Abstract
The typical genome of chronic lymphocytic leukemia (CLL) carries ∼2000 molecular lesions. Few mutations recur across patients at a frequency >5%, whereas a large number of biologically and clinically uncharacterized genes are mutated at lower frequency. Approximately 80% of CLL patients carry at least 1 of 4 common chromosomal alterations, namely deletion 13q14, deletion 11q22-23, deletion 17p12, and trisomy 12. Knowledge of the CLL genome has translated into the availability of molecular biomarkers for prognosis and treatment prediction. Prognostic biomarkers do not affect treatment choice, and can be integrated into prognostic scores that are based on both clinical and biological variables. Molecular predictive biomarkers affect treatment choice, and currently include TP53 disruption by mutation and/or deletion and IGHV mutation status. TP53 disruption by gene mutation and/or deletion associates with chemoimmunotherapy failure and mandates treatment with innovative drugs, including ibrutinib, idelalisib, or venetoclax. The mutation status of IGHV genes represents a predictive biomarker for identifying patients that may benefit the most from chemoimmunotherapy with fludarabine, cyclophosphamide, and rituximab. Assessment of these biomarkers at the time of treatment requirement is recommended by most current guidelines for CLL management. Other molecular predictors are under investigation, but their application in clinical practice is premature.
Learning Objectives
Understand that the major advances in the understanding of CLL genomics has led to the clarification of the mutational landscape of the disease and of its clonal evolution over time and under the selection pressure of treatment
Learn that such molecular knowledge has expanded the availability of prognostic and predictive biomarkers that can be used for patient counseling, treatment tailoring, and clinical trial design
Molecular genetics of chronic lymphocytic leukemia
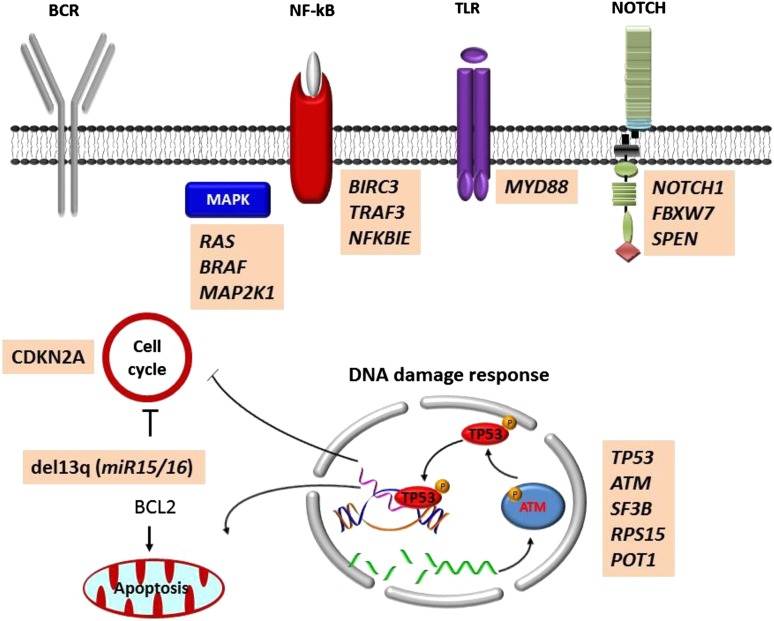
The typical genome of unselected chronic lymphocytic leukemia (CLL) carries ∼2000 molecular lesions, of which, however, only ∼20 are nonsynonymous mutations and only ∼5 are gross structural abnormalities.1,2 Few mutations recur across patients at a frequency >5%, whereas a large number of biologically and clinically uncharacterized genes are mutated at lower frequency. In total, >40 recurrently mutated driver genes have been identified in CLL (Figure 1).1,2 Recurrent mutations are not homogeneously spread across the CLL genome, but, rather, affect genes that can be integrated into a small set of pathways (Figure 2). These include microenvironment-dependent signaling through NOTCH (NOTCH1, FBXW7), inflammatory receptors (MYD88), MAPK–extracellular signal-regulated kinase (BRAF, KRAS, NRAS, MAP2K1) and NF-κB pathways (BIRC3, TRAF3, NFKBIE), as well as intracellular programs such as DNA damage and cell cycle control (ATM, TP53, SAMHD1, POT1), chromatin modification (HIST1H1E, CHD2, ZMYM3), transcription (EGR2, IRF4, BCOR, MED12), and ribosomal processing (XPO1, SF3B1, RPS15).1,2
Figure 1.
Genes and chromosomal regions affected by molecular lesions in CLL. The word cloud shows the genes that are reported as mutated in CLL by the Catalogue of Somatic Mutations in Cancer (COSMIC; v77) and the gross chromosomal abnormalities that are recurrent in CLL. The size of the font is proportional to the frequency of the molecular lesion.
Figure 2.
Mutated pathways in CLL. Cellular programs that are affected by the most recurrent molecular lesions are represented. Boxes show the genes that are recurrently mutated in each cellular program.
Approximately 80% of CLL patients carry at least 1 of 4 common chromosomal alterations, namely deletion 13q14, deletion 11q22-23, deletion 17p12, and trisomy 12. Deletion 13q14 is the most frequent genetic lesion of CLL occurring in 50% to 60% of cases.3 The minimal deleted region on 13q14 contains the miR15A and miR16A microRNAs.4 In normal cells, miR15A and miR16A inhibit the expression of key regulators of apoptosis and cell cycle. Deletion of miR15A and miR16A abrogates this inhibitory effect, favors the constitutive survival and cycling of tumor B cells, and leads to CLL development in mouse models.5 B-cell lymphoma 2 (BCL2) codes for an antiapoptotic protein and is one of the genes upregulated in CLL as a consequence of miR15A/miR16A deletion. Consistent with the central contribution of BCL2 activation in the pathogenesis of CLL, selective inhibition of BCL2 through the BH3 mimetic venetoclax results into high response rates and deep remissions.6
Although genes encoding for components of the B-cell receptor (BCR) signaling machinery are usually not targeted by somatic mutations in CLL, the critical role of this pathway in disease pathogenesis is supported by the therapeutic success of targeted agents that interfere with the BCR. These agents are commonly termed as BCR inhibitors, although they should be more properly defined as kinase inhibitors because they inhibit many different pathways. Functional evidence indicates that BCR pathway activation in CLL results from contacts between tumor cells and antigens, which is influenced, among others, by the somatic hypermutation (SHM) load of the rearranged immunoglobulin heavy-chain variable (IGHV) genes.7 CLLs carrying unmutated IGHV genes display enhanced BCR signaling compared with CLLs carrying mutated IGHV genes, reflecting functional differences in the type of antigenic interaction through the BCR. The BCRs of CLLs carrying unmutated IGHV genes maintain polyreactivity because of the lack of SHM, and thus are capable of binding multiple epitopes and are more prone to sustained signaling.7 Conversely, CLLs carrying mutated IGHV genes fail to respond to BCR stimulation because of anergy, or because the antigen specificity has been extremely narrowed by the accumulation of mutations through the SHM process.7
Definition and use of prognostic and predictive biomarkers in CLL
Biomarkers are variables that can be measured and associate with treatment or disease outcome. Biomarkers can be single variables or classifiers composed of multiple variables (ie, scores).
