Summary
The role of the host extracellular matrix (ECM) in infection tends to be neglected. However, the complex interactions between invading pathogens, host tissues and immune cells occur in the context of the ECM. On the pathogen side, a variety of surface and secreted molecules, including microbial surface components recognizing adhesive matrix molecules and tissue‐degrading enzymes, are employed that interact with different ECM proteins to effectively establish an infection at specific sites. Microbial pathogens can also hijack or misuse host proteolytic systems to modify the ECM, evade immune responses or process biologically active molecules such as cell surface receptors and cytokines that direct cell behaviour and immune defence. On the host side, the ECM composition and three‐dimensional ultrastructure undergo significant modifications, which have a profound impact on the specific signals that the ECM conveys to immune cells at the forefront of infection. Unexpectedly, activated immune cells participate in the remodelling of the local ECM by synthesizing ECM glycoproteins, proteoglycans and collagen molecules. The close interplay between the ECM and the innate immune response to microbial pathogens ultimately affects the outcome of infection. This review explores and discusses recent data that implicate an active role for the ECM in the immune response to infection, encompassing antimicrobial activities, microbial recognition, macrophage activation, phagocytosis, leucocyte population balance, and transcriptional and post‐transcriptional regulation of inflammatory networks, and may foster novel antimicrobial approaches.
Keywords: extracellular matrix, immunity, infection
Introduction
To successfully establish an infectious disease, most pathogenic bacteria adhere to the host, invade underlying tissues, and multiply and grow while evading or overcoming host immune defences.1 The underlying host–pathogen interactions are remarkably specific and involve a variety of substrates within the host. The interactions of pathogens with eukaryotic cells have been intensively studied in microbiology, immunology, cell biology, genetics and ecology research. Less attention has been given to the host extracellular matrix (ECM). However, the individual components of the ECM are substrates that pathogens can directly bind to or degrade, facilitating adhesion and penetration into the host; evidence for this started to accumulate 40 years ago.2, 3 Furthermore, the ECM is a fundamental component of the host cellular microenvironment where most of the events leading to infection, disease and tissue repair take place and is also a reservoir of diverse and tissue‐specific signals that feed into immunological pathways.4, 5 Recently, evidence has emerged that the ECM conveys specific signals to cells, thereby modulating essential immune functions, immune cell migration into and within infected tissues, and immune cell activation, proliferation and differentiation. Moreover, the ECM composition undergoes significant alterations during infection and, notably, specific immune cell types appear to contribute to that. Hence, regulatory circuits closely link the ECM and the immune system.
Important questions to emerge from these studies include (i) does the ECM play an active role in infection rather than simply providing a scaffold for bacterial adhesion or being a barrier to breach; (ii) which ECM–pathogen interactions significantly impact the ability of pathogens to colonize and invade host tissues, and/or bypass host defences, and influence how host cells respond to pathogens; and (iii) can we interfere with these interactions to develop new antimicrobial strategies or improve existing ones. Here, we briefly review the ECM and its interactions with microbial pathogens, and discuss the evidence for a direct implication of the ECM in the immune response to infection.
The ECM: an immunological perspective
Most infections and the resulting immune responses are tissue‐specific. Each tissue has distinct ECM signatures that arise from complex and dynamic combinations of up to ~ 300 different proteins in varying concentrations and geometries within the three‐dimensional extracellular space. This ever increasing number of proteins that contribute to matrices (the ‘matrisome’) include approximately 43 collagen subunits, 36 proteoglycans and 200 glycoproteins.6 ECM molecular multiplicity and complexity are amplified by post‐translational modifications, which can unveil cryptic epitopes or generate new ones capable of triggering immune reactions, and alternative splicing, which generates several isoforms with distinct functions.7, 8 For instance, laminin α4 and α5 isoforms in basement membranes contribute to immune cell subtype selectivity during leucocyte recruitment to sites of inflammation.9, 10
Interstitial ECMs, loose fibril‐like matrices that fill the tissue stroma, and basement membranes, laminar sheets that anchor cell layers to underlying tissues, are basic forms of ECM. Additionally, specialized reticular fibre networks combining features of these two ECM structures are found in secondary lymphoid organs.11, 12
The ECM is a highly dynamic yet strictly regulated tissue component. Its composition and normal function are determined and maintained by a fine balance between ECM synthesis, orchestrated by cytokines such as transforming growth factor‐β (TGF‐β), and turnover, accomplished by matrix metalloproteinases (MMPs), a disintegrin and metalloproteinases (ADAMs) and a disintegrin and metalloproteinases with thrombospondin motifs (ADAMTSs), whose activity is controlled by tissue inhibitors of metalloproteinases (TIMPs).13 During injury and infection, these enzymes are produced by activated immune cells such as monocytes/macrophages and promote immune cell migration into sites of infection and tissue damage, and affect their ability to mount inflammatory responses.14
The central dogma of matrix biology describes how the ECM provides structural support for cells and contributes to the unique structure of a tissue. However, this is only one function of the ECM. Host immune responses are carried out in the context of the ECM, so when immune cells contact the ECM, they receive vital instructions for their survival, proliferation, differentiation and activation, in addition to support for adhesion and guidance for migration. Several mechanisms are responsible for the communication between the ECM and cells. One such mechanism is signalling through cell surface adhesion molecules and receptors, including integrins and discoidin‐domain receptors (reviewed in refs 15, 16). Others involve binding, storage, activation and release of secreted molecules with potent immunomodulatory activity, including cytokines, chemokines and growth factors.17, 18 For instance, TGF‐β has been shown in vitro and in vivo to affect activation, proliferation and differentiation of most immune cell types, modulating nearly all stages of the immune response.19 Moreover, bioactive ECM fragments generated by tissue destruction (e.g. hyaluronan and heparan sulphate oligosaccharides) and ECM molecules whose expression is specifically induced upon tissue injury (e.g. fibronectin isoforms containing extra domain A, versican and biglycan) promote inflammation by inducing pro‐inflammatory gene expression or exhibiting chemoattractant properties (reviewed in refs 20, 21). These molecules form a class of endogenous damage‐associated molecular patterns,22 which, by activating pattern recognition receptors (PRRs) such as toll‐like receptors (TLRs) in immune cells (e.g. macrophages and dendritic cells) and non‐immune cells (e.g. fibroblasts and epithelial cells) alert the immune system to tissue damage and infection, which initiates not only pathogen clearance, but also tissue repair.23 Hence, the ECM and the immune system are intertwined: signals from the ECM help to coordinate immune responses and, in turn, immune cells promote ECM repair and regeneration through the release of cytokines such as tumour necrosis factor (TNF), interferon‐γ (IFN‐γ) and TGF‐β, which regulate the expression of many ECM molecules.
ECM–pathogen interactions
A tale of adhesion and colonization of the host
Tissue adherence represents the first essential mechanism that bacteria adopt to colonize the host. Failure to do so results in the organism being removed by physiological cleansing systems at sites of entry. The surface of host cells and the ECM are negatively charged and microbes employ a number of physico‐chemical forces to overcome these repulsive forces and establish interactions between a microbial ligand or ‘adhesin’ and a complementary molecule or ‘receptor’ on the host tissue. These interactions are highly specific, contributing to tissue tropism, species specificity and genetic specificity within a species. Bacterial adhesins are components of capsules, cell walls, pili or fimbriae, and those that bind to ECM components are called ‘microbial surface components recognizing adhesive matrix molecules’. Host receptors are usually glycoproteins found on the cell membrane and ECM components.24
The binding of microbial pathogens to specific ECM proteins has been extensively reviewed elsewhere1, 3, 25, 26 and will be briefly discussed here. The first report of a host ECM–pathogen interaction was published in 1978 by Kuusela et al.2 who studied the binding of Staphylococcus aureus to fibronectin and, a few years later, revealed two separate binding sites on fibronectin, in the N‐terminus and C‐terminus, respectively.27 Other groups further characterized the interaction of this ubiquitous and promiscuous ECM glycoprotein with Staphylococcus aureus and Streptococcus pyogenes, which has a remarkably large number of fibronectin binding adhesins.28, 29, 30, 31, 32, 33 Several other ECM proteins such as laminin, collagen, heparan and chondroitin sulphate, vitronectin, thrombospondin, elastin, bone sialoprotein and tenascin‐C, have since been implicated in specific interactions with pathogenic bacteria26, 34 (Table 1).
Table 1.
