Abstract
In the interspecies lager yeast hybrid there are MAL loci involved in maltose and maltotriose utilization derived from each parent (Saccharomyces cerevisiae and Saccharomyces eubayanus). We show that trans-regulation across hybrid subgenomes occurs for MAL genes. However, gene expression is less efficient with non-native activators (trans-activation) compared to native activators (cis-activation). MAL genes were induced by maltose and repressed by glucose irrespective of host. Despite the strong expression of S. cerevisiae-type genes in the S. eubayanus host, a very low amount of transporter protein was actually observed in cells. This suggests that proper formation and configuration of the S. cerevisiae transporters is not efficient in S. eubayanus. The S. eubayanus-type Malx1 transporter was present in the plasma membrane in high amounts in all hosts (S. cerevisiae, S. eubayanus and Saccharomyces pastorianus) at all times. However, the S. cerevisiae-type transporters appeared sequentially in the plasma membrane; scMalx1 was localized in the plasma membrane during early to late linear growth and subsequently withdrawn to intracellular compartments. In contrast, the scAgt1 transporter was found in the plasma membrane mainly in the stationary phase of growth. Different localization patterns may explain why certain transporter orthologues in natural S. pastorianus strains were lost to mutation.
Keywords: Agt1, Malx1, α-glucoside transporters, brewer's yeast, Saccharomyces eubayanus, hybrid
Gene transcription regulation across subgenomes of hybrid yeast.
INTRODUCTION
Maltose and maltotriose comprise the majority of the fermentable sugar of wort, and utilization of these two sugars influences overall fermentation performance of brewer's yeast (Rautio and Londesborough 2003; Stambuk et al.2006). Genes for maltose and maltotriose transporters are located in MAL loci, each of which typically contains three genes, one coding for a transmembrane maltose/maltotriose transporter, a second for maltase and a third for the activator of the two other genes of the locus. The lager yeast Saccharomyces pastorianus is a natural hybrid of Saccharomyces cerevisiae and Saccharomyces eubayanus (Libkind et al.2011), and contains MAL loci from both parents. There is at least one complete MAL locus from the S. eubayanus parent found in lager yeast that contains all three genes (Nakao et al.2009; Baker et al.2015). Whereas from the S. cerevisiae parent, lager yeast has inherited several MAL loci which are located on different chromosomes, e.g. in the A15 lager yeast strain there are intact MAL1, MAL2, MAL3 and MAL4 loci (Vidgren, Ruohonen and Londesborough 2005; Magalhães et al.2016). It is not known if S. cerevisiae-type (sc-type) activators can activate S. eubayanus-type (se-type) MAL genes and vice versa. Maltose induction is controlled by the MAL-activator elements binding to MAL sites in the promoters of MAL genes, whereas glucose repression is mediated through Mig1 transcription factors which also bind to promoters of MAL genes (Levine, Tanouye and Michels 1992; Hu et al.1995). Mig1 and MAL-activators compete for binding sites at MAL loci, and it has been suggested that this competition determines the balance between repression and induction (Wang, Sirenko and Needleman 1997). It is suggested that the MAL-activators require intracellular maltose to obtain active conformation (Wang, Sirenko and Needleman 1997) but there exist also mutated MAL-activators that are active in the absence of maltose, e.g. the MAL activator in the CEN.PK2-1D laboratory strain (Rodicio and Zimmerman 1985).
Trans-activation influences the phenotype of hybrid organisms (Tirosh et al.2009). In the present study, cross-genome regulation was investigated in relation to MAL genes. Two orthologous gene pairs AGT1 and MALx1 were studied in the S. eubayanus, S. cerevisiae and lager yeast S. pastorianus backgrounds. In addition to measuring gene expression, transporter-encoding genes were tagged with either Gfp or mCherry fluorescent tags to assess the amount and localization of each transporter in the different hosts. In this way, the localization pattern of transporter proteins in foreign and native hosts could be compared. Also, the effects of foreign and native transporter expression on growth, sugar utilization and ethanol production of each host were studied. Objectives of the study were (1) to determine if MAL activators can operate across genome boundaries, (2) to study the amount and localization of Agt1 and Malx1 transporters in different host backgrounds and (3) to ascertain why certain orthologues gained loss-of-function mutations during lager yeast evolution. It is known from earlier studies that in the lager hybrid, one gene of each orthologous pair, AGT1 and MALx1, is non-functional. In regard to the AGT1 gene, the mutated version is scAGT1 (Vidgren, Ruohonen and Londesborough 2005) whereas for the MALx1 gene pair the mutated form is the seMALx1 (Nakao et al.2009; Baker et al.2015).
MATERIALS AND METHODS
Strains
The S. eubayanus type strain VTT-C12902, here called C902, was from the VTT Culture Collection, Finland; deposited as CBS12357 at CBS-KNAW Fungal Biodiversity Centre. Two industrial brewer's yeasts, the lager strain A-63015 and ale strain A-75060, here called A15 and A60, respectively, were also obtained from the VTT Culture Collection. The S. cerevisiae laboratory strain were CEN.PK2-1D (VW-1B) (MATα, leu2-3/112 ura3-52 trp1-289 his3Δ1 MAL2-8c SUC2) and FY834 (MATα, his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63 GAL2+)
Strain construction
Four different hosts were transformed with a plasmid bearing a single copy of either sc- or se-type AGT1 or MALx1 genes. Genes were under the control of their own promoters and tagged either with mCherry or Gfp fluorophores. Those genes of orthologous gene pairs that were functional in the lager hybrid were amplified by PCR from the A15 strain, while those genes that had gained loss-of-function mutations in lager yeast were amplified from the corresponding parent strain. Thus, functional scMALx1 and seAGT1 were from the lager yeast A15, whereas scAGT1 was from the S. cerevisiae parent (A60 ale yeast was used here as the S. cerevisiae reference) and seMALx1 from S. eubayanus. Table 1 describes plasmids constructed in the present study and shows the location of amplified transporter genes in the original hosts. It has been shown earlier for Malx1 and Agt1 transporters that transformants containing tagged and untagged forms of the same transporter exhibited similar patterns of maltose transport activity through different growth phases. This suggests that the specific catalytic activities of the transporters were little changed by tagging and that the tagged and non-tagged transporters were processed similarly during intracellular trafficking (unpublished data). In addition, Agt1 transporters tagged either with mCherry or Gfp tags were studied in different host backgrounds. The amount of fluorescent signal measured was proportional for the Gfp and mCherry tags (unpublished data).
Table 1.
Plasmids constructed for the present study.
| Plasmid short name | Plasmid composition | Origin of transporter | Location of transporter gene in original species |
|---|---|---|---|
| Pl_scMALx1-mCherry | YEplac195-scMALx1-mCherry-KanMX | S. pastorianus A15 | In sequenced lager strain WS34/70 in sc-type Chr II contig208.1a |
| Pl_seMALx1-Gfp | YEplac195-seMALx1-Gfp-KanMX | S. eubayanus type strain CBS12357T | In sequenced S. eubayanus type strain CBS12357T in Chr XVIc |
| Pl_scAGT1-mCherry | YEplac195-scAGT1-mCherry-KanMX | S. cerevisiae ale A60 | In sequenced ale GSY2239 in Contig00198b; in A60 ale in Chr VIId |
| Pl_seAGT1-Gfp | YEplac195-seAGT1-Gfp-KanMX | S. pastorianus A15 | In sequenced lager strain WS34/70 in se-type Chr VIIa |
| Pl_empty plasmid | YEplac195-KanMX |
Strong constitutive promoter constructs of MAL genes prepared earlier were used as a starting material for constructs in this study. These constructs contained PGK1prom-scMALx1-mCherry-PGK1term, PGK1prom-seAGT1-GFP-PGK1term, TDH3prom-seMALx1-GFP-TDH3term and TDH3prom-scAGT1-mCherry-TDH3term. The GFP used in the constructs was a variant of GFP called enhanced green fluorescence protein and was described in Pöggeler et al. (2003). Red fluorescent protein used was the mCherry variant described in Shaner et al. (2004). By using the earlier construct TDH3prom-seMALx1-GFP-TDH3term as a template, seMALx1-GFP-TDH3term fragment was obtained by PCR. The seMALx1 promoter fragment (840 bp) was synthesized by PCR using S. eubayanus as a template. Each fragment was provided with short overlapping ends to help homologous recombination. Primers used for PCR reactions are listed in Table 2. Fragments generated were linked to each other and to the YEplac195 multicopy plasmid by homologous recombination near the SphI site previously cut with this enzyme. Yeast strain FY834 was used as a host in all recombination cloning steps of the present study similarly as in Vidgren et al. (2011). The PGK1prom-scMALx1-mCherry-PGK1 construct was used as a template to obtain a scMALx1-mCherry-PGK1 fragment. The scMALx1 promoter fragment (899 bp) was synthesized by PCR using A15 as a template. Fragments generated were linked to each other and to the YEplac195 plasmid by homologous recombination near the HindIII site previously cut with this enzyme. The TDH3prom-scAGT1-mCherry-TDH3term construct was used as a template to obtain scAGT1-mCherry-TDH3term fragment. The scAGT1 promoter fragment (1899 bp) was synthesized by PCR using A60 as a template. Fragments generated were linked to each other and to the YEplac195 plasmid containing an ura marker by homologous recombination near the SalI site previously cut with this enzyme. Similarly, the seAGT1-GFP-PGK1term fragment was obtained by using the earlier PGK1prom-seAGT1-GFP-PGK1t construct as a template. The seAGT1 promoter fragment (1898 bp) was synthesized by PCR using A15 as a template. Fragments generated were linked to each other and to the YEplac195 plasmid by homologous recombination near the HindIII site previously cut with this enzyme. All fragments were synthesized by PCR using standard methods and all homologous recombination reactions were performed as described in Vidgren et al. (2011). After verification of the sequences of each construct, transformation of the CEN.PK2-1D strain was performed using the lithium acetate transformation protocol (Gietz et al.1992). Transformants were identified using uracil selection. For growth in rich medium under G418 selection, a KanMX cassette which contains the kanamycin marker gene and confers resistance to the antibiotic G418 (Wach et al.1994) was used. The cassette was introduced to the seAGT1-GFP-Yeplac195 and scAGT1-mCherry-YEplac195 constructs as a NarI-XbaI fragment, to the scMALx1-mCherry-YEplac195 construct as SacI-XbaI fragment and to the seMALx1-GFP-YEplac195 construct as SalI-EcoRI fragment. The constructs obtained were transformed to the C902, A15 and A60 hosts using the lithium acetate transformation protocol, and transformants were selected using G418 (200 mg l−1). Table 3 describes all strains used in the present study.
