Abstract
The aging hematopoietic system undergoes numerous changes, including reduced production of red blood cells and lymphocytes as well as a relative increase in the production of myeloid cells. Emerging evidence indicates that many of these changes are due to selection pressures from cell-intrinsic and cell-extrinsic factors that result in clonal shifts in the hematopoietic stem cell (HSC) pool, resulting in predominant HSC clones that exhibit the functional characteristics associated with HSC aging. Given the recent descriptions of clonal hematopoiesis in aged populations, the increased risk of developing hematologic malignancies in individuals with clonal hematopoiesis, and the many similarities in hematopoietic aging and acquired bone marrow failure (BMF) syndromes, such as myelodysplastic syndromes (MDS), this raises significant questions regarding the relationship between aging hematopoiesis and MDS, including the factors that regulate HSC aging, whether clonal hematopoiesis is required for the development of MDS, and even whether BMF is an inevitable consequence of aging. In this article, we will review our current understanding of these processes and the potential intersections among them.
Learning Objectives
To understand the evidence for, and limitations of, the clonal selection model of hematopoietic stem cell aging
To understand the potential impact of clonal hematopoiesis during aging on the development of bone marrow failure syndromes, such as MDS
Introduction
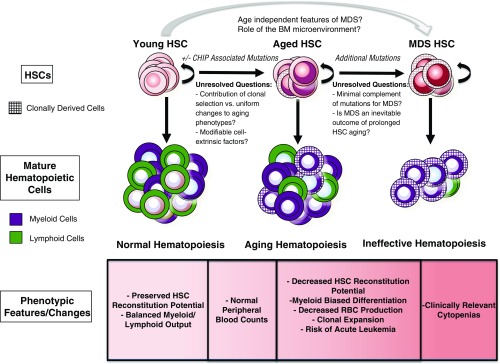
Alterations in the aging hematopoietic system have long been the subject of intense investigation. These studies have cataloged numerous changes that occur in both humans and mice, including reduced production of red blood cells and lymphocytes as well as a relative increase in the production of myeloid cells.1 These changes in hematopoietic potential are associated with numerous alterations in the bone marrow (BM), including decreasing cellularity, altered chemokine/cytokine levels, and alterations in the composition and architecture of the nonhematopoietic cells that make up the BM microenvironment.2 In addition, immunophenotypically defined hematopoietic stem cells (HSCs) are increased in number, presumably to compensate for a concomitant decline in their output on a per-cell basis to maintain blood production.3,4 The characterization of HSC cellular and transcriptomal changes during aging has revealed alterations in cell cycle distribution,5 response to DNA damage,6,7 and gene expression.3,4 Intriguingly, the changes observed during hematopoietic aging are qualitatively similar to those that occur in the setting of the acquired bone marrow failure (BMF) syndromes that also arise in the elderly, in particular the myelodysplastic syndromes (MDS). However, unlike in MDS, normal hematopoietic aging is not associated with changes that are sufficient to produce overt clinical manifestations (Figure 1).
Figure 1.
The features of HSCs in the context of aging and MDS are shown. With normal aging, there is an increase in the risk of the development of CHIP, but the contributions of and requirement for CHIP in aging phenotypes remain incompletely understood. Many functional characteristics of aged HSCs are accentuated in MDS, but the molecular events necessary to transition from CHIP to the clinically significant cytopenias seen in MDS remain unclear.