Examples of specific host ECM–pathogen interactions that facilitate microbial adhesion to tissues and pathogenesis (* indicates that animal models of infection were used in the study)
| ECM molecule | Pathogen | Adhesin | Effect of interaction | Disease | References |
|---|---|---|---|---|---|
| Fibronectin | Enterohaemorrhagic Escherichia coli O157:H7 | Lpf fimbriae (LpfA1 major subunit) | Colonization of GI tract | Acute diarrhoea; bloody diarrhoea; haemolytic uraemic syndrome | 130 |
| Streptococcus pyogenes (group A streptococci, GAS) | Protein F1 (functional upstream domain) | Fibronectin links F1 to integrin receptors, helping bacterial uptake | Tonsillopharyngitis; necrotizing fasciitis; myositis; streptococcal toxic shock syndrome | 131, 132, 133 | |
| Protein F2 (C‐terminal domains) | Host cell adhesion and internalization | 33 | |||
| SfbII (C‐terminal domain) | Host cell adhesion and internalization | 134 | |||
| Fba | Host cell adhesion and internalization | 31* | |||
| Staphylococcus aureus | MntC | Mucosal colonization | Nosocomial infections, septicaemia, osteomyelitis, endocarditis, etc. | 135 | |
| Salmonella typhimurium | MisL (N‐terminal non‐conserved region) | Intestinal colonization | Gastroenteritis | 136* | |
| ShdA | 137 | ||||
| Plasma fibronectin | Borrelia burgdorferi | BBK32 | Binding to fibronectin 13FnIII repeat module and intestinal colonization | Lyme disease | 35 |
| Fibronectin N‐terminal proteolytic fragments (30 and 70 kDa) | Streptococcus pyogenes | Protein F2(C‐terminal domains) | Fibronectin polymerization and colonization of vascular surfaces | Tonsillopharyngitis; necrotizing fasciitis; myositis; streptococcal toxic shock syndrome | 33 |
| 130 | |||||
| Laminin | EnterohaemorrhagicEscherichia coli O157:H7 | Lpf fimbriae (LpfA1 major subunit) | Eukaryotic cell adhesion and internalization | Acute diarrhoea; bloody diarrhoea; haemolytic uraemic syndrome | 130 |
| EnterohaemorrhagicEscherichia coli O157:H7 | Lpf fimbriae (LpfA1 major subunit) | Colonization of GI tract | Acute diarrhoea; bloody diarrhoea; haemolytic uraemic syndrome | 135 | |
| Staphylococcus aureus | MntC | Colonization of GI tract | Nosocomial infections, septicaemia, osteomyelitis, endocarditis, etc. | 138 | |
| Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Mucosal colonization | Infective endocarditis | 139 | |
| Collagen I | Borrelia burgdorferi | ErpX | Adhesion and invasion of endothelial cells | Lyme disease | 138 |
| Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Long‐term host tissue colonization | Infective endocarditis | 137 | |
| Salmonella typhimurium | ShdA | Adhesion and invasion of endothelial cells | Gastroenteritis | 140* | |
| Collagen II | Enterococcus faecium | Pilus subunits EmpA and EmpB | Intestinal colonization and persistence | UTIs, bacteraemia, and infective endocarditis | 138 |
| Collagen IV | Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Adherence to host tissue and biofilm formation | Infective endocarditis | 130 |
| EnterohaemorrhagicEscherichia coli O157:H7 | Lpf fimbriae (LpfA1 major subunit) | Adhesion and invasion of endothelial cells | Acute diarrhoea; bloody diarrhoea; haemolytic uraemic syndrome | 135 | |
| Staphylococcus aureus | MntC | 138 | |||
| Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Colonization of GI tract | Nosocomial infections, septicaemia, osteomyelitis, endocarditis, etc. | 136* | |
| Collagen V | Salmonella typhimurium | MisL (N‐terminal non‐conserved region) | Mucosal colonization | Infective endocarditis | 141 |
| Collagen VI | Enterococcus faecium | EcbA | Adhesion and invasion of endothelial cells | Gastroenteritis | 142* |
| Tenascin‐C | Legionella pneumophila | Mip | Intestinal colonization | UTIs, bacteraemia, and infective endocarditis | 138 |
| Vitronectin | Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Host tissue adhesion and biofilm formation | Legionellosis | 138 |
| Streptococcus gallolyticus (gallolyticus endocarditis isolates) | FimB, gtf and pilB | Adhesion to lung tissue and bacterial dissemination | Infective endocarditis | 143* | |
| Yersinia enterocolitica | YadA | Adhesion and invasion of endothelial cells | Infective endocarditis | 46 | |
| Thrombospondin | Trypanosoma cruzi | TcCRT | Adhesion and invasion of endothelial cells | Enteric and systemic diseases | 144* |
| Decorin | Borrelia burgdorferi | DbpA and DbpB | Adhesion to host cells and tissue; improved bacterial survival | Chagas disease | 141 |
| Nidogen 1 and 2 | Enterococcus faecium | SgrA | Enhancement of cellular infection | Lyme disease | 145, 146 |
| Soluble and immobilized fibrinogen(α‐ and β‐chains) | Staphylococcus aureus | ClfA, ClfB | Specific localization to decorin‐rich niches in the tunica adventitia and myocardial connective tissue; persistence of infection | UTIs, bacteraemia, and infective endocarditis | |
| Host tissue adhesion and biofilm formation | Nosocomial infections, septicaemia, osteomyelitis, endocarditis, etc. | ||||
| Colonization of biomaterial implants; bacterial spread |
GI, gastrointestinal; UTI, urinary tract infection; LpfAI, long polar fimbrae subunit 1; SfbII, fibronectin surface binding protein II; Fba, fibronectin binding protein; MntC, manganese transport protein C; MisL, autotransport protein MisL; FimB, type I fimbriae regulatory protein FimB; gtf, glucosyltransferase; pilB, type 4 fimbrial assembly protein; EcbA, E. faecium collagen binding protein A; MIP, macrophage infectivity potentiator; YadA, adhesin YadA; TcCRT, Trypanosoma cruzi calreticulin; DdpA/ DdpB, decorin binding protein A and B; SgrA, serine‐glutamate repeat containing protein A; ClfA/ClfB, clumping factor A and B.
Early studies were mostly limited by the use of binding assays, involving bacterial cells or recombinant adhesins and ECM molecules, and inhibition experiments to demonstrate these interactions. Cutting‐edge technologies, including live cell imaging and particle‐tracking methods, have now started to reveal the implications of these interactions in pathogenesis. For example, Niddam et al. showed that the Lyme disease spirochaete Borrelia burgdorferi exploits fibronectin to interact with vascular surfaces under physiological shear stress. Specifically, it recruits and induces polymerization of soluble plasma fibronectin that strengthens and stabilizes bacterial interactions with endothelia by a catch–bond mechanism.35
Breaking down barriers to invade the host
Having colonized the host, most pathogens need to invade tissues to cause disease. This requires the breakdown of primary and/or secondary defences of the host, involving the crossing of basement membranes and interstitial matrices. Pathogens have developed distinct ways to modify the ECM.1, 36 They can directly degrade ECM components using ‘invasins’ or bacterial tissue‐degrading enzymes such as hyaluronidases and collagenases, causing local tissue damage. Specific examples of these interactions are reported in Table 2 and reviewed elsewhere.26, 36 Degradation of the ECM not only facilitates the spread of pathogens, but can also favour tissue necrosis, bacterial toxin diffusion and host cell adhesion, migration and survival. Pathogens can also indirectly modify the ECM by altering the synthesis and turnover of ECM components by host cells in response to their presence (discussed later in this review). Moreover, pathogens can hijack or misuse host proteolytic systems. For instance, Staphylococcus aureus,37 Haemophilus influenzae 38 and Pseudomonas aeruginosa,39 among other common pathogenic bacteria, manipulate the plasminogen–plasmin system thereby degrading laminin and fibronectin and activating MMP zymogens that not only degrade all types of ECM proteins, but also process many biologically active molecules (e.g. cell surface receptors and cytokines) that direct cell behaviour and host defence.
Table 2.
Examples of specific ECM–pathogen interactions that facilitate host invasion through direct degradation of ECM components (# indicates that ex vivo mammalian tissue degradation models, were used in the study)
| ECM molecule | Pathogen | Microbial enzyme | Effect of ECM cleavage | Disease | References |
|---|---|---|---|---|---|
| Laminin | Psedomonas aeruginosa | Elastase; alkaline protease | Tissue invasion and necrosis | Necrotizing pneumonia, septic shock, UTI, skin and soft‐tissue infections | 147 # |
| Clostridium difficile | Cwp84 | Tissue integrity loss; facilitation of toxin diffusion | Pseudomembranous colitis and nosocomial diarrhoea | 148 | |
| Collagen I | Porphyromonas gingivalis | Gingipains in P. gingivalis supernatant | Tissue degradation | Periodontal disease | 149 |
| Vibrio parahaemolyticus | Metalloprotease VppC | Tissue damage | Acute gastroenteritis | 150 | |
| Collagen I, II, III, IV, V and VI | Clostridium histolyticum | Class I and II collagenases (ColG, ColH) | Necrotic tissue degradation; promote keratinocyte migration | Gas gangrene, infective endocarditis | 151, 152 |
| Collagen IV | Streptococcus gordonii | Serine protease | Basement membrane breakdown | Infective endocarditis | 153 |
| Fibronectin | Clostridium difficile | Cwp84 | Tissue integrity loss; facilitation of toxin diffusion | Pseudomembranous colitis and nosocomial diarrhoea | 148 |
| Porphyromonas gingivalis | Gingipains in P. gingivalis supernatant; HRgpA and RgpB gingipains | Cleavage and inactivation of cell‐binding region of fibronectin; gingival fibroblast detachment and death; tissue destruction | Periodontal disease | 149, 154 # | |
| Vitronectin | Clostridium difficile | Cwp84 | Tissue integrity loss; facilitation of toxin diffusion | Pseudomembranous colitis and nosocomial diarrhoea | 148 |
| Tenascin‐C (large isoforms) | Porphyromonas gingivalis | HRgpA, RgpB and Kgp gingipains | Enhanced anti‐adhesive activity of tenascin‐C; gingival fibroblasts detachment, apoptosis and tissue destruction | Periodontal disease | 154 # |
Cwp84, putative cell surface‐associated cysteine protease; HRgpA and RgpB, arginine‐gingipains; Kgp, lysine‐gingipains; UTI, urinary tract infection.
Strategies to evade the host immune response?
Although numerous pathogens undoubtedly exploit ECM components and ECM‐associated molecules to adhere to and degrade tissues for efficient host colonization and invasion, it is not always clear whether pathogens interact with these ECM molecules also to evade immune responses. For instance, Borrelia burgdorferi can avoid antibody‐mediated clearance and this may be partly explained by its specific interaction with decorin.36 The Haemophilus influenzae surface protein E binds to plasminogen, which is converted to plasmin, and uses plasmin for complement evasion and innate immune escape.37 Several other pathogenic bacteria take advantage of the host plasminogen system to facilitate their own spread and invasion through tissues.40, 41 Notably, Helicobacter pylori benefits from the complement regulatory property and the plasminogen‐binding ability of vitronectin to protect itself from innate immune responses.42 Furthermore, Group B streptococci and other Gram‐positive bacteria secrete hyaluronidases, whose activity allows immune evasion besides tissue invasion. Specifically, their hyaluronidases process pro‐inflammatory hyaluronan fragments into disaccharides, which block TLR2/4 signalling triggered by host‐derived hyaluronan fragments and pathogenic ligands, including lipopolysaccharide (LPS), thereby evading immune detection.43
Altered ECM dynamics in infection and its implications
The ECM undergoes significant alterations upon infection that promote or inhibit the establishment of infection and the host response to it. Below is an overview of the changes in ECM synthesis, degradation and post‐translational modification during infection and a discussion of their implications in pathogenesis.
ECM synthesis and deposition
Signal transduction pathways activated upon pathogen entry and recognition, and by mediators of inflammation and tissue repair during infection all contribute to ECM synthesis and deposition. This involves structural ECM proteins (e.g. collagens, laminins and proteoglycans) as well as non‐structural ECM components termed ‘matricellular proteins’, which are normally absent or scarcely expressed in healthy tissues (e.g. osteopontin, thrombospondins, galectins, tenascins). For instance, rhinovirus activates TLR3 and TLR7/8 signalling in airway smooth muscle cells, which leads to increased deposition of fibronectin, perlecan and collagen IV, contributing to airway remodelling and facilitating the migration of airway smooth muscle cells to the infection site.44 Interleukin‐33 (IL‐33) signalling induced by Staphylococcus aureus enhances fibronectin and collagen IIIa expression and deposition, accelerating wound repair.45 Systems biology approaches elucidating the ECM interactome network regulated by Trypanosoma cruzi and its gp83 ligand, which mediates trypanosome attachment and entry, have shown that activation of gp83 receptors in the cell via extracellular signal‐regulated kinase 1/2 results in up‐regulation of laminin γ1 and thrombospondin expression to facilitate trypomastigote recruitment, enhancing cellular infection.46 In mice, Citrobacter rodentium infection induces osteopontin and fibronectin expression through integrin‐linked kinase activation, facilitating bacterial colonization of the intestine.47 High expression levels of osteopontin have also been shown to be induced in murine acute and chronic coxsackievirus B3‐myocarditis together with those of MMP‐3, TIMP1, urokinase‐type plasminogen activator and TGF‐β 1 and, in turn, procollagen‐1α mRNA expression and fibrosis. Accordingly, osteopontin‐null mice are protected from viral myocarditis, and inhibition of osteopontin transcription by vitamin D decreases cardiac fibrosis in wild‐type animals.48 Viral myocarditis also results in up‐regulation of tenascin‐C before immune cell infiltration occurs and until scar tissue is formed.49 Elevated tenascin‐C levels have also been reported in patients with sepsis, parapneumonic infection, tuberculosis and Staphylococcus aureus infection.50, 51, 52, 53, 54 Furthermore, the lungs of mice infected with Gram‐negative bacteria show accumulation of versican, a chondroitin sulphate proteoglycan with multiple cytokine, chemokine, adhesion molecule and growth factor‐binding domains, that is implicated in the innate immune response.55 Similarly, when primary human lung fibroblasts are treated with the viral mimetic Poly I:C, they deposit a higher‐order structured ECM, rich in versican and hyaluronan, to which T cells avidly adhere and cease migration, an effect reversed by hyaluronidase treatment or versican antibody during matrix formation.56 Importantly, besides expression levels, infection can also alter the spatial distribution of ECM components as it emerges from influenza‐infected lungs, which show distinct regions that are enriched with either fibronectin or collagen.57 Hence, specific infectious diseases seem to generate distinct compositional changes of the ECM, inevitably influencing its biophysical structure and presentation of bioactive compounds that impact bacterial colonization and invasion, immune cell response and tissue repair.