Table 2.
PCR primers used in the homologous recombination cloning.
| Namea | Primer sequence | Sequence detectedb |
|---|---|---|
| seAGT1prom_F | 5΄-GCTATGACCATGATTACGCCAGAATTTAAAGGCGCGCGT -3΄ | YEplac195 flanking sequence 5΄ of HindIII underlined, −1900 to −1878 of seAGT1 promoter |
| seAGT1promR | 5΄- AGCGAAAGTATATTTTTCATATTCTGATATTTCGTAACTCTTTCTGATTA -3΄ | seAGT1 gene 1–20 underlined, seAGT1 promoter from −1 to −30 |
| seAGT1_F | 5΄- TATCAGAATATGAAAAATATACTTTCGCTGGTAGGAAG -3΄ | SeAGT1 prom from −1 to −10 underlined, seAGT1 gene from 1 to 29 |
| PGKterm_R | 5΄-TAGAGTCGACCTGCAGGC -’3΄ | YEplac195 flanking sequence 3΄ of HindIII |
| scMALx1prom_F | 5΄- GCTATGACCATGATTACGCC ATCAGAAATAGTCATTTATGTATTTTAGTTACGCT -3΄ | YEplac195 flanking sequence 5΄ of HindIII underlined, −800 to −765 of seMALx1 promoter |
| scMALx1_prom_R | 5΄- ATGAGGATAATCCCTTCAT AGTTAATTAATAGTCTTGGATGTAATTCTTATTGT -3΄ | scMALx1 gene underlined, scMALx1prom from −1 to −36 |
| scMALx1-mCherry-PGKterm_F | 5΄- TTAATTAACT ATGAAGGGATTATCCTCAT TAATAAACAGA -3΄ | scMALx1 promoter from −1 to −10, scMALx1 gene |
| scAGT1prom_F | 5΄- GATCCTCTAGAGTCGAGTTCTAGAAGCCTCCGGCAAA -3΄ | YEplac195 flanking sequence underlined (SalI site), scAGT1prom |
| scAGT1_prom_R | 5΄- CAATGAAATGATATTTTTCATATTATAATATTTTTTTAGTT GTTTGATGTTCTTCTATGTA -3΄ | scAGT1 gene underlined from 1 to 21,scAGT1prom from −1 to −38 |
| scAGT1_F | 5΄- CAACTAAAAAAATATTATAATATGAAAAATATCATTTCATTGGTAAGCAAG -3΄ | scAGT1prom from −1 to −21, scAGT1 gene from 1 to 30 |
| TDH3_R | 5΄-AAAGCTTGCATGCCTGCAGGTCGACATGTAGATGATACTGACTGCACG-3΄ | Yeplac195 flanking sequence underlined (SalI), TDH3 term |
| seMALx1_prom_F | 5΄-CCTCTAGAGTCGACCTGCAGG TTCTGTACGCGATTTGGTTATGTG-3΄ | YEplac195 flanking sequence underlined (SphI site), seMALx1prom |
| seMALx1_prom_R | 5΄-CATTGAGGATAGACCCTTCATAGTTGCCTAATAGCTTTTGAAATTTTTT-3΄ | seMALx1 gene underlined from 1 to 21, seMALx1 promoter from −1 to −28 |
| seMALx1_F | 5΄-AAATTTCAAAAGCTATTAGGCAACTATGAAGGGTCTATCCTCAATGATAAAT-3΄ | seMALx1 prom underlined from −1 to −25, seMALx1 from 1 to 27 |
| seMALx1_R | 5΄-AAATTCTGCGTTCGTTAAAGCTTG CATGTAGATGATACTGACTGCACG-3΄ | YEplac195 flanking sequence underlined (SphI site), TDH3 terminator |
| TDH3prom_FRW(SalI) | 5΄-ACCCGGGGATCCTCTAGAGTCGA CACGCTTTTTCAGTTCGAGT-3΄ | Yeplac195 flanking SalI sequence underlined, TDH3promoter sequence |
| TDH3promREV_AGT1 | 5΄-AATGAAATGATATTTTTCATTTTGTTTGTTTATGTGTGTTTATTCGA-3΄ | scAGT1 sequence underlined from 1 to 20, TDH3 prom |
| ScAGT1mCherry_FRW_TDH3p | 5΄-TAAACAAACAAAATGAAAAATATCATTTCATTGGTAAG-3΄ | TDH3 prom underlined, scAGT1 from 1 to 26 |
| ScAGT1mCherry_REV_TDH3t | 5΄-AAGATTTAAAGTAAATTCACTACAGCTCGTCCATGCC -3΄ | TDH3term underlined, mCherry |
| TDH3term_FRW_mCherry | 5΄-ATGGACGAGCTGTAGTGAATTTACTTTAAATCTTGCATTTAAAT-3΄ | mCherry underlined, TDH3 term |
| TDH3term_REV_plasmidi (SalI site) | 5΄- AAAGCTTGCATGCCTGCAGG CATGTAGATGATACTGACTGCACG-3΄ | Plasmid 3΄ SalI site underlined, TDH3 term |
| TDH3p_FRW_SphI_site | 5΄-CCTCTAGAGTCGACCTGCAGGCACGCTTTTTCAGTTCGAGT -3΄ | Plasmid 5΄ SphI site underlined, TDH3 prom |
| TDH3p_REV_SeMALx1 | 5΄-TAGACCCTTCATTTTGTTTGTTTATGTGTGTTTATTCGA-3΄ | seMALx1, TDH3prom |
| SeMALx1_FRW_TDH3prom | 5΄-TAAACAAACAAA ATGAAGGGTCTATCCTCAATG-3΄ | TDH3prom underlined, seMALx1 |
| SbMALx1_REV_GFP | 5΄-CGCCGCACTAGTTCATCTAATCATGAGAGATGGG-3΄ | linker underlined, seMALx1 |
| GFP_FRW(linker_SbMALx1) | 5΄-TGAACTAGTGCGGCGATGGTGAGCAAGGGCGA-3΄ | linker_seMALx1, GFP |
| GFP_REV (TDH3term) | 5΄-AAGTAAATTCACTTACTTGTACAGCTCGTCCATGC-3΄ | TDH3term, GFP |
| TDH3Term-FRW(Gfp) | 5΄-CTGTACAAGTAA GTGAATTTACTTTAAATCTTGCATTTAAAT -3΄ | GFP underlined, TDH3 term |
| TDH3term-REV(plasm) | 5΄- TCTGCGTTCGTTAAAGCTT GCATGTAGATGATACTGACTGCACG-3΄ | Yeplac195 flanking sequence underlined (SphI), TDH3 term |
aF, forward; R, reverse.
bThe numbering is from the first nucleotide of the translational start.
Table 3.
