Abstract
Introduction:
Newcastle Disease (ND), caused by Avian avulavirus 1 (AAvV 1, avulaviruses), is a notifiable disease throughout the world due to the economic impact on trading restrictions and its embargoes placed in endemic regions. The feral birds including aquatic/migratory birds and other wild birds may act as natural reservoir hosts of ND Viruses (NDVs) and may play a remarkable role in the spread of the virus in environment. In addition, other 19 avulaviruses namely: AAvV 2 to 20, have been potentially recognized from feral avian species.
Expalantion:
Many previous studies have investigated the field prevailing NDVs to adapt a wide range of susceptible host. Still the available data is not enough to declare the potential role of feral birds in transmission of the virus to poultry and/or other avian birds. In view of the latest evidence related to incidences of AAvVs in susceptible avian species, it is increasingly important to understand the potential of viruses to transmit within the domestic poultry and other avian hosts. Genomic and phylogenomic analysis of several investigations has shown the same (RK/RQRR↓F) motif cleavage site among NDV isolates with same genotypes from domestic poultry and other wild hosts. So, the insight of this, various semi-captive/free-ranging wild avian species could play a vital role in the dissemination of the virus, which is an important consideration to control the disease outbreaks. Insufficient data on AAvV 1 transmission from wild birds to poultry and vice versa is the main constraint to understand about its molecular biology and genomic potential to cause infection in all susceptible hosts.
Conclusion:
The current review details the pertinent features of several historical and contemporary aspects of NDVs and the vital role of feral birds in its molecular epidemiology and ecology.
Keywords: Avian avulavirus 1 (AAvV 1), Wild and Migratory birds, Epidemiology, Phylogeny, Newcastle Disease (ND), Fusion gene
1. INTRODUCTION
Newcastle Disease Virus (NDV) causes wide-spread mortality in poultry but clinical and subclinical presentations have also been observed in other avian species. Its causative agent belongs to Avulavirus genus under Paramyxoviridae family [1]. This family has the varied type of members which have been responsible to cause infections in several avian species including, domestic, captive/free-ranging wild, terrestrial, and aquatic birds globally [2-6]. Genus Avulavirus contains all the Avian avulaviruses (AAvVs) specie-types, causing clinical and subclinical infection in all avian species including poultry with interspecies transmission except for avian metapneumovirus. Based on Hemagglutination Inhibition (HI) and Neuraminidase Inhibition (NI) tests [7], NDVs have been identified and classified into two different subgroups, AAvV 2 and 6 among 20 specie-types (AAvV 1 to 20) [8]. Of these, one antigenic variant of AAvV 1, causes severe infection and is responsible for epidemics in pigeon termed as Pigeon Paramyxovirus type 1 (PPMV-1) [9]. Since NDV (AAvV 1) is a highly contagious disease of poultry and other birds, it can create an alarming situation in developing countries.
So, to elucidate the interspecies transmission, attention on molecular epidemiology of AAvV 1 is the need of time. Alongside to this, the data about the molecular biology and pathogenesis of different avulaviruses 2-20 [Rahman et al., 2018] is not enough to evaluate the virus mutation rate. AAvV 1 (or ND) is an OIE (Office International Epizootics; world organization for animal health) notifiable avian infection implicated with significant economic losses as well as natural genetic depletion of diverse hosts [5]. In the above context, the intention of this review is to provide a summary of overall pertinent evidence related to geographical distribution and susceptible host density of AAvV 1 with its temporal, terrestrial, and host density. However, up to date and comprehensive overview of the genetic diversity is the main consideration to assess the evolutionary analysis of NDVs among avian species. This review summarises, compares and discusses the available literature on NDVs genetic diversity and may be helpful to investigate the mutable aspects of NDVs in all genes particularly fusion (F) gene.
1.1. Geographical Distribution
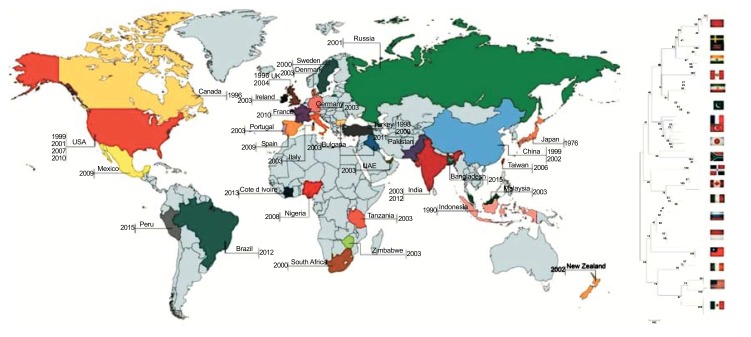
Several avian species can spread the numerous microbes across the globe, which are harmful to captive and free-ranging birds including poultry [10, 11]. During the last few years, NDV and influenza virus have been found to be transmitted by migratory birds across the globe [12, 13]. It provided the strong evidence that wild avian species have been implicated in the transferring of NDV to domestic and wild bird population as biological carriers/ natural reservoir [14]. ND was first described in 1926 in Newcastle-on-Tyne, England (from where it got its name) and on the island of Java, now part of Indonesia, although there have been some suggestions that there may have been earlier outbreaks. It appears that initially the disease spreads rapidly in Asia [15] but slowly reported a series of devastating outbreaks around the globe including Africa, Central America and parts of South America [16-18]. A lot of outbreaks in different avian species associated with virulent NDV were reported from North America (USA, Canada and Costa Rica), Europe (France, Italy, England, Scotland, Spain and Russia), Africa (Kenya) and Asia (China, Japan, India, Israel, Saudi Arabia and Kazakhstan) [19] (Fig. 1).
Fig. (1).
NDV from wild bird origin isolated in different regions/countries, also showed the geographical evolutionary relationship among these countries where it is endemic. For example, NDV from Pakistan, Iran, China and India grouped in same cluster.
Alongside poultry, NDV has been isolated from wild birds with incidences of virus transmission from wild psittacine/ cormorants to chickens in the United State during the 1970s and 1990s [20, 21]. The findings from these studies warrant the consistent monitoring of wild birds in a pause of virus dissemination. In India, many bird’s species including chicken, turkey, quail, emu, duck, peacock, pigeon, duck and guinea fowl have been found as potential susceptible host for NDV infection [22-29]. The current NDV epidemics in feral birds in Asian countries especially neighbours of India like Pakistan [17, 30] and Bangladesh [31] pose the threats to disease dissemination by sharing countries boundaries and the movement of migratory birds from one to another country. Regards to epidemiological context, different NDV strains are circulating in the field to infect the domestic and wild birds [32] that highlight the importance of the characterization of current prevailing NDV isolates from indigenous captive/semi-captive bird and from clinical or subclinical infection in free-ranging feral birds [33]. However, the interspecies transmission propensity of field circulating viruses may raise concerns about the efficacy of vaccine, used in the field. Various outbreaks have revealed the existence of vaccine strains of domestic poultry in wild birds from Mexico [34]. Moreover, vaccine NDV strain was confirmed in Australian wild bird [35] including feral birds in Japan [36], Finland [37], China [38], America [39] and in central Nigeria [40].