ECM degradation
The catabolic machinery that breaks down and remodels the ECM is also altered upon infection, affecting the supportive, barrier and biological functions of the ECM. Tissue degrading enzymes such as MMPs play a crucial role in regulating immune cell recruitment: they cleave the basement membrane ECM; expose cryptic pro‐migratory sites of ECM components (e.g. γ2 chain of laminin 5); target non‐ECM proteins such as adhesion molecules (e.g. E‐cadherin); activate, deactivate or regulate the bioavailability of chemokines (e.g. monocyte cheomattractant protein‐1 and IL‐8) and cytokines (e.g. IL‐1β and TNF‐α); and shed cell surface receptors associated with cell migration (e.g. CD44), modulating inflammation (reviewed in refs 58, 59, 60). With the exception of neutrophils, tissue‐degrading enzymes are not stored, but require de novo synthesis that is strictly regulated and can be induced by pro‐inflammatory cytokines (e.g. TNF‐α and IL‐1) and also by bacterial products (e.g. LPS and chlamydial heat‐shock proteins).61
Several pathogens, including Mycobacterium bovis, Mycobacterium tuberculosis, Streptococcus pyogenes and Helicobacter pylori, induce expression and activity of a number of MMPs, including MMP‐1, MMP‐2, MMP‐7, MMP‐9 and MMP‐13.62, 63, 64 In particular, the extensive production of MMP‐2 and MMP‐9 during mycobacterial infection is regulated by macrophage‐derived and T‐cell‐derived cytokines and causes ECM breakdown. This may be necessary for cell recruitment and granuloma formation, both protective immune responses to mycobacteria. However, dysregulation of MMP production at late stages of the infection could contribute to tissue damage and, by compromising tissue integrity, may facilitate bacterial dissemination and persistence of infection.62 Systemic Escherichia coli infection, acute Lyme neuroborreliosis and pneumococcal meningitis can all lead to secretion of high amounts of MMP‐9. In meningitis, MMP‐9 has been suggested to contribute to blood–brain barrier destruction and neuronal injury.65, 66, 67 Similarly, gastrin‐dependent induction of MMP‐7 upon Helicobacter pylori infection has been implicated in the development of gastric cancer through the release of heparin‐binding epidermal growth factor.64 Chlamydia trachomatis, which causes trachoma and blindness, up‐regulates MMP‐7, MMP‐9, MMP‐12 and TIMP‐1 expression, while it down‐regulates MMP‐10 and SPARC (secreted protein, acidic, cysteine‐rich)‐like 1, a matricellular protein that regulates decorin production and collagen assembly.68 In this study the expression pattern of these ECM‐modifying enzymes was correlated with the clinical scarring grade and inflammation. Interestingly, a dual function has been shown for MMP‐12 in viral myocarditis caused by coxsackievirus type B3. While intracellular MMP‐12 causes IFN‐α secretion and host protection, extracellular MMP‐12 cleaves the IFN‐α receptor 2 binding site of IFN‐α, preventing an uncontrolled immune response.69 In the same model of viral myocarditis, MMP‐9 exerts a protective role by inactivating IFN‐β/γ. Indeed, MMP‐9‐null mice display higher viral load, infiltration of CD3+ cells and tissue damage.70 However, murine MMP‐9 can also enhance susceptibility to infection and increase morbidity and mortality. This is the case in Francisella tularensis pulmonary infection where MMP‐9 generates pro‐inflammatory ECM‐derived peptides (i.e. Pro‐Gly‐Pro peptide from collagen I), enhancing neutrophil infiltration to lungs.71
In addition to inducing MMP expression, pathogens can also activate pro‐MMPs by secreting their own activating enzymes, as in the case of Pseudomonas aeruginosa, which activates pro‐MMP‐2 using LasB, a thermolysin‐like metalloprotease.39 The role of dysregulated tissue‐degrading enzymes, individually or in combination, is not well understood in every infectious disease. However, it is clear that the MMP/TIMP system changes the ECM composition and biophysics and its presentation of bioactive molecules, generating environmental cues that are detected and processed by immune cells into signalling events that direct their behaviour and response to infection.
ECM post‐translational modification
Post‐translational modifications (PTMs) add complexity to the ECM. Some are generated by proteolytic cleavage, while others are generated by citrullination of arginine, glycosylation, cross‐linking, hydroxylation of prolines, nitrosylation of tyrosines and aspartate isomerization (reviewed in ref. 7). Given the ability of pathogens to hijack host enzymes or secrete enzymes targeting host molecules, it is tempting to ask whether PTMs in the ECM are altered in infection. Indeed, the Porphyromonas gingivalis enzyme peptidylarginine deiminase can convert arginine residues to citrulline in mammalian ECM proteins, including fibrinogen, collagen II, fibronectin and tenascin‐C.72, 73, 74, 75, 76 Although arginine is positively charged at neutral pH, citrulline is uncharged, increasing protein hydrophobicity and, hence, altering protein three‐dimensional structure and function. Notably, Porphyromonas gingivalis periodontal infection has been linked to rheumatoid arthritis, an autoimmune disease of the joints characterized by high levels of citrullinated proteins and anti‐citrullinated ECM protein antibodies.77 Trypanosoma cruzi, the aetiological agent of Chagas disease, which features extensive inflammation and fibrosis of the heart, has been reported to increase the expression of lysyl oxidase (LOX).78 This enzyme carries out cross‐linking of collagen fibres thereby altering matrix stiffness, which has been linked to cancer metastasis.79 By irreversibly altering collagen structure and function, LOX has been proposed to cause dysfunction of cardiomyocytes and, in turn, of the heart in Chagas disease. Similarly, dengue virus infection has been shown to suppress the expression of cartilage‐associated protein (CRTAP), alongside the protein associated to tight junctions (PATJ). CRTAP associates with the proteoglycan leprecan, which has collagen prolyl 3‐hydroxylase activity, and cyclophilin B in the endoplasmic reticulum. This trimeric complex is required for proline 3‐hydroxylation of collagen and, hence, collagen assembly. As the levels of CRTAP mRNA negatively correlate with viral replication, the authors of the study have speculated that CRTAP, together with PATJ, restrict dengue infection by influencing cell–cell adhesion.80
While evidence supporting a link between specific PTMs of ECM proteins and diseases such as arthritis and cancer is accumulating, the implications of altered PTMs of ECM components during infection are only beginning to emerge. For example, collagen fibre cross‐linking mediated by LOX can significantly alter tissue structure and ECM mechanics. Mammoto et al. recently showed that LOX‐dependent changes in ECM mechanics control vascular permeability and pulmonary oedema. In vivo, mouse lungs treated with LPS, which contributes to pulmonary oedema and acute respiratory distress syndrome in patients with sepsis,81 become much stiffer than untreated lungs and exhibit enhanced vascular permeability. Increased LOX expression and LOX and LOX1 protein isoform activity control alveolar architecture and vascular permeability, which are restored by LOX activity inhibition.82
Macrophages: not just destroyers of the ECM
In bacterial and viral infection, activated macrophages [e.g. M(IFN‐γ), M(LPS)] have long been implicated in ECM destabilization and destruction through the secretion of tissue‐degrading enzymes, including MMP‐9. Similarly, parasitic infections involve activated macrophages [i.e. M(IL‐4)], which release proteases such as MMP‐1 and MMP‐12.83 However, macrophages can also produce a number of ECM proteins. Increasing evidence of this and its implications are discussed below.
Fibronectin is the first ECM glycoprotein reported to be produced by human macrophages and IFN‐γ‐stimulated mouse peritoneal macrophages.84, 85 Moreover, bacterial components induce tenascin‐C expression in human monocyte‐derived and mouse bone‐marrow‐derived macrophages.53, 86 Lipopolysaccharide from Porphyromonas gingivalis induces thrombospondin‐1 production in THP‐1 cells,87 while LPS from Escherichia coli and IFN‐γ up‐regulate galectin‐1 and galectin‐3, including five galectin‐3 truncated forms, in primary human macrophages.88 In mice with viral myocarditis, macrophages infiltrating the heart are the main producers of osteopontin.48 Notably, in murine macrophages, LPS causes the formation of chromosomal loops in the osteopontin promoter by bridging nuclear factor‐κB (NF‐κB) and activator protein‐1 together, leading to osteopontin transcription,89 which is negatively regulated by GSK3b.90
Macrophage expression of proteoglycans has also been demonstrated. Treatment of bone marrow‐derived and alveolar macrophages with Escherichia coli LPS results in rapid induction of versican and hyaluronan synthase 1, and simultaneous inhibition of the major hyaluronan‐degrading enzymes (hyaluronidases 1/2).91 Serglycin, decorin and biglycan are also secreted by LPS‐activated mouse peritoneal macrophages.92, 93, 94 By using inhibition, chromatin immunoprecipitation and NF‐κB reporter gene assays, some of these studies have demonstrated that pathogenic activation of the NF‐κB signalling pathway downstream of TLR4 leads to ECM molecule transcription.86, 89, 93, 94, 95 Importantly, in the case of tenascin‐C, biglycan and decorin, which can activate TLR4, this can promote autocrine loops of inflammation.86, 93, 94 Hence, the infected cellular microenvironment influences TLR function and, in turn, TLR activation affects the microenvironment.