Strains used in the present study.
| Host | Species | Transformed plasmid |
|---|---|---|
| CEN.PK2-1D | S. cerevisiae | Pl_scMALx1-mCherry |
| CEN.PK2-1D | Pl_seMALx1-Gfp | |
| CEN.PK2-1D | pl_scAGT1-mCherry | |
| CEN.PK2-1D | pl_seAGT1-Gfp | |
| CEN.PK2-1D | pl_empty plasmid | |
| A60 ale | S. cerevisiae | Pl_scMALx1-mCherry |
| A60 ale | Pl_seMALx1-Gfp | |
| A60 ale | pl_scAGT1-mCherry | |
| A60 ale | pl_empty plasmid | |
| C902 | S. eubayanus | Pl_scMALx1-mCherry |
| C902 | Pl_seMALx1-Gfp | |
| C902 | pl_scAGT1-mCherry | |
| C902 | pl_empty plasmid | |
| A15 lager | S. pastorianus | Pl_scMALx1-mCherry |
| A15 lager | Pl_seMALx1-Gfp | |
| A15 lager | pl_scAGT1-mCherry | |
| A15 lager | pl_empty plasmid |
Cultivations
Native, non-laboratory yeasts bearing the constructs were grown in YP (10 g yeast extract and 20 g peptone l−1) containing 20 g l−1 maltose or glucose supplemented with G418 (200 mg l−1) at 24°C with 200 rpm shaking. Strains of the laboratory yeast CEN.PK2-1D bearing the constructs were grown in synthetic complete medium (Sherman, Fink and Hicks 1983) lacking uracil (SC-Ura) and containing either 20 g l−1 glucose or maltose at 30°C with 200 rpm shaking. Host strains transformed with empty plasmids were used as controls. Samples were collected from cultivations for OD600 determination, HPLC, RNA extraction and confocal microscopic studies.
Total RNA extraction
Frozen cells were mechanically disrupted using a Precellys24 homogenizer (Bertin Technologies). Total RNA was extracted using the RNeasy mini kit (Qiagen). To eliminate genomic DNA, an additional DNase treatment was performed with RNase-free DNaseI (Thermo scientific) according to instructions. The extracted RNA was quantified using the ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies). As another preliminary quality control assay, the absence of contaminant genomic DNA in RNA preparations was verified using RNA as a template in real-time PCR assays (RNA not reverse-transcribed to cDNA).
Quantitative RT-qPCR
One microgram of total RNA from each sample was reverse transcribed to cDNA with the Transcriptor First Strand cDNA Synthesis kit (Roche) according to kit's instructions. A total of 5 μl of cDNA (1:100 diluted) was PCR amplified from each of the above reverse transcription reactions.
Primers were designed for specific binding to GFP, mCherry and the reference gene IPP1 (Table 4) with the PerlPrimer v 1.1.21 program. The PCR efficiency of each primer pair (E) was evaluated by the dilution series method using a mix of sample cDNAs as a template. The efficiencies (E) of the qPCR assays (ranging from 1.94 to 2.00) for each primer pair were calculated using the formula 10(−1/m), where m is the slope of the line of the threshold cycle (CT)-versus-log dilution plot of the DNA template (5 pg–50 ng input DNA) (Pfaffl 2001). Amplification efficiencies were for mCherry primer pair 94%, for Gfp 98% and for IPP1 pair 96%. The PCRs were performed using a LightCycler® 480 SYBR Green I Master mix (Roche Diagnostics) on a LightCycler® 480 II instrument (Roche Diagnostics, Switzerland) in two independent replicates. The following program was used: pre-incubation (95°C for 5 min), amplification cycle repeated 45 times (95°C for 10 s, 60°C for 10 s, 72°C for 10 s with a single fluorescence measurement), melting curve program (65–97°C with continuous fluorescence measurement), and finally a cooling step to 40°C. Data analysis was performed using the supplied LightCycler® 480 Software, version 1.5 (Roche Diagnostics) by the 2−ΔΔCT method.
Table 4.
Primers for RT-qPCR.
| Name | Primer sequence | Sequence detected |
|---|---|---|
| mCherry_F | 5΄-GCGTGATGAACTTCGAGGAC-3΄ | 299–318 of mCherry gene |
| mCherry_R | 5΄-TTCAGCCTCTGCTTGATCTC-3΄ | 512–493 of mCherry gene |
| GFP_F | 5΄-CACCATCTTCTTCAAGGACGA-3΄ | 291–311 of GFP gene |
| GFP_R | 5΄-GGCTGTTGTAGTTGTACTCC-3΄ | 445–426 of GFP gene |
| IPP1_F | 5΄-TTGCTTACACTGGTCAAGTC-3΄ | 385–405 of IPP1 gene |
| IPP1_R | 5΄-AACCATTCGTTAGTAGCCCTC-3΄ | 549–579 of IPP1 gene |
Confocal microscopic studies
Cells containing GFP- or mCherry-tagged transporters were examined on Ibidi Ibitreat μ-Slides (Integrated Biodiagnostics). Confocal images were taken using a Zeiss Axiovert 200M with a spinning disc confocal unit Yokogawa CSU22 and Zeiss Plan-Neofluar 100xOil/1.4 NA objective. Image analysis was performed with SlideBook 4.2.0.7 software and cell-associated fluorescence was quantified using the National Institutes of Health (NIH) ImageJ program (http://rsbweb.nih.gov/ij/). A possible disadvantage of this method is that kinetics of the degradation of reporter proteins might be slower compared to transporter proteins. There is the possibility that fluorescent signal might be observed in cells when the transporters have already been degraded. The average fluorescence was calculated as total cell-associated signal intensity in 150–200 cells. Samples were collected for confocal microscopic analysis at the following time points, early linear phase (OD600 2-4), late linear phase (OD600 15-20) and stationary phase (after 40 h cultivation; OD600 >20) for native yeasts grown in rich media. For laboratory yeast grown in minimal media, samples were taken at the early linear phase (OD600 2-4), late linear phase (OD600 7-10) and stationary phase (after 40 h cultivation; OD600 >10).
HPLC
The culture supernatants were analyzed by HPLC for maltose, glucose and ethanol using a Waters 2690 Separation Module and Waters System Interphase Module liquid chromatography coupled with a Waters 2414 differential refractometer and a Waters 2487 dual λ absorbance detector (Waters, Milford, MA). A Fast Juice Column (50 × 7.8 mm, Phenomenex, Torrance, CA) and a Fast Acid Analysis Column (100 × 7.8 mm, Bio-Rad, Hercules, CA) and an Aminex HPX-87H Organic Acid Analysis Column (300 × 7.8 mm, Bio-Rad) were equilibrated with 2.5 mM H2SO4 in water at 60°C and samples were eluted with 2.5 mM H2SO4 in water at a 0.5 ml/min flow rate. Data were acquired with Waters Millennium software.
RESULTS
Sequence comparisons
The gene encoding the se-type MAL activator is located in Chr XVI in S. eubayanus type strain CBS12357T genome (Libkind et al.2011). This is the same locus from which the seMALx1 transporter used in the present study as a transporter was amplified. There are no other MAL activator genes found in the genome of the sequenced S. eubayanus type strain. The sc-type MAL activator used in the sequence comparison is encoded by a gene located in contig208.1 of the sequenced WS34/70 lager yeast (Nakao et al.2009) in the same locus from which scMALx1 used in the present study as a transporter was obtained. In the lager WS34/70 genome there are three other sc-type MAL activators present and they have 95%, 97% and 98% sequence identities to the contig 208.1 activator.
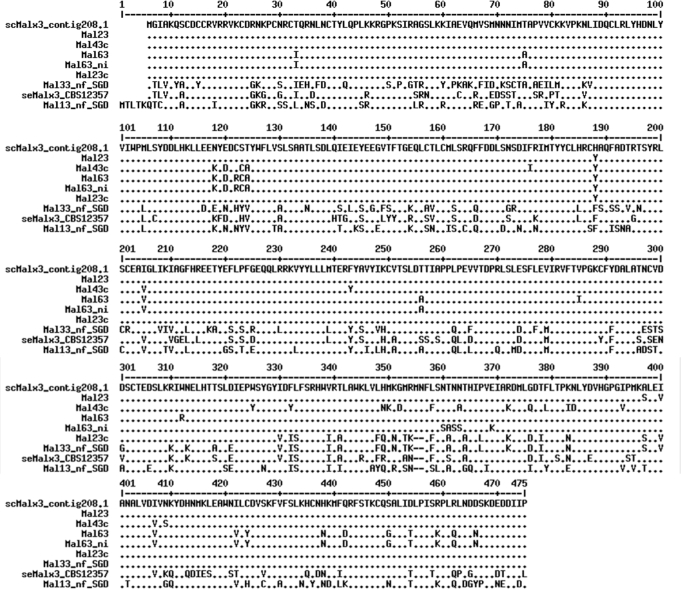
A comparison of the S. eubayanus and S. cerevisiae MAL activators is shown in Fig. 1. Sequence identity of se-type activator to different sc-type activators varied from 69% to 74% at the protein level (Fig. 1). Regulatory domains of the MAL activator have been studied. It has been reported that the sequence-specific DNA binding domain of the MAL activator is contained in residues 1-100, residues 60-283 constitute a functional core region, including the transactivation domain, residues 251-299 are required to inhibit the activation function of MAL activator and the rest of the C-terminal region of the protein contains maltose responsive domains that act to relieve the inhibitory effect on the activation domain (Levine, Tanouye and Michels 1992; Hu et al.1999). Our comparison of se and sc-type MAL activators showed that changes are distributed evenly along the protein sequence and they are not concentrated in specific functional domains.
