Abstract
Myelodysplastic syndrome (MDS) is a clinically heterogeneous disease characterized by functional impairment of hematopoiesis and abnormal bone marrow morphology. The type and severity of hematopoietic dysfunction in MDS are highly variable, and the kinetics of disease progression are difficult to predict. Genomic studies have shown that MDS is typically driven by a multistep somatic genetic process affecting a core set of genes. By definition, recurrent MDS driver mutations all drive clonal dominance, although they can have stereotyped positions in the clonal hierarchy or patterns of comutation association and exclusivity. Furthermore, environmental context, such as exposures to cytotoxic chemotherapy or the presence of germ-line predisposition, can influence disease pathogenesis and clinical outcomes. This review will address how an enhanced understanding of MDS genetics may enable refinement of current diagnostic schema, improve understanding of the pathogenesis of therapy-related MDS, and identify germ-line predispositions to development of MDS that are more common than recognized by standard clinical evaluation.
Learning Objectives
Understand that genetic characteristics may enable refinement of current classification schema by improving the distinction between MDS and ICUS and between MDS and AML
Understand that leukemic transformation of MDS is characterized by clonal genetic evolution, often affecting genes involved in the RAS pathway
Understand that therapy-related MDS is highly associated with TP53 and PPM1D mutations, which impair the cellular stress response, and that t-MDS without TP53 or PPM1D mutations is genetically similar to de novo MDS
Understand that inherited mutations that predispose to development of MDS, such as GATA2, SBDS, TERT, and TERC, are more common than recognized by standard clinical evaluation and may have prognostic significance
Introduction
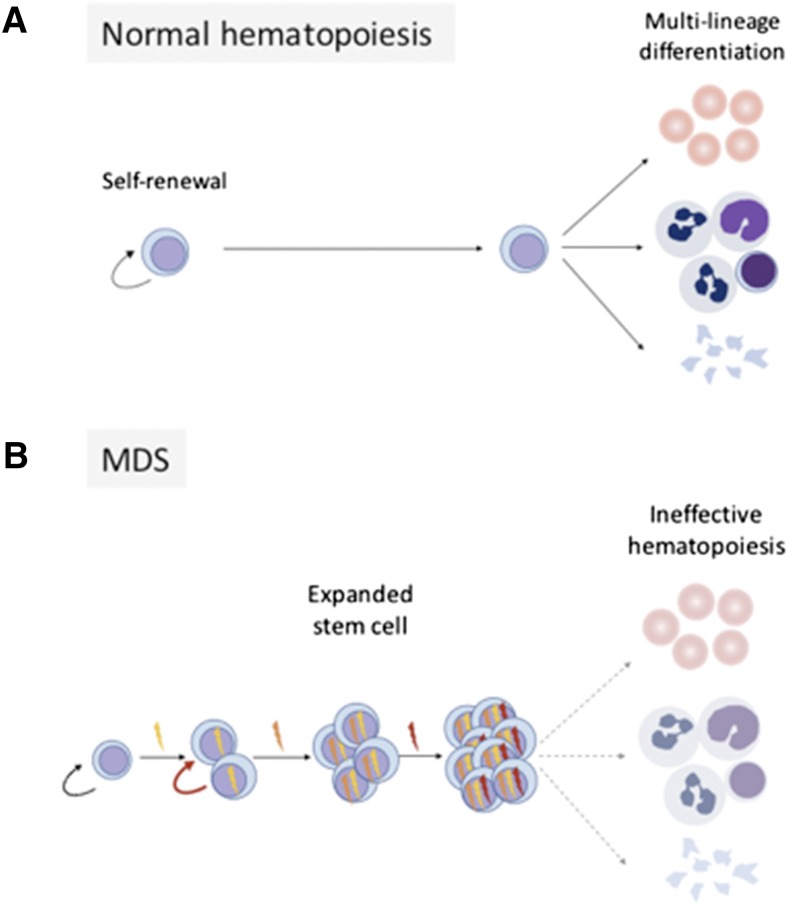
Peripheral blood derives from the hematopoietic stem cell (HSC), which is defined by its dual capacities for sustained self-renewal and multilineage differentiation (Figure 1). Normally, a diverse pool of HSCs is maintained in homeostatic balance and contributes to polyclonal hematopoiesis. A pathogenic mutation in one of a small subset of genes can endow a single stem cell with a competitive advantage over neighboring cells and drive clonal expansion. However, clonality alone is not sufficient to cause or diagnose disease, because even densely clonal hematopoiesis can remain functionally intact.1,2 Instead, a diagnosis of myelodysplastic syndrome (MDS) requires identification of peripheral cytopenias and characteristic bone marrow morphologic abnormalities.