Prognostic biomarkers usually reflect the underlying biology and natural history of CLL, and thus define prognosis in the absence of treatment, or, for patients requiring treatment, independent of the treatment received.8,9 Clinical endpoints captured by prognostic biomarkers include: (1) death; (2) treatment requirement, if “watch and wait” is the proposed management; and (3) other clinically relevant events such as Richter syndrome. In clinical practice, the impact of CLL prognostic biomarkers is limited. In the field of clinical research, prognostic biomarkers allow for identification of those CLL patients appropriate for early intervention studies. Predictive biomarkers, on the other hand, provide information on the likely benefit from a specific treatment. Consistently, the efficacy of a given treatment is different in biomarker-positive vs biomarker-negative patients. In clinical practice, predictive CLL biomarkers can significantly impact treatment decisions for a particular patient.
Variables can be defined as predictive biomarkers on statistical grounds or on biological grounds.8,9 On statistical grounds, at least 2 comparison groups are needed (ie, 2 different treatment arms in a randomized trial) to determine whether a biomarker is potentially predictive, and a formal test for an interaction between the biomarker and the treatment group needs to be performed. Conversely, to establish whether a biomarker is purely prognostic requires the demonstration that there is a significant association between the biomarker and outcome, regardless of treatment, and that treatment effects do not depend on the biomarker. A biomarker may have both predictive and prognostic implications. On biological grounds, a predictive biomarker is one whose presence is required for the treatment to work. Thus, in this context, the term predictive should be reserved for situations in which the biomarker is the target of the therapy and needs to be present for the treatment response to occur (eg, BCR/ABL1 transcript in chronic myeloid leukemia treated with tyrosine kinase inhibitors).
A number of molecular biomarkers have been proposed in CLL that fulfill the definition of prognostic factors, though only a few of them have been consistently validated. Conversely, a few molecular biomarkers meet the statistical definition of predictive factors, and none the biological definition (Table 1).
Table 1.
Prognostic and predictive biomarkers commonly used in clinical practice
| Prognostic | Predictive | |
|---|---|---|
| Age |  |
 |
| CIRS |  |
 |
| Stage |  |
 |
| β2-microglobulin |  |
 |
| CD49d |  |
 |
| CD38 |  |
 |
| ZAP70 |  |
 |
| IGHV mutation |  |
 |
| 17p13 deletion |  |
 |
| 11q22-23 deletion |  |
 |
| Trisomy 12 |  |
 |
| 13q14 deletion |  |
 |
| TP53 mutation |  |
 |
| SF3B1 mutation |  |
 |
| NOTCH1 mutation |  |
 |
A green circle indicates yes, a red circle indicates no, and an orange circle indicates not yet fully validated. CIRS, Cumulative Illness Rating Scale; IGHV, immunoglobulin heavy variable gene.
Molecular prognostic biomarkers in CLL
Immunoglobulin gene mutations
The IGHV genes of CLL can accumulate variations as a consequence of the SHM process. The prevalence of mutated IGHV genes (defined as a <98% identity compared with the germ line nucleotide sequence) is higher among newly diagnosed and asymptomatic CLL patients (∼60%), whereas the prevalence of unmutated IGHV genes (defined as ≥98% identity compared with the germ line nucleotide sequence) is higher among progressive (∼50%-60%) and relapsed/refractory (∼70%-80%) CLL patients. Patients with mutated IGHV genes, when compared with patients with unmutated IGHV genes, experience longer time to first treatment when managed with “watch and wait,” longer progression-free survival (PFS) when treated with chemoimmunotherapy, lower risk of transformation, and thus, in the end, better overall survival (OS) (Table 2).10-29 The robust and consistent prognostic value of IGHV mutation status has been formally validated by a systematic review and meta-analysis.30 Apart from the mutation status, the usage of specific IGHV genes may also affect prognosis, including progression (ie, IGHV3-21 in stereotyped subset 2) and transformation to Richter syndrome (IGHV4-39 in stereotyped subset 8).17,19
Table 2.
Clinical outcome of unselected, previously untreated CLL from study cohorts
| Ref. | Time to first treatment | OS | ||
|---|---|---|---|---|
| TP53mut | TP53wt | TP53mut | TP53wt | |
| 37 | 4.8 y (median) | 7.5 y (median) | 60.9% (at 5 y) | 86.8% (at 5 y) |
| 19 | — | — | 4.6 y (median) | 13.9 y (median) |
| 31 | — | — | 50.7% (at 5 y) | 76.5% (at 5 y) |
| 28 | — | — | 54.6% (at 5 y) | 81.8% (at 5 y) |
| 23 | 0.3 y (median) | 5.3 y (median) | 4.6 y (median) | 12.5 y (median) |
| 39 | 1.5 y (median) | 3.5 y (median) | — | — |
| 26 | — | — | 31.9% (at 5 y) | 74.6% (at 5 y) |
| 45 | — | — | 66.2% (at 5 y) | 85.2% (at 5 y) |
| 27 | — | — | 43.5% (at 5 y) | 68.0% (at 5 y) |
| 13 | 1.8 y (median) | Not reached | 2.5 y (median) | Not reached |
| NOTCH1mut | NOTCH1wt | NOTCH1mut | NOTCH1wt | |
| 37 | 3.5 y (median) | 7.6 y (median) | 75.7% (at 5 y) | 85.1% (at 5 y) |
| 19 | — | — | 3.5 y (median) | 13.9 y (median) |
| 31 | — | — | 56.3% (at 5 y) | 75.2% at 5 y) |
| 28 | 0.2 y (median) | 3.4 y (median) | 63.6% (at 5 y) | 85.5% (at 5 y) |
| 23 | 0.4 y (median) | 5.3 y (median) | 5.5 y (median) | 12.8 y (median) |
| 39 | 3.1 y (median) | 3.1 y (median) | — | — |
| 26 | — | — | 36.3% (at 5 y) | 72.5% (at 5 y) |
| 27 | — | — | 69.9% (at 5 y) | 76.1% (at 5 y) |
| 35 | 3.6 y (median) | 7.5 y (median) | — | — |
| 34 | 1.8 y (median) | 6.0 y (median) | 64% (at 10 y) | 35% (at 10 y) |
| 36 | 0.2 y (median) | 1.2 y (median) | 55% (at 5 y) | 75% (at 5 y) |
| 38 | 45% (at 5 y) | 55% (at 5 y) | 75% (at 5 y) | 95% (at 5 y) |
| 33 | — | — | 10.4 y (median) | Not reached |
| 21 | 0.4 y (median) | 4.9 y (median) | 5.5 y (median) | Not reached |
| 40 | — | — | 38% (at 5 y) | 70% (at 5 y) |
