + | + | + | + | + |
| R135 | + | + | + | + | + |
| E138 | + | + | + | +a | + |
| V139 | + | + | + | + | + |
| H142 | + | + | + | + | + |
| R145 | + | + | + | + | + |
| D147 | + | + | + | + | + |
| R149 | + | + | + | + | + |
| R150 | + | + | + | + | + |
A soluble form of CD6 was mutated in domain 1, immobilized on a BIAcore chip and tested for binding to CD6 domain 1 monoclonal antibodies. +;binding. −; no or reduced binding.
Increased off rate for E138A.
Itolizumab has a faster dissociation rate compared with other CD6 domain 1 mAbs
To further differentiate between the properties of itolizumab and MEM98 we compared their binding kinetics and compared them with CD6 domain 1 mAbs specific for the other epitopes. In preliminary SPR kinetic analysis at 25°, injections of itolizumab, MEM98, MT605 and UMCD6 (100 nm at 100 μl/min) over CD6–CD4d3+4 immobilized via a CD4d3+4 mAb revealed a pattern comparable with that shown in Fig. 2.This pattern was observed in three measurements in two independent experiments over different levels of immobilized CD6 for itolizumab, MEM98 and UMCD6 and one measurement for MT605. The most striking differences among the mAbs was that itolizumab had a faster dissociation rate. The three measurements for dissociation rates that are concentration independent for itolizumab (K off = 2·3, 4·2 and 4·3 × 10−2 s−1) were similar.
Figure 2.
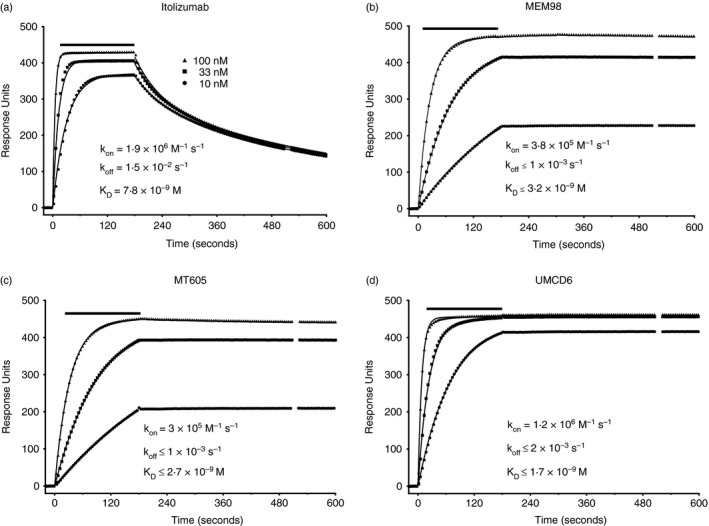
Itolizumab has a faster dissociation rate compared with other CD6 domain 1 monoclonal antibodies (mAbs). (a‐d) Surface plasmon resonance sensograms show binding traces for itolizumab, MEM98, MT605 and UMCD6 injection (bars) at 10, 33 and 100 nm (symbols) and curve fits (lines) over CD6‐CD4d3+4 chimeric protein immobilized via streptavidin at 37°. The background signal over CD4d3+4 (control surface) is subtracted from each trace. For clarity only every 10th data point is shown.
We then measured kinetics at physiological temperature, 37°. The same concentrations of each mAb were injected over the same level of immobilized CD6 with a fast flow rate to minimize rebinding. It is clear from the data shown in Fig. 2 that compared with the other three CD6 domain 1 mAbs, MEM98, MT605 and UMCD6, itolizumab dissociated more rapidly from CD6, K off = 0·015 s−1 for the majority. For the timescale of the experiment there was minimal dissociation of MEM98, MT605 and UMCD6; rates were calculated as being at least an order of magnitude lower compared with itolizumab. Itolizumab also bound more rapidly to CD6 (K on = 1·9 × 106 m −1 s−1) compared with MEM98 (K on = 3·8 × 105 m −1 s), the mAb with the overlapping epitope. Analysis of these kinetic data indicated that itolizumab (K D = 7·8 × 10−9 m) has a lower affinity compared with three other CD6 domain 1 mAbs, which all have similar binding characteristics, MEM98 (K D ≤ 3·2 × 10−9 m), MT605 (K D ≤ 2·7 × 10−9 m) and UMCD6 (K D ≤ 1·7 × 10−9 m), i.e. in the range expected for most mAbs, with the same hierarchy that we observed at 25°.