Figure 1.
MDS gene mutations corrupt normal hematopoiesis. Panel A shows normal hematopoiesis, where HSCs possess the capacity for self-renewal and multilineage differentation. Panel B shows that serial acquisition of somatic mutations causes clonal stem cell expansion and impaired differentation.
Recent MDS-focused sequencing studies have shown concordant core findings.3-6 MDS is typically driven by a multistep genetic process characterized by recurrent mutations affecting basic cellular pathways, including RNA splicing, epigenome regulation, myeloid transcriptional coordination, DNA damage and stress responses, and growth factor signaling. Despite the high number of recurrently mutated genes and heterogeneity of affected pathways, myeloid driver mutations share a fundamental biological property: they all have the potential to cause clonal dominance at the stem cell level. The diversity of clinical MDS phenotypes associated with specific mutations may be attributable to differential coregulation of the HSC self-renewal program and lineage-specific differentiation programs.
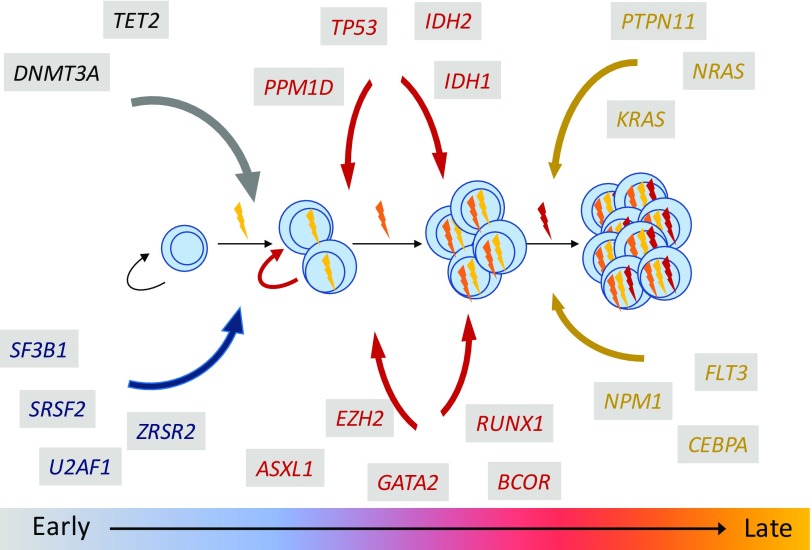
The impact of gene mutations on MDS pathobiology can be informed by genetic, temporal, or environmental contexts. Mutations do not manifest in random order or random combinations. Instead, individual mutations can have highly stereotyped positions in the clonal hierarchy and strong patterns of comutation association and exclusivity (Figure 2). Moreover, environmental context, such as leukemogenic exposures or germ-line predisposition syndromes, can influence disease pathogenesis and clinical outcomes. Because clinical sequencing platforms are deployed more broadly in practice, it is imperative to define the core regulatory logic of MDS genetics; to integrate clinical, morphologic, and genetic characteristics into a new MDS diagnostic paradigm; and to understand how MDS clonal genetic architecture may predict clinical evolution and therapeutic response. Recent MDS genetic studies have yielded key insights into our understanding of how gene mutations cooperate to initiate and propagate disease.
Figure 2.
Gene mutations have stereotyped positions in the MDS clonal hierarchy.
Genetics and the evolving definition of MDS
The evolving understanding of myeloid genetics has challenged our historical definition of MDS.
Does a diagnosis of MDS require morphologic dysplasia? Where does MDS end and acute myeloid leukemia (AML) begin? Genetic studies have begun to delineate the boundaries of MDS more precisely.