Except for Asian countries, zoo birds in Israel and Mexico have become infected with virulent NDV strains similar to those causing infection in domestic birds, which indicated the spill-over of virus in the environment [41]. In this account, the similar findings have been observed in Africa [42] and other continents [43, 44] which raised the concerns regarding the potential role of wild birds in shedding of NDVs in environment and transmission to poultry. Moreover, the existence of anti-NDV antibodies in feral birds of South Africa [45], Burkina Faso [46], and Nigeria [47] has also exposed the susceptibility of a wide range of avian hosts. The molecular detection, evolutionary dynamics and viral genetic manipulation will be helpful to control the epidemics and thus, we need to focus on the molecular and geographically epidemiology of NDV strains and should be perceived as an epidemiological alert to re-assess the control and quarantine measures against NDV [48, 49].
1.2. Genetic Variability of Newcastle Disease Virus
Avian avulaviruses are RNA viruses, having non-segmented, a single-stranded negative-sense genome with helical capsid symmetry. It has a molecular weight of about 5 × 106 Da, with approximately 15.2 kb in length and encodes six structural and two non-structural proteins in the order 3'-NP–V/W/P-M-F–HN-L-'5 [50]. The surface projections on envelope are approximately 8 nm long, present on the HN molecule, whilst F molecules form smaller projections. The F protein has significance as a type-I integral membrane protein with the trans-membrane domain located in the carboxyl-terminal region followed by a short cytoplasmic domain [51], responsible for virulence. During the infection, the first HN protein of virus attaches to a cell and then fusion protein makes linkage for the invasion of viral genetic material into the host cell [52]. The virulence of NDV depends on the primary molecular determinant which is known as F protein cleavage site having specific amino acid sequence pattern and position [53]. The susceptible host got an infection when the cellular proteases cleave an inactive F0 precursor protein into F1 and F2 subunits [51]. The NDV pathotype totally depends on the F protein cleavage site having dibasic amino acids in velogenic and mesogenic strains, while the F protein of lentogenic NDV isolates lack this motif [51, 54]. The NDV isolates from migratory birds have shared the similar specific dibasic amino acid at the cleavage site of F gene which has been detected in chickens from the same area during an outbreak [55]. Usually, the virulent strain has RQK/RRF residue pattern whereas K/RQG/ERL residues [53] have been observed at specific position 112-117 in the NDV strain of low virulence [56] (Table 1). In comparison, a mutative NDV isolate has also been detected from a dove having a K for Q substitution at residue 114; evidence of adaptation of NDV to the environment [57, 58].
Table 1.
NDV susceptible host range with geographical distribution.
| Accession No. | Host | Scientific Names | Year | Country | Genotype | CSP* | Virulence | References |
|---|---|---|---|---|---|---|---|---|
| FJ436303 | Chicken | Gallus gallus domesticus | 1986 | China | IX | RRQRRF | virulence | [123] |
| FJ872531 | Muscovy duck | Cairina moschata | 2002 | China | VII | RRQKRF | virulence | [124] |
| GQ288389 | Mallard | Anas platyrhynchos | 1999 | USA | IIa | EKQGRL | avirulence | [48] |
| AY865652 | Sterna | S. hirundo | 2001 | Russia | Vb | RRQRRF | virulence | [125] |
| JQ013039 | Teal | Anas crecca | 2010 | France | Class I | ERQERL | avirulence | [126] |
| KJ627773 | Crane | Gruidae | 1992 | India | - | GKQGRL | avirulence | [127] |
| KJ398400 | Peafowl | Pavo cristatus | 2012 | India | VII | RRQRRF | virulence | [127] |
| AY581302 | Guinea fowl | Numididae | 2000 | India | - | RRQRRF | virulence | [128] |
| GQ288391 | Mottled duck | Anas fulvigula | 2001 | USA | IIa | GKQGRL | avirulence | [48] |
| GQ288378 | Northern pintail | Anas acuta | 1987 | USA | IIa | GKQGRL | avirulence | [48] |
| EF564816 | Red knot | Calidris canutus | 2001 | USA | Ia | GKQGRL | avirulence | [13] |
| AY289194 | Turkey | Meleagris gallopavo | 2003 | Canada | - | GKQGRL | avirulence | [129] |
| JX854452 | Pheasant | Phasianus colchicus | 2011 | Pakistan | VII | RRQKRF | virulence | [67] |
| KC934169 | Blackbird | Turdus merula | 2008 | China | IX | RRQRRF | virulence | [130] |
| KC934170 | Spotted dove | Spilopelia chinensis | 2008 | China | IX | RRQRRF | virulence | [130] |
| KC424431 | White-cheeked starling | Sturnus cineraceus | 2008 | China | IX | RRQRRF | virulence | [131] |
| JN255774 | Double-crested cormorant | Phalacrocorax auritus | 2010 | USA | VII | KRQKRF | virulence | [62] |
| JN255778 | Black-backed gull | Larus marinus | 2010 | USA | VII | KRQKRF | virulence | [62] |
| JN255779 | Herring gull | Larus argentatus | 2010 | USA | VII | KRQKRF | virulence | [62] |