Figure 1.
Comparison of the deduced amino acid sequences of se and sc-type MAL activators. Se-type MAL activator is encoded by MALx3 located in Chr XVI in S. eubayanus type strain CBS12357T (Libkind et al.2011). Sc-type MAL activator encoded by scMALx1 is present in the same locus as transporter gene used at the present study and is located in Chr II in WS34/70 (Contig 208.1) (Nakao et al.2009). In addition, Mal13 non-functional activator (accession number 853205), Mal23 activator (accession number AF002704), Mal23 constitutive activator (accession number AF002703) and Mal33 non-functional activator (accession number 852600). Mal43c constitutive activator (accession number M81157), Mal63 activator (accession number M6537) and Mal63 non-inducible activator (accession number AF003003) are included in the comparison. Multiple sequence alignment with hierarchical clustering (Corpet 1988).
A phylogeny of the MAL activators shows three distinct subgroups of regulators (Brown, Murray and Verstrepen 2010). These are MAL63-like subgroup responding to several sugars (including maltose, maltotriose and palatinose), MAL13-like subgroup which is responsible only for palatinose; non-functional Mal13 and Mal33 activators of S288c belong to this group, even if they are ‘non-functional’ for maltose they respond to palatinose (Brown, Murray and Verstrepen 2010). A third subgroup contains putative maltose-responsive transcription factors whose sequences are not close to activators of the present study and for this reason were excluded from the sequence comparison shown in Fig. 1. Sequence analysis shows that se-type MAL activator and MAL13 subgroup activators (Mal13 and Mal33) share several of the amino acid changes in relation to MAL63-like subgroup. Thus, the seMALx1 activator is closer to Mal13-like subgroup than MAL63-like subgroup and due to this might be more responsive to palatinose than maltose.
Pougach et al. (2014) have studied amino acid changes in MAL activators responsible for the maltose to palatinose change, i.e. altered DNA-binding ability from CGC(9N)CGN to CGG(9N)CGG. They have identified Arg12Cys and Val13Ile amino acid changes to be responsible for binding ability change. However, there must be also other not yet identified changes as only Val13Ile change can be found in MAL13 subgroup promoters of the present study responding only to palatinose (in Mal13 activator). MAL63-type activators are promiscuous and can bind both CGC and CGG motifs (respond to both maltose and palatinose). The se-type MAL activator does not possess Arg12Cys and Val13Ile changes and, thus, there is so far no evidence that the se-type MAL activator is not a promiscuous activator able to bind both CGC and CGG motifs.
Relevant promoters seMALx1prom, scMALx1prom, seAGT1prom and scAGT1prom, were analyzed for elements responsible for MAL-activator and Mig1 repressor binding. Figure S1 (Supporting Information) shows sequence comparison of MALx1 promoters and the divergent MALx1-MALx2 promoter studied earlier (Levine, Tanouye and Michels 1992; Bell et al.1997), where two Mig1 binding sites and three MAL activator binding sites have been found. Table 5 shows sequence identities in promoter elements between sc and se-type promoters, i.e. MAL activator binding sites were 56%–84% identical whereas Mig1 repressor binding sites were only 56%–58% identical. AGT1 promoters differentiated even more between sc and se-type (Table 6; Fig. S2, Supporting Information). MAL activator and Mig1 binding sites were only 33%–53% and 33%–58% identical, respectively. Sequence analysis indicated that trans-regulation of transporters would be unlikely, especially with the AGT1 gene. However, based on current knowledge three-nucleotide CG-rich motifs separated by a spacer of fixed length are sufficient for a MAL activator to bind Pougach et al. (2014). Binding sites described in Pougach et al. were found in promoters in the present study. Figure S1 (Supporting Information) includes CGC(9N)CGN and CGG(9N)CGG binding sites in MALx1 promoters and Fig. S2 (Supporting Information) those of AGT1 promoters in addition to other described MAL activator binding sites described in Levine, Tanouye and Michels (1992), Bell et al. (1997) and Vidgren et al. (2011). There were two CGC(9N)CGN and CGG(9N)CGG binding sites in each of the sc and se type MALx1 promoters. There were also two such sites in the seAGT1 promoter and one site in A15 and A60 AGT1 promoters.
Table 5.
MALx1 promoter element sequence identities between sc and se-type promoters. Location of elements in aligned sequences (Fig. S1, Supporting Information) is shown in parenthesis.
| Element | se-type promoter element identity to sc-type (identical nucleotides/total nucleotides in element) | % Identity |
|---|---|---|
| Mig1 (283–300) | 11/19 | 58 |
| MAL activator (465–496) | 27/32 | 84 |
| MAL activator (542–559) | 14/19 | 74 |
| MAL activator (574–598) | 14/25 | 56 |
| Mig1 (599–614) | 9/16 | 56 |
Table 6.
AGT1 promoter element sequence identities between sc and se-type promoters. Location of element in aligned sequences (Fig. S2, Supporting Information) is shown in parenthesis.
| Element | se-type promoter element identity to sc-type (identical nucleotides/total nucleotides in element) | % Identity |
|---|---|---|
| MAL activator (812–827) | 5/15 | 33 |
| MAL activator (907–921) | 4/15 | 27 |
| MAL activator (943–957) | 5/14 | 36 |
| Mig1 (1088–1099) | 4/12 | 33 |
| Mig1 (1098–1110) | 7/12 | 58 |
| Mig1 (1109–1120) | 7/12 | 58 |
| Mig1 (1119–1130) | 7/12 | 58 |
| Mig1 (1128–1140) | 6/12 | 50 |
| Mig1 (1184–1195) | 5/12 | 42 |
| Mig1 (1195–1205) | 5/12 | 42 |
| MAL activator (1703–1717) | 8/15 | 53 |
| MAL activator (1882–1895) | 6/14 | 43 |
Gene transcription in the laboratory yeast CEN.PK2-1D
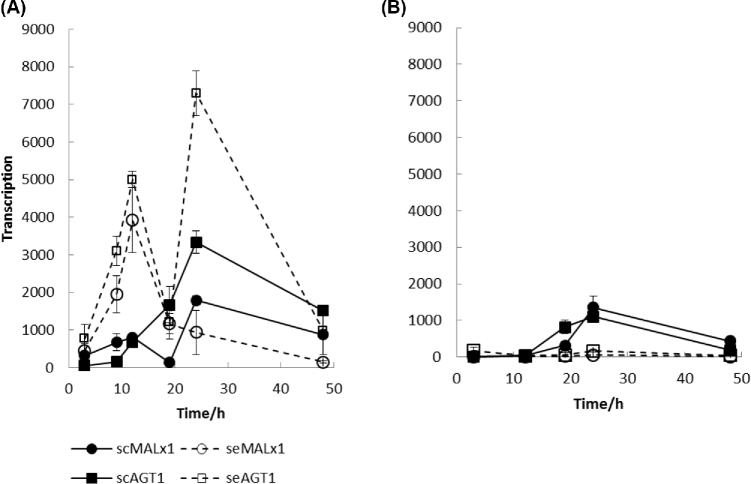
Initially, transporter constructs were studied in the laboratory yeast CEN.PK2-1D containing a strong sc-type MAL activator in a MAL2-8c locus (Rodicio and Zimmermann 1985). Under inducing conditions (on maltose), the MAL activator present in CEN.PK2-1D was able to induce both sc- and se-type genes (both MALx1 and AGT1). However, the se-type genes were transcribed at levels up to 10-fold higher than those of the sc-type genes (Fig. 2). There were two peaks in expression of the scMAL genes in the CEN.PK2-1D host. The first peak was at early linear growth phase, when transcription of both se-type genes peaked, while transcription of sc-type genes remained at a low level. A second peak took place at late linear growth phase when both sc-type genes were induced as well as seAGT1, but not seMALx1. Under repressive conditions (on glucose) repression was very strong with se-type gene transcription being very weak, even after glucose exhaustion. In contrast, sc-type genes were transcribed relatively strongly after glucose exhaustion.
Figure 2.
Gene transcription in the CEN.PK2-1D host transformed with sc and se-type transporters and grown on (A) maltose and (B) glucose. Results are averages ± ranges of data from two replicate growths of each yeast strain on each sugar.