MDS and its distinction from idiopathic cytopenia of undetermined significance and clonal cytopenia of undetermined signficance
At present, a definitive diagnosis of MDS in a patient with cytopenias requires demonstration of characteristic morphologic changes or MDS-defining cytogenetic abnormalities. However, many such patients have a normal karyotype and lack the most distinctive pathologic features of MDS, such as ring sideroblasts or an excess of myeloid blasts. In these cases, it may be difficult to exclude or establish a diagnosis of MDS, and pathologists rely on more subjective identification and quantitation of dysplastic bone marrow elements. The term idiopathic cytopenia of undetermined significance (ICUS) was established to describe patients with persistent unexplained cytopenias that do not meet diagnostic criteria for MDS.
ICUS clearly represents a heterogeneous clinical category, including many patients who will never develop a myeloid neoplasm and other patients who may rapidly manifest frank disease. Indeed, it was recognized that a subset of patients with ICUS has MDS-associated somatic mutations and that these patients may share genetic and clinical characteristics with bona fide MDS.7,8 The distinct natural history of patients with clonal vs nonclonal cytopenias has recently been explored in a large cohort of patients presenting for clinical evaluation of unexplained cytopenias.9 Using gene panel testing, it was shown that ICUS could be segregated by molecular genetic profile into subgroups with distinct outcomes or likelihood of clinical progression. Not surprisingly, patients with clonal ICUS had a much higher rate of progression than patients with nonclonal ICUS. However, not all clonality was equivalent. The group of patients with mutations affecting RNA splicing (SF3B1, SRSF2, and U2AF1) and those with TET2, DNMT3A, or ASXL1 mutations plus additional gene mutations had clinical characteristics that were very similar to those of low-risk MDS patients, including older age, male bias, worse overall survival, and increased risk of disease progression. By contrast, those with mutations affecting TET2, DNMT3A, or ASXL1 alone had slower kinetics of progression unless paired with additional myeloid driver mutations. These results are consistent with studies showing that somatic TET2, DNMT3A, and ASXL1 mutations are commonly found in the blood of aging individuals without overt hematologic abnormalities or clinically apparent myeloid malignancies.1,2
Somatic mutation status has not yet been fully integrated into the World Health Organization MDS diagnostic classification scheme.10 As additional longitudinal cohorts with comprehensive genetic annotation are reported, MDS may be more precisely defined by the presence of specific mutation patterns in patients with cytopenias, even in the absence of definitive morphologic findings.
MDS and its distinction from AML
Medullary blast count has long been recognized as one of the most important prognostic parameters in MDS. Based on a series of studies that evaluated the association between the proportion of bone marrow blasts and overall survival or risk of AML transformation, a formal distinction between MDS and AML was defined. Irrespective of other clinicopathologic features, a patient is characterized as having MDS if bone marrow blasts are measured below 20% and AML if bone marrow blasts are 20% or higher.10,11 Although this quantitative threshold is useful for disease classification, clinical trial enrollment, and population-based studies, it does not provide resolution of clinically relevant heterogeneity at the level of an individual patient. For example, some patients have a low bone marrow blast count but progress rapidly to AML, suggesting that a biological transition to more aggressive AML can precede development of a leukemic blast count. Other patients can have a relatively high blast count that remains stable for a prolonged period of time, suggesting that even profound defects in hematopoietic differentiation do not always cause florid decompensation.12
Accurate prediction of disease kinetics and dynamic monitoring of disease status are central challenges in the clinical care of MDS patients. In individual patients, it can be difficult at the time of initial diagnosis to predict the pace of disease progression or identify incipient transformation, even with serial clinical monitoring. Can an understanding of the stereotyped position of gene mutations in the MDS clonal hierarchy improve prognostic models? Specifically, can a point assessment of MDS genetics be used to predict the kinetics of disease progression in individual patients? By defining the most relevant associations between genetic characteristics and disease status, it may be possible to more precisely identify clinically actionable disease transitions.
Serial genetic analysis of patients with MDS before and after transformation to secondary acute myeloid leukemia (s-AML) has revealed a key principle of disease progression: the transition from MDS to s-AML is typically marked by a change in clone constituency rather than a change in overall clone abundance. Walter et al13 used whole-genome sequencing to compare the genetic characteristics of samples obtained from MDS patients at 2 time points. They showed that the proportion of bone marrow involved by a genetically defined clone remains the same in MDS and s-AML, irrespective of blast count. In each case, mutations associated with the founding clone persisted at high variant allele fraction, but there was selective outgrowth or emergence of at least one genetically distinct subclone. This study established the basic paradigm that clinical transformation of MDS is associated with clonal genetic evolution.