Intriguingly, there is mounting evidence that macrophages can synthesize collagen molecules. In Drosophila, phagocytes (haemocytes) produce functional collagen IV, which controls key signalling events in the germline stem cell niche.96 The first reports of collagen synthesis by mammalian macrophages were published in the 1990s. The first showed collagen I synthesis in mouse peritoneal macrophages and the second demonstrated collagen VIII synthesis and secretion in human macrophages, which was decreased by IFN‐γ and LPS treatment.97, 98 Collagen VIII is a short‐chain, non‐fibrillar collagen that forms unique hexagonal lattice structures and possesses both structural and signalling properties. Later, human macrophages were shown to secrete collagen VI, which forms beaded filaments with a multidomain structure that interact with ECMs and cell surface receptors to anchor interstitial structures and cells within tissues. Expression of collagen VI was decreased by IFN‐γ and LPS, but increased upon IL‐4 treatment.99 Recently, the expression of all 28 collagen‐encoding mRNAs was quantified in steady‐state and LPS‐activated primary human macrophages and compared with that of human dermal fibroblasts, an abundant matrix source.95 Steady‐state macrophages expressed basal levels (lower than those in dermal fibroblasts) of collagen mRNAs with the exception of collagen III, X, XI, XVI, XX and XXVI. However, LPS specifically increased the expression of fibril‐associated collagens with interrupted triple helices (FACITs; collagen VII, XII, XV, XIX and XXI), the collagenase‐resistant collagen V, the collagen‐containing von Willebrand factor collagen XXVIII and collagen I, IV, XVIII, XXV and XXVII. Collagen II, VIII, IX, XIII, XIV, XXIII and XXIV expression was not increased. LPS also down‐regulated the expression of collagen VI, confirming previous studies,99 and collagen XIII and XVII.95 By secreting collagens, depending upon their mode of activation, macrophages may contribute to the ECM, and therefore to tissue stabilization and repair, and to cell–cell and cell–matrix interactions (e.g. in vitro, monocytes adhere strongly to collagen VI99). Furthermore, macrophages may bind to their secreted collagen molecules as they express several receptors known to interact with collagen (e.g. integrins and proteoglycans). However, collagen secretion by macrophages and its potential role in macrophage adhesion and the immune response in vivo remain to be clarified.
The ECM: an integral part of the innate immune response to infection?
Antimicrobial activity of the ECM
The innate immune system employs endogenous peptides like α‐defensin and LL‐37, which bind to heparin and dermatan sulphate glycosaminoglycans and have antimicrobial properties. During the inflammatory response to infection, tissue‐degrading enzymes generate bioactive ECM fragments. A number of studies showed that heparin‐binding peptides derived from laminin isoforms, vitronectin, thrombospondin and fibronectin, exert antimicrobial activities against Gram‐positive and Gram‐negative bacteria, and the fungus Candida albicans.100, 101, 102
Certain ECM proteins are also found in biological fluids. One such protein is tenascin‐C, which has been found in human breast milk (2·2–671 μg/ml) where it acts as an innate broad‐spectrum human immunodeficiency virus 1 (HIV‐1) ‐neutralizing protein.103 Tenascin‐C directly captures HIV‐1 virions by binding to the HIV‐1 envelope gp120 protein at a CD4‐inducible epitope that overlaps the chemokine co‐receptor binding site. Accordingly, tenascin‐C depletion abolishes the HIV‐1‐neutralizing activity of milk.103
Direct antimicrobial activity has also been reported for the ECM‐associated protein MMP‐12, which is abundantly expressed in mature tissue macrophages and mobilized to macrophage phagolysosomes after the ingestion of bacteria. Inside phagolysosomes, MMP‐12 adheres to bacterial cell walls and disrupts cellular membranes, killing the bacteria. Notably, the C‐terminal domain of MMP‐12, but not its catalytic domain, contains a four‐amino‐acid sequence on an exposed β loop of the protein that is unique in nature and confers antimicrobial activity.104 Together, these data may help the search for safe, endogenous antimicrobial molecules from complex biological mixtures. Furthermore, binding epitopes could serve as templates for de novo synthesis of novel antimicrobial molecules.
ECM‐mediated recognition of microbial pathogens, macrophage activation and phagocytosis
The innate immune system employs highly conserved receptors, namely PRRs, to recognize conserved motifs in microbial pathogens, called pathogen‐associated molecular patterns. Mindin, a member of the mindin‐F‐spondin family of secreted ECM proteins, has emerged as a unique pattern recognition molecule in the ECM for microbial pathogens and has been proposed to function as an integral part of the innate immune response.105 He et al., showed that genetic ablation of mindin confers resistance to LPS‐induced shock and systemic Salmonella typhimurium and Streptococcus pneumoniae infections in vivo. Moreover, mindin‐null mice feature impaired bacterial clearance in lungs infected with Gram‐positive group B streptococcus or Haemophilus influenzae. In vitro, macrophages and mast cells lacking mindin display impaired TNF‐α and IL‐6 production and defective phagocytosis. By using recombinant mindin, the authors showed that this ECM protein recognizes carbohydrate moieties of Gram‐positive and Gram‐negative bacterial components and, by binding to them, it agglutinates bacteria. As glucose inhibited not only mindin binding to pathogens but also macrophage activation by LPS, the authors concluded that mindin‐mediated carbohydrate recognition of microbial pathogens is a secondary stimulation necessary for the activation of macrophages and mast cells105 (Fig. 1). Notably, mindin is not a universal, but is a specific pattern recognition molecule as it only recognizes and opsonizes certain bacteria. Following this, the innate immune function of mindin has been extended to include promoting influenza virus clearance from the nasal cavity by allowing efficient macrophage activation.106 Furthermore, the proteoglycan lumican, whose core protein contains tandem repeats of leucine‐rich motifs similarly to PRRs, interacts with CD14 on the surface of macrophages and neutrophils, promoting CD14‐TLR4‐mediated responses to LPS (Fig. 1). Hence, lumican‐null mice are hyporesponsive to LPS‐induced septic shock.107 In a Pseudomonas aeruginosa model of lung infection, mortality of lumican‐deficient mice is increased as animals fail to clear bacteria from tissues. This study showed that CD14‐mediated phagocytosis of Escherichia coli and Pseudomonas aeruginosa bacteria by macrophages is impaired in the absence of lumican and identified Tyr‐20 as a vital residue for CD14 binding and phagocytosis.108 Finally, infection of the cornea with Pseudomonas aeruginosa readily increases lumican expression before inflammatory cell infiltration, and lumican‐null mice display poor resolution of bacterial keratitis and sustained production of pro‐inflammatory cytokines.109 Another ECM protein that senses a number of microbial pathogens is galectin‐3, which binds to carbohydrate structures on glycoproteins and glycolipids (e.g. N‐acetyl‐d‐lactosamine and LPS) from (myco)bacteria, protozoan parasites and yeast.110 Although the in vivo function of galectin‐3 during infection has not been fully investigated, upon microorganism recognition, it contributes to macrophage‐mediated phagocytosis, at least in vitro 111 (Fig. 1). Whether these ECM components exert the same essential immune functions in humans represents an outstanding, important question.
Figure 1.
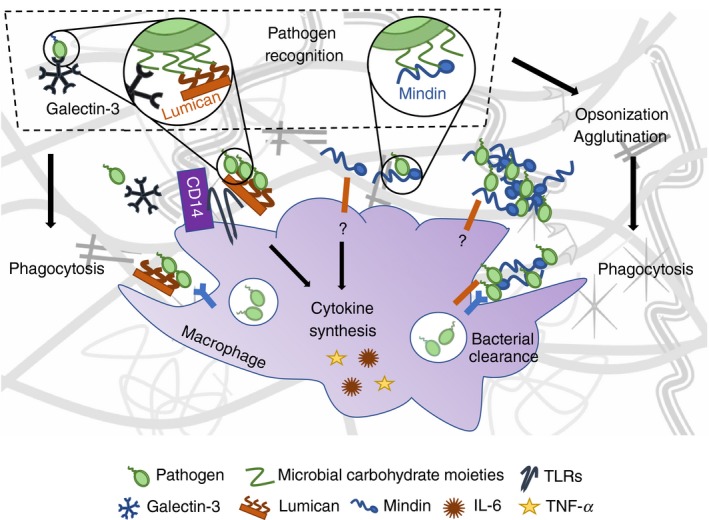
Mindin, lumican and galectin‐3: three extracellular sentinels. Mindin, lumican and galectin‐3 recognize and bind to sugar moieties found in the cell wall of several types of bacteria. All of them promote phagocytosis of bacteria by macrophages. Mindin binds to bacteria, causes their opsonization and agglutination, and facilitates their phagocytosis by macrophages. Mindin also induces the synthesis of pro‐inflammatory cytokines by these cells. Lumican instead interacts with CD14 on the surface of macrophages, promoting CD14‐TLR4‐mediated responses to lipopolysaccharide (LPS) and CD14‐mediated phagocytosis.
Leucocyte population balance in infection: emerging roles for ECM components
Severe infection demands large numbers of leucocytes that are compensated by the emergency myelopoiesis response, which is initiated by activated PRRs and cytokines, including IL‐6, granulocyte–macrophage colony‐stimulating factor and granulocyte colony‐stimulating factor. This protects the host from systemic infection by quickly generating the required leucocyte population. Kanayama et al.112 have recently found that osteopontin skewed the balance of myeloid and lymphoid cell populations during systemic infection with Candida albicans. Specifically, the authors showed that osteopontin limited the supply of neutrophils and Ly6C+ monocytes–macrophages by enhancing the apoptosis of common myeloid and granulocyte–macrophage progenitors through a down‐regulation of the expression of the apoptosis inhibitor survivin. This resulted in greater fungal load in kidneys and significantly higher mortality of wild‐type mice with systemic fungal infection compared with osteopontin‐deficient littermates. The detrimental effect of osteopontin was observed early, 24 hr after infection.112 However, it is unclear if this effect is maintained, exacerbated or reversed at later stages of infection.
Dendritic cells (DCs) are the main antigen‐presenting cells and, upon capturing microbial pathogens, mature and migrate to lymphoid tissues where they activate naive T cells. Distinct classes of microbes elicit lineage‐specific responses from the effector T‐cell repertoire. Type 1 helper T (Th1) cells are involved in infection by intracellular bacteria and viruses, Th2 cells in parasitic infection, and Th17 cells in infection by extracellular pathogens and facultative and obligate intracellular bacteria and fungi. Regulatory T cells hold the inflammatory response in check. Specific ECM proteins have been shown to contribute to the T‐cell polarizing function of DCs without affecting DC development. For instance, generation of Th17 cells by Escherichia coli LPS or Mycobacterium tuberculosis stimulated bone marrow‐derived DCs from tenascin‐C‐null mice is significantly impaired.113 Furthermore, the expression of galectin‐3 in DCs controls the magnitude of T‐cell priming in vitro and in vivo during helminthic infection with Schistosoma mansoni. Galectin‐3‐deficient mice have significantly fewer T cells in their spleen and higher cellular and humoral Th1 responses.114 Although this study shows that galectin‐3 expression by DCs modulates the proliferation and cytokine release by T cells, it does not explain the mechanism responsible for the biased Th1 response. Efficient T‐cell priming by DCs has also been shown to depend on mindin.115 When DCs from mindin‐null mice are activated with bacterial components, including LPS from Salmonella typhosa or Escherichia coli and lipoteichoic acid from Staphylococcus aureus, CD4+ T‐cell priming is 60–70% lower than that of wild‐type mice. Investigation of this demonstrated that DCs interact with mindin via integrins α 4 β 1 and α 5 β 1, leading to up‐regulated expression of the Rho GTPases Rac1/2, which are known to regulate DC priming of T cells.115 As DCs link innate to adaptive immunity and unbalanced effector T‐cell populations lead to pathological inflammation, understanding how the ECM can regulate T‐cell responses is crucial.