| SF3B1mut | SF3B1wt | SF3B1mut | SF3B1wt | |
| 37 | 3.8 y (median) | 8.0 y (median) | 64.7% (at 5 y) | 86.7% (at 5 y) |
| 31 | — | — | 60.3% (at 5 y) | 73.85 (at 5 y) |
| 28 | 0.2 y (median) | 2.0 y (median) | — | — |
| 23 | 0.1 y (median) | 4.8 y (median) | 5.3 y (median) | 12.2 y (median) |
| 39 | 1.9 y (median) | 3.0 y (median) | — | — |
| 26 | — | — | 53% (at 5 y) | 70.8% (at 5 y) |
| 27 | — | — | 64.9% (at 5 y) | 77.7% (at 5 y) |
| 21 | 0.2 y (median) | 4.7 y (median) | 5.3 y (median) | Not reached |
| 40 | — | — | 46% (at 5 y) | 68% (at 5 y) |
| 29 | 0.8 y (median) | 1.1 y (median) | — | — |
| 32 | — | — | 30.0% (at 10 y) | 77% (at 10 y) |
| 41 | 1.2 y (median) | 5.0 y (median) | — | — |
| IGHVunmut | IGHVmut | IGHVunmut | IGHVmut | |
| 19 | — | — | 11.7 y (median) | Not reached |
| 28 | 0.3 y | 7 y (median) | 74.6% (at 5 y) | 87.3% (at 5 y) |
| 23 | 0.9 y (median) | 9.9 y (median) | 6.7 y (median) | Not reached |
| 26 | — | — | 55.6% (at 5 y) | 76.4% (at 5 y) |
| 27 | — | — | 6.4 y (median) | Not reached |
| 13 | 1.5 y (median) | 9.2 y (median) | 81% (at 5 y) | 88% (at 5 y) |
| 21 | 0.8 y (median) | 9.9 y (median) | 6.4 y (median) | Not reached |
| 29 | 0.3 y (median) | 5.4 y (median) | — | — |
| 10 | — | — | 9.7 y (median) | 24.5 y (median) |
| 11 | — | — | 9.0 y (median) | — |
| 22 | — | — | 7.0 y (median) | Not reached |
| 12 | — | — | 8.0 y (median) | Not reached |
| 25 | — | — | 43% (at 10 y) | 87% (at 10 y) |
| 16 | 5.0 y (median) | Not reached | 3.2 y (median) | — |
| 17 | — | — | 7.0 y (median) | Not reached |
| 20 | — | — | 41% (at 10 y) | 75% (at 10 y) |
| 24 | 2.4 y (median) | 19 y (median) | 9.8 y (median) | 17.9 y (median) |
| 18 | 4.6 y (median) | Not reached | 13.0 y (median) | 23.0 y (median) |
| 15 | 1.1 y (median) | Not reached | 7.0 y (median) | Not reached |
| 14 | 2.8 y (median) | 11.0 y (median) | 9.7 y (median) | Not reached |
Clinical outcome of unselected, previously untreated CLL from study cohorts including >200 patients stratified according to the TP53, NOTCH1, SF3B1, and IGHV mutation status. —, not available; Ref., reference.
Genetic lesions
Over the last few years, several genetic prognostic biomarkers have been identified that are significantly associated with CLL OS, time to first treatment in cases managed with “watch and wait,”, or PFS in treated cases.
In a relevant fraction of CLL patients (∼25%), deletion of 13q14 occurs in the absence of any concomitant driver genetic lesion. Patients harboring solely 13q14 deletion have an excellent clinical outcome with a progression rate of <1% per year, longer PFS after therapy, and low risk of transformation, which overall translate into an expected survival only slightly lower than that of the general population.31
Deletion of 11q22-23 always includes ATM and occurs in <10% of newly diagnosed CLL, whereas its prevalence rises to ∼20% at the time of first treatment and 30% at disease relapse. Mutations of the SF3B1 and NOTCH1 genes are observed in 10% to 15% of newly diagnosed CLL, whereas their prevalence rises to ∼20% in progressive or relapsing CLL. Deletion of 11q22-23, SF3B1 mutations and NOTCH1 mutations identify patients with intermediate-risk disease (∼40% of cases are alive at 10 years) whose survival is ∼50% less than that expected for the general population.31 Patients with deletion of 11q22-23, SF3B1 mutations, or NOTCH1 mutations have short time to progression if managed with “watch and wait,” and faster relapse if treated with chemoimmunotherapy, but not if treated with ibrutinib (Table 2).3,19,21,23,26-29,31-44 The risk of transformation is specifically affected by NOTCH1 mutations, but not by 11q22-23 deletion or by SF3B1 mutations.19,34,42
At diagnosis, the TP53 gene is disrupted in 4% to 8% of unselected CLL by deletions, mutations, or a combination of both. The incidence rises to 10% at the time of first-line treatment, 30% to 40% in relapsed/refractory CLL, and 50% to 60% in Richter syndrome because TP53 abnormalities can be acquired/selected during the course of the disease.13,19,23,26-28,31,37,39,43 TP53 abnormalities mark the worst outcome in CLL, with an estimated median OS of 3 to 5 years, which is ∼70% less than that expected for the general population.31 Patients having TP53 abnormalities have short time to progression if managed with “watch and wait,” and generally fail chemoimmunotherapy (Table 2).13,19,23,26-28,31,37,39,45 The risk of transformation is also increased among patients with TP53 abnormalities.42 The robust and consistent prognostic value of TP53 abnormalities in CLL has been formally validated by a systematic review and meta-analysis.30 The negative impact of TP53 abnormalities is only partly mitigated, but not overcome, by ibrutinib, idelalisib, or venetoclax, at least in the relapsed/refractory setting.6,44,46,47
A number of other gene mutations recurring at lower frequency have been signaled for their association with CLL outcome, including BIRC3, SAMHD1, RPS15, NFKBIE, EGR2, KRAS, and POT1. However, their annotation as CLL prognostic biomarkers is controversial or awaits validation studies.47
Score systems
Hierarchical prognostic models integrating together multiple genetic features of CLL and partitioning the risk according to the prognostic weight of the genetic biomarkers effectively stratify CLL survival. In 2000, this notion was unequivocally documented by the seminal study of Döhner et al who established a hierarchical prognostic model for CLL based on 5 risk categories.3 CLL cases harboring 17p13 deletion independent of concomitant abnormalities had the worst prognosis (median survival, 32 months), followed by cases carrying 11q22-23 deletion (median survival, 79 months), trisomy 12 (median survival, 114 months), normal fluorescence in situ hybridization (FISH) (median survival, 111 months), and 13q14 deletion (median survival, 133 months).3 Though the FISH hierarchical model was developed in the chemotherapy era, it stands also when applied to stratify the survival of patients treated with ibrutinib.48