However, clonal evolution itself does not always mean clinical transformation. Comparisons of serial samples from individual patients and genetic analyses of MDS and s-AML cohorts have shown that specific gene mutations have highly stereotyped positions in MDS clonal hierarchy.4,6,14 For example, mutations affecting genes that encode epigenetic modifiers (DNMT3A, TET2, ASXL1, EH2, etc.) or RNA spliceosome components (SF3B1, SRSF2, and U2AF1) tend to arise in the MDS phase of disease and rarely occur at the time of transformation. Acquisition of mutations in these pathways characterizes the initiation and early progression of MDS. By contrast, mutations that drive activated growth factor signaling pathways (NRAS, KRAS, PTPN11, FLT3, etc.) are rarely identified early in disease, and instead, they are frequently gained or expanded in subclones at time of progression to high-grade MDS or transformation to acute leukemia.14-17 Aberrant activation of rat sarcoma/mitogen-activated protein kinase (RAS/MAPK) signaling thus seems to represent a discrete biological transition in MDS, although the precise molecular consequences of individual RAS pathway gene mutations on intracellular signaling can be distinct.
Deriving clinical utility from genetic knowledge: examples from stereotyped clonal hierarchies and comutation associations
The stereotyped subclonal position of RAS pathway mutations in the MDS clonal hierarchy may inform interpretation of clinical genetic results. For example, we recently showed that the presence of RAS pathway mutations at the time of allogeneic HSC transplantation for MDS is associated with poor survival.3 Notably, the adverse prognostic significance of RAS pathway mutations was only manifest in patients receiving reduced intensity conditioning regimens and driven by a markedly increased risk of early relapse. We suggest that RAS pathway mutations that persist at the time of transplantation reflect low-volume but biologically transformed disease that outpaces the development of effective graft-versus-leukemia activity, unless patients receive intensive cytoreduction before transplantation. These data are consistent with another transplant study, where persistence of RAS pathway mutations in s-AML patients after induction chemotherapy was associated with poor posttransplant survival,18 although this study was not powered to evaluate RAS pathway mutations in MDS patients receiving reduced intensity conditioning vs myeloablative regimens. In the nontransplant setting, patients with RAS-mutated or FLT3 internal tandem duplication s-AML have inferior survival compared with those with wild-type RAS or FLT3, suggesting that mutations causing activated signaling drive a biologically distinct transformation.16 In aggregate, these data suggest that diagnostic sequencing or serial genetic monitoring for mutations that cause aberrant activation of growth factor signaling (NRAS, KRAS, FLT3, PTPN11, CBL, NF1, RIT1, and KIT) may enable more accurate and clinically meaningful assessment of biological disease status.
Point mutations in RNA splicing factors (SF3B1, U2AF1, and SRSF2) are the most common class of genetic alterations in MDS patients.4,5 Splicing mutations are always heterozygous and display a striking mutual exclusivity, where mutations very rarely cooccur in the same patient.19 Experimental models have shown that the genetic characteristics of human MDS reflect a cellular intolerance of severe spliceosome impairment and showed proof of concept that it may be possible to leverage this vulnerability to therapeutic benefit using pharmacologic modulators of spliceosome function.20 These studies suggest an exciting path forward: ongoing genomic discovery efforts or analysis of large datasets may stimulate biological hypotheses or novel therapeutic approaches. For example, mutations in the cohesin pathway (STAG2, RAD21, SMC3, and SMC1A) display a mutual exclusivity similar to spliceosome mutations. Alternatively, there may be powerful, albeit unexpected, genetic associations and exclusivities that are apparent only in specific clinical or genetic contexts.
Pathogenetic context: distinct MDS biology with cytotoxic therapy or germ-line predisposition syndromes
Genetics of therapy-related MDS.
MDS that develops after chemotherapy or radiation for a nonmyeloid disease is categorized as therapy-related myelodysplastic syndrome (t-MDS), and it is recognized as a distinct group because of its association with poor clinical outcome.10 The definition of t-MDS is based solely on clinical history, and it is independent of the type or intensity of exposure and of the latency between exposure and t-MDS diagnosis. All patients who are at risk of having biological t-MDS are thus encompassed within the category. However, whether a specific exposure is truly linked to disease pathogenesis and the extent to which therapy-related pathogenesis directly drives poor clinical outcomes have not been clearly established.