Transcriptional and post‐transcriptional roles for ECM molecules in host defence signalling pathways
Pathogen recognition via PRRs initiates inflammatory signalling pathways that are tightly regulated to allow microbial clearance with minimal damage to the host. Recent research implicates a role for ECM and ECM‐associated proteins in regulating inflammatory networks at the transcriptional and post‐transcriptional levels during the immune response to infection.
An elegant study by Marchant et al.69 found a transcriptional role for MMP‐12 (macrophage elastase) in immunity against viral infection. During coxsackievirus type B3 and respiratory syncytial virus infections, MMP‐12‐null mice display increased viral load, mortality and lower levels of IFN‐α, which is essential for viral immunity. Mechanistically, secreted MMP‐12 is taken up by virus‐infected cells and traffics to the nucleus, where it binds to the NFKBIA promoter, driving its transcription, which is essential for optimal IFN‐α secretion and host protection. Additionally, MMP‐12 regulates specific substrates by two distinct mechanisms: (i) through DNA binding of gene exons (e.g. exons encoding PSME3, the immunoproteasome cap protein, and SPARC‐like protein 1, which decreases their mRNA and protein levels in MMP‐12‐null mice); and (ii) extracellularly, through substrate protein cleavage (e.g. IFN‐α receptor 2 binding site). Hence, MMP‐12 clears systemic IFN‐α and, accordingly, selective inhibition of extracellular MMP‐12 in infected wild‐type mice elevates systemic IFN‐α levels and reduces viral replication.69
At the post‐transcriptional level, a role for decorin and tenascin‐C has been found in regulating microRNAs in LPS‐induced sepsis.53, 93, 116 Merline et al. detected increased decorin levels in patients with sepsis and mice with LPS‐induced sepsis, and elevated IL‐10 amounts in decorin‐null mice. They demonstrated that, in the presence of LPS, decorin reduces the levels of the anti‐inflammatory cytokine IL‐10 through two mechanisms. First, it activates extracellular signal‐regulated kinase and p38 mitogen‐activated protein kinases downstream of TLR2 and TLR4, thereby inducing the expression of the pro‐inflammatory modulator programmed cell death 4 (PDCD4), which translationally represses IL‐10. Second, decorin inhibits TGF‐β 1 signalling, leading to lower levels of the microRNA miR‐21, which represses PDCD4 expression and so decreases IL‐10 levels.93 Soon after, tenascin‐C was shown to orchestrate the secretion of specific cytokine subsets in response to LPS. Specifically, tenascin‐C regulates the biosynthesis of the LPS‐responsive miRNA miR‐155, allowing optimal TNF‐α production in macrophages and effective immune response to LPS in vivo.53 Hence, specific ECM and ECM‐associated proteins possess previously unknown transcriptional and post‐transcriptional regulatory activities, which fine‐tune the innate immune response to infection.
The ECM in infectious granulomas
Tuberculosis, syphilis, toxoplasmosis, infectious mononucleosis and measles are a few examples of infectious diseases characterized by the formation of granulomas. Traditionally considered host‐protective structures, infectious granulomas are compact, organized immune cell clusters that are generated in response to specific pathogens. Granulomas contain large numbers of mature macrophages, which can fuse into multinucleated giant cells or differentiate into foam cells. Neutrophils, dendritic cells, B, T and natural killer cells, and fibroblasts are also found in granulomas, which are surrounded by epithelial cells. Granulomas also contain ECM proteins, whose expression and function in infectious granulomatous disease pathogenesis has only recently been investigated.
In humans, osteopontin has been detected in granulomas of diverse aetiology;117 in those caused by Paracoccidioides brasiliensis, it localizes in ECM, macrophages and multinucleated giant cells at the centre of lesions in the early phase of infection.118 In tuberculosis granulomas, osteopontin is markedly expressed within lymphocytes, macrophages, epithelioid cells and multinucleated giant cells, but not in the central necrotic core.54 These granulomas also stain strongly for tenascin‐C in the surrounding fibrotic rings and weakly inside, in the ECM. A diffuse ECM‐associated staining of galectin‐9 is detected in the granuloma and in epithelioid and multinucleated giant cells. Notably, osteopontin, tenascin‐C and galectin‐9 are absent in non‐infectious Crohn disease granulomas.54 Analysis of human lung biopsies from patients with atypical mycobacteriosis and tuberculosis revealed expression of tenascin‐C and precursor proteins of collagens I and III around granulomas. Precursor proteins of collagen I were also found within granulomas that co‐localized with myofibroblasts.119 In mice infected with Paracoccidioides brasiliensis, Gonzalez et al. detected increased expression of laminin, fibronectin, fibrinogen, collagen I, collagen III, elastic fibres and proteoglycans during granuloma formation.120 Analysis of their arrangement showed that they were mostly surrounding the granuloma, but sporadically inside it. Initially arranged in a disorganized manner, ECM fibres later acquired a compact, concentric arrangement, which originated from an anchorage point that may contribute to tissue integrity and enhance distribution of growth factors and cytokines.120 Notably, during chronic mycobacterial infection, the fibrinolytic system has been shown to limit progressive fibrosis with plasminogen regulating the turnover of ECM proteins within the granuloma.121
Recent studies on ECM turnover in tuberculosis granuloma have helped to rewrite tuberculosis immunopathology. Traditionally considered a host‐protective structure that ‘quarantines’ the infecting mycobacteria, tuberculosis granuloma has been implicated in the expansion and dissemination of infection. Specifically, caseous necrosis leading to ECM destruction and bacterial dissemination was thought to be the cornerstone of tuberculosis pathogenesis. Conversely, collagen destruction has now been proposed to initiate caseous necrosis. Elkington et al.122 have first shown that MMP‐1‐expressing mice develop collagen destruction within granulomas upon infection with Mycobacterium tuberculosis in the absence of caseous necrosis. They then demonstrated that proteolytic collagen destruction of the lung ECM is the initial pathological event that reduces the survival of Mycobacterium tuberculosis‐infected cells, resulting in caseous necrosis and cavitation, and so diverting the immune response in favour of the pathogen. Conversely, intact collagen fibrils increase survival of infected cells.123, 124 In line with this, Parasa et al. reported up‐regulation of MMP‐1, MMP‐3, MMP‐9 and MMP‐12 in a human lung‐tissue model and in biopsies from patients with non‐cavitary tuberculosis. Global MMP inhibition via marimastat reduced granuloma formation and bacterial load.125
Future work should take into account the hypoxic conditions in infectious granulomas and include unbiased global analyses of ECM and ECM‐associated molecules in granulomas at various disease stages and gain‐ and loss‐of‐function experiments in three‐dimensional systems and/or genetically modified animals to understand the role of the ECM in granuloma formation and function.
Concluding remarks and future perspectives
The ECM and the innate immune response to infection are inextricably linked. Largely ignored or overlooked, the diverse yet specific functions of the ECM in infection influence the establishment and dissemination of microbial pathogens in host tissues as well as the outcome of the immune response to the infection (Fig. 2). In addition to the studies discussed here, ECM immunological research, which is expanding into the areas of prion disease,126 virus transmission,127 exosomes in viral pathogenesis128 and vaccine development,129 shows the breadth and complexity of ECM activity and regulation in infection.
Figure 2.
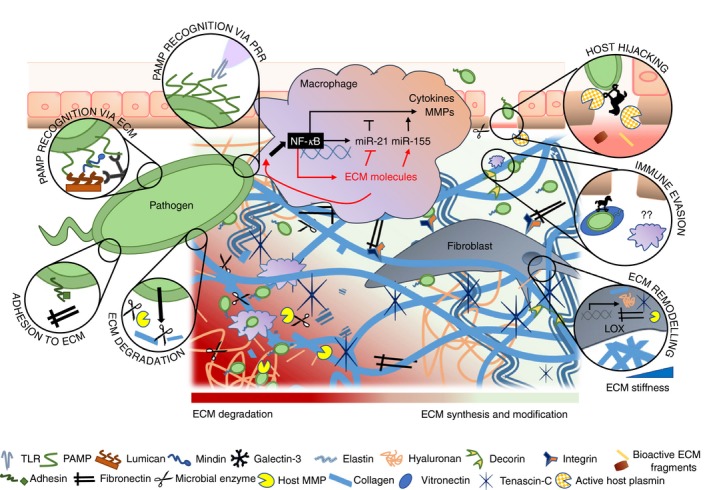
Multiple functions of the extracellular matrix (ECM) in the immune response to infection. Recognition of pathogen‐associated molecular patterns by macrophages through pattern recognition receptors (PRRs) and by ECM components is shown. Pathogen binding to ECM molecules such as fibronectin helps host colonization. Degradation of the ECM through microbial tissue‐degrading enzymes or host matrix metalloproteases (MMPs) activated by pathogen facilitates host invasion. To establish the infection, pathogens can also hijack host proteolytic systems such as the plasminogen–plasmin system and evade innate immune responses by binding to ECM components such as vitronectin. Fibroblasts in the interstitial ECM produce and secrete ECM proteins, MMPs and higher levels of lysyl oxidase (LOX), which cross‐links collagen fibres, increasing ECM stiffness. Pathogen‐mediated activation of macrophages triggers inflammatory signalling pathways such as the nuclear factor‐κB (NF‐κB) pathway, which culminates in the synthesis of cytokines, MMPs and microRNAs. Activated macrophages synthesize also ECM components such as decorin and tenascin‐C, which regulate the biosynthesis of miR‐21 and miR‐155 and generate positive feedback loops that propagate inflammation.
The ECM undergoes significant changes upon microbial invasion, magnifying the complexity of the cellular microenvironment at sites of infection. This presents challenges in setting up appropriate model systems and identifying the ECM and immunological pathways that are directly responsible for the outcome of individual infectious diseases. Unbiased omics, systems biology and genome editing approaches are promising resources for defining these pathways, designing mechanistic studies, and, in the longer‐term, elucidating them in the context of the microbiota.
As antimicrobial resistance is of global concern and tackling it is becoming increasingly challenging, investigating the now evident role of the ECM in infection may reveal novel therapeutic strategies or improve existing ones. It may also inform biomaterials and tissue engineering in their effort to prolong the lifespan and integrity of medical devices that are compromised by infection.
Disclosures
None to declare.
Acknowledgements
The authors are supported by an Anne McLaren Fellowship (University of Nottingham; awarded to AMP) and a BBSRC‐DTP studentship (HT). We thank E.P. Ling for assistance with the literature search and N. Zordan for help with the figures.