Integration of the most recurrent mutations onto the backbone of the FISH hierarchical model has allowed an improvement in outcome discrimination.31 Four groups of patients are hierarchically classified by the integrated mutational-cytogenetic model: (1) high-risk patients, harboring TP53 and/or BIRC3 abnormalities independent of co-occurring genetic lesions, who account for ∼15% to 20% of newly diagnosed CLL and show a 10-year survival of 29%; (2) intermediate-risk patients, harboring NOTCH1 and/or SF3B1 mutations and/or del11q22-23 in the absence of BIRC3 and TP53 abnormalities, who account for ∼15% to 20% of newly diagnosed CLL and show a 10-year survival of 37%; (3) low-risk patients, harboring +12 or normal genetics, who account for ∼40% of newly diagnosed CLL and show a 10-year survival of 57%; and (4) very-low-risk patients, harboring del13q14 only in the absence of any additional abnormality, who account for ∼25% of newly diagnosed CLL and have a nearly normal life expectancy with a 10-year survival of 69%.31
With the broadening of the number of prognostic biomarkers, it has become difficult for the practicing clinician to determine which factors are most important to counsel individual patients, and how to combine the results of multiple prognostic tests when some suggest an aggressive clinical course and others a more indolent one in the same patient. Systematic reviews and meta-analyses illustrated the robust and consistent prognostic value of IGHV mutation status and FISH results for the 11q22-23 and 17p13 chromosomal abnormalities in patients with previously untreated CLL.30 These studies also indicated that IGHV mutation status and FISH karyotyping provide complementary information with respect to outcome anticipation. Accordingly, recent efforts have focused on combining IGHV mutation status and genetic biomarkers into scoring systems that may anticipate CLL survival.
The German CLL Study Group (GCLLSG) developed a comprehensive prognostic index by taking advantage of a large population of prospectively collected patients within its clinical trials.49 The prognostic index score is calculated by giving points to 17p13 deletion, 11q22-q23 deletion, and IGHV mutation status among molecular biomarkers, and sex, age, ECOG performance status, serum thymidine kinase, and β2-microglobulin among clinical factors (Table 3). Patients are then grouped into low (5-year survival, 95%), intermediate (5-year survival, 86%), high (5-year survival, 67%), and very-high (5-year survival, 18%) risk groups.49 Among biomarkers, the GCLLSG score includes serum thymidine kinase. Thymidine kinase is a key cellular enzyme of the salvage pathway of DNA synthesis whose activity is cell cycle dependent, and therefore it represents a general proliferation marker. Serum thymidine kinase is available as a routine clinical assay in some European countries, whereas it is less used in the United States. Therefore, a slightly modified version of the model based on 7 prognostic variables, instead of 8, and not including serum thymidine kinase has been proposed.50
Table 3.
GCLLSG Prognostic Index
| Score | 5-y survival, % | |
|---|---|---|
| Biomarker | ||
| Age, y | ||
| ≤60 | 0 | |
| >60 | 1 | |
| Sex | ||
| Female | 0 | |
| Male | 1 | |
| ECOG PS | ||
| 0 | 0 | |
| >0 | 1 | |
| β2-microglobulin, mg/L | ||
| ≤1.7 | 0 | |
| 1.7-3.5 | 1 | |
| >3.5 | 2 | |
| Serum thymidine kinase, U/L | ||
| ≤10 | 0 | |
| >10 | 2 | |
| IGHV | ||
| Mutated | 0 | |
| Unmutated | 1 | |
| 11q22-q23 | ||
| Nondeleted | 0 | |
| Deleted | 1 | |
| 17p13 | ||
| Nondeleted | 0 | |
| Deleted | 6 | |
| Risk group | ||
| Low risk | 1-2 | 95 |
| Intermediate risk | 3-5 | 86 |
| High risk | 6-10 | 67 |
| Very-high risk | 11-14 | 18 |
ECOG PS, Eastern Cooperative Oncology Group performance status; GCLLSG, German CLL Study Group.
More recently, an international collaboration used information from ∼3500 patients enrolled in clinical trials or institutional/population-based cohorts to develop a comprehensive CLL-International Prognostic Index (IPI), which was subsequently validated in independent CLL populations (Table 4).27,51,52 The CLL-IPI score incorporates both molecular and clinical CLL aspects, and is based on 5 robust and widely used prognostic biomarkers (age, clinical stage, 17p13 deletion and/or TP53 mutation, IGHV mutation status, and β2-microglobulin), and documents that TP53 disruption is the sole recurrent genetic abnormality that shows independent prognostic information after adjusting for IGHV mutation status and for the most important clinical variables. Conversely, 11q22-q23 deletion and mutations of NOTCH1 and SF3B1 do not carry independent prognostic value in this model. The CLL-IPI identifies low-, intermediate-, high-, and very-high-risk patients showing significantly different survival at 5 years (93%, 79%, 63%, 23%, respectively). Although the CLL-IPI has been primarily developed to prognosticate CLL survival, it also anticipates time to first treatment in “watch and wait” patients.27 A prognostic system based only on IGHV mutation status and FISH cytogenetics that simplifies the CLL-IPI has also been proposed.53
Table 4.
CLL-IPI
| Score | 5-y survival, % | |
|---|---|---|
| Biomarker | ||
| Age, y | ||
| ≤65 | 0 | |
| >65 | 1 | |
| Stage | ||
| Rai 0/Binet A | 0 | |
| Rai I-IV/Binet B-C | 1 | |
| β2-microglobulin, mg/L | ||
| ≤3.5 | 0 | |
| >3.5 | 2 | |
| IGHV | ||
| Mutated | 0 | |
| Unmutated | 2 | |
| TP53 | ||
| No abnormalities | 0 | |
| Deletion and/or mutation | 4 | |
| Risk group | ||
| Low risk | 0-1 | 93 |
| Intermediate risk | 2-3 | 79 |
| High risk | 4-6 | 63 |
| Very-high risk | 7-10 | 23 |
Importantly, these indices were developed in patients in the chemoimmunotherapy era, and will need to be validated in the novel agent era. The use of molecular biomarkers in CLL clinical practice will be discussed below in “Recommendations for clinical practice.”