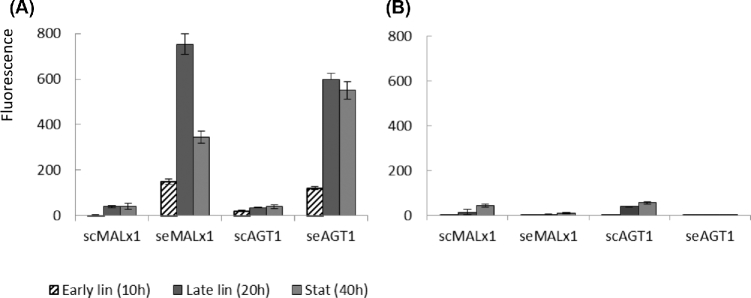
Next, the amount of fluorescence from each transporter was determined in cells. This was done by confocal microscopic analysis and signal quantification of mCherry or Gfp tagged transporters. The amount of tagged transporter (fluorescence) was determined for whole cells and thus included transporters present on the plasma membrane as well as in the intracellular compartments (Fig. 3). On maltose, the amount of fluorescence for both se-type transporters in cells was relatively high. For example, at stationary growth phase the se-type transporter fluorescence was 9–15 times higher than the corresponding fluorescence of sc-type transporters. Under repressive conditions (on glucose) there was a similar amount of fluorescence for sc-type transporter proteins as when grown under inducing conditions (on maltose), correlating to the observed transcription levels in both conditions; se-type transporter fluorescence was present at a very low basal level under repressive conditions, again correlating with transcription.
Figure 3.
Amount of fluorescent signal in the CEN.PK2-1D host transformed with sc and se-type transporters and grown on (A) maltose and (B) glucose. Samples were collected at early logarithmic growth phase at 10 h, at late logarithmic growth phase at 20 h and at stationary growth phase at 40 h. Results are averages ± ranges of data from two replicate growths of each yeast strain on each sugar.
The division of transporters between plasma membrane and intracellular compartments was also studied by confocal microscopy. Figure 3 shows average total fluorescence in cells but it did not differentiate between fluorescence in the plasma membrane and intracellular compartments. In Table 7 fluorescence detected either in plasma membrane or in intracellular compartments, determined by visual inspection of fluorescent images, is shown. Different localization patterns were observed depending on transporter type. Under inducing conditions, seMalx1 transporter fluorescence was found consistently in the plasma membrane as well as in intracellular compartments during each growth phase. The sc-type version, scMalx1, was not present, or was below detection limit on the plasma membrane, but found only in intracellular compartments. For Agt1 transporters, a similar localization pattern was found for both the sc and se-type, with transporters in the plasma membrane from late linear until stationary phase. Under repressive conditions there were faint signals for scAgt1 and scMalx1 transporters in the plasma membrane and in intracellular compartments. The scMalx1 transporter signals were found in the plasma membrane only during late linear growth phase, while the scAgt1 transporter signals were found in the plasma membrane only in the stationary phase.
Table 7.
Localization of tagged transporters in the CEN.PK2-1D host when grown on maltose or glucose in minimal media. Localization determined at early linear phase (OD600 2-4), late linear phase (OD600 7-10) and stationary phase (after 40 h cultivation; OD600 >10). Determined from fluorescent images by visual inspection.
| On maltose | On plasma membrane | In intracellular compartments | ||||
|---|---|---|---|---|---|---|
| Early lin | Late lin | Stat | Early lin | Late lin | Stat | |
| scMalx1 | − | − | − | + | + | + |
| seMalx1 | ++ | ++ | ++ | ++ | ++ | ++ |
| scAgt1 | − | + | + | + | + | + |
| seAgt1 | − | ++ | ++ | ++ | ++ | ++ |
| Early lin | Late lin | Stat | Early lin | Late lin | Stat | |
| scMalx1 | − | + | − | − | + | − |
| seMalx1 | − | − | − | − | − | − |
| scAgt1 | − | − | + | − | + | + |
| seAgt1 | − | - | - | − | - | - |
− Not visible.
+ Fluorescent signals.
++ Strong fluorescent signals.
On maltose, contrary to what was expected, for strains possessing transporter constructs, utilization of maltose was not improved, but was rather slower (Fig. S3, Supporting Information). In particular, the scMALx1 construct caused a significant delay in growth, maltose utilization and ethanol production. On glucose, strains possessing a transporter construct had in many cases slightly slower glucose utilization capacity.
Gene transcription in non-laboratory yeasts
Constructs were studied in native, non-laboratory yeast hosts, S. cerevisiae A60, S. eubayanus C902 and S. pastorianus A15 (hybrid of the first two species). In this manner, transregulation across genomes could be studied. Yeasts were transformed with the same sc and se transporter constructs that were used earlier to transform CEN.PK2-1D. However, the seAGT1 construct could not be successfully transformed to any of these three yeasts. The reason for this is not known but possibly overexpression of seAGT1 from the multicopy plasmid caused non-bearable effects for these yeasts, as no viable transformants were found. Results were therefore obtained only for scAGT1, and the se- and sc-type MALx1 constructs.
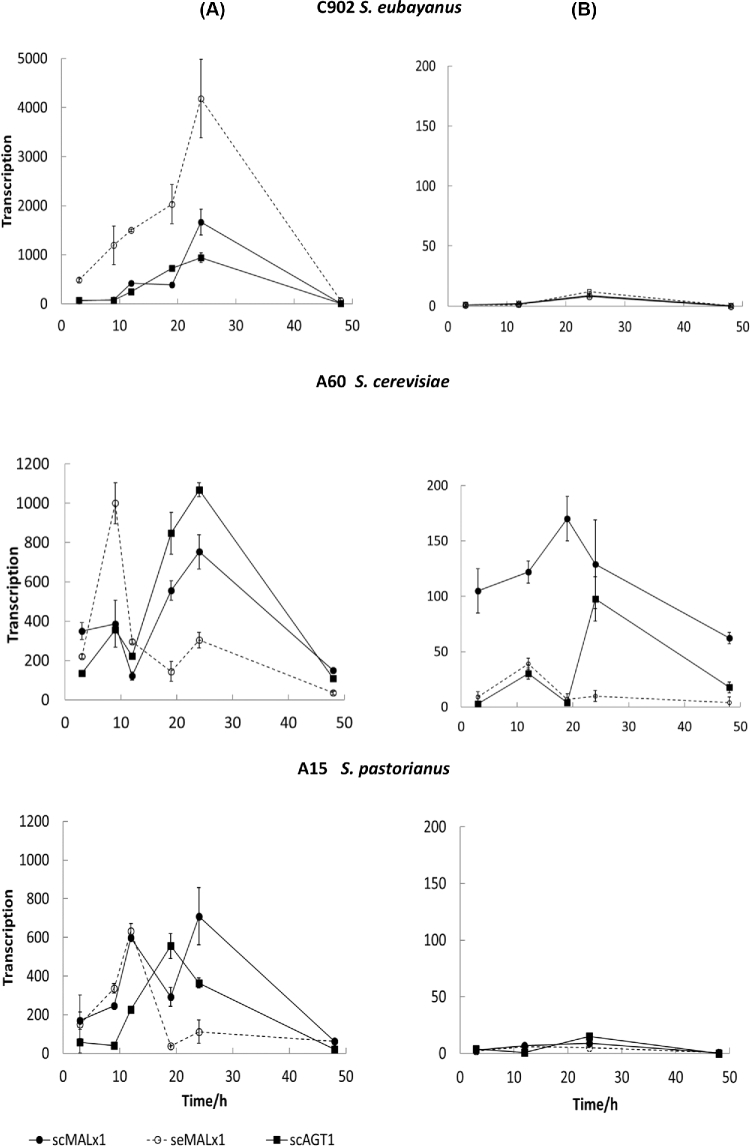
All maltose transporter genes studied, both se- and sc-type, were induced strongly by maltose in all hosts (Fig. 4A), showing clearly that transregulation across S. eubayanus and S. cerevisiae genomes occurs for MAL genes. However, in the C902 host its ‘own gene’ seMALx1, i.e. gene originally cloned from its own genome, had higher transcription levels than the sc-type genes. Reciprocally, in the S. cerevisiae host, transcription of sc-type genes was more efficient compared to the se-type gene during most of the growth period. A similar response was seen with the lager yeast hybrid. It seems that trans-activation was less efficient compared to cis-activation.
Figure 4.
Gene transcription in S. eubayanus C902, S. cerevisiae A60 and S. pastorianus A15 hosts transformed with sc- and se-type transporters and grown on (A) maltose and (B) glucose. Results are averages ± ranges of data from two replicate growths of each yeast strain on each sugar. Due to great difference in transcription level between maltose and glucose ordinate scales are different.