References
- 1. Pizarro‐Cerda J, Cossart P. Bacterial adhesion and entry into host cells. Cell 2006; 124:715–27. [DOI] [PubMed] [Google Scholar]
- 2. Kuusela P. Fibronectin binds to Staphylococcus aureus . Nature 1978; 276:718–20. [DOI] [PubMed] [Google Scholar]
- 3. Chagnot C, Listrat A, Astruc T, Desvaux M. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol 2012; 14:1687–96. [DOI] [PubMed] [Google Scholar]
- 4. Sorokin L. The impact of the extracellular matrix on inflammation. Nat Rev Immunol 2010; 10:712–23. [DOI] [PubMed] [Google Scholar]
- 5. Boyd DF, Thomas PG. Towards integrating extracellular matrix and immunological pathways. Cytokine 2017; 98:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hynes RO, Naba A. Overview of the matrisome – an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol 2012; 4:a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leeming DJ, Bay‐Jensen AC, Vassiliadis E, Larsen MR, Henriksen K, Karsdal MA. Post‐translational modifications of the extracellular matrix are key events in cancer progression: opportunities for biochemical marker development. Biomarkers 2011; 16:193–205. [DOI] [PubMed] [Google Scholar]
- 8. Boyd CD, Pierce RA, Schwarzbauer JE, Doege K, Sandell LJ. Alternate exon usage is a commonly used mechanism for increasing coding diversity within genes coding for extracellular matrix proteins. Matrix. 1993; 13:457–69. [DOI] [PubMed] [Google Scholar]
- 9. Kenne E, Soehnlein O, Genove G, Rotzius P, Eriksson EE, Lindbom L. Immune cell recruitment to inflammatory loci is impaired in mice deficient in basement membrane protein laminin α4. J Leukoc Biol 2010; 88:523–8. [DOI] [PubMed] [Google Scholar]
- 10. Wu C, Ivars F, Anderson P, Hallmann R, Vestweber D, Nilsson P, et al Endothelial basement membrane laminin α5 selectively inhibits T lymphocyte extravasation into the brain. Nat Med 2009; 15:519–27. [DOI] [PubMed] [Google Scholar]
- 11. Lokmic Z, Lammermann T, Sixt M, Cardell S, Hallmann R, Sorokin L. The extracellular matrix of the spleen as a potential organizer of immune cell compartments. Semin Immunol 2008; 20:4–13. [DOI] [PubMed] [Google Scholar]
- 12. Sixt M, Kanazawa N, Selg M, Samson T, Roos G, Reinhardt DP, et al The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity 2005; 22:19–29. [DOI] [PubMed] [Google Scholar]
- 13. Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 2011; 3:a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chou J, Chan MF, Werb Z. Metalloproteinases: a functional pathway for myeloid cells. Microbiol Spectr 2016; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leitinger B. Transmembrane collagen receptors. Annu Rev Cell Dev Biol 2011; 27:265–90. [DOI] [PubMed] [Google Scholar]
- 16. Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol 2011; 3:a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doyle JJ, Gerber EE, Dietz HC. Matrix‐dependent perturbation of TGFβ signaling and disease. FEBS Lett 2012; 586:2003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 2009; 1:a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Travis MA, Sheppard D. TGF‐β activation and function in immunity. Annu Rev Immunol 2014; 32:51–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol 2010; 10:826–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piccinini AM, Midwood KS. DAMPening inflammation by modulating TLR signalling. Mediators Inflamm 2010; 2010:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matzinger P. The danger model: a renewed sense of self. Science 2002; 296:301–5. [DOI] [PubMed] [Google Scholar]
- 23. Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell 2002; 111:927–30. [DOI] [PubMed] [Google Scholar]
- 24. Kline KA, Falker S, Dahlberg S, Normark S, Henriques‐Normark B. Bacterial adhesins in host–microbe interactions. Cell Host Microbe 2009; 5:580–92. [DOI] [PubMed] [Google Scholar]
- 25. Westerlund B, Korhonen TK. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol 1993; 9:687–94. [DOI] [PubMed] [Google Scholar]
- 26. Singh B, Fleury C, Jalalvand F, Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev 2012; 36:1122–80. [DOI] [PubMed] [Google Scholar]
- 27. Kuusela P, Vartio T, Vuento M, Myhre EB. Binding sites for streptococci and staphylococci in fibronectin. Infect Immun 1984; 45:433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kreikemeyer B, Klenk M, Podbielski A. The intracellular status of Streptococcus pyogenes: role of extracellular matrix‐binding proteins and their regulation. Int J Med Microbiol 2004; 294:177–88. [DOI] [PubMed] [Google Scholar]
- 29. Schwarz‐Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, et al Pathogenic bacteria attach to human fibronectin through a tandem β‐zipper. Nature 2003; 423:177–81. [DOI] [PubMed] [Google Scholar]
- 30. Joh D, Wann ER, Kreikemeyer B, Speziale P, Hook M. Role of fibronectin‐binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol 1999; 18:211–23. [DOI] [PubMed] [Google Scholar]
- 31. Terao Y, Kawabata S, Kunitomo E, Murakami J, Nakagawa I, Hamada S. Fba, a novel fibronectin‐binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol Microbiol 2001; 42:75–86. [DOI] [PubMed] [Google Scholar]
- 32. Hanski E, Caparon M. Protein‐F, a fibronectin‐binding protein, is an adhesin of the group a streptococcus Streptococcus pyogenes . Proc Natl Acad Sci U S A 1992; 89:6172–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kreikemeyer B, Oehmcke S, Nakata M, Hoffrogge R, Podbielski A. Streptococcus pyogenes fibronectin‐binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J Biol Chem 2004; 279:15850–9. [DOI] [PubMed] [Google Scholar]
- 34. Ljungh A, Moran AP, Wadstrom T. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol Med Microbiol 1996; 16:117–26. [DOI] [PubMed] [Google Scholar]
- 35. Niddam AF, Ebady R, Bansal A, Koehler A, Hinz B, Moriarty TJ. Plasma fibronectin stabilizes Borrelia burgdorferi–endothelial interactions under vascular shear stress by a catch‐bond mechanism. Proc Natl Acad Sci U S A 2017; 114:E3490–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steukers L, Glorieux S, Vandekerckhove AP, Favoreel HW, Nauwynck HJ. Diverse microbial interactions with the basement membrane barrier. Trends Microbiol 2012; 20:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beaufort N, Wojciechowski P, Sommerhoff CP, Szmyd G, Dubin G, Eick S, et al The human fibrinolytic system is a target for the staphylococcal metalloprotease aureolysin. Biochem J 2008; 410:157–65. [DOI] [PubMed] [Google Scholar]
- 38. Barthel D, Singh B, Riesbeck K, Zipfel PF. Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity. J Immunol 2012; 188:379–85. [DOI] [PubMed] [Google Scholar]
- 39. Beaufort N, Seweryn P, de Bentzmann S, Tang A, Kellermann J, Grebenchtchikov N, et al Activation of human pro‐urokinase by unrelated proteases secreted by Pseudomonas aeruginosa . Biochem J 2010; 428:473–82. [DOI] [PubMed] [Google Scholar]
- 40. Bergmann S, Hammerschmidt S. Fibrinolysis and host response in bacterial infections. Thromb Haemost 2007; 98:512–20. [PubMed] [Google Scholar]
- 41. Lahteenmaki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol 2005; 13:79–85. [DOI] [PubMed] [Google Scholar]
- 42. Ringner M, Valkonen KH, Wadstrom T. Binding of vitronectin and plasminogen to Helicobacter pylori . FEMS Immunol Med Microbiol 1994; 9:29–34. [DOI] [PubMed] [Google Scholar]
- 43. Kolar SL, Kyme P, Tseng CW, Soliman A, Kaplan A, Liang J, et al Group B Streptococcus evades host immunity by degrading hyaluronan. Cell Host Microbe 2015; 18:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuo C, Lim S, King NJ, Johnston SL, Burgess JK, Black JL, et al Rhinovirus infection induces extracellular matrix protein deposition in asthmatic and nonasthmatic airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2011; 300:L951–7. [DOI] [PubMed] [Google Scholar]
- 45. Yin H, Li X, Hu S, Liu T, Yuan B, Ni Q, et al IL‐33 promotes Staphylococcus aureus‐infected wound healing in mice. Int Immunopharmacol 2013; 17:432–8. [DOI] [PubMed] [Google Scholar]
- 46. Nde PN, Lima MF, Johnson CA, Pratap S, Villalta F. Regulation and use of the extracellular matrix by Trypanosoma cruzi during early infection. Front Immunol 2012; 3:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Assi K, Bergstrom K, Vallance B, Owen D, Salh B. Requirement of epithelial integrin‐linked kinase for facilitation of Citrobacter rodentium‐induced colitis. BMC Gastroenterol 2013; 13:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szalay G, Sauter M, Haberland M, Zuegel U, Steinmeyer A, Kandolf R, et al Osteopontin: a fibrosis‐related marker molecule in cardiac remodeling of enterovirus myocarditis in the susceptible host. Circ Res 2009; 104:851–9. [DOI] [PubMed] [Google Scholar]
- 49. Imanaka‐Yoshida K, Hiroe M, Yasutomi Y, Toyozaki T, Tsuchiya T, Noda N, et al Tenascin‐C is a useful marker for disease activity in myocarditis. J Pathol 2002; 197:388–94. [DOI] [PubMed] [Google Scholar]
- 50. Schenk S, Muser J, Vollmer G, Chiquet‐Ehrismann R. Tenascin‐C in serum: a questionable tumor marker. Int J Cancer 1995; 61:443–9. [DOI] [PubMed] [Google Scholar]
- 51. Kaarteenaho‐Wiik R, Lakari E, Soini Y, Pollanen R, Kinnula VL, Paakko P. Tenascin expression and distribution in pleural inflammatory and fibrotic diseases. J Histochem Cytochem 2000; 48:1257–68. [DOI] [PubMed] [Google Scholar]
- 52. Paallysaho T, Tervo K, Kivela T, Virtanen I, Tarkkanen A, Tervo T. Cellular fibronectin and tenascin in an orbital nylon prosthesis removed because of infection caused by Staphylococcus aureus . Graefes Arch Clin Exp Ophthalmol 1993; 231:61–5. [DOI] [PubMed] [Google Scholar]
- 53. Piccinini AM, Midwood KS. Endogenous control of immunity against infection: tenascin‐C regulates TLR4‐mediated inflammation via microRNA‐155. Cell Rep 2012; 2:914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hasibuan FM, Shiratori B, Senoputra MA, Chagan‐Yasutan H, Koesoemadinata RC, Apriani L, et al Evaluation of matricellular proteins in systemic and local immune response to Mycobacterium tuberculosis infection. Microbiol Immunol 2015; 59:623–32. [DOI] [PubMed] [Google Scholar]
- 55. Wight TN, Frevert CW, Debley JS, Reeves SR, Parks WC, Ziegler SF. Interplay of extracellular matrix and leukocytes in lung inflammation. Cell Immunol 2017; 312:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Evanko SP, Potter‐Perigo S, Bollyky PL, Nepom GT, Wight TN. Hyaluronan and versican in the control of human T‐lymphocyte adhesion and migration. Matrix Biol 2012; 31:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Overstreet MG, Gaylo A, Angermann BR, Hughson A, Hyun YM, Lambert K, et al Inflammation‐induced interstitial migration of effector CD4+ T cells is dependent on integrin αV. Nat Immunol 2013; 14:949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 2001; 17:463–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore!. Curr Opin Cell Biol 2001; 13:534–40. [DOI] [PubMed] [Google Scholar]