| JN941993 | Pelican | Pelecanus | 2008 | USA | - | KRQKRF | virulence | [132] |
| KC808510 | Scarlet macaw | Ara macao | 2009 | Mexico | Vb | RRQKRF | virulence | [34] |
| KC503422 | Spectacled eider | Somateria fischeri | 2007 | USA | Ib | GKQGRL | avirulence | [133] |
| KC503482 | Slaty-backed gull | Larus schistisagus | 2007 | USA | Class I | GKQGRL | avirulence | [133] |
| AY562985 | Cockatoo | Cacatuidae | 1990 | Indonesia | VIId | RRQKRF | virulence | [134] |
| KF740478 | Japanese quail | Coturnix japonica | 2003 | India | VII | RRQKRF | virulence | [135] |
| JN599167 | Penguin | Spheniscidae | 1999 | China | VII | RRQKRF | virulence | [136] |
| AY471773 | Budgerigar | Melopsittacus undulatus | 1995 | Turkey | VIId | RRQKRF | virulence | [137] |
| HG326606 | Spur-winged goose | Plectropterus gambensis | 2008 | Nigeria | I | GKQGRL | avirulence | [42] |
| HM063424 | Water rail | Rallus aquaticus | 2005 | China | I | GKQGRL | avirulence | [138] |
| HM063425 | Wild pigeon | Columba livia | 2003 | China | VI | RRQKRF | virulence | [138] |
| AF109886 | Finches | Fringillidae | 1997 | UK | Ve | RRQKRF | virulence | [139] |
| AF091623 | Goosander | Mergus merganser | 1996 | UK | Vb | RRQRRF | virulence | [139] |
| AY471818 | Fantail | Rhipidura | 1996 | Turkey | VIIc | GRQKRF | avirulence | [137] |
| AY471721 | Falcon | Falconiforme | 2000 | Turkey | VIIf | RRQKRF | virulence | [137] |
| AY471785 | Kestrel | Falco tinnunculus | 1999 | Turkey | VIId | RRQKRF | virulence | [137] |
| AY471843 | Turtle dove | Streptopelia turtur | 1996 | Turkey | VIIb | GRQKRF | avirulence | [137] |
| KC808497 | Egret | Ardea alba | 2009 | Mexico | II | GRQGRL | avirulence | [34] |
| KC808491 | Robin | Turdus grayi | 2009 | Mexico | II | GRQGRL | avirulence | [34] |
| KC808490 | Yellow-napped parrot | Amazona auropalliata | 2009 | Mexico | II | GRQGRL | avirulence | [34] |
| KC808489 | Hawk | Buteo brachyurus | 2009 | Mexico | II | GRQGRL | avirulence | [34] |
| KC808487 | Oropendola | Zacua spp. | 2009 | Mexico | II | GRQGRL | avirulence | [34] |
| KC808493 | Amazon parrot | Amazona autumnalis | 2009 | Mexico | Ia | GRQGRL | avirulence | [34] |
| KC808494 | Chachalaca | Ortalis vetula | 2009 | Mexico | Ia | GRQGRL | avirulence | [34] |
| KC808498 | Guan | Penelopina nigra | 2009 | Mexico | Ia | GRQGRL | avirulence | [34] |
| FJ938175 | Sparrow | Passer domesticus | 2005–2007 | China | VII | RRQKRF | virulence | [140] |
| EF564817 | Ruddy turnstone | Arenaria interpres | 2002 | USA | Ia | GKQGRL | avirulence | [13] |
| JX855036 | Crested Ibis | Nipponia nippon | 2013 | China | VIId | RRQKRF | virulence | 116 |
| JF820295 | Ostrich | Struthio camelus | 2011 | Iran | VII | RRQKRF | virulence | 141 |
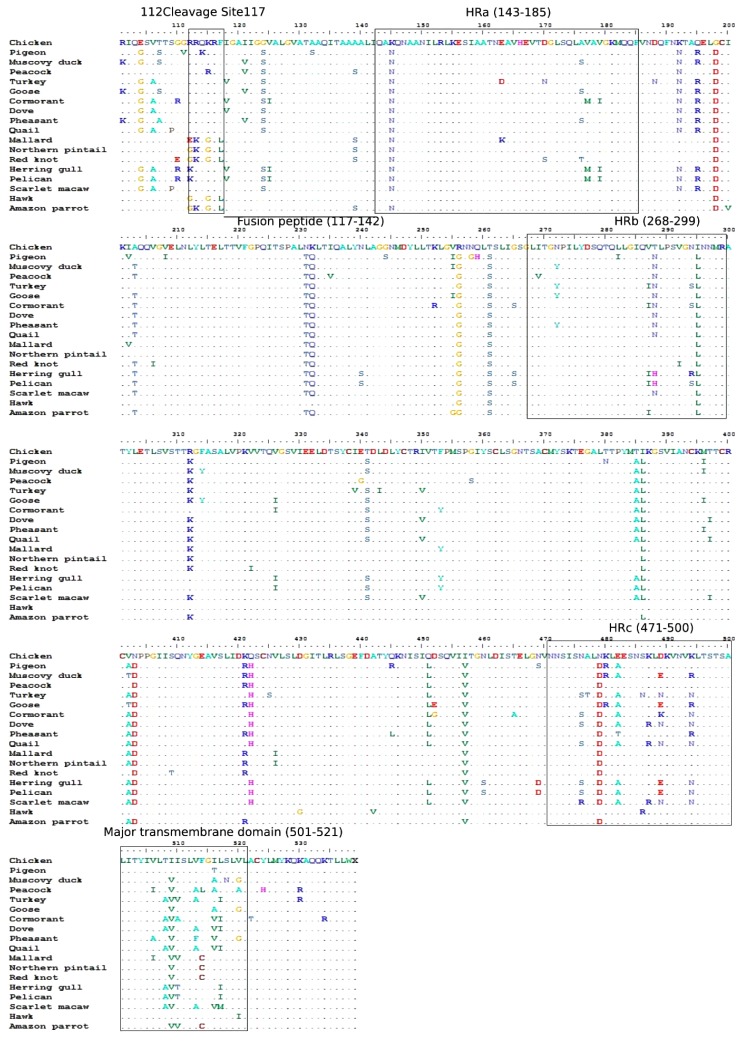
Although, F protein is highly conserved among all isolates consisting of a series of Heptad Repeat (HR) regions [51], some sort of mutations have also been observed in it such as the mutation of the L at position 154 in the HR1 region from residues 130 to 170 interferes in cell fusion [59]. In addition, substitutions in the other conserved regions like HR2 region from N-terminal to the TM domain, a helical region HR3 domain from residues 263 to 289 and another helical domain HR4 from residues 81 to 102 [60, 61] have been observed (Fig. 2). Based on the evolutionary estimation, the F gene divergences of 1-18% between NDVs isolates from chicken-origin and wild-origin [62], and it was highly similar to the hawk origin NDV strain with 2% divergence. Although, NDVs from wild bird origin had nucleotide homologies of up to 99.8% and low homologies of 82% with chicken origin NDVs [63].
Fig. (2).
Alignment of deduced amino acid sequence of complete F gene of NDV isolate from different wild birds with reference of chicken isolate with accession numbers AY562991, JX901111, FJ872531, HQ011508, JN942045, HG326605, GQ288385, GQ429293, JX854452, KC808512, KJ920203, EF612277, EF564816, JN255779, JN941993, KC808510, KC808489, KC808493 from chicken to amazon parrot, respectively. Structurally and functionally important residues are boxed and highlighted based on residue profile to explore similarities among them.
In addition, NDVs originated from quail and dove showed a minor nucleotide difference (15%) when compared to isolates from chicken, but these did not exhibit any significant antigenic differences (1%) between them. Similarly, duck and goose shared only 1% nucleotide dissimilates (Table 2). To date, the complete genome sequences of a number of strains of AAvV 1-20 [64, 65] have been published. The genetic information was available only from Nigeria [66], Pakistan [17, 67], India [29, 33, 68, 69], China [70] and South Africa [71], but still, a wide range of birds remains uncovered to investigate the potential of NDV to transmit and infect diverse host range.
Table 2.