CD6 domain 1 and domain 3 mAbs are equally effective in triggering signalling through the extracellular region of CD6
We compared CD6 mAbs for their ability to bind to the CD6 extracellular region in a manner that leads to signalling in a simplified assay where the mode of action was unambiguous. CD6 is a non‐catalytic tyrosine‐phosphorylated receptor, which are all proposed to signal by kinetic segregation of receptors at the cell surface.25 As the complex nature of activating and inhibitory signalling of CD6 makes interpretation of receptor triggering more difficult, to compare the CD6 mAbs, we substituted the native cytoplasmic region of CD6 with the CD3 ζ‐chain, a well characterized activating non‐catalytic tyrosine‐phosphorylated receptor.5, 25 We used a mouse hybridoma cell line expressing a chimeric antigen receptor consisting of the extracellular and transmembrane regions of human CD6 fused to the cytoplasmic region of the mouse CD3 ζ‐chain.19 The chimera was well expressed, at the same levels as full‐length CD6 in the same hybridoma.6 We measured IL‐2 production by these cells in response to binding to the extracellular region of CD6 with immobilized CD6 mAbs (Fig. 3).
Figure 3.

CD6 domain 1 and 3 monoclonal antibodies (mAbs) are equally effective at cross‐linking the extracellular region of CD6 to trigger interleukin‐2 (IL‐2) production. IL‐2 production by 2B4 hybridoma cells expressing CD6‐ζ in response to immobilized CD6 mAbs was measured by ELISA. Of five CD6 domain 1 (a,b) and three CD6 domain 3 (c) mAbs, itolizumab, MEM98 and T12.1, which were further titrated (b) and OX124 and OX125 (c) were the most effective at triggering CD6‐ζ. (d) Itolizumab and OX125 were equally effective in triggering CD6‐ζ. Negativeve control; CAMPATH‐1H.
Five CD6 domain 1 mAbs were compared at three concentrations establishing a trend in the hierarchy of efficacy in triggering IL‐2 production under conditions where the mode of action is clearly defined: itolizumab ≥ T12.1 ≥ MEM98 > UMCD6 ≥ MT605 (Fig. 3a). Interleukin‐2 production depended on binding CD6 as none was triggered by the non‐binding CAMPATH‐1H (CD52) mAb. Further titration of itolizumab, MEM98 and T12.1 showed that they produced similar levels of IL‐2 at saturation (Fig. 3b).
We tested three CD6 mAbs, OX124, OX125 and OX126 specific for different epitopes on domain 3 for efficacy in triggering through the extracellular region of CD6.6 OX125 and OX126 were comparable and both superior to OX124 in triggering through the extracellular region of CD6 to produce IL‐2 (Fig. 3c). Direct comparison between a CD6 domain 1 mAb, itolizumab and a CD6 domain 3 mAb, OX125 showed both were equally effective in triggering a receptor by binding to the extracellular region of CD6 (Fig. 3d).