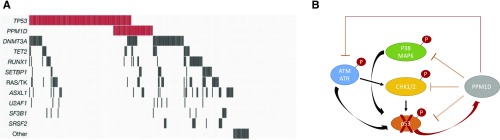
Several mechanisms have been proposed to explain the pathogenesis of t-MDS, including direct genotoxic stress to HSCs, the selective outgrowth HSC clones with impaired DNA damage response or MDS-associated driver mutations, or manifestation of an inherited cancer susceptibility.21-23 Recent genetic studies of t-MDS and therapy-related AML cases have sought to better define a subgroup of patients who have true therapy-driven disease.14,24-26 In our recent analysis of 1514 MDS patients receiving allogeneic stem cell transplantation, we compared the mutation profile of 311 patients with t-MDS with that of 1203 patients with primary MDS. Mutations in PPM1D or TP53 were present in 46% of the patients with t-MDS, and they were the only gene mutations that were significantly associated with t-MDS. By contrast, the group of t-MDS cases without TP53 or PPM1D mutations appeared genetically similar to primary MDS and did not have adverse outcomes (Figure 3A).
Figure 3.
The genetics of t-MDS. Panel A shows the spectrum of recurrent gene mutations in a cohort of 311 t-MDS cases. Each column represents one patient, and colored bars are mutations. TP53 and PPM1D mutations are found in one-half of cases, and they frequently cooccur in individual patients. Panel B represents key components of the DNA damage response. Red circles with “P” represent phosphates that are regulated by PPM1D phosphatase activity.
PPM1D is a serine-threonine protein phosphatase that negatively regulates the cellular stress response (Figure 3B). Cells that encounter replicative stress through exposure to chemotherapy rapidly activate DNA repair and cell cycle checkpoint pathways via phosphorylation of specific residues on various proteins, including ATM, CHK1, and P53. P53, which represents the central coordinating node of these pathways, induces expression of PPM1D, which in turn triggers a return to steady state through its phosphatase activity. Mutations in PPM1D are localized to exon 6 and cause C-terminal truncations that may cause an increase in phosphatase activity that aberrantly inhibits checkpoint and DNA damage response pathways.27
TP53 mutations have long been recognized to occur more frequently in therapy-related myeloid neoplasms than in de novo myeloid neoplasms.28 This epidemiological observation is consistent with the established consequences of TP53 dysfunction during the DNA damage response after exposure to cytotoxic agents.29 Moreover, TP53 mutations can be identified in the peripheral blood of some patients who develop t-MDS/therapy-related AML, even before exposure to chemotherapy, suggesting that cytotoxic therapies can select for preexisting clones with acquired DNA damage response dysfunction.24 Experimental evidence in a mouse model of Tp53 insufficiency supports this clinical observation, showing that Tp53+/− hematopoietic stem/progenitor cells preferentially expand after exposure to the alkylating agent N-ethyl-N-nitrosurea.24
The strong statistical association between t-MDS and TP53 or PPM1D mutations as well as the shared biological role of TP53 or PPM1D in the DNA damage response suggest that these mutations mark a subset of t-MDS patients whose disease is directly driven or selected by therapeutic exposure. The potent selective pressures exerted by such therapies are further evidenced by the significant cooccurrence of PPM1D and TP53 mutations in individual patients. This association suggests either convergent evolution of distinct subclones under conditions of environmental selection or cooperative biological activity in the same clone. Together, these findings show that somatic genetic analysis can reveal the biological basis of clinically relevant heterogeneity in disease pathogenesis, even within a World Health Organization–defined subcategory. Furthermore, genetic annotation enables interrogation of alternative biological hypotheses. For example, poor outcomes in t-MDS without TP53 pathway alterations may be independent of disease biology and reflect global end organ dysfunction, alteration of the bone marrow microenvironment, or poor hematopoietic reserve.
Age and MDS genetics.