The nucleotide based genome homology of different NDVs isolated from a wide range of wild/feral birds
| % Nucleotide Divergence and Homology Between Each Pair of Isolates | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | Pigeon |
Muscovy
duck |
Peacock | Turkey | Goose | Cormorant | Dove | Pheasant | Quail | Mallard | Sterna |
Northern
pintail |
Red
knot |
Herring
gull |
Pelican |
Scarlet
macaw |
Hawk |
Amazon
parrot |
|
| Chicken | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | |
| Pigeon | 0.17 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
|
Muscovy duck |
0.17 | 0.11 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Peacock | 0.14 | 0.14 | 0.13 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Turkey | 0.18 | 0.15 | 0.14 | 0.15 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Goose | 0.17 | 0.12 | 0.01 | 0.14 | 0.15 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Cormorant | 0.18 | 0.15 | 0.13 | 0.16 | 0.10 | 0.14 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.01 | |
| Dove | 0.15 | 0.12 | 0.11 | 0.12 | 0.06 | 0.11 | 0.07 | 0.01 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | |
| Pheasant | 0.18 | 0.13 | 0.08 | 0.15 | 0.15 | 0.08 | 0.16 | 0.13 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Quail | 0.15 | 0.13 | 0.11 | 0.13 | 0.07 | 0.11 | 0.08 | 0.01 | 0.14 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | |
| Mallard | 0.11 | 0.16 | 0.16 | 0.13 | 0.18 | 0.16 | 0.18 | 0.15 | 0.17 | 0.16 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
| Sterna | 0.18 | 0.12 | 0.09 | 0.14 | 0.14 | 0.09 | 0.15 | 0.12 | 0.11 | 0.12 | 0.17 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
|
Northern pintail |
0.10 | 0.16 | 0.15 | 0.14 | 0.17 | 0.15 | 0.17 | 0.15 | 0.16 | 0.15 | 0.04 | 0.16 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
|
Red knot |
0.14 | 0.17 | 0.16 | 0.15 | 0.17 | 0.17 | 0.19 | 0.16 | 0.17 | 0.16 | 0.12 | 0.16 | 0.12 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | |
|
Herring gull |
0.18 | 0.15 | 0.14 | 0.16 | 0.11 | 0.14 | 0.04 | 0.08 | 0.16 | 0.09 | 0.18 | 0.15 | 0.18 | 0.19 | 0.00 | 0.01 | 0.01 | 0.01 | |
| Pelican | 0.18 | 0.15 | 0.14 | 0.16 | 0.10 | 0.14 | 0.04 | 0.08 | 0.16 | 0.08 | 0.18 | 0.15 | 0.17 | 0.18 | 0.01 | 0.01 | 0.01 | 0.01 | |
|
Scarlet macaw |
0.16 | 0.13 | 0.11 | 0.13 | 0.07 | 0.12 | 0.08 | 0.02 | 0.14 | 0.01 | 0.16 | 0.13 | 0.16 | 0.17 | 0.09 | 0.08 | 0.01 | 0.01 | |
| Hawk | 0.02 | 0.17 | 0.17 | 0.14 | 0.18 | 0.17 | 0.19 | 0.15 | 0.18 | 0.15 | 0.11 | 0.17 | 0.10 | 0.14 | 0.18 | 0.18 | 0.16 | 0.01 | |
|
Amazon parrot |
0.11 | 0.15 | 0.14 | 0.13 | 0.16 | 0.15 | 0.17 | 0.13 | 0.14 | 0.14 | 0.10 | 0.14 | 0.09 | 0.08 | 0.17 | 0.16 | 0.15 | 0.11 | |
1.3. Host Range
According to the world organization for animal health (WHO), ND is a contagious disease affecting more than 250 avian species around the globe [72]. In this context, a lot of free-ranging aquatic fowls are considered as a potential carrier of AAvV 1 [7, 73]. In 1946, after the first evidence of virulent strain of NDV in United States of America and Mexico, wild migratory avian and white storks were found to be susceptible for virulent NDV strain [74, 75]. Such kind of circumstantial evidence revealed the potential of virus and mode of transmission AAvV 1 by implicating wild birds to poultry [14]. It is not only about migratory birds, some recreational birds and captive wild birds also seemed to be as a natural reservoir/ carrier and susceptible hosts of NDVs [76]. Previously, the incidences of NDVs in wild and migratory aquatic birds [37], pigeons [36, 40], Peacock (17, 29, 33, 68], pheasant [67], duck and geese [70] have been reported.
NDV as a mutable RNA virus has the ability to cross interspecies barrier by mutating itself. Related evidence has been observed in Luxembourg, where lentogenic viruses highly similar to the LaSota strain were isolated from waterfowl subsequent to spill over and interspecies adaptation [42]. In another previous study related to Turkey, the causative virus was identified in genotype Ia along with several cases of NDVs in wild birds which were kept in live bird’s market [77]. Taken together, it was suggested that viruses from wild birds may spill over into poultry. Based on AAvVs genetic diversity analysis, the viral transmission between wild birds and poultry was proved, previously [13, 42, 78, 79]. Furthermore, the NDV strains isolated from wild birds could cause outbreaks in chickens. For example, NDV isolates from migrating cormorants were identified as the likely source of epidemic in poultry. The interaction between wild birds and poultry happens frequently; the wild birds possibly play a pivotal role in the evolution of NDV for the adaptation of environment [78, 80].
Interestingly, it has been shown that double-crested cormorant is an important reservoir of NDVs among all feral bird population [81]. Similarly, pigeons and mallards have been suggested to be the reservoir of NDVs [82, 83]. In New Zealand, NDV has been isolated from a red-breasted musk parrot following its seizure after illegal importation forms Fiji [84]. Specifically, waterfowls are considered important reservoirs of NDVs and may act as a carrier for NDV transfer to poultry, causing outbreaks of different pathogenicity [12, 85, 86]. In recent years, a disease resembling those of ND has been reported in ducks and geese in the regions of China [87]. Thus, it could be inferred that feral birds are carriers of virulent strains but transmission pattern of virulent NDV strains is not yet fully understood.
Small wild birds mainly Passeriformes can also transmit NDVs due to their peri-domestic habits and some avian species are playing a significant role in epizootiology of NDVs [4]. NDV had also been recovered from domestic duck farms [88] and wild birds [89], which raise concerns regarding NDV transmission among several avian species. So, few reports on mortality in free-living birds other than feral pigeons (Columba livia), recently captured teal (Anas crecca) [90], double-crested cormorants (Phalacrocorax auritus), white pelicans (Pelecanus erythrorhynchos), and gulls (Larus spp.), raised the concerns about apparent epizootic nature of the virus [91]. In North America, during ND outbreaks, virulent strains of NDV of non-poultry origin have been reported from cormorants, gulls and pelicans [62, 92], and NDV strain of low virulence has been isolated from gulls, shorebirds, and waterfowl as part of avian influenza surveillance [12, 93]. NDV strains of variable virulence have also been isolated from wild pigeons and doves [94, 95]. In view of host susceptibility, NDVs have been reported from a quite wide range of various feral birds, including wild, aquatic and recreational birds (Table 1). Although a variety of wild birds are susceptible to ND, the information available is limited due to the non-availability of clinical/necropsy samples and poor disease surveillance in feral avian population. Thus, there is a need for NDV surveillance in all susceptible wild birds to insight the evolution and adaptation of virus for controlling ND outbreaks worldwide.
1.4. Clinico-pathological Presentation of NDV
Based on pathogenicity, NDVs are classified into different four stains [19]. First, the Velogenic Viscerotropic NDV (VVNDV) formerly Dolye form, responsible for acute and lethal infections causing hemorrhagic lesions in visceral organs. Second, the Velogenic neurotrophic NDV (VNNDV) formerly Beach form, responsible for high mortality involve in respiratory and neurologic signs (gut lesions are absent). Third, the Mesogenic NDV formerly Beaudette form causes low mortality with acute respiratory disease and nervous signs. At four, the Lentogenic or asymptomatic enteric NDV formerly Hitchner form is a virulent virus that appears to replicate primarily in the gut with mild or inn-apparent respiratory infections [96].
Wild and migratory birds were considered the natural reservoirs of NDV and harboured mainly Lentogenic strains but occasionally Velogenic strains have potential to shed in the environment [42, 89, 97]. Mortality in wild birds such as pigeon, juvenile double-crested cormorants (Phalacrocorax auritus) and teal (Anas crecca) has also been reported due to the infection caused by Velogenic strains [25, 29, 62, 74, 92]. Highly virulent viruses from psittacines and in a variety of zoo and live market birds have also been isolated [42, 98-100]. Depend on infection, strain, dose rate, route of exposure, hostage, immunological status and environmental conditions, signs can range from no clinical presentation to neurologic signs, paralysis, and/or acute death [19]. However, few pathotypes are responsible for peracute infection with almost 100% mortality to subclinical disease with no lesions [93].