Timing of MAL gene transcription was also different between hosts. In the C902 host, transcription of all three genes followed a similar pattern, reaching their peak at the 24 h time point. However, for A60 and A15 hosts, timing between transcription of sc- and se-type transporters varied. In the A60 host seMALx1 transcription peaked early in the growth, at the 10–12 h time point, whereas sc-type genes peaked only at 20–24 h time point. In the A15 host both MALx1 genes peaked early, both at the 12 h time point. In addition, scMALx1 peaked again at the 24 h time point but its se-version did not show this second peak. In the A15 host, scAGT1 peaked at the 20 h time point. It seems that the transcription pattern in the A15 host (hybrid) was different to patterns seen in the C902 and A60 hosts and timing of transcription in the hybrid is not directly inherited from either of the parental species, S. eubayanus or S. cerevisiae, but formed its own distinct pattern. This seems reasonable as there is a mix of sc- and se-type MAL activators and Mig1 repressors that generate a hybrid-specific expression pattern, which does not directly follow that of the parents. All maltose transporter genes studied, with the possible exception of scMALx1 in the A60 host, were repressed in the presence of glucose (Fig. 4B). In the C902 and A15 hosts, glucose repression was strong and even after glucose exhaustion only very low transcription was observed. In the A60 host there was a basal level of scMALx1 transcription from the beginning of the growth when glucose concentration was high (20 g l−1) and expression remained at this constant basal level during the linear growth phase. Contrarily, the scAGT1 gene in the same host was apparently subject to glucose repression and weak expression was observed only after glucose had been exhausted, at around the 15 h time point. The third gene, seMALx1, was strictly under glucose repression regardless of host strain.
In the present study, the multicopy plasmid YEplac195 was used. For this there are multiple promoter regions present in the cell and there was some risk that transcription factors, Mig1 repressors and MAL activators, would be titered out within the cell. Results obtained showed that at least Mig1 factors were not titered out (Fig. 4). In the C902 and A15 hosts, glucose repression of all genes investigated was very strong during the whole growth period. This means that there were enough Mig1 factors to bind all promoters present. In the A60 and CEN.PK2 hosts (Figs. 4 and 2) grown on glucose there was a low level of expression of sc-type genes but not those of se-type. Mig1 factors were not titered out in these hosts either, as otherwise continuous repression of se-type genes would not be possible. The YEplac195 plasmid number is expected to be similar in the same host independent of the transporter it overexpresses. This means that there would have been enough Mig1 repressors also for sc-type genes overexpressed in A60 and CEN.PK2 hosts but for some other reason repression was alleviated. If MAL activators were titered out, it would lead to a situation where maximum expression peaks would have been higher, but it would not otherwise affect to expression pattern seen as Mig1 repressors were not titered out. In the present study, the maximum level of expression was limited to what can be obtained with native MAL activators present in the cell.
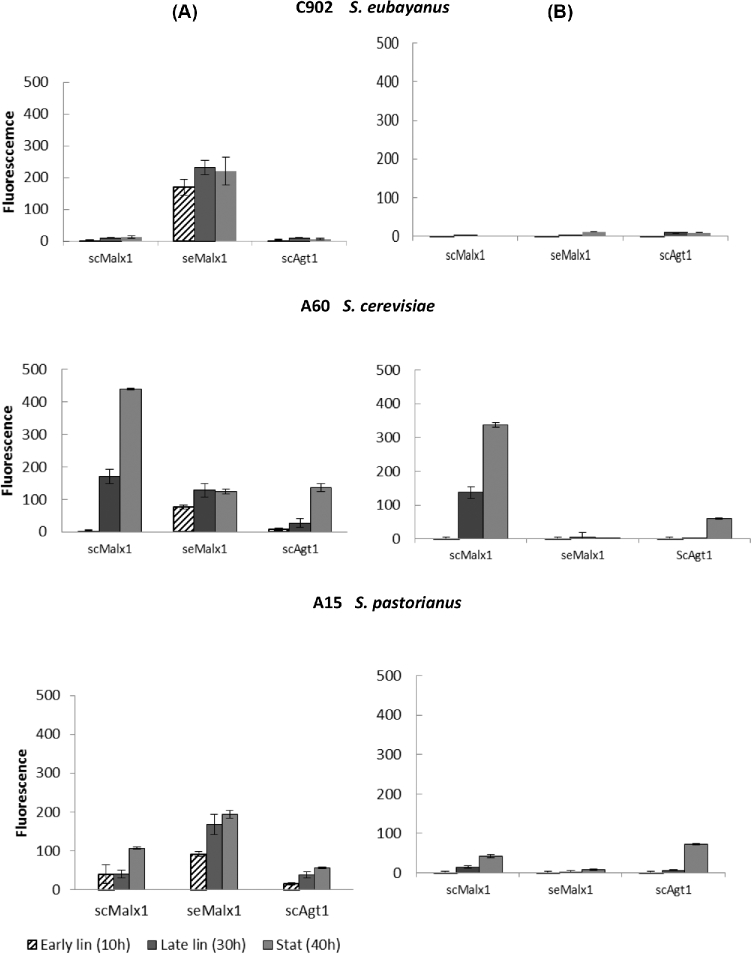
The amount of fluorescence from each transporter, as determined by confocal microscopy, showed that, despite strong expression of foreign sc-type transporters in the C902 host, there were only low amounts of fluorescence for the actual transporters (Fig. 5A). This would suggest that sc-type transporter mRNAs detected by RT-qPCR were not translated efficiently to protein products, or these proteins were rapidly removed in S. eubayanus. Conversely, fluorescence form the native transporter of C902, seMalx1, was present in relatively high amounts during different growth phases of C902. This was consistent with strong expression of the gene. In the A60 and A15 hosts an early transcriptional peak for seMALx1 was, apparently, sufficient to produce seMalx1 transporters for the whole period of growth as the amount of fluorescence for seMalx1 transporters remained quite stable during growth. For all strains, late transcription of sc-type genes correlated with an increasing fluorescence amount towards the stationary phase. In C902 and A15, glucose repression was strong and even after glucose exhaustion there was only a low transcription level observed. However, even this moderate transcription was enough to produce a small amount of transporter signals in cells (Fig. 5B). In the A60 host, even a low basal level of expression was enough to produce a significant amount of fluorescence for transporter protein.
Figure 5.
Amount of fluorescent signal in S. eubayanus C902, S. cerevisiae A60 and S. pastorianus A15 hosts transformed with sc and se-type transporters and grown on (A) maltose and (B) glucose. Samples were collected at early logarithmic growth phase at 10 h, at late logarithmic growth phase at 30 h and at stationary growth phase at 40 h. Results are averages ± ranges of data from two replicate growths of each yeast strain on each sugar.
Confocal microscopic studies showed that there was temporal variation in the localization of transporters on the plasma membrane depending on the transporter type. As an example, Fig. 6 shows localization of different transporters between plasma membrane and intracellular compartments in the A15 host. In Table 8 division of fluorescence between plasma membrane and intracellular compartments, determined by visual analysis of fluorescent images, in different hosts is shown. The seMalx1 transporter fluorescence was the most stable as it was found on the plasma membrane at a constant level during all growth phases in all hosts. In hosts where scAgt1 and scMalx1 transporters were present they were observed to have a certain pattern of localization on the plasma membrane, i.e. in both A15 and A60 hosts, signals for scMalx1 transporters were present in the plasma membrane during the linear growth phase, but were missing in the plasma membrane in the stationary growth phase. Possibly, they were already recycled back to intracellular compartments, as there were high amounts of fluorescence present in the intracellular space. Unfortunately, distinguishing between vacuoles and other intracellular compartments was not possible to do reliably based on data available. In the yeast cell the vacuole is a dynamic structure that can rapidly modify its morphology depending on needs of the cell and cell cycle stage and, as such, it does not have any basic size or shape. There can also be more than one vacuole per yeast cell. Staining of vacuoles with an organelle specific dye would have been needed to make this kind of distinction between vacuoles and other intracellular compartments. In contrast, signals for scAgt1 transporters were not found in the plasma membrane until stationary phase, even if they were abundant in intracellular compartments earlier. There seems, therefore, to be a mechanism excluding scAgt1 transporters from the plasma membrane until later phases of growth. In the C902 host, signals for seMalx1 transporters were present in the plasma membrane in high amounts during all growth phases, whereas very faint signals of sc-type transporters were found only in intracellular compartments. When yeast was grown on glucose, signals started to emerge in some of the hosts only in the late linear growth phase (Table 8). Also under repressive conditions signals for scMalx1 transporters appeared earlier in the plasma membrane.
Figure 6.