Molecular predictive biomarkers in CLL
Immunoglobulin gene mutations
Among patients receiving chemoimmunotherapy, the IGHV mutation status affects the kinetics of relapse and thus PFS, which is longer in IGHV-mutated vs IGHV-unmutated patients. Accordingly, ∼50% to 60% of patients with mutated IGHV genes who receive potent chemoimmunotherapy regimens maintain disease remission long-term, including persistent negativity of minimal residual disease in some instances, which translates into a plateau on the PFS curve, no relapses beyond 10 years, and an OS similar to the one expected in normal healthy subjects. Conversely, long-term, almost all IGHV unmutated CLL patients are projected to progress after chemoimmunotherapy.54-56
In contrast to chemoimmunotherapy, CLL patients benefit from ibrutinib and idelalisib independent of IGHV mutation status. Indeed, upon treatment with ibrutinib or idelalisib, the PFS of IGHV-unmutated patients is similar to that of IGHV-mutated cases.44,48,57 The lymphocytosis tends to resolve more quickly in the IGHV-unmutated group. On these bases, though the interaction between different treatments (chemoimmunotherapy vs novel targeted agents), IGHV mutation status, and outcome has never been tested in a clinical trial, cross-trial comparisons suggest that unmutated IGHV genes might fulfill the statistical definition of predictive biomarker for lack of benefit from chemoimmunotherapy.
Genetic lesions
TP53 codes for a central regulator of the DNA damage response pathway and is the target of the genotoxic effect of chemotherapy. Upon chemotherapy-induced DNA damage, TP53 is activated and CLL cells undergo apoptosis. Conversely, when TP53 is nonfunctional because of deletion or mutation, the apoptosis of CLL cells cannot be triggered in response to chemotherapy. Consistently, CLL patients with 17p13 deletion or TP53 mutation have a very poor response to chemoimmunotherapy regimens, including fludarabine and cyclophosphamide (FC) and rituximab (FCR), bendamustine and rituximab (BR), obinutuzumab and chlorambucil (G-Clb), ofatumumab and chlorambucil (O-Clb), or rituximab and chlorambucil (R-Clb).58-61 Ibrutinib, idelalisib, and venetoclax do not exert their antileukemic activity through genotoxic mechanisms, and are therefore active irrespective of TP53 dysfunction.62 Consistently, the response rate in TP53-disrupted patients is superimposable to that observed in patients with a wild-type TP53 gene, and better than that observed in every previous historical control treated with chemoimmunotherapy.63-65 Notably, the interaction between treatment (chemoimmunotherapy vs novel targeted agents), TP53 status, and outcome has never been formally tested in a clinical trial. However, cross-trial comparison of patients’ outcome according to TP53 status allows for indirectly suggesting that TP53 disruption might fulfill the statistical definition of predictive biomarker for lack of benefit from chemoimmunotherapy in CLL.
Among new CLL genetic lesions, only NOTCH1 mutations behave as a candidate predictive biomarker. Indeed, among CLL harboring NOTCH1 mutations, treatment with FCR, R-Clb, or O-Clb does not result in the expected increase in PFS compared with treatment with FC or with chlorambucil alone.66-68 These observations point to NOTCH1 mutations as a biomarker of resistance to the anti-CD20 monoclonal antibodies rituximab and ofatumumab in CLL. The outcome of CLL patients treated with obinutuzumab combined to chlorambucil is improved independent of NOTCH1 mutation status, suggesting that the augmented cytotoxicity of obinutuzumab or the increased dose of the anti-CD20 antibody used in the obinutuzumab-chlorambucil schema overcomes NOTCH1 mutation–associated resistance to rituximab.67 The mechanism underlying the anti-CD20 refractoriness associated with NOTCH1 mutations remains obscure.
Resistance to ibrutinib, idelalisib, or venetoclax is not fully captured by predictive biomarkers that are otherwise used in the setting of chemoimmunotherapy. On this basis, novel biomarkers are required to redefine high-risk CLL patients in the era of novel agents. Development of resistance to ibrutinib is due to acquired mutations in Bruton tyrosine kinase (BTK) at the binding site of ibrutinib, and gain-of-function mutations in phospholipase C γ 2 (PLCG2), a direct downstream target of BTK.69 The reason why some CLL cases are prone to develop mutation-driven resistance to ibrutinib, whereas other cases persist in long-lasting remission, is unknown. The notion that BTK and PLCG2 mutations associate with complex karyotype and with defects of the DNA damage response pathway (ie, TP53 and ATM abnormalities) suggests that genetic instability may be involved in the development of resistance.
Recommendations for clinical practice
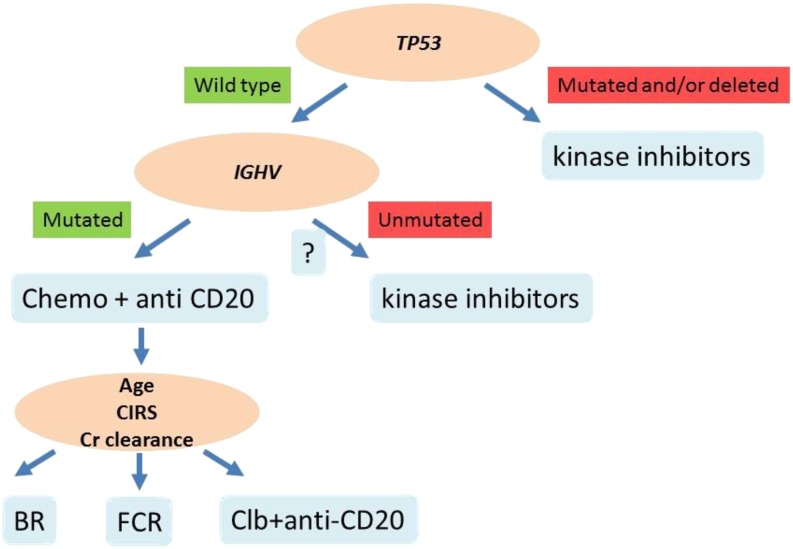
The great activity of FCR in patients with mutated IGHV, and the improvement of outcome among IGHV-unmutated patients when treated with ibrutinib, may announce the emergence of a therapeutic approach that is guided by the IGHV mutational status (Figure 3). Accordingly, the most recent guidelines support IGHV mutation analysis as desirable at the time of treatment requirement, and a consensus has been reached on the minimal technical requirements for a reliable and reproducible analysis of the rearranged IGHV sequences.70-72 Because the IGHV mutation status does not change over time, there is no need to repeat its analysis during disease course.