CD6 domain 1 mAbs interfere with binding to CD6 domain 3
We undertook a systematic analysis of the ability of CD6 mAbs to interfere with binding of CD6 to its best characterized ligand, CD166. We compared a panel of CD6 domain 1 mAbs for their ability to interfere with interactions of CD6 domain 3. We began by asking whether CD6 domain 1 mAbs hinder binding of a CD6 domain 3 mAb well characterized at the biochemical level as blocking ligand binding between purified proteins, CD6 and CD166.6 There was a hierarchy of effects of the CD6 domain 1 mAbs, UMCD6 > itolizumab > MT605 > MEM98 > T12.1, UMCD6 reaching up to 100% inhibition (Fig. 4, top panel). The effect was titratable (see Supplementary material, Fig. S2A) and the hierarchy of hindrance reproducible (see Supplementary material, Fig. S2B). It was clear that CD6 domain 1 mAbs could effectively interfere with interactions of domain 3.
Figure 4.
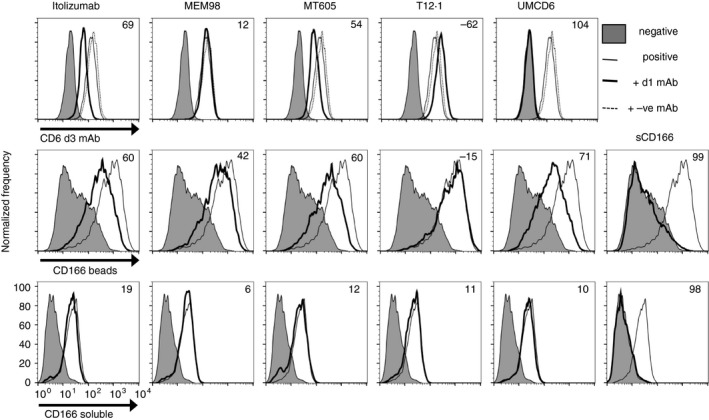
CD6 domain 1 monoclonal antibodies (mAbs) interfere with interactions of CD6 domain 3. Flow cytometry analysis of mean fluorescence intensity showing the hierarchy of inhibition by saturating concentrations of CD6 domain (d) 1 mAbs UMCD6 > itolizumab>MT605 > MEM98 > T12.1 on binding by a CD6 domain 3 mAb, OX126 (top row), CD166 immobilized on beads (middle row) or soluble multivalent CD166 (bottom row) binding to 2B4 hybridoma cells expressing human CD6. (Top row) CD6 domain 1 mAbs (40 μg/ml; heavy solid line) or relevant isotype control, CAMPATH or OX21 (dotted line), OX126‐FITC (solid line), negative staining control, OX21‐FITC (filled histogram). (Middle and bottom rows) Pre‐incubation with CD6 domain 1 mAbs interfered with CD166 immobilized on beads (middle panel) but was ineffectual at blocking soluble multivalent CD166 (lower panel). CD6 domain 1 mAbs (5 μg/ml; heavy solid line), CD166 (solid line), negative staining control, CD166d2‐5 (filled histogram). Soluble CD166 VVC (sCD166; 10 μm) (solid line) effectively blocked immobilized and soluble CD166 binding.
CD6 domain 1 mAbs interfere with binding of CD166 on beads
We next tested the ability of the CD6 mAbs to interfere with ligand binding. It has been reported that CD6 domain 1 mAbs do not block interactions of soluble forms of CD166 but do block interactions between cells.24, 26 To mimic presentation of CD166 at the cell surface, we immobilized CD166 on beads and tested the ability of saturating amounts (≥ 5 μg/ml) of CD6 domain 1 mAbs to perturb binding. The same hierarchy as with hindering the CD6 domain 3 mAb (OX126) binding was observed; UMCD6 > itolizumab > MT605 > MEM98 > T12.1 (Fig. 4, middle panel). None of the CD6 domain 1 mAbs was as effective as 10 μm soluble CD166 protein at blocking binding of CD166 beads (Fig. 4, middle panel, extreme right). Control mAbs, CAMPATH and OX21 and signal regulatory protein α did not perturb the interaction of CD166 beads with the cells (data not shown). Comparatively, the CD6 domain 1 mAbs had marginal or no effect on binding of soluble multivalent CD166 (Fig. 4 lower panel) whereas soluble CD166 blocked binding entirely (Fig. 4 lower panel, extreme right). These data confirm and extend previous reports that CD6 domain 1 mAbs including UMCD6 and T12.1 were ineffective at blocking binding of soluble multivalent forms of CD166.24
CD6 domain 1 mAbs are less effective compared with a domain 3 mAb at preventing binding of CD166
We compared the two CD6 domain 1 mAbs, UMCD6 and itolizumab which were most effective at hindering CD6/CD166 interactions with the effect of the CD6 domain 3 mAb, OX126. OX126 was superior in inhibiting CD6/CD166 interactions compared with the CD6 domain 1 mAbs (Fig. 5, upper panel). In contrast to the CD6 domain 1 mAbs, OX126 also markedly inhibited binding of soluble multivalent CD166 (Fig. 5, lower panel).