This discussion regarding the relationship between aging hematopoiesis and BMF will focus on MDS, which is nearly universally associated with alterations in HSC function, lineage bias, and reductions in the production of red blood cells, lymphocytes, platelets, and/or neutrophils, with qualitative changes in HSC function, lineage bias, and red blood cell and lymphocyte production also observed in aging individuals without MDS.8 Acquired aplastic anemia and most of the inherited aplastic anemia syndromes, such as Fanconi anemia (FA) are not typically associated with significant lineage skewing and variably feature increased apoptosis and decreased numbers of hematopoietic stem and progenitor cells (HSPCs). They are also associated with unique cell-intrinsic defects that affect DNA damage responses (eg, FA),9 telomere attrition (eg, dyskeratosis congenita),10 aberrant immune recognition of HSPCs (eg, acquired aplastic anemia),11 or decreased ribosomal biogenesis (eg, Diamond-Blackfan anemia and Shwachman-Diamond syndrome).12 Notably, in contrast to MDS, these disorders present clinically in younger individuals. One might speculate that the different phenotypes of BMF disorders that present in younger vs older individuals may reflect age-dependent differences in HSC function, because HSCs from children and young adults typically show more balanced lymphomyeloid differentiation, whereas HSCs from older individuals show more myeloid-biased differentiation. Thus, perhaps it is not surprising that inherited BMF syndromes may affect all lineages, whereas age-related BMFs may preferentially affect the myeloid lineage. Given the diversity of molecular mechanisms that drive the various BMF disorders, we will focus our discussion on those most associated with aging, in particular MDS.
Current concepts in HSC aging
Current thinking about the relationships among aging, hematopoiesis, and MDS is influenced by recent advances in our understanding of the mechanisms of HSC aging. One of the most important ideas to emerge recently is that aging hematopoiesis is the result of clonal selection at the level of the HSC. Previous views of hematopoietic aging held that HSCs undergo uniform changes in lineage output with age, but more recent studies support an alternative model, in which a pool of HSCs that are heterogeneous with respect to their self-renewal and differentiation capacities at birth are clonally selected over time.13 Although these selection pressures are many and incompletely understood, it is clear that they must regulate HSC-intrinsic and -extrinsic mechanisms of self-renewal and responses to stress. Under these selective pressures, the composition of HSCs changes with age, resulting in a net change in hematopoietic output. This model is supported by single-cell and limiting dilution experiments that have shown that HSCs may exhibit myeloid-biased (My-HSCs), lymphoid-biased, or balanced lymphomyeloid differentiation, as has been shown in the mouse and human hematopoietic system.14 Thus, aging hematopoiesis may be viewed as the net result of many selection pressures that promote alterations in the composition of HSC clones endowed with relatively stable functional characteristics.
But what mechanisms drive clonal selection in HSCs? One of the best-known molecular phenomena associated with reduced HSC function in older individuals is telomere attrition,10 but the degree of telomere shortening is not uniform in all humans, which suggests a possible explanation for individual differences in hematopoietic aging. Aging HSCs in both mice and humans also exhibit broad changes in their methylomes, and loss-of-function mutations in the regulators of DNA methylation, such as DNMT3A and TET2, which are also observed in clonal hematopoiesis in otherwise healthy aging individuals, are associated with enhanced HSC self-renewal and/or myeloid lineage bias,15-17 thereby providing a potential mechanism of clonal selection and age-associated lineage skewing. Although all of these molecular changes are likely to contribute to HSC aging, their described phenotypes do not explain all the changes observed in the aging hematopoietic system, indicating that more diverse and complex mechanisms are involved. Indeed, gene expression profiling studies of old HSCs have demonstrated altered expression of transcripts that encode components of the epigenetic machinery as well as an enrichment for transcripts that promote myeloid differentiation,18 likely reflecting an expansion of the My-HSC pool and a shift in the clonal composition of HSCs. Because these studies evaluated pools of HSCs, however, it is not clear whether the altered gene expression reflects the expansion of a preexisting clone, the appearance of a novel clone with unique self-renewal and differentiation properties, or a qualitative change that occurs more broadly in all aging HSCs.