1.4.1. Galliformes, Anseriformes and Columbiformes Birds
Turkey is a susceptible host to vNDV with clinical signs of depression, nasal discharge, blood-tinged diarrhea and nervous incoordination [101]. Partridges and pheasants are considered to be extremely sensitive to vNDV [102] with similar clinical signs as observed in chickens from the acute onset with high mortality, severe nervous signs to inapparent infection [7]. In a study, neurological signs in experimentally infected ducks with a mesogenic NDV strain were also observed [103]. Similarly, the goose was also reported as a susceptible host for NDV [104, 105] with moderate to severe depression, anorexia, diarrhea, ocular and nasal discharges, and swelling of the eyelids [87]. Histopathologically, the susceptibility of birds was characterized by multifocal areas of ulceration and hemorrhages in the esophagus, gizzard, and multifocal necrosis of the intestinal mucosa [106]. Ulceration and fibrin deposition in the intestinal mucosa and over the cecal tonsils, severe atrophy of lymphoid organs, and lymphoid depletion, multifocal areas of necrosis in the pancreas and less frequently in the liver were also observed. In a few cases, the brain was affected by neuronal degeneration present [106]. These types of investigation have clear evidence about the interspecies transmission prosperity of NDV.
In pigeons, ND is caused by pigeon specific variant of ND virus known as pigeon paramyxovirus-1 (PPMV-1) [58]. The outbreak in pigeon was reported first time in the Middle East during 1970 and spread to Europe during 1980 and now become endemic around the globe [81]. Neurological signs and diarrhea are major clinical signs seen mainly in young birds [107]. Gross lesions in pigeons infected with PPMV-1 from natural outbreaks consist of pancreatic necrosis, enteritis, and proventricular hemorrhages [108]. Histologically, lesions consist of non-suppurative encephalitis, multifocal necrosis in spleen, bursa, liver, larynx, and pancreas, and multifocal accumulation of lymphocytes in several organs [108] with spleen enlargement and perivascular cuffing in the cerebellum and brainstem [101].
1.4.2. Psittaciformes, Passeriformes and Suliformes Birds
NDVs were also isolated from non-domesticated species of Psittaciformes and Passeriformes [4] with neurological signs in psittacine and Passeriformes birds [109]. Different cases of ND were also reported in six different states of the United States in 1991 [110]. Clinical signs included tremors, lateral recumbency, respiratory distress, greenish diarrhea, ruffled plumage, and head drawn back between the shoulders, and eventual death. The VVND viruses were isolated from affected birds, including yellow-headed Amazon parrots (Amazona ochrocephala oratrix), yellow-naped Amazon parrots (Amazona ochrocephala auropalliata), cockatiels (Nymphicus hollandicus), and Canaries (Serinus canarius). In another study, VVNDV was isolated from experimentally infected birds including budgerigars (Melopsittacus undulates), Amazon parrots (Amazona ochrocephala auropalliata), and Canaries (Serinus canaries) with neurological signs consisting of tremors, ataxia, wing droop, and uni or bilateral leg paralysis [111].
Histopathology following exposure to VVNDV results in haemorrhages and necrosis of the intestinal mucosa, haemorrhages on the skullcap and around the orbit, fibrinous peritonitis, hepatosplenomegaly, focal hepatic necrosis, airsacculitis, and hemorrhagic tracheitis in the Canaries, Amazon parrots, and budgerigars. These birds were also able to spread the virus and infect cage-mates. Shedding of VVNDV has been observed for more than 1 year in Amazon parrots and for more than 80 days in budgerigars, both enabled in the spread of virus in the environment [111]. NDV outbreaks in double-crested cormorant (Phalacrocorax auritus) populations [112] were also reported with neurological signs including prominent gross lesions of enlarged and mottled spleen associated with bursal atrophy and multifocal hemorrhagic foci in the meninges [112]. Histologically, lesions having multifocal nonsuppurative encephalitis with areas of gliosis appeared more prominent in the cerebellar white matter, interstitial nephritis and multifocal myocarditis [113]. Isolation of NDVs from commercial or feral birds reported no associated clinical signs [114, 115]. So, based on previous studies, existence and transmission of NDVs among different avian species including domesticated and wild birds with a variable clinical infection impose the continuous surveillance of wild and migratory birds.
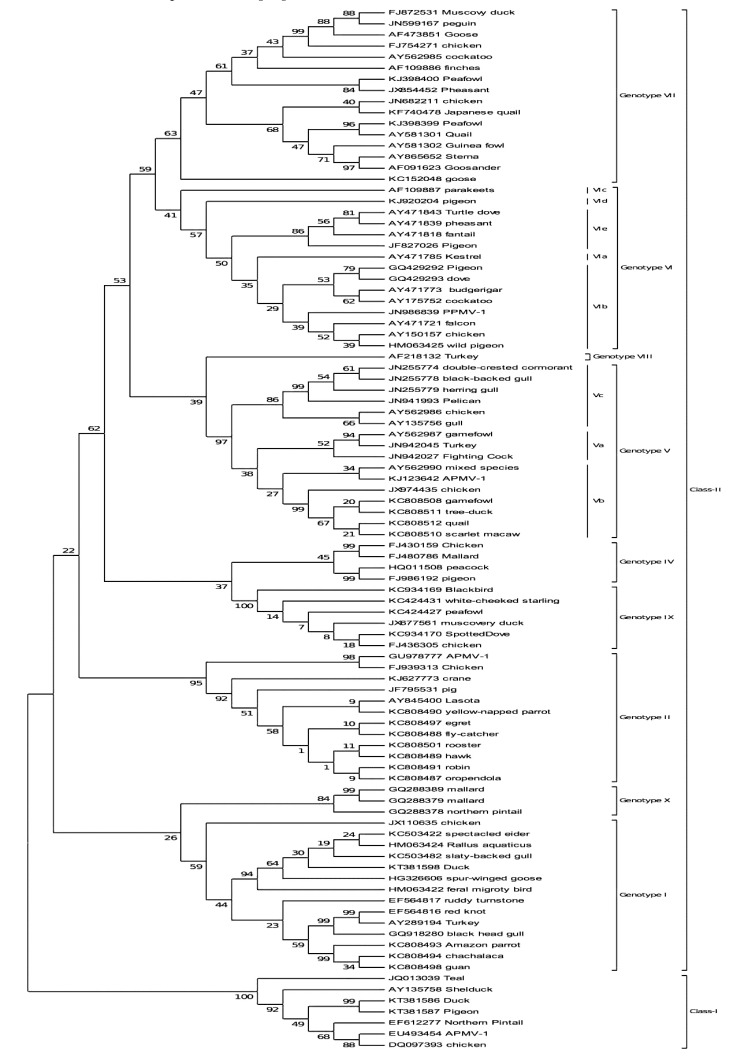
1.5. Phylogenetic Analysis of Wild Originated NDVs
Two major classes (class I and II) of NDVs were premeditated according to genome length-based phylogenetic analysis [74]. In class I, there are atleast nine different genotypes, while in class II comprised of eighteen genotypes (I–XVIII) (Maminiaina et al., 2010; Meng et al., 2012). In class II, the genotypes VI and VII were further divided into nine (a-i) and five (a-e) sub-genotypes, respectively [116]. Noteworthy, Munir et al. [17] concluded that the isolates can be divided into six broadly distinct lineages (1 to 6). Of these, lineage 3 and 4 further subdivided into four (a-d) and five (a-e) sub-lineages, respectively. According to the phylogenetic analysis, most of NDV strains from pigeon or dove were classified into genotype VI [117] often considered as mesogenic strains, and a little information about other genotypes NDV from Columbidae was recorded [32, 92]. The diversity was found in amino acid sequences of F genes of NDVs, all publicly available isolates at GenBank®, indicating the evolutionary and mutative potential of NDVs. Dissimilarities among vaccine strain and field isolates from genotype V of class II have also been reported [74, 118], that aid in viral shedding, further, the persistence of NDVs in backyard poultry and free-living wild birds, explaining why vNDV caused sporadic outbreaks in the poultry industry until recently [119].