Figure 5.

CD6 domain 1 monoclonal antibodies (mAbs) are less effective compared with a domain 3 mAb at preventing binding of CD166 on beads. Flow cytometry analysis showing comparison with CD6 domain 1 mAbs, pre‐incubation with a CD6 domain 3 mAb was more effective, (OX126 > UMCD6 > itolizumab) at inhibiting CD166 immobilized on beads (upper panel) and soluble multivalent CD166 (lower panel) binding to 2B4 hybridoma cells expressing human CD6. CD6 domain 1 mAbs (5 μg/ml; heavy solid line), CD166 (solid line), negative staining control, CD166d2‐5 (filled histogram).
Discussion
Mapping epitopes for CD6 domain 1 mAbs onto the structure of CD6 identified two separate binding sites on CD6 domain 1.17 Classification of CD6 domain 1 mAbs according to their epitopes groups MEM98 and itolizumab as a pair and MT605 and T12.1 as another. These two sites containing E63 or R77 lie on different faces of domain 1. Itolizumab and MEM98 competed for binding to recombinant and native cell surface CD627 but were distinguishable from each other in dependence of the former on R61 and binding kinetics. Itolizumab dissociated more rapidly compared with MEM98 from CD6 and was more effective at triggering through the extracellular region of CD6. Greater dependence on bivalent binding of low‐affinity mAbs may be important for clustering surface receptors to trigger signal transduction.28 Characterization of ior t1 mAb from which the humanized itolizumab was derived indicated it was marginally better compared with other CD6 domain 1 mAbs in enhancing co‐stimulation of purified T cells triggered through the T‐cell receptor by CD3 mAbs.13 Humanization of ior t1 indicated that it retained a similar affinity for CD6.23 MEM98 with the overlapping epitope to itolizumab was more effective at triggering signal transduction compared with MT605 specific for a different epitope but similar affinity, which supports the conclusion that E63 is the optimal epitope to target on CD6 domain 1 for triggering through CD6.
The alanine screen did not define a residue that affected UMCD6 binding. A previous study showed that MEM98 blocks binding of itolizumab to cells.27 Displacement of itolizumab by UMCD6 but not the other way round in a binding assay could be explained by overlapping epitopes together with UMCD6 having a higher affinity for CD6.23 The antibody blocking data (see supplementary material, Fig. S1) provide further evidence that the epitope of UMCD6 may involve the same residues as the epitopes of itolizumab and MEM98 and more drastic mutations are needed to reveal dependency.