The events leading to the establishment of functionally diverse HSCs are poorly understood, but it has been suggested that the number of HSC clones is limited by the number of HSCs that expand from birth to adolescence.19 Thus, selection of HSC clones with specific functional properties may be a common event during the lifetime of most individuals. However, it is unclear if HSC clone properties are fixed or if they diversify as the number of HSCs expands. Although this is not an easy question to address, emerging technologies, such as genetic barcoding (eg, lentivirus or transposon-based), and single-cell genomic and transcriptomic studies have demonstrated the potential to capture the transcriptional diversification that occurs in individual HSCs as they differentiate,20 and these approaches may also allow for the evaluation of individual clones and their functional and molecular changes during aging. At this time, however, most data support the clonal shift model of hematopoietic aging, namely, that selection allows HSC clones with fixed characteristics to dominate over time. Although studies of mouse HSCs indicate that individual HSCs can retain their engraftment and lineage potentials even with serial transplantation, such studies of HSCs from various developmental stages in mice (eg, fetal liver, newborn, or old adult) and humans (eg, cord blood and young and old adults) have only been performed at the population and not the single-cell level. Performing such studies by using human HSCs will represent a significant challenge due to both technical and resource limitations for performing single-cell HSC xenotransplants. With the advent of improved models of xenografting, including increasingly “humanized” mice harboring ectopic human marrow niches21 or human hematopoietic cytokines and growth factors,22 such studies may be possible in the near future.
Role of the bone marrow microenvironment
It is becoming increasingly clear that alterations in the aging BM microenvironment can induce alterations in hematopoiesis. Mice with genetically engineered stromal components that mimic the features of aging exhibit features of hematopoiesis reminiscent of aging, such as decreased HSC self-renewal and increased cell cycling.2 Additionally, numerous studies have described alterations in chemokine/cytokine levels with age,23 many of which are thought to regulate HSPC lineage skewing, predominantly toward the myeloid lineage, as well as HSC self-renewal. Changes in the overall function of the aging HSC pool may be the result of clonal shifts due to relative loss of self-renewal in normal HSCs or positive selection for mutant and/or lineage-biased HSCs during aging. Intriguingly, changes in the BM microenvironment may be intrinsic to BM stromal cells in MDS patients through the acquisition of genetic abnormalities,24,25 whereas in other cases, such changes may be the result of intercommunication between hematopoietic cells derived from neoplastic clones and the microenvironment. Recent evidence from both mouse models as well as the use of primary human cells derived from MDS patients also support the role of hematopoietic cells derived from neoplastic HSPCs in modifying the niche by inducing changes in the stroma, thus completing a positive feedback loop enforcing selection pressures that favor the expansion of neoplastic HSC clones.26,27
In addition to the increased risk of MDS and hematologic malignancies with age, another phenomenon associated with HSC aging is the shift to myeloid biased differentiation. An important consequence of this lineage shift is an increase in myeloid-derived cells mediating innate immunity at the expense of lymphoid-derived cells that mediate the adaptive immune response, leading to reduced B- and T-cell diversity and decreased memory immune responses.28 The outcome of this process, also referred to as immunosenescence or immune remodeling, is decreased cellular immunity and increased systemic inflammation, which both have major implications for human diseases with significant inflammatory components, such as cancer, atherosclerosis, autoimmune disease, and neurodegenerative disease.29 Such alterations in hematopoietic cell differentiation/activation may also feedback to alter hematopoietic differentiation in the BM, as some have suggested.30 Although promoting HSC differentiation into innate immune cells might provide an evolutionary advantage by providing protection against bacterial infections, the long-term deleterious effects of increased inflammation and impaired cellular immunity would be expected to increase the risk of developing chronic inflammatory disorders. Indeed, this hypothesis is supported by recent studies in which patients with clonal hematopoiesis with mutations in genes, including DNMT3A, TET2, ASXL1, and JAK2, exhibit increased all-cause mortality largely attributable to vascular disease,31-33 and hypercholesteremia-prone mice engrafted with BM from Tet2-deleted mice exhibit increased atherosclerosis.32,34 The drive to form myeloid cells may not only arise from exposure to infectious organisms, but may also be due to alterations in the gut and skin microbiome that occur with age, much of which is influenced by modifiable behaviors, such as diet and exercise.35 Thus, the possible extrinsic selection pressures on HSCs are many, but it remains unclear in humans whether these pressures predominantly act to drive the expansion of My-HSC clones or to promote myeloid-biased differentiation in all HSCs.