- 60. Van Lint P, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol 2007; 82:1375–81. [DOI] [PubMed] [Google Scholar]
- 61. Gaffney J, Solomonov I, Zehorai E, Sagi I. Multilevel regulation of matrix metalloproteinases in tissue homeostasis indicates their molecular specificity in vivo . Matrix Biol 2015; 44–46:191–9. [DOI] [PubMed] [Google Scholar]
- 62. Quiding‐Jarbrink M, Smith DA, Bancroft GJ. Production of matrix metalloproteinases in response to mycobacterial infection. Infect Immun 2001; 69:5661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakurai A, Okahashi N, Maruyama F, Ooshima T, Hamada S, Nakagawa I. Streptococcus pyogenes degrades extracellular matrix in chondrocytes via MMP‐13. Biochem Biophys Res Commun 2008; 373:450–4. [DOI] [PubMed] [Google Scholar]
- 64. Yin Y, Grabowska AM, Clarke PA, Whelband E, Robinson K, Argent RH, et al Helicobacter pylori potentiates epithelial:mesenchymal transition in gastric cancer: links to soluble HB‐EGF, gastrin and matrix metalloproteinase‐7. Gut 2010; 59:1037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kirchner A, Koedel U, Fingerle V, Paul R, Wilske B, Pfister HW. Upregulation of matrix metalloproteinase‐9 in the cerebrospinal fluid of patients with acute Lyme neuroborreliosis. J Neurol Neurosurg Psychiatry 2000; 68:368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leib SL, Leppert D, Clements J, Tauber MG. Matrix metalloproteinases contribute to brain damage in experimental pneumococcal meningitis. Infect Immun 2000; 68:615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Paemen L, Jansen PM, Proost P, Van Damme J, Opdenakker G, Hack E, et al Induction of gelatinase B and MCP‐2 in baboons during sublethal and lethal bacteraemia. Cytokine 1997; 9:412–5. [DOI] [PubMed] [Google Scholar]
- 68. Hu VH, Weiss HA, Ramadhani AM, Tolbert SB, Massae P, Mabey DC, et al Innate immune responses and modified extracellular matrix regulation characterize bacterial infection and cellular/connective tissue changes in scarring trachoma. Infect Immun 2012; 80:121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marchant DJ, Bellac CL, Moraes TJ, Wadsworth SJ, Dufour A, Butler GS, et al A new transcriptional role for matrix metalloproteinase‐12 in antiviral immunity. Nat Med 2014; 20:493–502. [DOI] [PubMed] [Google Scholar]
- 70. Cheung C, Marchant D, Walker EK, Luo Z, Zhang J, Yanagawa B, et al Ablation of matrix metalloproteinase‐9 increases severity of viral myocarditis in mice. Circulation 2008; 117:1574–82. [DOI] [PubMed] [Google Scholar]
- 71. Malik M, Bakshi CS, McCabe K, Catlett SV, Shah A, Singh R, et al Matrix metalloproteinase 9 activity enhances host susceptibility to pulmonary infection with type A and B strains of Francisella tularensis . J Immunol 2007; 178:1013–20. [DOI] [PubMed] [Google Scholar]
- 72. Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, et al Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α‐enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum 2010; 62:2662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van der Woude D, Rantapaa‐Dahlqvist S, Ioan‐Facsinay A, Onnekink C, Schwarte CM, Verpoort KN, et al Epitope spreading of the anti‐citrullinated protein antibody response occurs before disease onset and is associated with the disease course of early arthritis. Ann Rheum Dis 2010; 69:1554–61. [DOI] [PubMed] [Google Scholar]
- 74. Tutturen AE, Fleckenstein B, de Souza GA. Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res 2014; 13:2867–73. [DOI] [PubMed] [Google Scholar]
- 75. Chang X, Yamada R, Suzuki A, Kochi Y, Sawada T, Yamamoto K. Citrullination of fibronectin in rheumatoid arthritis synovial tissue. Rheumatology (Oxford) 2005; 44:1374–82. [DOI] [PubMed] [Google Scholar]
- 76. Anzilotti C, Pratesi F, Tommasi C, Migliorini P. Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun Rev 2010; 9:158–60. [DOI] [PubMed] [Google Scholar]
- 77. Wegner N, Lundberg K, Kinloch A, Fisher B, Malmstrom V, Feldmann M, et al Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol Rev 2010; 233:34–54. [DOI] [PubMed] [Google Scholar]
- 78. Soares MB, de Lima RS, Rocha LL, Vasconcelos JF, Rogatto SR, dos Santos RR, et al Gene expression changes associated with myocarditis and fibrosis in hearts of mice with chronic chagasic cardiomyopathy. J Infect Dis 2010; 202:416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, et al Lysyl oxidase is essential for hypoxia‐induced metastasis. Nature 2006; 440:1222–6. [DOI] [PubMed] [Google Scholar]
- 80. Afroz S, Giddaluru J, Abbas MM, Khan N. Transcriptome meta‐analysis reveals a dysregulation in extra cellular matrix and cell junction associated gene signatures during Dengue virus infection. Sci Rep 2016; 6:33752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mammoto T, Parikh SM, Mammoto A, Gallagher D, Chan B, Mostoslavsky G, et al Angiopoietin‐1 requires p190 RhoGAP to protect against vascular leakage in vivo . J Biol Chem 2007; 282:23910–8. [DOI] [PubMed] [Google Scholar]
- 82. Mammoto A, Mammoto T, Kanapathipillai M, Wing Yung C, Jiang E, Jiang A, et al Control of lung vascular permeability and endotoxin‐induced pulmonary oedema by changes in extracellular matrix mechanics. Nat Commun 2013; 4:1759. [DOI] [PubMed] [Google Scholar]
- 83. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Alitalo K, Hovi T, Vaheri A. Fibronectin is produced by human macrophages. J Exp Med 1980; 151:602–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cofano F, Comoglio PM, Landolfo S, Tarone G. Mouse immune interferon enhances fibronectin production of elicited macrophages. J Immunol 1984; 133:3102–6. [PubMed] [Google Scholar]
- 86. Goh FG, Piccinini AM, Krausgruber T, Udalova IA, Midwood KS. Transcriptional regulation of the endogenous danger signal tenascin‐C: a novel autocrine loop in inflammation. J Immunol 2010; 184:2655–62. [DOI] [PubMed] [Google Scholar]
- 87. Gokyu M, Kobayashi H, Nanbara H, Sudo T, Ikeda Y, Suda T, et al Thrombospondin‐1 production is enhanced by Porphyromonas gingivalis lipopolysaccharide in THP‐1 cells. PLoS One 2014; 9:e115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Novak R, Dabelic S, Dumic J. Galectin‐1 and galectin‐3 expression profiles in classically and alternatively activated human macrophages. Biochim Biophys Acta 2012; 1820:1383–90. [DOI] [PubMed] [Google Scholar]
- 89. Zhao W, Wang L, Zhang M, Wang P, Zhang L, Yuan C, et al NF‐κB‐ and AP‐1‐mediated DNA looping regulates osteopontin transcription in endotoxin‐stimulated murine macrophages. J Immunol 2011; 186:3173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Song H, Deng B, Zou C, Huai W, Zhao R, Zhao W. GSK3β negatively regulates LPS‐induced osteopontin expression via inhibiting its transcription. Scand J Immunol 2015; 81:186–91. [DOI] [PubMed] [Google Scholar]
- 91. Chang MY, Tanino Y, Vidova V, Kinsella MG, Chan CK, Johnson PY, et al Reprint of: A rapid increase in macrophage‐derived versican and hyaluronan in infectious lung disease. Matrix Biol 2014; 35:162–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zernichow L, Abrink M, Hallgren J, Grujic M, Pejler G, Kolset SO. Serglycin is the major secreted proteoglycan in macrophages and has a role in the regulation of macrophage tumor necrosis factor‐α secretion in response to lipopolysaccharide. J Biol Chem 2006; 281:26792–801. [DOI] [PubMed] [Google Scholar]
- 93. Merline R, Moreth K, Beckmann J, Nastase MV, Zeng‐Brouwers J, Tralhao JG, et al Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA‐21. Sci Signal 2011; 4:ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zeng‐Brouwers J, Beckmann J, Nastase MV, Iozzo RV, Schaefer L. De novo expression of circulating biglycan evokes an innate inflammatory tissue response via MyD88/TRIF pathways. Matrix Biol 2014; 35:132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Piccinini AM, Zuliani‐Alvarez L, Lim JM, Midwood KS. Distinct microenvironmental cues stimulate divergent TLR4‐mediated signaling pathways in macrophages. Sci Signal 2016; 9:ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Van De Bor V, Zimniak G, Papone L, Cerezo D, Malbouyres M, Juan T, et al Companion blood cells control ovarian stem cell niche microenvironment and homeostasis. Cell Rep 2015; 13:546–60. [DOI] [PubMed] [Google Scholar]
- 97. Vaage J, Lindblad WJ. Production of collagen type I by mouse peritoneal macrophages. J Leukoc Biol 1990; 48:274–80. [DOI] [PubMed] [Google Scholar]
- 98. Weitkamp B, Cullen P, Plenz G, Robenek H, Rauterberg J. Human macrophages synthesize type VIII collagen in vitro and in the atherosclerotic plaque. FASEB J 1999; 13:1445–57. [DOI] [PubMed] [Google Scholar]
- 99. Schnoor M, Cullen P, Lorkowski J, Stolle K, Robenek H, Troyer D, et al Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J Immunol 2008; 180:5707–19. [DOI] [PubMed] [Google Scholar]
- 100. Andersson E, Rydengard V, Sonesson A, Morgelin M, Bjorck L, Schmidtchen A. Antimicrobial activities of heparin‐binding peptides. Eur J Biochem 2004; 271:1219–26. [DOI] [PubMed] [Google Scholar]
- 101. Malmsten M, Davoudi M, Schmidtchen A. Bacterial killing by heparin‐binding peptides from PRELP and thrombospondin. Matrix Biol 2006; 25:294–300. [DOI] [PubMed] [Google Scholar]
- 102. Kobayashi N, Yoshida T. Binding sites on laminin receptors as components for antibiotics. Protein Pept Lett 2007; 14:33–6. [DOI] [PubMed] [Google Scholar]
- 103. Fouda GG, Jaeger FH, Amos JD, Ho C, Kunz EL, Anasti K, et al Tenascin‐C is an innate broad‐spectrum, HIV‐1‐neutralizing protein in breast milk. Proc Natl Acad Sci U S A 2013; 110:18220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Houghton AM, Hartzell WO, Robbins CS, Gomis‐Ruth FX, Shapiro SD. Macrophage elastase kills bacteria within murine macrophages. Nature 2009; 460:637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. He YW, Li H, Zhang J, Hsu CL, Lin E, Zhang N, et al The extracellular matrix protein mindin is a pattern‐recognition molecule for microbial pathogens. Nat Immunol 2004; 5:88–97. [DOI] [PubMed] [Google Scholar]
- 106. Jia W, Li H, He YW. Pattern recognition molecule mindin promotes intranasal clearance of influenza viruses. J Immunol 2008; 180:6255–61. [DOI] [PubMed] [Google Scholar]
- 107. Wu F, Vij N, Roberts L, Lopez‐Briones S, Joyce S, Chakravarti S. A novel role of the lumican core protein in bacterial lipopolysaccharide‐induced innate immune response. J Biol Chem 2007; 282:26409–17. [DOI] [PubMed] [Google Scholar]