Aging is associated with the development of clonally restricted hematopoiesis.1,2,30 At least 10% of individuals over age 70 years old have detectable mutations in canonical drivers of myeloid malignancies in their peripheral blood, whereas others exhibit clonal skewing without driver mutations. The presence of clonal hematopoiesis with myeloid driver mutations is associated with an elevated risk of developing hematologic cancers.1,2 Accordingly, the age-dependent accumulation of somatic mutations is thought to underlie the increasing prevalence of MDS with increasing age. In children and young adults, MDS is rare and more commonly arises in the context of predisposing conditions, such as leukomogenic exposures, familial MDS/AML syndromes, or inherited or acquired bone marrow failure syndromes.31 The spectrum of genetic alterations in young MDS patients is different than that of older MDS patients, likely reflecting these distinct pathogenetic mechanisms.3 Whereas older patients more frequently harbor somatic mutations in genes encoding epigenetic modifiers (TET2 and DNMT3A) or RNA splicing (SRSF2 and SF3B1), younger patients have much higher frequency of genes associated with germ-line conditions (GATA2 and SBDS) and acquired predispositions (PIGA).
By uniformly interrogating genetic loci associated with IBMF and MDS predisposition syndromes in a large cohort of MDS patients, we made several observations: some MDS patients had germ-line mutations in IBMF genes, most patients with IBMF mutations did not have a documented clinical history of the associated syndrome, the latency of MDS development in patients with IBMF gene mutations was highly variable, and MDS transformation in specific IBMF syndromes was associated with characteristic somatic progression events.
For example, we found compound heterozygous SBDS mutations in 4% of adults with MDS under the age of 40 years old.3 SBDS is the causative gene for Shwachman–Diamond Syndrome, an autosomal recessive congenital bone marrow failure syndrome associated with short stature, exocrine pancreatic dysfunction, and a strong predisposition to MDS/AML transformation.32 Surprisingly, most patients in this cohort with biallelic SBDS mutations had no clinical history of Shwachman–Diamond Syndrome, although clinical and genetic evidence suggested that these were bona fide cases of clinically unrecognized disease. First, SBDS mutations were present at the heterozygous variant allele fraction and were previously reported as pathogenic in affected patients. Second, patients with biallelic SBDS mutations were significantly and uniformly younger than carriers who harbored a single mutated allele. Third, patients with biallelic SBDS mutations had significantly shorter stature than carriers. These results were consistent with the increasing recognition that the clinical presentation of Shwachman–Diamond Syndrome can be highly variable and that patients can escape clinical diagnosis into young adulthood.33
Not all MDS cases with genetically identifiable inherited predispositions were restricted to young patients. We found that almost 1% of the total MDS cohort had germ-line pathogenic mutations in TERC or TERT. These patients had highly variable latency of MDS transformation, with a median age of diagnosis in the sixth decade of life. Despite harboring canonical TERC or TERT mutations known to impair telomere maintenance, only one patient had a clinical history of dyskeratosis congenita. This suggests that germ-line defects in telomere maintenance have variable clinical presentation, where the most severely affected individuals present early in life with syndromic characteristics of dyskeratosis congenita, whereas less severely affected individuals present later in life with MDS or other end organ manifestations. The variable clinical presentation and disease latency are consistent with the genetic anticipation that is seen with inherited defects in telomere maintenance, where each successive generation has increasingly shortened telomeres and a more severe phenotype.34
These examples show that inherited predispositions are more common in MDS patients than recognized by standard clinical evaluation. Identification of MDS patients with cryptic germ-line predisposition mutations may influence several aspects of clinical management. For example, in the setting of stem cell transplantation, patients with SBDS, TERC, or TERT mutations all had poor survival, but the apparent causes of poor clinical outcomes were different. In patients with SBDS mutations, relapse was common. In contrast, those with cryptic TERC or TERT mutations had low relapse but high risk of nonrelapse mortality, consistent with clinical characteristics of patients with bona fide dyskeratosis congenita.35 Developing strategies for improving transplantation outcomes in these patients may thus depend on understanding the biological differences that govern their distinct susceptibility to transplant- or conditioning regimen–specific toxicities. Similarly, objective identification of cryptic IBMF/predisposition mutations will impact transplant donor selection and family counseling and screening recommendations.