Clonal hematopoiesis in aging
Several recent studies have strongly suggested that a significant proportion of HSCs in healthy aging individuals are likely to be derived from HSC clones that have acquired somatic mutations that also are associated with hematologic malignancies, such as MDS and acute myeloid leukemia (AML).36 In these studies, whole-exome sequencing analyses of normal human blood samples revealed evidence for clonally derived hematopoiesis, as evidenced by the presence of detectable somatic mutations, at low frequencies among patients <45 years of age, with a significant increase among patients >70 years of age. Although clonal hematopoiesis during aging was previously appreciated based on nonrandom inactivation of the X chromosome,36 the frequent observation of clonal hematopoiesis in the general population not only raises the question of how such mutations relate to the development of age-related hematologic cancers, such as AML and MDS, but also whether such mutations are sufficient or necessary to promote hematopoietic aging.37 These uncertainties are captured in the proposed term for this phenomenon, clonal hematopoiesis of indeterminate potential (CHIP).38 It is important to note that although clonal hematopoiesis in the healthy aging population likely originates from HSCs, this has not been formally demonstrated by direct evaluation of functionally characterized purified HSCs. Interestingly, the presence of a detectable clone in the peripheral blood was associated with an increased risk of developing a hematologic cancer on the order of 1% per year,31,33 which is similar to the risk of monoclonal B-cell lymphocytosis and monoclonal gammopathy of undetermined significance giving rise to B-cell or plasma cell neoplasms, respectively, but overall, the appearance of a clone was infrequently linked to clinically relevant alterations in hematopoiesis. Thus, BMF, if defined as the inability to maintain normal blood counts, does not appear to be an inevitable consequence of aging.
Although these studies do not exclude the possibility that acquired somatic mutations may contribute to hematopoietic aging in some individuals, they do not address why some individuals without mutations may exhibit hematopoietic aging phenotypes, or why individuals with clonal hematopoiesis may not exhibit aging phenotypes. Ultimately, these studies were unlikely to capture the full clonal composition of the HSC pool, because they relied on exome sequencing of limited depth performed on peripheral blood cells, and they could only discriminate a limited number of mutations associated with myeloid malignancies due to a lack of germ line controls. Furthermore, the correlation of these mutations with steady-state blood counts was unlikely to reflect subtler functional changes in HSPCs, including number, self-renewal, or lineage potential. Future studies that use whole-exome sequencing, or even whole-genome sequencing, with nonhematopoietic tissue germ line controls (eg, buccal mucosa or fingernails) may improve our understanding of the contribution of genetic changes to hematopoietic aging, and studies directly comparing the lineage potential and self-renewal potential of mutation-bearing HSPCs to nonmutant HSPCs will provide critical information regarding the possible differences and similarities in HSCs. Such studies might include clonal assays, especially single-cell in vivo transplantation assays, coupled with direct evaluation of the genetic status of such clones. Methods that allow prospective separation of mutation-bearing HSPCs, similar to that developed for the purification of preleukemic HSCs from AML blasts in AML patients,39-41 might significantly expedite such studies by circumventing the need for single-cell assays. Finally, understanding these differences may have significant implications for human health beyond MDS, because clonal hematopoiesis is associated with an increase in all-cause mortality,31,33 which may be driven in large part by an increased risk for cardiovascular atherosclerotic disease.32,34
An important observation has been that the vast majority of individuals with clonal hematopoiesis will not develop MDS or other hematologic malignancies. Thus, the genetic makeup of an individual plays a major modifying role in the aging hematopoietic phenotype as well as the risk of developing MDS, with the latter likely due to the random acquisition of additional somatic mutations in HSCs over time, as has been proposed for human cancers in general.42 This speculation is congruous with investigations of age-related changes in various mouse strains, such as C57BL/J, CBA, DBA/2, and BALB/c, with studies showing dramatic differences in HSC number and function between strains.13 Thus, frequently cited changes in the aging hematopoietic system, including the reduced frequency of functional HSCs, as determined by limiting dilution reconstitution studies in C57BL/6J mice may not faithfully capture the heterogeneity in human hematopoietic aging phenotypes. Although the physiologic basis of differences in HSC aging and the influence of clonal hematopoiesis is unclear at this time, important factors likely include differences in HSC self-renewal, lineage commitment, and/or responses to environmental stimuli and cellular stress. Understanding the contributions of each of these factors will be the focus of many future investigations.