Similarly, phylogenetic relationships of non-virulent NDV strains in shorebirds and waterfowl provide support for the spread of viruses among different avian species and geography (12, 94). Virulent NDVs of identical genetic make-up were isolated from wild pigeons and doves in the United States, providing the evidence of virus spread from the different origin within different avian species and geography of North America [95, 96]. However, intercontinental transport of viruses may be attributed to the trade of racing pigeons. Thus, dissemination of NDVs within North America is supported, but evidence for intercontinental virus spread by migratory birds is limited. Thus, previous studies on genetic diversity among strains of NDVs revealed that some strains from wild birds were phylogenetically related with NDVs isolated from live-bird markets (12, 42). One previous study from Africa revealed the different host adaptation of NDVs where, the domestic pigeons originated NDVs of genotype VI have been found responsible for causing infection in different birds, such as feral pigeons and doves [57]. Such type of transmission has also been reported with high mortality rates in other studies [96, 120] This actually points out the viral evolution for adaptation of hosts leads to constrain in its eradication.
So far, among wild birds, virulent NDVs seem enzootic not only in cormorants in North America [62] but also in pigeons worldwide [87, 95]. Additionally, viruses from genotype XVIII have been reported from wild birds in the United States. Phylogenetically, the viruses originated from wild birds showed a close relationship to strains isolated from live birds markets [121], raise the questions about interspecies transmission of NDVs for the conservation of endangered wild birds. Such findings highlight the potential of NDVs to transmit via migration of wild birds, which are natural reservoirs of NDVs [55]. Furthermore, isolation of virulent NDVs from wild aquatic birds raise the concern that wild and migratory waterfowls could play a role as a long-distance vector of virulent NDV, highlighting the need for increased NDV surveillance in aquatic fowls [89].
To estimate the evolutionary distances between wild birds originated NDV strains, complete F gene-based phylogenetic analysis was performed by the maximum composite likelihood method in MEGA6 software [122]. The evolutionary analysis of already reported wild bird originated NDVs revealed the strong relationship among all NDVs originated from wild birds, domestic birds and poultry (Fig. 3).
Fig. (3).
Molecular phylogenetic analysis of fusion (F) gene by a maximum likelihood method conducted in MEGA6. All NDV isolates showed an evolutionary relationship with chicken origin NDVs and also with other wild or captive bird's origin NDVS.
In this figure, NDVs from different feral birds showed a strong evolutionary relationship with the poultry originated strains as mentioned in class I and II and also clustered within different genotypes. Generally, the velogenic NDV strains from poultry origin have been clustered in genotype VII but NDVs from wild bird’s origin are also found in genotype IX, V and class I beyond its pathotype properties. Such type of grouping of NDV strain in different genotype points out the continuous evolution of field circulating viruses, which may be due to the vaccine failure because, in many countries, NDV epidemics have been observed in vaccinated birds. Therefore, the active surveillance and monitoring of wild birds can help us prevent the disease transmission in all susceptible host and evolution of NDVs.
CONCLUDING REMARKS
Based on the previous molecular investigations, it can be concluded that poultry and non-poultry host cohabiting raise NDV epidemics around the globe particularly in endemic countries. Phylogenetic analysis of wild and domestic originated NDVs provided such evidence, which supports that wild birds are contributing to the global redistribution of NDVs alongside the regular vaccination but only in poultry. The cleavage site in F protein of the NDVs has been found to be an important factor contributing to the pathogenesis in hosts. However, few mutations have been observed in different isolates originated from different bird’s species, result in variability of virulence. So, the investigation of genetic variation and isolate’s evolutionary analysis can provide valuable data on characterization, epidemiology among different avian species and diagnosis of NDVs circulating in the environment. In future, the attention of the global scientific community towards these aspects would help to elucidate the complete epidemiology trends of NDVs and to validate the robustness of diagnostic screening, particularly in endemic countries. In addition, a continuous surveillance of wild and migratory/aquatic fowls will share the information to design the appropriate control strategies.
ACKNOWLEDGEMENTS
Declared none
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
REFERENCES
- 1.ICTV. International Committee on Taxonomy of Viruses. Virus Taxonomy. https:// talk.ictvonline.org/ ictv-reports/ https:// talk.ictvonline.org/ taxonomy/ p/ taxonomy- history?taxnode_id=20161031. 2017. Accessed July 2017.
- 2.Wang L.F., Eaton B.T. Emerging paramyxoviruses. Infect. Dis. Rep. 2001;3:52–69. [Google Scholar]
- 3.Lamb R.A., Parks G.D. Paramyxoviridae: The viruses and their replication. In: Knipe, D.M., Howley, P.M. (Eds.), Fields Virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, 2007; 1449-96. [Google Scholar]
- 4.Alexander D.J., Aldous E.W., Fuller C.M. The long view: A selective review of 40 years of Newcastle disease research. Avian Pathol. 2012;41(4):329–335. doi: 10.1080/03079457.2012.697991. [DOI] [PubMed] [Google Scholar]
- 5.OIE. Newcastle disease. In Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France. . http:// www.oie.int/ fileadmin / Home/eng/ Health_standards/ tahm/2.03.14_NEWCASTLE_DIS.pdf. 2012.
- 6.Dimitrov K.M., Ramey A.M., Qiu X., Bahl J., Afonso C.L. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect. Genet. Evol. 2016;39:22–34. doi: 10.1016/j.meegid.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Alexander D.J. Newcastle disease, other avian paramyxoviruses, and pneumovirus infection. In: Disease of poultry, ed. Shaif YM, Barnes HJ, Glisson JR, et al., 12th ed., pp. 75-100. Blackwell, Oxford, UK. 2003. [Google Scholar]
- 8.Aziz-ul-Rahman Munir M, Shabbir MZ. Comparative evolutionary and phylogenomic analysis of Avian avulaviruses 1 to 20. Mol. Phylogenet. Evol. doi: 10.1016/j.ympev.2018.06.040. In press. [DOI] [PubMed] [Google Scholar]
- 9.Lipkind M., Shihmanter E. Antigenic relationships between avian paramyxoviruses. I. Quantitative characteristics based on hemagglutination and neuraminidase inhibition tests. Arch. Virol. 1986;89(1-4):89–111. doi: 10.1007/BF01309882. [DOI] [PubMed] [Google Scholar]
- 10.Hubalek Z. Pathogenic microorganisms associated with free-living birds (a review). Acta Scientiarum Naturalium Brno. 1994;28:174. [Google Scholar]