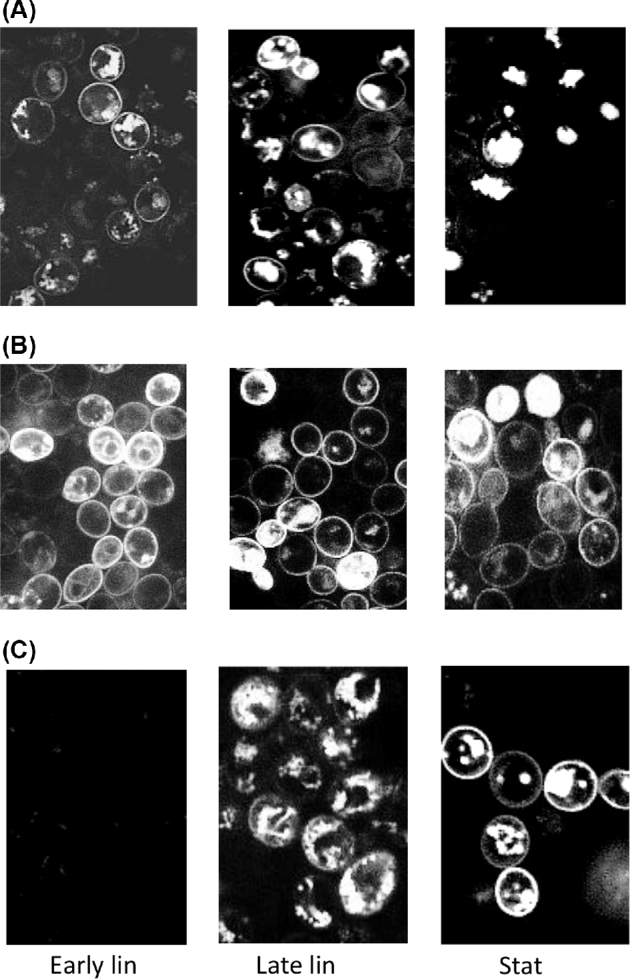
Fluorescent signals in A15 host for (A) scMalx1-mCherry, (B) seMalx1-Gfp and (C) scAgt1-mCherry in early linear, late linear and stationary growth phases on glucose, respectively.
Table 8.
Localization of tagged transporters when grown on maltose and glucose in rich media. Localization determined at early linear phase (OD600 2-4), late linear phase (OD600 15-20) and stationary phase (after 40 h cultivation; OD600 >20). Determined from fluorescent images by visual inspection.
| On maltose | On plasma membrane | In intracellular compartments | |||||
|---|---|---|---|---|---|---|---|
| Early lin | Late lin | Stat | Early lin | Late lin | Stat | ||
| C902 | ScMalx1 | − | − | − | + | + | + |
| SeMalx1 | ++ | ++ | ++ | + | + | + | |
| ScAgt1 | − | − | − | − | + | + | |
| A60 | ScMalx1 | − | ++ | − | ++ | ++ | ++ |
| SeMalx1 | ++ | ++ | ++ | − | − | − | |
| ScAgt1 | − | − | ++ | ++ | ++ | ++ | |
| A15 | ScMalx1 | + | + | − | ++ | ++ | ++ |
| SeMalx1 | ++ | ++ | ++ | + | + | + | |
| ScAgt1 | − | − | ++ | - | ++ | ++ | |
| Early lin | Late lin | Stat | Early lin | Late lin | Stat | ||
| C902 | ScMalx1 | − | − | − | − | − | − |
| SeMalx1 | − | + | + | − | + | + | |
| ScAgt1 | − | − | − | − | − | − | |
| A60 | ScMalx1 | − | + | + | − | + | + |
| SeMalx1 | − | − | − | − | − | − | |
| ScAgt1 | − | − | + | − | + | + | |
| A15 | ScMalx1 | − | + | + | − | + | + |
| SeMalx1 | − | − | − | − | − | − | |
| ScAgt1 | − | − | + | − | + | + | |
− Not visible.
+ Fluorescent signals.
++ Strong fluorescent signals.
When grown on glucose, no significant effect was seen in growth, ethanol production and glucose utilization in strains transformed with constructs (Fig. S4, Supporting Information), whereas on maltose differences were observed. In the A15 host, both se- and sc-type maltose transporter expression had beneficial effects, as a slightly higher ethanol production compared to the control strain was observed (Fig. S5, Supporting Information). In the A60 host, however, expression of seMalx1 did not have an effect compared to the control but both sc-type transporters, scAgt1 and scMalx1, decreased growth rate and ethanol utilization. In the C902 host, utilization of maltose as well as growth and ethanol production was delayed in all strains possessing the construct compared to host. This was especially apparent with the scAGT1 construct, where it was seen that maltose was not consumed efficiently and only approximately 2 g l−1 ethanol was produced.
DISCUSSION
Results of the present study showed for the first time that MAL activators work beyond genome boundaries. Newly introduced sc- and se-type transporter genes were transcribed at a high level in both S. cerevisiae and S. eubayanus backgrounds, thus demonstrating trans-regulation across subgenomes. Sequence analysis showed low sequence conservation in the Mig1 and MAL activator binding sites described by Levine, Tanouye and Michels (1992), Bell et al. (1997) and Vidgren et al. (2011) between sc and se-type promoters. This was observed especially for AGT1 promoters. However, CG-rich motifs forming binding sites for MAL activators described in Pougach et al. (2014) were found in all promoters of the present study. These CGC(9N)CGN and CGG(9N)CGG binding sites were most probably responsible for much of the MAL gene activation observed. Response to maltose required the presence of more than one MAL activator binding site and an increase in the number of such sites in the promoter from two to three increases the expression significantly (Pougach et al.2014). All other promoters in this study contained a sufficient number of CGC(9N)CGN and CGG(9N)CGG binding sites (two) for expression. The exception were the sc-type AGT1 promoters, which contained only one site. Since sc-type AGT1 promoters possessed other types of MAL activator binding sites (Vidgren et al.2011), activation might have occurred in combination with other MAL activator binding elements and this one CGC(9N)CGN binding site. It is possible that also in other promoters, where other MAL activator binding sites are present, they intensify the effect conferred with two CGC(9N)CGN and CGG(9N)CGG binding sites. However, sc- and se-type activators were not completely interchangeable, as stronger activation of the gene was observed in the native host with native MAL activators. Also, timing of transcription varied depending on activator type. While the se-type MAL activator induced both orthologous genes simultaneously, the sc-type activator caused different timing in transcription, so that se-type genes were induced earlier than sc-type genes. These results are similar to those of an earlier investigation on sc- and se-type ARO10 (2-oxo-acid decarboxylase) genes, where it was shown that trans-regulation across subgenomes occurs in lager yeast but, correspondingly, that se- and sc-type activators were not wholly interchangeable, as a basal level of se-type ARO10 gene expression required the se-type activator, even if under inducing conditions both activator types were similarly efficient for both orthologous genes (Bolat et al.2013).
Comparison of transcription patterns in different hosts showed that in the lager hybrid the pattern was not directly inherited from either of the parents as such but instead was a combination of characteristics derived from each parent, i.e. strong glucose repression of MAL genes was similar to that found in S. eubayanus, whereas the transcription pattern of MAL genes under inducing conditions was more similar to that seen in the S. cerevisiae parent. The observation that the MAL gene activation pattern followed that of S. cerevisiae suggests that MAL activators which are operational in this context are mostly of the sc-type. This seems reasonable as there are more sc-type MAL loci present in the lager hybrid (Nakao et al.2009) and it is expected that there are more sc-type activators present relative to se-type. Hybrid-specific gene expression has been found in other studies also. Tirosh et al. (2009) performed allele-specific expression profiling of two yeast species, S. cerevisiae and S. paradoxus, and an artificially made hybrid of these two. For many genes they observed hybrid-specific expression deviating from the parental pattern. They explained most of these changes by novel cis-trans interactions or through modified trans-regulation associated with environmental sensing and signalling.
In general, it seems to be common that, once the hybrid has formed, the transcription pattern of orthologous genes is co-regulated. This means that timing of transcription for orthologous genes follows the same pattern (Horinouchi et al.2010). This seems reasonable as promoters of orthologous genes possess similar activator and repressor binding elements and are exposed to the same mixture of cis- and trans-factors in the hybrid cell. However, even if the timing of transcription followed the same pattern, differences in the level of gene transcription between orthologous genes have been found (He et al.2014; Gibson et al.2015). Nevertheless, in the present study sc- and se-type MAL genes were not found to be co-regulated. Early induction of se-type MAL genes compared to sc-type genes was observed in all other hosts except C902. Studies by Gibson et al. (2013) had already shown some examples where in the lager hybrid there are differences in timing of transcription of orthologous maltose metabolism genes and possibly this is the characteristic of MAL genes.
In the C902 host, high transcription levels of both sc-type transporter genes did not correlate with transporter amount detected in cells. It seems that the sc-type transporter mRNAs are not translated to mature, folded transporter proteins efficiently or, if they are, they are unstable and degraded rapidly. Nevertheless, a small amount of both scAgt1 and scMalx1 transporter proteins present in C902 cells showed that protein translation is not totally defective. Poor conversion of mRNA to sc-type transporters could be due to, for example, a missing lipid component in C902 membranes. As transporters are in effect protein-lipid complexes, they need certain lipids for formation of a functional structure. Lipid composition is important for determining the correct topological organization of transporters, either during the initial membrane assembly or dynamically after membrane insertion has occurred (Bogdanov, Heacock and Dowhan 2002). In general, in cold tolerant strains, such as S. eubayanus, lipid composition of cells differs from non-cold tolerant strains (Krogerus et al.2017). Changes in the lipid composition may be an adaptive response to low temperature fermentations (Torija et al.2003). A study with the Fur4 transporter has shown that conformational alterations in the transporter caused its rapid degradation. When different versions of the Fur4 transporter tagged with Gfp were studied with fluorescence microscopy, it was observed that versions with non-folding mutations were barely detectable by fluorescence microscopy because of rapid degradation (Keener and Babst 2013). Fluorescent images in the present study concerning sc-type transporters in the C902 host were remarkably similar to those obtained with Fur4 non-folding mutants.