Based on work in AML and the development of methods to prospectively isolate preleukemic HSCs,39-41 we can envision a model in which the most competitive HSC clones may be predisposed to oncogenic transformation because they have been selected for their ability to self-renew under genotoxic stress or other stress conditions that occur with aging. These features may be acquired at the expense of the ability to differentiate and may thus explain the increasing incidence of clinically relevant BMF syndromes with age. Patients with clonal hematopoiesis do appear to have an increased risk for developing a therapy-related myeloid neoplasm following chemotherapy,43-45 suggesting that clonal hematopoiesis may contribute to disease initiation under exogenous stressors that perhaps mimic an accelerated form of aging. Indeed, HSCs appear to use unique DNA repair pathways as compared with more committed progenitors in response to DNA damage,46 and these mechanisms also appear to differ by age.6,7
Considerations for the future
Although the mechanisms that regulate HSC aging are complex, much attention is currently being devoted to determining whether the development of clonal hematopoiesis is necessary to promote age-related hematopoietic changes. It is important to note that although clonal hematopoiesis is observed with increasing frequency with age, not all aging individuals develop BMF or other hematologic malignancies. The reasons for this are not clear, but likely involve interactions between cell-intrinsic and microenvironmental mechanisms as well as the genetic/epigenetic factors that regulate them. For example, given the emerging evidence for the microenvironment as a regulator of many aspects of HSC function, it would be interesting to determine the extent to which the microenvironment plays a modifying role in hematopoietic aging. Although these studies could certainly be performed in mice, it will be important to conduct similar studies in humans to interpret the relevance of mouse studies to human health. Such studies might include a closer examination of hematopoiesis in commonly encountered clinical contexts, such as in patients with chronic inflammatory illnesses or after bone marrow transplantation using donors and/or recipients of varying ages. Finally, it should be noted that the changes in the aging hematopoietic system and the MDS marrow are likely interdependent. For example, the decline in the production of mature T and B lymphocytes during aging is likely to alter the inflammatory environment observed in the BM microenvironment in the context of both aging and MDS.28,47
Although much of our discussion and the scientific literature view hematopoietic aging from the viewpoint of the HSC, which represents the most obvious unit of selection for evolutionary pressure, it is not entirely clear whether the phenotypic manifestations of hematopoietic aging might also reflect alterations in other more differentiated hematopoietic cell types. For example, in the evaluation of primary MDS BM HSPCs, we and others have shown that they exhibit consistent alterations in myeloid progenitor composition.48-50 It may even be the case that, in some situations, aging and MDS phenotypes are not the result of changes in HSCs, but rather changes in committed progenitors that exhibit altered function, whether intrinsic or in response to environmental cues. For example, although the decreased red blood cell mass observed in MDS patients or normal elderly individuals may be expected to promote erythroid development through the production of erythropoietin (EPO), this feedback mechanism may not exert significant effects on HSCs, because they may not sense and respond to EPO directly.51 Similarly, particular HSC clones or their progeny may exhibit an increased ability to resist genomic insults, the ability to expand in response to systemic inflammatory signals, or the ability to compete against other HSCs for finite numbers of specialized niches. Such clones might be selected for over time, and such a shift may also result in the expansion of HSC clones that exhibit alterations in differentiation or proliferation capacities. Based on this model, it is not difficult to envision a direct ancestral relationship between aging HSCs and the clones observed in CHIP. Given the recent demonstrations that early driver mutations in mature non-Hodgkin lymphomas can arise in HSCs,52 this suggests that similar selection pressures on HSCs during aging may promote the development of many hematologic cancers in addition to the myeloid malignancies, a speculation that is supported by the development of both myeloid and lymphoid malignancies after the development of CHIP.31,33
Given the potential health benefits of inhibiting or reversing HSC aging, numerous investigators have explored this possibility. Proof-of-concept studies for such therapeutic interventions have shown partial reversal of aging HSC phenotypes by pharmacological targeting of dysregulated genes and elevated reactive oxygen species that characterize aging HSCs.53 Thus, it would be interesting to test whether such interventions might similarly inhibit the development or severity of MDS phenotypes or aging hematopoiesis, both in the context of somatic mutations and without. It is unclear whether such interventions would exert direct or indirect selective effects on mutant or nonmutant aged HSPCs, or if such interventions would temporarily or durably inhibit aging in the hematopoietic system. These therapeutic strategies, even if only partially effective, would hold the promise of improving the health of large numbers of people by increasing red blood cell production, improving cellular immunity, and decreasing the sequelae of the systemic inflammation that accompanies aging.