- 108. Shao H, Lee S, Gae‐Scott S, Nakata C, Chen S, Hamad AR, et al Extracellular matrix lumican promotes bacterial phagocytosis, and Lum–/– mice show increased Pseudomonas aeruginosa lung infection severity. J Biol Chem 2012; 287:35860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shao H, Scott SG, Nakata C, Hamad AR, Chakravarti S. Extracellular matrix protein lumican promotes clearance and resolution of Pseudomonas aeruginosa keratitis in a mouse model. PLoS One 2013; 8:e54765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. van den Berg TK, Honing H, Franke N, van Remoortere A, Schiphorst WE, Liu FT, et al LacdiNAc‐glycans constitute a parasite pattern for galectin‐3‐mediated immune recognition. J Immunol 2004; 173:1902–7. [DOI] [PubMed] [Google Scholar]
- 111. Sano H, Hsu DK, Apgar JR, Yu L, Sharma BB, Kuwabara I, et al Critical role of galectin‐3 in phagocytosis by macrophages. J Clin Invest 2003; 112:389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kanayama M, Xu SJ, Danzaki K, Gibson JR, Inoue M, Gregory SG, et al Skewing of the population balance of lymphoid and myeloid cells by secreted and intracellular osteopontin. Nat Immunol 2017; 18:973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ruhmann M, Piccinini AM, Kong PL, Midwood KS. Endogenous activation of adaptive immunity: tenascin‐C drives interleukin‐17 synthesis in murine arthritic joint disease. Arthritis Rheum 2012; 64:2179–90. [DOI] [PubMed] [Google Scholar]
- 114. Breuilh L, Vanhoutte F, Fontaine J, van Stijn CM, Tillie‐Leblond I, Capron M, et al Galectin‐3 modulates immune and inflammatory responses during helminthic infection: impact of galectin‐3 deficiency on the functions of dendritic cells. Infect Immun 2007; 75:5148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li H, Oliver T, Jia W, He YW. Efficient dendritic cell priming of T lymphocytes depends on the extracellular matrix protein mindin. EMBO J 2006; 25:4097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Piccinini AM, Midwood KS. Illustrating the interplay between the extracellular matrix and microRNAs. Int J Exp Pathol 2014; 95:158–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Carlson I, Tognazzi K, Manseau EJ, Dvorak HF, Brown LF. Osteopontin is strongly expressed by histiocytes in granulomas of diverse etiology. Lab Invest 1997; 77:103–8. [PubMed] [Google Scholar]
- 118. Nishikaku AS, Scavone R, Molina RF, Albe BP, Cunha Cda S, Burger E. Osteopontin involvement in granuloma formation and in the severity of Paracoccidioides brasiliensis infection. Med Mycol 2009; 47:495–507. [DOI] [PubMed] [Google Scholar]
- 119. Kaarteenaho‐Wiik R, Sademies O, Paakko P, Risteli J, Soini Y. Extracellular matrix proteins and myofibroblasts in granulomas of sarcoidosis, atypical mycobacteriosis, and tuberculosis of the lung. Hum Pathol 2007; 38:147–53. [DOI] [PubMed] [Google Scholar]
- 120. Gonzalez A, Lenzi HL, Motta EM, Caputo L, Restrepo A, Cano LE. Expression and arrangement of extracellular matrix proteins in the lungs of mice infected with Paracoccidioides brasiliensis conidia. Int J Exp Pathol 2008; 89:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sato J, Schorey J, Ploplis VA, Haalboom E, Krahule L, Castellino FJ. The fibrinolytic system in dissemination and matrix protein deposition during a mycobacterium infection. Am J Pathol 2003; 163:517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Elkington P, Shiomi T, Breen R, Nuttall RK, Ugarte‐Gil CA, Walker NF, et al MMP‐1 drives immunopathology in human tuberculosis and transgenic mice. J Clin Invest 2011; 121:1827–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Al Shammari B, Shiomi T, Tezera L, Bielecka MK, Workman V, Sathyamoorthy T, et al The extracellular matrix regulates granuloma necrosis in tuberculosis. J Infect Dis 2015; 212:463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tezera LB, Bielecka MK, Chancellor A, Reichmann MT, Shammari BA, Brace P, et al Dissection of the host‐pathogen interaction in human tuberculosis using a bioengineered 3‐dimensional model. Elife. 2017; 6:e21283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Parasa VR, Muvva JR, Rose JF, Braian C, Brighenti S, Lerm M. Inhibition of tissue matrix metalloproteinases interferes with Mycobacterium tuberculosis‐induced granuloma formation and reduces bacterial load in a human lung tissue model. Front Microbiol 2017; 8:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Marbiah MM, Harvey A, West BT, Louzolo A, Banerjee P, Alden J, et al Identification of a gene regulatory network associated with prion replication. EMBO J 2014; 33:1527–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pais‐Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, et al Biofilm‐like extracellular viral assemblies mediate HTLV‐1 cell‐to‐cell transmission at virological synapses. Nat Med 2010; 16:83–9. [DOI] [PubMed] [Google Scholar]
- 128. Kulkarni R, Prasad A. Exosomes derived from HIV‐1 infected DCs mediate viral trans‐infection via fibronectin and galectin‐3. Sci Rep 2017; 7:14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sun X, Mei M, Zhang X, Han F, Jia B, Wei X, et al The extracellular matrix protein mindin as a novel adjuvant elicits stronger immune responses for rBAG1, rSRS4 and rSRS9 antigens of Toxoplasma gondii in BALB/c mice. BMC Infect Dis 2014; 14:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Farfan MJ, Cantero L, Vidal R, Botkin DJ, Torres AG. Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect Immun 2011; 79:3744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ensenberger MG, Tomasini‐Johansson BR, Sottile J, Ozeri V, Hanski E, Mosher DF. Specific interactions between F1 adhesin of Streptococcus pyogenes and N‐terminal modules of fibronectin. J Biol Chem 2001; 276:35606–13. [DOI] [PubMed] [Google Scholar]
- 132. Ozeri V, Tovi A, Burstein I, Natanson‐Yaron S, Caparon MG, Yamada KM, et al A two‐domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J 1996; 15:989–98. [PMC free article] [PubMed] [Google Scholar]
- 133. Cue D, Southern SO, Southern PJ, Prabhakar J, Lorelli W, Smallheer JM, et al A nonpeptide integrin antagonist can inhibit epithelial cell ingestion of Streptococcus pyogenes by blocking formation of integrin α 5 β 1‐fibronectin‐M1 protein complexes. Proc Natl Acad Sci U S A 2000; 97:2858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kreikemeyer B, Talay SR, Chhatwal GS. Characterization of a novel fibronectin‐binding surface protein in group A streptococci. Mol Microbiol 1995; 17:137–45. [DOI] [PubMed] [Google Scholar]
- 135. Salazar N, Castiblanco‐Valencia MM, da Silva LB, de Castro I, Monaris D, Masuda HP, et al Staphylococcus aureus manganese transport protein C (MntC) is an extracellular matrix‐ and plasminogen‐binding protein. PLoS One 2014; 9:e112730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 2005; 57:196–211. [DOI] [PubMed] [Google Scholar]
- 137. Kingsley RA, Keestra AM, de Zoete MR, Baumler AJ. The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: evidence for molecular mimicry of heparin binding. Mol Microbiol 2004; 52:345–55. [DOI] [PubMed] [Google Scholar]
- 138. Vollmer T, Hinse D, Kleesiek K, Dreier J. Interactions between endocarditis‐derived Streptococcus gallolyticus subsp. gallolyticus isolates and human endothelial cells. BMC Microbiol 2010; 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Brissette CA, Verma A, Bowman A, Cooley AE, Stevenson B. The Borrelia burgdorferi outer‐surface protein ErpX binds mammalian laminin. Microbiology 2009; 155:863–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Montealegre MC, Singh KV, Somarajan SR, Yadav P, Chang C, Spencer R, et al Role of the Emp Pilus subunits of Enterococcus faecium in biofilm formation, adherence to host extracellular matrix components, and experimental infection. Infect Immun 2016; 84:1491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hendrickx AP, van Luit‐Asbroek M, Schapendonk CM, van Wamel WJ, Braat JC, Wijnands LM, et al SgrA, a nidogen‐binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital‐acquired Enterococcus faecium . Infect Immun 2009; 77:5097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, Lorenz U, et al Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI‐H292 lung epithelial cells and extracellular matrix. Cell Microbiol 2007; 9:450–62. [DOI] [PubMed] [Google Scholar]
- 143. Muhlenkamp MC, Hallstrom T, Autenrieth IB, Bohn E, Linke D, Rinker J, et al Vitronectin binds to a specific stretch within the head region of Yersinia Adhesin A and thereby modulates Yersinia enterocolitica host interaction. J Innate Immun 2017; 9:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Imai DM, Feng S, Hodzic E, Barthold SW. Dynamics of connective‐tissue localization during chronic Borrelia burgdorferi infection. Lab Invest 2013; 93:900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Eidhin DN, Perkins S, Francois P, Vaudaux P, Hook M, Foster TJ. Clumping factor B (ClfB), a new surface‐located fibrinogen‐binding adhesin of Staphylococcus aureus . Mol Microbiol 1998; 30:245–57. [DOI] [PubMed] [Google Scholar]
- 146. McDevitt D, Nanavaty T, House‐Pompeo K, Bell E, Turner N, McIntire L, et al Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem 1997; 247:416–24. [DOI] [PubMed] [Google Scholar]
- 147. Heck LW, Morihara K, Abrahamson DR. Degradation of soluble laminin and depletion of tissue‐associated basement membrane laminin by Pseudomonas aeruginosa elastase and alkaline protease. Infect Immun 1986; 54:149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Janoir C, Pechine S, Grosdidier C, Collignon A. Cwp84, a surface‐associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J Bacteriol 2007; 189:7174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sofat N, Wait R, Robertson SD, Baines DL, Baker EH. Interaction between extracellular matrix molecules and microbial pathogens: evidence for the missing link in autoimmunity with rheumatoid arthritis as a disease model. Front Microbiol 2014; 5:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kim SK, Yang JY, Cha J. Cloning and sequence analysis of a novel metalloprotease gene from Vibrio parahaemolyticus 04. Gene 2002; 283:277–86. [DOI] [PubMed] [Google Scholar]
- 151. Shi L, Ermis R, Garcia A, Telgenhoff D, Aust D. Degradation of human collagen isoforms by Clostridium collagenase and the effects of degradation products on cell migration. Int Wound J 2010; 7:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Watanabe K. Collagenolytic proteases from bacteria. Appl Microbiol Biotechnol 2004; 63:520–6. [DOI] [PubMed] [Google Scholar]
- 153. Juarez ZE, Stinson MW. An extracellular protease of Streptococcus gordonii hydrolyzes type IV collagen and collagen analogues. Infect Immun 1999; 67:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ruggiero S, Cosgarea R, Potempa J, Potempa B, Eick S, Chiquet M. Cleavage of extracellular matrix in periodontitis: gingipains differentially affect cell adhesion activities of fibronectin and tenascin‐C. Biochim Biophys Acta 2013; 1832:517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]