- 11.Hubálek Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J. Wildl. Dis. 2004;40(4):639–659. doi: 10.7589/0090-3558-40.4.639. [DOI] [PubMed] [Google Scholar]
- 12.Stroud DA, Davidson NC, West R, Scott DA, Hanstra L, Thorup O, Ganter B, Delany S. Status of migratory wader population in Africa and Western Eurasia in the 1990s. . Int water studies. 2004;15:1–259. [Google Scholar]
- 13.Dhama K., Mahendran M., Tomar S. Pathogens transmitted by migratory birds: Threat perceptions to poultry health and production. Int. J. Poult. Sci. 2008;7:516–525. doi: 10.3923/ijps.2008.516.525. [DOI] [Google Scholar]
- 14.Kim L.M., King D.J., Suarez D.L., Wong C.W., Afonso C.L. Characterization of class I Newcastle disease virus isolates from Hong Kong live bird markets and detection using real-time reverse transcription-PCR. J. Clin. Microbiol. 2007;45(4):1310–1314. doi: 10.1128/JCM.02594-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle T.M. A hitherto unrecorded disease of fowls due to a filter-passing virus. J. Comp. Pathol. Ther. 1927;40:144–169. [Google Scholar]
- 16.Alexander DJ. Orthomyxoviridae–avian influenza. Poultry diseases. 2008:317. doi: 10.1016/B978-0-7020-2862-5.50031-3. [DOI] [Google Scholar]
- 17.Munir M., Abbas M., Khan M.T., Zohari S., Berg M. Genomic and biological characterization of a velogenic Newcastle disease virus isolated from a healthy backyard poultry flock in 2010. Virol. J. 2012;9:46. doi: 10.1186/1743-422X-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen K., Marks D.R., Arsnoe D.M., Afonso C.L., Bevins S.N., Miller P.J., Randall A.R., DeLiberto T.J. Avian paramyxovirus serotype 1 (Newcastle disease virus), avian influenza virus, and Salmonella spp. in mute swans (Cygnus olor) in the Great Lakes region and Atlantic Coast of the United States. Avian Dis. 2014;58(1):129–136. doi: 10.1637/10638-081413-Reg.1. [DOI] [PubMed] [Google Scholar]
- 19.Alexander D.J., Senne D.A. Newcastle disease, other avian paramyxoviruses and pneumovirus infections, p 75-100. In Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE (ed), Diseases of poultry, 12th ed. Blackwell Publishing, Ames, IA. 2008. [Google Scholar]
- 20.Walker J.W., Heron B.R., Mixson M.A. Exotic Newcastle disease eradication program in the United States. Avian Dis. 1973;17(3):486–503. doi: 10.2307/1589147. [DOI] [PubMed] [Google Scholar]
- 21.Wobeser G., Leighton F.A., Norman R., Myers D.J., Onderka D., Pybus M.J., Neufeld J.L., Fox G.A., Alexander D.J. Newcastle disease in wild water birds in western Canada, 1990. Can. Vet. J. 1993;34(6):353–359. [PMC free article] [PubMed] [Google Scholar]
- 22.Roy P., Venugopalan A.T., Manvell R. Characterization of Newcastle disease viruses isolated from chickens and ducks in Tamilnadu, India. Vet. Res. Commun. 2000;24(2):135–142. doi: 10.1023/A:1006416724050. [DOI] [PubMed] [Google Scholar]
- 24.Chang P.C., Hsieh M.L., Shien J.H., Graham D.A., Lee M.S., Shieh H.K. Complete nucleotide sequence of avian paramyxovirus type 6 isolated from ducks. J. Gen. Virol. 2001;82(Pt 9):2157–2168. doi: 10.1099/0022-1317-82-9-2157. [DOI] [PubMed] [Google Scholar]
- 25.Kataria J.M., Dhama K. Sohini Dey, Madhan Mohan C. Ranikhet Disease in Pigeon. Poultry World. 2006;1(6):16–18. [Google Scholar]
- 26.Naveen K.A., Singh S.D., Kataria J.M., Barathidasan R., Dhama K. Detection and differentiation of pigeon paramyxovirus serotype-1 (PPMV-1) isolates by RT-PCR and restriction enzyme analysis. Trop. Anim. Health Prod. 2013;45(5):1231–1236. doi: 10.1007/s11250-013-0352-0. [DOI] [PubMed] [Google Scholar]
- 27.Gowthaman V., Singh S.D., Barathidasan R., Ayanur A., Dhama K. Natural Outbreak of Newcastle Disease in Turkeys and Japanese Quails Housed Along with Chicken in a Multi-Species Poultry Farm in Northern India. Adv Anim Vet Sci. 2013;1(3S):17–20. [Google Scholar]
- 28.Gowthaman V., Singh S.D., Dhama K., Desingu P.A., Kumar A., Malik Y.S., Munir M. Isolation and characterization of genotype XIII Newcastle disease virus from Emu in India. Virusdisease. 2016;27(3):315–318. doi: 10.1007/s13337-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desingu P.A., Singh S.D., Dhama K., Vinodhkumar O.R., Barathidasan R., Malik Y.S., Singh R., Singh R.K. Molecular characterization, isolation, pathology and pathotyping of peafowl (Pavo cristatus) origin Newcastle disease virus isolates recovered from disease outbreaks in three states of India. Avian Pathol. 2016;45(6):674–682. doi: 10.1080/03079457.2016.1198005. [DOI] [PubMed] [Google Scholar]
- 30.Shabbir M.Z., Zohari S., Yaqub T., Nazir J., Shabbir M.A.B., Mukhtar N., Shafee M., Sajid M., Anees M., Abbas M., Khan M.T., Ali A.A., Ghafoor A., Ahad A., Channa A.A., Anjum A.A., Hussain N., Ahmad A., Goraya M.U., Iqbal Z., Khan S.A., Aslam H.B., Zehra K., Sohail M.U., Yaqub W., Ahmad N., Berg M., Munir M. Genetic diversity of Newcastle disease virus in Pakistan: A countrywide perspective. Virol. J. 2013;10(1):170. doi: 10.1186/1743-422X-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nooruzzaman M., Mazumder A.C., Khatun S., Chowdhury E.H., Das P.M., Islam M.R. Pathotypic and genotypic characterization of two Bangladeshi isolates of Newcastle disease virus of chicken and pigeon origin. Transbound. Emerg. Dis. 2015;62(1):102–107. doi: 10.1111/tbed.12086. [DOI] [PubMed] [Google Scholar]
- 32.Tirumurugaan K.G., Vinupriya M.K., Vijayarani K., Kumanan K. Analysis of the fusion protein cleavage site of Newcastle disease virus isolates from India reveals preliminary evidence for the existence of II, VI and VII genotypes. Indian J. Virol. 2011;22(2):131–137. doi: 10.1007/s13337-011-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A., Maan S., Mahajan N.K., Rana V.P., Jindal N., Batra K., Ghosh A., Mishra S.K., Kapoor S., Maan N.S. Detection and molecular characterization of Newcastle disease virus in peafowl (Pavo cristatus) in Haryana State, India. Indian J. Virol. 2013;24(3):380–385. doi: 10.1007/s13337-013-0169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardenas Garcia S., Navarro Lopez R., Morales R., Olvera M.A., Marquez M.A., Merino R., Miller P.J., Afonso C.L. Molecular epidemiology of Newcastle disease in Mexico and the potential spillover of viruses from poultry into wild bird species. Appl. Environ. Microbiol. 2013;79(16):4985–4992. doi: 10.1128/AEM.00993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackenzie J., Britten D., Hinshaw V., Wood J. Isolation of avian influenza and paramyxoviruses from wild birds in Western Australia, in Veterinary Viral Diseases: Their significance in South-east Asia and the Western Pacific. In: Della-Porta A.J., editor. Veterinary Viral Diseases. 1985. pp. 336–339. [Google Scholar]