This is an exciting time in hematopoietic aging and MDS research. Understanding the connections between CHIP and MDS pathogenesis will have significant ramifications for the development of novel approaches to predict which patients may be at risk of developing MDS as well as other age-related hematologic diseases or other chronic diseases associated with inflammation. Similar to normal aging, MDS is multigenic and multifactorial, with both cell-intrinsic and cell-extrinsic contributions to pathogenesis. We expect the application of novel genomic and experimental genetic techniques will for the first time allow us to track individual HSC clones and to define the molecular basis of their different functional properties, and hopefully more investigations will be performed in normal aging individuals and MDS patients to better understand the ramifications of the results generated in inbred mouse strains. We expect these investigations will further reveal the mechanistic bases of HSC aging, identify those factors that are necessary and/or sufficient to drive clonal selection in the context of acquired somatic mutations, and provide new opportunities to both diagnose and treat MDS and other age-associated disorders.
Acknowledgments
This work was supported by grants from the EvansMDS Foundation (C.Y.P.), the Taub Foundation Grants Program for MDS Research (C.Y.P.), the Starr Cancer Consortium (C.Y.P.), the Tri-Institutional Stem Cell Initiative (C.Y.P.), a Leukemia and Lymphoma Society Scholar Award (C.Y.P.), an American Society of Hematology Fellow Scholar Award (S.S.C.), National Institutes of Health K08 Clinical Investigator Award 1K08CA194275 (S.S.C.), and an Early Career Award from the Dresner Foundation (S.S.C.).
References
- 1.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13(5):376-389. [DOI] [PubMed] [Google Scholar]
- 2.Latchney SE, Calvi LM. The aging hematopoietic stem cell niche: Phenotypic and functional changes and mechanisms that contribute to hematopoietic aging. Semin Hematol. 2017;54(1):25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi DJ, Bryder D, Zahn JM, et al. . Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102(26):9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang WW, Price EA, Sahoo D, et al. . Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA. 2011;108(50):20012-20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996;2(9):1011-1016. [DOI] [PubMed] [Google Scholar]
- 6.Flach J, Bakker ST, Mohrin M, et al. . Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15(1):37-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makipour S, Kanapuru B, Ershler WB. Unexplained anemia in the elderly. Semin Hematol. 2008;45(4):250-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23-34. [DOI] [PubMed] [Google Scholar]
- 10.Townsley DM, Dumitriu B, Young NS. Bone marrow failure and the telomeropathies. Blood. 2014;124(18):2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young NS. Current concepts in the pathophysiology and treatment of aplastic anemia. Hematology Am Soc Hematol Educ Program. 2013;2013:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park CY. Hematopoiesis in aging: Current concepts and challenges. Semin Hematol. 2017;54(1):1-3. [DOI] [PubMed] [Google Scholar]
- 14.Biasco L, Pellin D, Scala S, et al. . In vivo tracking of human hematopoiesis reveals patterns of clonal dynamics during early and steady-state reconstitution phases. Cell Stem Cell. 2016;19(1):107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran-Crusio K, Reavie L, Shih A, et al. . Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Challen GA, Sun D, Mayle A, et al. . Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15(3):350-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Luo M, Jeong M, et al. . Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14(5):673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elias HK, Bryder D, Park CY. Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin Hematol. 2017;54(1):4-11. [DOI] [PubMed] [Google Scholar]