Somatic clonal hematopoiesis has been observed in individuals with familial MDS and related asymptomatic carriers.36 Some such patients exhibit clonal skewing without recurrent driver mutation, whereas others have somatic mutations in myeloid driver genes. The clinical implications of detecting emergence of somatic clones have not been defined. However, development of clonal hematopoiesis seems to occur precociously in patients with IBMF and MDS/AML predisposition syndromes, suggesting that inherited alterations in hematopoiesis or bone marrow microenvironment favor the selection of clonally dominant HSCs and the development of myeloid malignancies. Furthermore, recent data suggest that the pathobiology of different germ-line syndromes affects the spectrum of somatic mutations that drive clinical progression. In MDS patients with genetically defined Shwachman–Diamond Syndrome, TP53 mutations are remarkably common.3 In contrast, TP53 mutations are rare in patients with germ-line GATA2 deficiency, and instead, somatic ASXL1 mutations are more common.37 Understanding the distinct pathways of clonal transformation in patients with germ-line predispositions may improve clinical monitoring and yield advances in development of disease-specific therapeutic strategies.
Conclusion
Genetic characteristics of MDS are powerfully associated with clinical phenotype. An enhanced understanding of the regulatory logic of MDS genetics may stimulate refined and biologically based diagnostic and monitoring schema that may be integrated into clinical practice.
References
- 1.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013; 122(22):3616-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makishima H, Yoshizato T, Yoshida K, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwok B, Hall JM, Witte JS, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cargo CA, Rowbotham N, Evans PA, et al. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126(21):2362-2365. [DOI] [PubMed] [Google Scholar]
- 9.Malcovati L, Gallì A, Travaglino E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129(25):3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 11.WHO. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008 [Google Scholar]
- 12.Germing U, Gattermann N, Strupp C, Aivado M, Aul C. Validation of the WHO proposals for a new classification of primary myelodysplastic syndromes: a retrospective analysis of 1600 patients. Leuk Res. 2000;24(12):983-992. [DOI] [PubMed] [Google Scholar]
- 13.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badar T, Patel KP, Thompson PA, et al. Detectable FLT3-ITD or RAS mutation at the time of transformation from MDS to AML predicts for very poor outcomes. Leuk Res. 2015;39(12):1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T, Tyndel MS, Kim HJ, et al. The clonal origins of leukemic progression of myelodysplasia. Leukemia. 2017;31(9):1928-1935. [DOI] [PubMed] [Google Scholar]
- 18.Yoshizato T, Nannya Y, Atsuta Y, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129(17):2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 20.Lee SC, Dvinge H, Kim E, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganser A, Heuser M. Therapy-related myeloid neoplasms. Curr Opin Hematol. 2017;24(2):152-158. [DOI] [PubMed] [Google Scholar]
- 22.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin oncol. 2017;35(14):1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K, Wang F, Kantarjian H, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18(1):100-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ok CY, Patel KP, Garcia-Manero G, et al. Mutational profiling of therapy-related myelodysplastic syndromes and acute myeloid leukemia by next generation sequencing, a comparison with de novo diseases. Leuk Res. 2015;39(3):348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih AH, Chung SS, Dolezal EK, et al. Mutational analysis of therapy-related myelodysplastic syndromes and acute myelogenous leukemia. Haematologica. 2013;98(6):908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleiblova P, Shaltiel IA, Benada J, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013;201(4):511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19(5):1405-1413. [DOI] [PubMed] [Google Scholar]
- 29.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babushok DV, Bessler M. Genetic predisposition syndromes: when should they be considered in the work-up of MDS? Best Pract Res Clin Haematol. 2015;28(1):55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boocock GRB, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97-101. [DOI] [PubMed] [Google Scholar]
- 33.Myers KC, Bolyard AA, Otto B, et al. Variable clinical presentation of Shwachman-Diamond syndrome: update from the North American Shwachman-Diamond Syndrome Registry. J Pediatr. 2014;164(4):866-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savage SA, Bertuch AA. The genetics and clinical manifestations of telomere biology disorders. Genet Med. 2010;12(12):753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadalla SM, Sales-Bonfim C, Carreras J, et al. Outcomes of allogeneic hematopoietic cell transplantation in patients with dyskeratosis congenita. Biol Blood Marrow Transplant. 2013;19(8):1238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churpek JE, Pyrtel K, Kanchi K-L, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West RR, Hsu AP, Holland SM, Cuellar-Rodriguez J, Hickstein DD. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica. 2014;99(2):276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]