There was no difference between binding to domains 1 and 3 in the efficacy of triggering with CD6 mAbs through the extracellular region of CD6. The way a superagonist CD28 mAb bound to CD28 at the surface was an important parameter in its ability to trigger CD28. The epitope for the superagonist was close to the cell surface.29 The arrangement of the three scavenger receptor cysteine‐rich domains in CD6 is not linear and the topology of CD6 at the cell surface has not been determined.17 CD6 contains a membrane proximal stalk region, which may introduce an element of flexibility. A complex of CD6 with a CD6 domain 1 mAb compared with a complex of CD6 with a domain 3 mAb may not differ in distance from the cell surface as if the domains of CD6 were in a strictly linear configuration. Binding to either of the two domain 1 epitopes on the extracellular region of CD6 triggered signal transduction. For both epitopes, the distance spanned by the extracellular region of CD6 at the surface in complex with immobilized mAbs did not impede segregation and rearrangement of cell surface receptors necessary to trigger signalling.19, 25
The CD6 domain 1 mAbs, UMCD6 and ior t1 were both originally characterized for their agonistic properties.12, 13 UMCD6 was not effective at triggering the T‐cell hybridoma cells, consistent with agonistic properties of UMCD6 being dependent on Fc‐mediated interactions with macrophages.12 The phenomenon of a co‐stimulatory effect of a CD6 mAb has been observed by others using similar assay conditions.9 Replacing immobilized CD6 mAb with immobilized Fc fusion protein of the CD6 ligand, CD166 (ALCAM)‐Fc also enhanced CD3‐mediated proliferation, strengthening the interpretation that triggering through CD6 either by its ligand or through an antibody was activating in vitro.30
The conclusions from these data are that CD6 mAbs mimicked ligand engagement and CD6 signalling is activating under the conditions of the assays. The corollary of these data is that blocking ligand engagement reduces responses. Several groups including ourselves have provided evidence that this is so, using soluble recombinant protein and antibodies.6, 9, 10, 21, 31 Reagents that perturb CD6/CD166 interactions include CD6 domain 1 mAbs. The significance of the latter statement is that CD6 domain 1 is not the ligand‐binding domain.17, 18 CD6 domain 1 mAbs do not prevent the biochemical interaction between CD6 and the soluble form of its ligand but can sterically hinder the CD6/CD166 interaction between the molecules displayed on a surface as on cells as we have confirmed in this study.24, 26 The large antibody molecules may cover the surface of the molecule and prevent CD166 expressed on antigen‐presenting cells from reaching the membrane proximal CD6 binding domain. The CD6 domain 1 mAbs differed in their ability to interfere with domain 3 interactions. The precise way a mAb binds to CD6 seems to be critical for steric hindrance as pairs of mAbs specific for the same epitope, itolizumab and MEM98 or MT605 and T12.1 differed in their ability to interfere with CD6 domain 3 interactions. The competition experiment with a CD6 domain 3 mAb established a clear hierarchy with UMCD6 being the most effective, followed by itolizumab, MT605, MEM98 and T12.1 the least. This hierarchy was observed throughout the series of experiments testing inhibition of CD166 binding.
The Fc region of CD6 mAbs appears to be important for maximum steric hindrance of interactions between cells. The Fc region of itolizumab was necessary for its inhibitory effects.27 As none of the mutants that prevented CD6/CD166 interactions compromised binding by the CD6 domain 3 mAb, OX126, the most effective CD6/CD166 blocking mAb characterized to date appears to block by steric hindrance.6 A recombinant form of OX126 with a human Fc region engineered to lack binding to Fc receptors (Absolute Antibody) had reduced ability to block CD6/CD166 interactions (Marion H. Brown and Andrew Hartland unpublished data). Fab fragments of OX126 had reduced efficacy in blocking interactions between CD6 and soluble CD166.6 Agonistic effects of UMCD6 were overcome by the addition of a secondary antibody consistent with steric hindrance of interactions between cells.12