In the lager hybrid, one of the two orthologous genes encoding α-glucoside transporters is mutated and non-functional (Vidgren, Ruohonen and Londesborough 2005). It has been suggested that orthologous transporters compete with each other for limited resources of delivery and stable localization of transporters into the plasma membrane. It can be hypothesized that the se-type Malx1 transporters, which are constantly present in the plasma membrane, limit membrane space for other transporters. During the fermentation process, wide-substrate-range transporters (especially for maltotriose) are important, and their presence in the plasma membrane is needed for efficient fermentation. The presence of se-type Malx1 transporters may not therefore be suitable for brewing yeast, which must contend with a dynamic substrate composition during fermentation. Consequently, loss-of-function mutations in the seMALx1 gene may actually have been beneficial for lager yeast in its typical environment and this may explain why only scMalx1 transporters are functional in present-day lager yeast.
As for MALx1, in the case of the AGT1 orthologous gene pair in lager yeast, one of the transporters (sc-type) is mutated and non-functional (Vidgren and Londesborough 2012; Cousseau et al.2013). However, both sc- and se-type Agt1 transporters seem to localize to the plasma membrane similarly at late stages of growth. Also, they are both maltose/maltotriose transporters having the same wide substrate range (Vidgren et al.2010). A possible reason for seAgt1 being more suitable in the hybrid background is that it functions better at the low temperatures typical of lager fermentations. The seAgt1 transporters retain more maltose and maltotriose uptake activity at low temperatures compared to scAgt1, especially below 10°C which is a temperature traditionally employed in lager fermentations (Vidgren et al.2010).
Thus, it seems that lager yeasts may have adapted to the brewing environment by accumulating loss-of-function mutations in genes that were less suited to brewing conditions. Also, these yeast appear to be capable of fine-tuning sugar uptake during fermentation by changing the main transporters present in the plasma membrane in response to changing substrate composition. Firstly, maltose, the most abundant sugar in the wort, is taken up by scMalx1 transporters, which are highly active and specific for maltose. In later phases of growth, scMalx1 transporters are withdrawn and seAgt1 transporters, which have a wider substrate range and operate well at low temperatures, are carried to the plasma membrane so that remaining sugars of wort, and in particular maltotriose, can be taken up by the cells.
In conclusion, with the MALx1 orthologous gene pair, differential timing of localization of the respective transporters to the plasma membrane feasibly confers a competitive advantage upon the scMalx1 transporter. With the AGT1 orthologous pair, the competitive advantage may be the greater suitability of the seAgt1 type transporter to low-temperature fermentations. Both conjectures could be tested by evolving de novo hybrid yeast in brewing-relevant conditions and determining the rate at which loss-of-function mutations arise in the respective transporter genes.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSYR online.
Acknowledgements
We thank Sirpa Holm for skilled technical assistance and John Londesborough for critical reading of the manuscript.
FUNDING
This work was supported by the Academy of Finland [grant number 276480].
Conflict of interest. None declared
REFERENCES
- Baker E, Wang B, Bellora N et al. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Yeast 2015;32:1793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell PJ, Higgins VJ, Dawes IW et al. Tandemly repeated 147 bp elements cause structural and functional variation in divergent MAL promoters of Saccharomyces cerevisiae. Yeast 1997;13:1135–44. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J 2002;21:2107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolat I, Romagnoli G, Zhu F et al. Functional analysis and transcriptional regulation of two orthologs of ARO10, encoding broad-substrate-specificity 2-oxo-acid decarboxylases, in the brewing yeast Saccharomyces pastorianus CBS1483. FEMS Yeast Res 2013;13:505–17. [DOI] [PubMed] [Google Scholar]
- Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol 2010; 20:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988;16:10881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousseau FE, Alves SL Jr, Trichez D et al. Characterization of maltotriose transporters from the Saccharomyces eubayanus subgenome of the hybrid Saccharomyces pastorianus lager brewing yeast strain Weihenstephan 34/70. Lett Appl Microbiol 2013;56:21–9. [DOI] [PubMed] [Google Scholar]
- Gibson BR, Londesborough J, Rautio J et al. Transcription of α-glucoside transport and metabolism genes in the hybrid brewing yeast Saccharomyces pastorianus with respect to gene provenance and fermentation temperature. J Inst Brew 2013;119:23–31. [Google Scholar]
- Gibson B, Krogerus K, Ekberg J et al. Variation in α-acetolactate production within the hybrid lager yeast group Saccharomyces pastorianus and affirmation of the central role of the ILV6 gene. Yeast 2015;32:301–16. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA et al. Improved method for high efficiency transformation of Intact yeast cells. Nucl Acids Res 1992;20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dong J, Yin H et al. Monitoring of the production of flavour compounds by analysis of the gene transcription involved in higher alcohol and ester formation by the brewer's yeast Saccharomyces pastorianus using a multiplex RT-qPCR assa. J Inst Brew 2014;120:119–26. [Google Scholar]
- Hu Z, Nehlin JO, Ronne H et al. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet 1995;28:258–66. [DOI] [PubMed] [Google Scholar]
- Hu Z, Gibson AW, Kim JH et al. Functional domain analysis of the Saccharomyces MAL-activator. Curr Genet 1999;1–2: 1–12. [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Yoshikawa K, Kawaide R et al. Genome-wide expression analysis of Saccharomyces pastorianus orthologous genes using oligonucleotide microarrays. J Biosci Bioeng 2010;110:602–7. [DOI] [PubMed] [Google Scholar]
- Keener JM, Babst M. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic 2013;14:412–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogerus K, Seppänen-Laakso T, Castillo S et al. Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb Cell Fact 2017;16:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine J, Tanouye L, Michels CA. The UAS(MAL) is a bidirectional promotor element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr Genet 1992;22:181–9. [DOI] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valério E et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci USA 2011;108:14539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães F, Vidgren V, Ruohonen L et al. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res 2016;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y, Kanamori T, Itoh T et al. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res 2009;16:115–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pougach K, Voet A, Kondrashov FA et al. Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nat Commun 2014;5:4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S, Masloff S, Hoff B et al. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr Genet 2003;43:54–61. [DOI] [PubMed] [Google Scholar]
- Rautio J, Londesborough J. Maltose transport by brewer's yeast in brewer's wort. J Inst Brew 109:251–61. [Google Scholar]
- Rodicio R, Zimmermann FK. Cloning of maltase regulatory genes in Saccharomyces cerevisiae isolation of the MAL2-8 c regulatory gene. Curr Genet 1985;9:539–45. [Google Scholar]
- Tirosh I, Reikhav S, Levy AA et al. Yeast hybrid provides insight into the evolution of gene expression regulation. Science 2009;324:659–62. [DOI] [PubMed] [Google Scholar]
- Torija MJ, Beltran G, Novo M et al. Effects of fermentation temperature and Saccharomyces species on the cell fatty acid composition and presence of volatile compounds in wine. Int J Food Microbiol 2003;85:127–36. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp.red fluorescent protein. Nat Biotechnol 2004;12:1524–5. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB. 1983. Methods in Yeast Genetics. A Laboratory Manual. Cold Springs Harbor Laboratory, Cold Springs Harbor, NY, USA. [Google Scholar]
- Stambuk BU, Alves SL Jr, Hollatz C et al. Improvement of maltotriose fermentation by Saccharomyces cerevisiae. Lett Appl Microbiol 200643:370–6. [DOI] [PubMed] [Google Scholar]
- U’Ren JM, Wisecaver JH, Paek AL et al. Draft genome sequence of the ale-fermenting Saccharomyces cerevisiae strain GSY2239. Genome Announc 2015;3:e00776–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sirenko O, Needleman R. 1997. Genomic footprinting of Mig1p in the MAL62 promoter. Binding is dependent upon carbon source and competitive with the Mal63p activator. J Biol Chem 1997;272:4613–22. [DOI] [PubMed] [Google Scholar]
- Vidgren V, Ruohonen L, Londesborough J. Characterization and functional analysis of the MAL and MPH Loci for maltose utilization in some ale and lager yeast strains. Appl Environ Microbiol 2005;71:7846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgren V, Multanen JP, Ruohonen L et al. The temperature dependence of maltose transport in ale and lager strains of brewer's yeast. FEMS Yeast Res 2010;10:402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgren V, Londesborough J. Characterization of the Saccharomyces bayanus-type AGT1 transporter of lager yeast. J Inst Brew 2012; 118;148–51. [Google Scholar]
- Vidgren V, Kankainen M, Londesborough J et al. Identification of regulatory elements in the AGT1 promoter of ale and lager strains of brewer's yeast. Yeast 2011;28:579–94. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R et al. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994;10:1793–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.