- 19.Crisan M, Dzierzak E. The many faces of hematopoietic stem cell heterogeneity. Development. 2016;143(24):4571-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson NK, Kent DG, Buettner F, et al. . Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell Populations. Cell Stem Cell. 2015;16(6):712-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinisch A, Thomas D, Corces MR, et al. . A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22(7):812-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theocharides AP, Rongvaux A, Fritsch K, Flavell RA, Manz MG. Humanized hemato-lymphoid system mice. Haematologica. 2016;101(1):5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ergen AV, Boles NC, Goodell MA. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 2012;119(11):2500-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores-Figueroa E, Arana-Trejo RM, Gutiérrez-Espíndola G, Pérez-Cabrera A, Mayani H. Mesenchymal stem cells in myelodysplastic syndromes: phenotypic and cytogenetic characterization. Leuk Res. 2005;29(2):215-224. [DOI] [PubMed] [Google Scholar]
- 25.Blau O, Baldus CD, Hofmann WK, et al. . Mesenchymal stromal cells of myelodysplastic syndrome and acute myeloid leukemia patients have distinct genetic abnormalities compared with leukemic blasts. Blood. 2011;118(20):5583-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvi LM, Adams GB, Weibrecht KW, et al. . Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841-846. [DOI] [PubMed] [Google Scholar]
- 27.Medyouf H, Mossner M, Jann JC, et al. . Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14(6):824-837. [DOI] [PubMed] [Google Scholar]
- 28.Yanes RE, Gustafson CE, Weyand CM, Goronzy JJ. Lymphocyte generation and population homeostasis throughout life. Semin Hematol. 2017;54(1):33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denkinger MD, Leins H, Schirmbeck R, Florian MC, Geiger H. HSC aging and senescent immune remodeling. Trends Immunol. 2015;36(12):815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11(10):685-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal S, Fontanillas P, Flannick J, et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaiswal S, Natarajan P, Silver AJ, et al. . Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genovese G, Kähler AK, Handsaker RE, et al. . Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. . Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156(3):408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jan M, Ebert BL, Jaiswal S. Clonal hematopoiesis. Semin Hematol. 2017;54(1):43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, Jasper H, Rudolph KL. Aging-induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell. 2015;16(6):601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jan M, Snyder TM, Corces-Zimmerman MR, et al. . Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung SS, Eng WS, Hu W, et al. . CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci Transl Med. 2017;9(374):eaaj2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jan M, Chao MP, Cha AC, et al. . Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci USA. 2011;108(12):5009-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillis NK, Ball M, Zhang Q, et al. . Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18(1):112-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong TN, Ramsingh G, Young AL, et al. . Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coombs CC, Zehir A, Devlin SM, et al. . Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374-382.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohrin M, Bourke E, Alexander D, et al. . Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovtonyuk LV, Fritsch K, Feng X, Manz MG, Takizawa H. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front Immunol. 2016;7:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang WW, Pluvinage JV, Price EA, et al. . Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci USA. 2013;110(8):3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Will B, Zhou L, Vogler TO, et al. . Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120(10):2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woll PS, Kjällquist U, Chowdhury O, et al. . Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25(6):794-808. [DOI] [PubMed] [Google Scholar]
- 51.Gibbs KD Jr, Gilbert PM, Sachs K, et al. . Single-cell phospho-specific flow cytometric analysis demonstrates biochemical and functional heterogeneity in human hematopoietic stem and progenitor compartments. Blood. 2011;117(16):4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damm F, Mylonas E, Cosson A, et al. . Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4(9):1088-1101. [DOI] [PubMed] [Google Scholar]
- 53.Guidi N, Geiger H. Rejuvenation of aged hematopoietic stem cells. Semin Hematol. 2017;54(1):51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]