CD6 domain 1 mAbs, UMCD6, itolizumab and MT605 have all been reported to suppress immune responses.9, 10, 11, 27, 32 Inhibitory effects of the CD6 domain 1 mAb MT605 have been interpreted as being caused by steric hindrance of CD6/CD166 interactions and preventing co‐stimulation by CD6.9, 10 In these two studies there was good evidence of productive engagement of CD6 by CD166 and this interpretation is supported by our cell‐binding studies showing MT605 can sterically hinder multivalent immobilized CD166 from binding to cell surface CD6. The two CD6 mAbs, itolizumab and UMCD6, that have recently been put forward for therapeutic exploitation2, 32 have different binding characteristics and efficacy in cross‐linking and blocking ligand binding. It has been claimed that neither of these mAbs exerts its effects by depleting cells.27, 32 Soluble humanized itolizumab inhibited immobilized CD3‐mediated activation of human peripheral blood mononuclear cells in vitro.30 Interpretations of these data include the immunosuppression being a consequence of steric hindrance of CD6/CD166 interactions between cells.27 Steric hindrance of CD6/CD166 interactions has been proposed as a mechanism of immunosuppressive action of itolizumab in patients.27 Itolizumab did interfere with domain 3 interactions including hindering binding of CD166 on the surface of beads. The high concentrations of itolizumab required to reveal the suppressive effects in vitro are consistent with the mAb behaving in a monovalent fashion that would favour blocking not co‐stimulatory triggering.27 That immunosuppressive effects of UMCD6 in a mouse model of autoimmune disease could be attributed to steric hindrance of CD6/CD166 interactions does need to be considered in addition to other proposed explanations.32 UMCD6 was the most effective CD6 domain 1 mAb at blocking CD6 domain 3 interactions. Partial blocking may be adequate to reduce occupancy in a low‐affinity interaction sufficiently to have a functional effect. To date it appears that development of CD6‐targeted immunotherapy has been based on empirical observations of the beneficial effects of a particular reagent to hand without a clear prediction as to the mode of action. Molecular characterization of different CD6 mAbs provides a more specific selection of properties to test and subsequently exploit, particularly if the proposed mode of action is to exert immunosuppression by blocking CD6/CD166 interactions. Of the available CD6 domain 1 mAbs, more effective blocking, a high affinity and reduced efficacy as an agonist in triggering CD6 may favour UMCD6 over itolizumab. However, as we have shown and commented on previously, UMCD6 is not as efficacious at blocking CD6/CD166 interactions compared with a CD6 domain 3 mAb.33, 34 Molecular characterization of mouse CD6 mAbs that are effective in suppressing mouse models of autoimmune disease will be very interesting as the approach to developing CD6 as a drug target becomes more rational.27, 35
Disclosures
The authors declare no commercial or financial conflict of interest.
Supporting information
Figure S1. Competitive and steric hindrance of binding among CD6 domain 1 monoclonal antibodies.
Figure S2. CD6 domain 1 monoclonal antibodies interfere with binding to CD6 domain 3.
Acknowledgements
This work was supported by a Medical Research Council UK Programme grant (G0400808) and the CIU Trust. We thank Omer Dushek for assistance with SPR kinetic analysis, and Ellie Denham and Duncan Howie for critically reading the manuscript. Mutagenesis and SPR were carried out by LIG, functional experiments by AH and JB, cell binding experiments by AH and MHB. MHB was responsible for overall supervision and wrote the manuscript.