Figure 3.
Biomarker-informed decision nodes in the management of newly presented CLL. At the time of treatment requirement, the presence of TP53 mutation/deletion represents an indication for treatment with kinase inhibitors, for example, ibrutinib or, alternatively, idelalisib. Patients who require first-line treatment and carry a wild-type TP53 gene may be candidate to chemoimmunotherapy with one of the available combinations, especially in the presence of mutated IGHV genes. In patients with wild-type, a TP53 gene, and unmutated IGHV genes, the role of chemoimmunotherapy is under debate, and kinase inhibitors may represent a valuable option. CIRS, Cumulative Illness Rating Scale.
Given the opportunity to effectively treat TP53-disrupted patients with novel targeted agents, CLL guidelines recommend testing for 17p13 deletion and TP53 mutations in patients requiring therapy (Figure 3). Mutations represent the most frequent form of TP53 inactivation in CLL, and frequently (∼70% of the cases harboring TP53 disruption) pair with the loss of the second TP53 allele through 17p13 deletion. A proportion of patients harbor the sole TP53 mutation without 17p13 deletion, whereas 17p13 deletion in the absence of TP53 mutation is less frequent. The direct clinical implications of such a complex and heterogeneous picture of TP53 defects in CLL are that both FISH cytogenetics and sequencing should be done to have a comprehensive assessment of the TP53 gene status.73 Because leukemic clones may evolve, 17p13 deletion and TP53 mutation analyses should be repeated at each disease progression requiring treatment if it was normal before. Sanger sequencing is the currently recommended approach for TP53 mutation analysis.
Prognostic scores allow very-low-risk patients to be sorted out; these patients have a life expectancy similar to that expected in the normal population and show a low likelihood of progression. On the other hand, prognostic scores also allow for identification of very-high-risk patients who generally progress and shortly succumb to the disease. However, prognostic scores are not 100% accurate (the outcome discrimination capacity is 73% for the CLL-IPI and 68% for the integrated mutational-cytogenetic model) because they all comprise intermediate-risk categories, which are a case mix containing both low- and high-risk patients whose outcome is not captured by the scoring system.27,31 Although scoring systems may be useful for patient counseling and for the definition of the timing of follow-up, they can turn a “watch-and-wait” strategy into a “watch-and-worry” situation in patients without treatment indication. Therefore, at the time of diagnosis, in asymptomatic patients who do not require treatment, current guidelines do not support the need for a full prognostic assessment also incorporating molecular biomarkers such as gene mutations, FISH, and IGHV analysis.
Perspectives
Conventional Sanger sequencing has limited sensitivity and allows for the identification of TP53 mutations represented in at least 10% to 20% of the alleles. Next-generation sequencing is more sensitive than Sanger sequencing for the identification of small TP53-mutated subclones (down to 1%), which occur in a significant fraction of CLL, have the same unfavorable prognostic impact as clonal TP53 defects, and anticipate the development of a chemorefractory phenotype among CLL patients requiring treatment.26,28 However, given the risk of false-positive calls when dealing with the identification of small subclonal mutations by next-generation sequencing, methodologies and analyses should be standardized and harmonized at the international level before incorporating this approach in the clinical setting.
The robustness of NOTCH1 mutations as a biomarker of resistance to rituximab and ofatumumab in CLL, as well as the possibility that obinutuzumab may overcome such resistance, requires independent clinical validation.66-68 Thus, it is premature to recommend NOTCH1 mutation testing routinely in clinical practice.
Treatment-emergent mutations of BTK and PLCG2 are found in 85% of ibrutinib-resistant CLLs and these mutations are detected up to 15 months before clinical relapse.69 Although these observations support BTK and PLCG2 mutations as a sensitive biomarker of ibrutinib resistance, the notion that such mutations can be detected even in CLL patients without clues of clinical relapse may suggest a limited specificity for ibrutinib resistance. Prospective investigations are thus needed before translating BTK and PLCG2 mutations as biomarkers to inform the decision to switch patients from ibrutinib treatment toward alternative options.
Conclusions
The genome of CLL has a strong impact on the clinical outcome of the disease and has revealed important molecular biomarkers. In clinical practice, predictive biomarkers heavily impact on clinician’s choice because they can inform CLL treatment decisions. Currently, the best example is represented by TP53 disruption by gene mutation and/or deletion that associates with chemoimmunotherapy failure and mandates treatment with innovative drugs, including ibrutinib, idelalisib, or venetoclax. The mutation status of IGHV genes represents an additional predictive biomarker, which helps identify patients who may benefit the most from chemoimmunotherapy with FCR. Assessment of these biomarkers at the time of treatment requirement is recommended by most current guidelines for the management of CLL. Other molecular predictors are under investigation, but their application in clinical practice is premature.
Acknowledgments
The authors are thankful to Adalgisa Condoluci for writing assistance.
This work was supported by Special Program Molecular Clinical Oncology 5 × 1000 (no. 10007), Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy; Progetto Ricerca Finalizzata (RF-2011-02349712), Ministero della Salute, Rome, Italy; Ministero dell’Istruzione, dell’Università e della Ricerca–Progetti di Ricerca di Interesse Nazionale (MIUR-PRIN; 2015ZMRFEA_004), Rome, Italy; Swiss Cancer League (grant no. KFS-3746-08-2015), Bern, Switzerland; and Swiss National Science Foundation (grant no. 320030_169670/1), Bern, Switzerland.