References
- 1. Simpson BS, Coles AJ. Rationale for cytotoxic monoclonal antibodies in MS. Int MS J 2007; 14:48–56. [PubMed] [Google Scholar]
- 2. Hernandez P, Moreno E, Aira LE, Rodriguez PC. Therapeutic targeting of CD6 in autoimmune diseases: a review of Cuban clinical studies with the antibodies IOR‐T1 and itolizumab. Curr Drug Targets 2016; 17:666–77. [DOI] [PubMed] [Google Scholar]
- 3. Menon R, David BG. Itolizumab – a humanized anti‐CD6 monoclonal antibody with a better side effects profile for the treatment of psoriasis. Clin Cosmet Investig Dermatol 2015; 8:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Montero E, Falcon L, Morera Y, Delgado J, Amador JF, Perez R. CD6 molecule may be important in the pathological mechanisms of lymphocytes adhesion to human skin in psoriasis and ior t1 MAb a possible new approach to treat this disease. Autoimmunity 1999; 29:155–6. [DOI] [PubMed] [Google Scholar]
- 5. Breuning J, Brown MH. T cell costimulation by CD6 is dependent on bivalent binding of a GADS/SLP‐76 complex. Mol Cell Biol 2017; 37:e00071–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hassan NJ, Simmonds SJ, Clarkson NG, Hanrahan S, Puklavec MJ, Bomb M et al CD6 regulates T‐cell responses through activation‐dependent recruitment of the positive regulator SLP‐76. Mol Cell Biol 2006; 26:6727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oliveira MI, Goncalves CM, Pinto M, Fabre S, Santos AM, Lee SF et al CD6 attenuates early and late signaling events, setting thresholds for T‐cell activation. Eur J Immunol 2012; 42:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orta‐Mascaro M, Consuegra‐Fernandez M, Carreras E, Roncagalli R, Carreras‐Sureda A, Alvarez P et al CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med 2016; 213:1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zimmerman AW, Joosten B, Torensma R, Parnes JR, van Leeuwen FN, Figdor CG. Long‐term engagement of CD6 and ALCAM is essential for T‐cell proliferation induced by dendritic cells. Blood 2006; 107:3212–20. [DOI] [PubMed] [Google Scholar]
- 10. Kofler DM, Severson CA, Mousissian N, De Jager PL, Hafler DA. The CD6 multiple sclerosis susceptibility allele is associated with alterations in CD4+ T cell proliferation. J Immunol 2011; 187:3286–91. [DOI] [PubMed] [Google Scholar]
- 11. Singer NG, Richardson BC, Powers D, Hooper F, Lialios F, Endres J et al Role of the CD6 glycoprotein in antigen‐specific and autoreactive responses of cloned human T lymphocytes. Immunology 1996; 88:537–43. [PMC free article] [PubMed] [Google Scholar]
- 12. Bott CM, Doshi JB, Morimoto C, Romain PL, Fox DA. Activation of human T cells through CD6: functional effects of a novel anti‐CD6 monoclonal antibody and definition of four epitopes of the CD6 glycoprotein. Int Immunol 1993; 5:783–92. [DOI] [PubMed] [Google Scholar]
- 13. Osorio LM, Garcia CA, Jondal M, Chow SC. The anti‐CD6 mAb, IOR‐T1, defined a new epitope on the human CD6 molecule that induces greater responsiveness in T cell receptor/CD3‐mediated T cell proliferation. Cell Immunol 1994; 154:123–33. [DOI] [PubMed] [Google Scholar]
- 14. Jayaraman K. Biocon's first‐in‐class anti‐CD6 mAb reaches the market. Nat Biotechnol 2013; 31:1062–3. [DOI] [PubMed] [Google Scholar]
- 15. Aira LE, Lopez‐Requena A, Fuentes D, Sanchez L, Perez T, Urquiza A et al Immunological and histological evaluation of clinical samples from psoriasis patients treated with anti‐CD6 itolizumab. MAbs 2014; 6:783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aira LE, Hernandez P, Prada D, Chico A, Gomez JA, Gonzalez Z et al Immunological evaluation of rheumatoid arthritis patients treated with itolizumab. MAbs 2016; 8:187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chappell PE, Garner LI, Yan J, Metcalfe C, Hatherley D, Johnson S et al Structures of CD6 and its ligand CD166 give insight into their interaction. Structure 2015; 23:1426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowen MA, Aruffo AA, Bajorath J. Cell surface receptors and their ligands: in vitro analysis of CD6–CD166 interactions. Proteins 2000; 40:420–8. [PubMed] [Google Scholar]