References
- 1.Landau DA, Tausch E, Taylor-Weiner AN, et al. . Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puente XS, Beà S, Valdés-Mas R, et al. . Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. [DOI] [PubMed] [Google Scholar]
- 3.Döhner H, Stilgenbauer S, Benner A, et al. . Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Dumitru CD, Shimizu M, et al. . Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein U, Lia M, Crespo M, et al. . The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28-40. [DOI] [PubMed] [Google Scholar]
- 6.Roberts AW, Davids MS, Pagel JM, et al. . Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374(4):311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vardi A, Agathangelidis A, Sutton LA, Ghia P, Rosenquist R, Stamatopoulos K. Immunogenetic studies of chronic lymphocytic leukemia: revelations and speculations about ontogeny and clinical evolution. Cancer Res. 2014;74(16):4211-4216. [DOI] [PubMed] [Google Scholar]
- 8.Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol. 2015;33(33):3968-3971. [DOI] [PubMed] [Google Scholar]
- 9.Minden M. Is it time to redefine prognostic and predictive in oncology? J Clin Oncol. 2016;34(14):1702-1703. [DOI] [PubMed] [Google Scholar]
- 10.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 11.Damle RN, Wasil T, Fais F, et al. . Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840-1847. [PubMed] [Google Scholar]
- 12.Delgado J, Pratt G, Phillips N, et al. . Beta2-microglobulin is a better predictor of treatment-free survival in patients with chronic lymphocytic leukaemia if adjusted according to glomerular filtration rate. Br J Haematol. 2009;145(6):801-805. [DOI] [PubMed] [Google Scholar]
- 13.Haferlach C, Dicker F, Weiss T, et al. . Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49(9):851-859. [DOI] [PubMed] [Google Scholar]
- 14.Shanafelt TD, Rabe KG, Kay NE, et al. . Age at diagnosis and the utility of prognostic testing in patients with chronic lymphocytic leukemia. Cancer. 2010;116(20):4777-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaderi MA, Kanduri M, Buhl AM, et al. . LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia. Haematologica. 2011;96(8):1153-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouillette P, Collins R, Shakhan S, et al. . Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118(11):3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahill N, Sutton LA, Jansson M, et al. . IGHV3-21 gene frequency in a Swedish cohort of patients with newly diagnosed chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2012;12(3):201-206. [DOI] [PubMed] [Google Scholar]
- 18.Pepper C, Majid A, Lin TT, et al. . Defining the prognosis of early stage chronic lymphocytic leukaemia patients. Br J Haematol. 2012;156(4):499-507. [DOI] [PubMed] [Google Scholar]
- 19.Rossi D, Rasi S, Fabbri G, et al. . Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119(2):521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lech-Maranda E, Mlynarski W, Grzybowska-Izydorczyk O, et al. . Polymorphisms of TNF and IL-10 genes and clinical outcome of patients with chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2013;52(3):287-296. [DOI] [PubMed] [Google Scholar]
- 21.Mansouri L, Cahill N, Gunnarsson R, et al. . NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic leukemia. Leukemia. 2013;27(2):512-514. [DOI] [PubMed] [Google Scholar]
- 22.Mansouri L, Grabowski P, Degerman S, et al. . Short telomere length is associated with NOTCH1/SF3B1/TP53 aberrations and poor outcome in newly diagnosed chronic lymphocytic leukemia patients. Am J Hematol. 2013;88(8):647-651. [DOI] [PubMed] [Google Scholar]
- 23.Cortese D, Sutton LA, Cahill N, et al. . On the way towards a ‘CLL prognostic index’: focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a population-based cohort. Leukemia. 2014;28(3):710-713. [DOI] [PubMed] [Google Scholar]
- 24.González-Gascón Y Marín I, Hernández JA, Martín A, et al. Mutation status and immunoglobulin gene rearrangements in patients from northwest and central region of Spain with chronic lymphocytic leukemia. BioMed Res Int. 2014;2014:257517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lionetti M, Fabris S, Cutrona G, et al. . High-throughput sequencing for the identification of NOTCH1 mutations in early stage chronic lymphocytic leukaemia: biological and clinical implications. Br J Haematol. 2014;165(5):629-639. [DOI] [PubMed] [Google Scholar]
- 26.Rossi D, Khiabanian H, Spina V, et al. . Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International CLL-IPI Working Group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17(6):779-790. [DOI] [PubMed] [Google Scholar]
- 28.Nadeu F, Delgado J, Royo C, et al. . Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putowski M, Podgórniak M, Piróg M, et al. . Prognostic impact of NOTCH1, MYD88, and SF3B1 mutations in Polish patients with chronic lymphocytic leukemia. Pol Arch Intern Med. 2017;127(4):238-244. [DOI] [PubMed] [Google Scholar]
- 30.Parikh SA, Strati P, Tsang M, West CP, Shanafelt TD. Should IGHV status and FISH testing be performed in all CLL patients at diagnosis? A systematic review and meta-analysis. Blood. 2016;127(14):1752-1760. [DOI] [PubMed] [Google Scholar]
- 31.Rossi D, Rasi S, Spina V, et al. . Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quesada V, Conde L, Villamor N, et al. . Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47-52. [DOI] [PubMed] [Google Scholar]
- 33.Del Poeta G, Dal Bo M, Del Principe MI, et al. . Clinical significance of c.7544-7545 delCT NOTCH1 mutation in chronic lymphocytic leukemia. Br J Haematol. 2013;160(3):404-418. [DOI] [PubMed] [Google Scholar]
- 34.Villamor N, Conde L, Martínez-Trillos A, et al. . NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia. 2013;27(5):1100-1106. [DOI] [PubMed] [Google Scholar]
- 35.Weissmann S, Roller A, Jeromin S, et al. . Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on 852 patients. Leukemia. 2013;27(12):2393-2396. [DOI] [PubMed] [Google Scholar]
- 36.Willander K, Dutta RK, Ungerbäck J, et al. . NOTCH1 mutations influence survival in chronic lymphocytic leukemia patients. BMC Cancer. 2013;13:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeromin S, Weissmann S, Haferlach C, et al. . SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28(1):108-117. [DOI] [PubMed] [Google Scholar]
- 38.Sportoletti P, Baldoni S, Del Papa B, et al. . A revised NOTCH1 mutation frequency still impacts survival while the allele burden predicts early progression in chronic lymphocytic leukemia. Leukemia. 2014;28(2):436-439. [DOI] [PubMed] [Google Scholar]
- 39.Baliakas P, Hadzidimitriou A, Sutton LA, et al. ; European Research Initiative on CLL (ERIC). Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329-336. [DOI] [PubMed] [Google Scholar]
- 40.Rasi S, Khiabanian H, Ciardullo C, et al. . Clinical impact of small subclones harboring NOTCH1, SF3B1 or BIRC3 mutations in chronic lymphocytic leukemia. Haematologica. 2016;101(4):e135-e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavallini C, Visco C, Putta S, et al. . Integration of B-cell receptor-induced ERK1/2 phosphorylation and mutations of SF3B1 gene refines prognosis in treatment-naïve chronic lymphocytic leukemia. Haematologica. 2017;102(4):e144-e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi D, Rasi S, Spina V, et al. . Different impact of NOTCH1 and SF3B1 mutations on the risk of chronic lymphocytic leukemia transformation to Richter syndrome. Br J Haematol. 2012;158(3):426-429. [DOI] [PubMed] [Google Scholar]
- 43.Stilgenbauer S, Jones JA, Coutre S, et al. . Outcome of ibrutinib treatment by baseline genetic features in patients with relapsed or refractory CLL/SLL with del17p in the Resonate-17 study [abstract]. Blood. 2015;126(23). Abstract 833. [Google Scholar]
- 44.Coutre SE, Leonard JP, Barrientos JC, et al. . Clinical activity of idelalisib (GS-1101), a selective inhibitor of PI3Kδ, in phase 1 and 2 trials in chronic lymphocytic leukemia (CLL): effect of del(17p)/TP53 mutation, del(11q), IGHV mutation, and NOTCH1 mutation [abstract]. Blood. 2013;122(21). Abstract 1632. [Google Scholar]
- 45.Rossi D, Cerri M, Deambrogi C, et al. . The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15(3):995-1004. [DOI] [PubMed] [Google Scholar]
- 46.Byrd JC, Furman RR, Coutre SE, et al. . Three-year follow-up of treatment-naïve and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125(16):2497-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amin NA, Malek SN. Gene mutations in chronic lymphocytic leukemia. Semin Oncol. 2016;43(2):215-221. [DOI] [PubMed] [Google Scholar]
- 48.O’Brien SM, Furman RR, Coutre SE, et al. . Five-year experience with single-agent ibrutinib in patients with previously untreated and relapsed/refractory chronic lymphocytic leukemia/small lymphocytic lymphoma. Blood. 2016;128(22). Abstract 233. [Google Scholar]
- 49.Pflug N, Bahlo J, Shanafelt TD, et al. . Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood. 2014;124(1):49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molica S, Giannarelli D, Mirabelli R, et al. . Unavailability of thymidine kinase does not preclude the use of German comprehensive prognostic index: results of an external validation analysis in early chronic lymphocytic leukemia and comparison with MD Anderson Cancer Center model. Eur J Haematol. 2016;96(1):72-77. [DOI] [PubMed] [Google Scholar]
- 51.Gentile M, Shanafelt TD, Rossi D, et al. . Validation of the CLL-IPI and comparison with the MDACC prognostic index in newly diagnosed patients. Blood. 2016;128(16):2093-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Cunha-Bang C, Christiansen I, Niemann CU. The CLL-IPI applied in a population-based cohort. Blood. 2016;128(17):2181-2183. [DOI] [PubMed] [Google Scholar]
- 53.Delgado J, Doubek M, Baumann T, et al. . Chronic lymphocytic leukemia: a prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am J Hematol. 2017;92(4):375-380. [DOI] [PubMed] [Google Scholar]
- 54.Rossi D, Terzi-di-Bergamo L, De Paoli L, et al. . Molecular prediction of durable remission after first-line fludarabine-cyclophosphamide-rituximab in chronic lymphocytic leukemia. Blood. 2015;126(16):1921-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer K, Bahlo J, Fink AM, et al. . Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127(2):208-215. [DOI] [PubMed] [Google Scholar]
- 56.Thompson PA, Tam CS, O’Brien SM, et al. . Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127(3):303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burger JA, Tedeschi A, Barr PM, et al. ; RESONATE-2 Investigators. Ibrutinib as initial therapy for patients with chronic lymphocytic leukemia. N Engl J Med. 2015;373(25):2425-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hallek M, Fischer K, Fingerle-Rowson G, et al. ; International Group of Investigators; German Chronic Lymphocytic Leukaemia Study Group. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376(9747):1164-1174. [DOI] [PubMed] [Google Scholar]
- 59.Fischer K, Cramer P, Busch R, et al. . Bendamustine in combination with rituximab for previously untreated patients with chronic lymphocytic leukemia: a multicenter phase II trial of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. 2012;30(26):3209-3216. [DOI] [PubMed] [Google Scholar]
- 60.Goede V, Fischer K, Busch R, et al. . Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med. 2014;370(12):1101-1110. [DOI] [PubMed] [Google Scholar]
- 61.Hillmen P, Robak T, Janssens A, et al. ; COMPLEMENT 1 Study Investigators. Chlorambucil plus ofatumumab versus chlorambucil alone in previously untreated patients with chronic lymphocytic leukaemia (COMPLEMENT 1): a randomised, multicentre, open-label phase 3 trial. Lancet. 2015;385(9980):1873-1883. [DOI] [PubMed] [Google Scholar]
- 62.Anderson MA, Deng J, Seymour JF, et al. . The BCL2 selective inhibitor venetoclax induces rapid onset apoptosis of CLL cells in patients via a TP53-independent mechanism. Blood. 2016;127(25):3215-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furman RR, Sharman JP, Coutre SE, et al. . Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;370(11):997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stilgenbauer S, Eichhorst B, Schetelig J, et al. . Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. Lancet Oncol. 2016;17(6):768-778. [DOI] [PubMed] [Google Scholar]
- 65.O’Brien S, Jones JA, Coutre SE, et al. . Ibrutinib for patients with relapsed or refractory chronic lymphocytic leukaemia with 17p deletion (RESONATE-17): a phase 2, open-label, multicentre study. Lancet Oncol. 2016;17(10):1409-1418. [DOI] [PubMed] [Google Scholar]
- 66.Stilgenbauer S, Schnaiter A, Paschka P, et al. . Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247-3254. [DOI] [PubMed] [Google Scholar]
- 67.Tausch E, Beck P, Schlenk RF, et al. . NOTCH1 mutation and treatment outcome in CLL patients treated with chlorambucil (Chl) or ofatumumab-Chl (O-Chl): results from the phase III study complement 1 (OMB110911) [abstract]. Blood. 2013;122(21). Abstract 527. [Google Scholar]
- 68.Estenfelder S, Tausch E, Robrecht S, et al. . Gene mutations and treatment outcome in the context of chlorambucil (Clb) without or with the addition of rituximab (R) or obinutuzumab (GA-101, G): results of an extensive analysis of the phase III study CLL11 of the German CLL Study Group [abstract]. Blood. 2016;128(22). Abstract 3227. [Google Scholar]
- 69.Woyach JA, Ruppert AS, Guinn D, et al. . BTK(C481S)-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35(13):1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eichhorst B, Robak T, Montserrat E, et al. ; ESMO Guidelines Committee. Chronic lymphocytic leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(suppl 5):v78-v84. [DOI] [PubMed] [Google Scholar]
- 71.Zelenetz AD, Gordon LI, Wierda WG, et al. ; National comprehension cancer network. Chronic lymphocytic leukemia/small lymphocytic lymphoma, version 1.2015. J Natl Compr Canc Netw. 2015;13(3):326-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenquist R, Ghia P, Hadzidimitriou A, et al. . Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia. 2017;31(7):1477-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pospisilova S, Sutton LA, Malcikova J, et al. ; European Research Initiative on CLL (ERIC). Innovation in the prognostication of chronic lymphocytic leukemia: how far beyond TP53 gene analysis can we go? Haematologica. 2016;101(3):263-265. [DOI] [PMC free article] [PubMed] [Google Scholar]