- 19. Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T‐cell receptor triggering is critically dependent on the dimensions of its peptide–MHC ligand. Nature 2005; 436:578–82. [DOI] [PubMed] [Google Scholar]
- 20. Aricescu AR, Lu W, Jones EY. A time‐ and cost‐efficient system for high‐level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 2006; 62:1243–50. [DOI] [PubMed] [Google Scholar]
- 21. Hassan NJ, Barclay AN, Brown MH. Frontline: optimal T cell activation requires the engagement of CD6 and CD166. Eur J Immunol 2004; 34:930–40. [DOI] [PubMed] [Google Scholar]
- 22. Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med 1998; 188:2083–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alonso R, Huerta V, de Leon J, Piedra P, Puchades Y, Guirola O et al Towards the definition of a chimpanzee and human conserved CD6 domain 1 epitope recognized by T1 monoclonal antibody. Hybridoma 2008; 27:291–301. [DOI] [PubMed] [Google Scholar]
- 24. Bowen MA, Bajorath J, Siadak AW, Modrell B, Malacko AR, Marquardt H et al The amino‐terminal immunoglobulin‐like domain of activated leukocyte cell adhesion molecule binds specifically to the membrane‐proximal scavenger receptor cysteine‐rich domain of CD6 with a 1:1 stoichiometry. J Biol Chem 1996; 271:17390–6. [DOI] [PubMed] [Google Scholar]
- 25. Dushek O, Goyette J, van der Merwe PA. Non‐catalytic tyrosine‐phosphorylated receptors. Immunol Rev 2012; 250:258–76. [DOI] [PubMed] [Google Scholar]
- 26. Bowen MA, Patel DD, Li X, Modrell B, Malacko AR, Wang WC et al Cloning, mapping, and characterization of activated leukocyte‐cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med 1995; 181:2213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bughani U, Saha A, Kuriakose A, Nair R, Sadashivarao RB, Venkataraman R et al T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS ONE 2017; 12:e0180088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koch C, Staffler G, Huttinger R, Hilgert I, Prager E, Cerny J et al T cell activation‐associated epitopes of CD147 in regulation of the T cell response, and their definition by antibody affinity and antigen density. Int Immunol 1999; 11:777–86. [DOI] [PubMed] [Google Scholar]
- 29. Evans EJ, Esnouf RM, Manso‐Sancho R, Gilbert RJ, James JR, Yu C et al Crystal structure of a soluble CD28‐Fab complex. Nat Immunol 2005; 6:271–9. [DOI] [PubMed] [Google Scholar]
- 30. Nair P, Melarkode R, Rajkumar D, Montero E. CD6 synergistic co‐stimulation promoting proinflammatory response is modulated without interfering with the activated leucocyte cell adhesion molecule interaction. Clin Exp Immunol 2010; 162:116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gimferrer I, Calvo M, Mittelbrunn M, Farnos M, Sarrias MR, Enrich C et al Relevance of CD6‐mediated interactions in T cell activation and proliferation. J Immunol 2004; 173:2262–70. [DOI] [PubMed] [Google Scholar]
- 32. Li Y, Singer NG, Whitbred J, Bowen MA, Fox DA, Lin F. CD6 as a potential target for treating multiple sclerosis. Proc Natl Acad Sci USA 2017; 114:2687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown MH. CD6 as a cell surface receptor and as a target for regulating immune responses. Curr Drug Targets 2016; 17:619–29. [DOI] [PubMed] [Google Scholar]
- 34. Castro MA, Oliveira MI, Nunes RJ, Fabre S, Barbosa R, Peixoto A et al Extracellular isoforms of CD6 generated by alternative splicing regulate targeting of CD6 to the immunological synapse. J Immunol 2007; 178:4351–61. [DOI] [PubMed] [Google Scholar]
- 35. Zhang L, Li Y, Qiu W, Bell BA, Dvorina N, Baldwin WM 3rd et al Targeting CD6 for the treatment of experimental autoimmune uveitis. J Autoimmun 2018; 90:84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Competitive and steric hindrance of binding among CD6 domain 1 monoclonal antibodies.
Figure S2. CD6 domain 1 monoclonal antibodies interfere with binding to CD6 domain 3.