- 36.Jahangir A., Ruenphet S., Ueda S., Ueno Y., Shoham D., Shindo J., Okamura M., Nakamura M., Takehara K. Avian influenza and Newcastle disease viruses from northern pintail in Japan: Isolation, characterization and inter-annual comparisons during 2006-2008. Virus Res. 2009;143(1):44–52. doi: 10.1016/j.virusres.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Lindh E., Huovilainen A., Rätti O., Ek-Kommonen C., Sironen T., Huhtamo E., Pöysä H., Vaheri A., Vapalahti O. Orthomyxo-, paramyxo- and flavivirus infections in wild waterfowl in Finland. Virol. J. 2008;5:35. doi: 10.1186/1743-422X-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng X., Hua Y., Li X., Zhang Z. Monitoring influenza A virus and Newcastle disease virus in migratory waterfowls in Sanjiang natural reserve of Heilongjiang Province. Wei Sheng Wu Xue Bao. 2008;48(10):1403–1407. [PubMed] [Google Scholar]
- 39.Krauss S., Walker D., Pryor S.P., Niles L., Chenghong L., Hinshaw V.S., Webster R.G. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 2004;4(3):177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- 40.Ibu O.J., Okoye J.O.A., Adulugba E.P., Chah K.F., Shoyinka S.V.O., Salihu E., Chukwuedo A.A., Baba S.S. Prevalence of Newcastle disease viruses in wild and captive birds in central Nigeria. Int. J. Poult. Sci. 2009;8:574–578. doi: 10.3923/ijps.2009.574.578. [DOI] [Google Scholar]
- 41.Haddas R., Meir R., Perk S., Horowitz I., Lapin E., Rosenbluth E., Lublin A. Newcastle disease virus in little owls (Athene noctua) and African penguins (Spheniscus demersus) in an Israeli zoo. Transbound. Emerg. Dis. 2014;61(6):e79–e82. doi: 10.1111/tbed.12064. [DOI] [PubMed] [Google Scholar]
- 42.Snoeck C.J., Marinelli M., Charpentier E., Sausy A., Conzemius T., Losch S., Muller C.P. Characterization of newcastle disease viruses in wild and domestic birds in Luxembourg from 2006 to 2008. Appl. Environ. Microbiol. 2013;79(2):639–645. doi: 10.1128/AEM.02437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaleta E.F. Avian paramyxovirus infections. Infect dis of Wild Mammal Bird Eur; 2012. pp. 59–66. [Google Scholar]
- 44.Yuan X., Wang Y., Li J., Yu K., Yang J., Xu H., Zhang Y., Ai H., Wang J. Surveillance and molecular characterization of Newcastle disease virus in seafowl from coastal areas of China in 2011. Virus Genes. 2013;46(2):377–382. doi: 10.1007/s11262-012-0863-1. [DOI] [PubMed] [Google Scholar]
- 45.Pfitzer S., Verwoerd D.J., Gerdes G.H., Labuschagne A.E., Erasmus A., Manvell R.J., Grund C. Newcastle disease and avian influenza A virus in wild waterfowl in South Africa. Avian Dis. 2000;44(3):655–660. doi: 10.2307/1593107. [DOI] [PubMed] [Google Scholar]
- 46.Tarnagda Z., Yougbare I., Kam A., Tahita M.C., Ouedraogo J.B. Prevalence of infectious bronchitis and Newcastle disease virus among domestic and wild birds in H5N1 outbreaks areas. J. Infect. Dev. Ctries. 2011;5(8):565–570. doi: 10.3855/jidc.1441. [DOI] [PubMed] [Google Scholar]
- 47.Oladele S.B., Enam S.J., Okubanjo O.O. Pathogenic haemoparasites and antibody to Newcastle disease virus from apparently healthy wild birds in Zaria, Nigeria. Vet. World. 2012;5(1):13–18. doi: 10.5455/vetworld.2012.13-18. [DOI] [Google Scholar]
- 48.Zhang G.Z., Zhao J.X., Wang M. Serological survey on prevalence of antibodies to avian paramyxovirus serotype 2 in China. Avian Dis. 2007;51(1):137–139. doi: 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 49.Desingu P.A., Singh S.D., Dhama K., Karthik K., Vinodh Kumar O.R., Malik Y.S. Phylogenetic analysis of Newcastle disease virus isolates occurring in India during 1989-2013. Virusdisease. 2016;27(2):203–206. doi: 10.1007/s13337-016-0320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan Y., Samal S.K. Role of intergenic sequences in newcastle disease virus RNA transcription and pathogenesis. J. Virol. 2008;82(3):1323–1331. doi: 10.1128/JVI.01989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison T.G. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta. 2003;1614(1):73–84. doi: 10.1016/S0005-2736(03)00164-0. [DOI] [PubMed] [Google Scholar]
- 52.Swanson K., Wen X., Leser G.P., Paterson R.G., Lamb R.A., Jardetzky T.S. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402(2):372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glickman R.L., Syddall R.J., Iorio R.M., Sheehan J.P., Bratt M.A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 1988;62(1):354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagai Y., Klenk H.D., Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976;72(2):494–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 55.Takakuwa H., Ito T., Takada A., Okazaki K., Kida H. Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations. Jpn. J. Vet. Res. 1998;45(4):207–215. [PubMed] [Google Scholar]
- 56.Wei D., Yang B., Li Y.L., Xue C.F., Chen Z.N., Bian H. Characterization of the genome sequence of an oncolytic Newcastle disease virus strain Italien. Virus Res. 2008;135(2):312–319. doi: 10.1016/j.virusres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Terregino C., Cattoli G., Grossele B., Bertoli E., Tisato E., Capua I. Characterization of Newcastle disease virus isolates obtained from Eurasian collared doves (Streptopelia decaocto) in Italy. Avian Pathol. 2003;32(1):63–68. doi: 10.1080/0307945021000070732. [DOI] [PubMed] [Google Scholar]
- 58.Ujvári D., Wehmann E., Kaleta E.F., Werner O., Savić V., Nagy E., Czifra G., Lomniczi B. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 2003;96(1-2):63–73. doi: 10.1016/S0168-1702(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 59.Sergel T.A., McGinnes L.W., Morrison T.G. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 2000;74(11):5101–5107. doi: 10.1128/JVI.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L., Colman P.M., Cosgrove L.J., Lawrence M.C., Lawrence L.J., Tulloch P.A., Gorman J.J. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology. 2001;290(2):290–299. doi: 10.1006/viro.2001.1172. [DOI] [PubMed] [Google Scholar]
- 61.Chen L., Gorman J.J., McKimm-Breschkin J., Lawrence L.J., Tulloch P.A., Smith B.J., Colman P.M., Lawrence M.C. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9(3):255–266. doi: 10.1016/S0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 62.Diel D.G., da Silva L.H., Liu H., Wang Z., Miller P.J., Afonso C.L. Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 2012;12(8):1770–1779. doi: 10.1016/j.meegid.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 63.Courtney S.C., Susta L., Gomez D., Hines N.L., Pedersen J.C., Brown C.C., Miller P.J., Afonso C.L. Highly divergent virulent isolates of Newcastle disease virus from the Dominican Republic are members of a new genotype that may have evolved unnoticed for over 2 decades. J. Clin. Microbiol. 2013;51(2):508–517. doi: 10.1128/JCM.02393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnamurthy S., Samal S.K. Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J. Gen. Virol. 1998;79(Pt 10):2419–2424. doi: 10.1099/0022-1317-79-10-2419. [DOI] [PubMed] [Google Scholar]
- 65.de Leeuw O., Peeters B. Complete nucleotide sequence of Newcastle disease virus: Evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 1999;80(Pt 1):131–136. doi: 10.1099/0022-1317-80-1-131. [DOI] [PubMed] [Google Scholar]
- 66.Van Borm S., Obishakin E., Joannis T., Lambrecht B., van den Berg T. Further evidence for the widespread co-circulation of lineages 4b and 7 velogenic Newcastle disease viruses in rural Nigeria. Avian Pathol. 2012;41:377–382. doi: 10.1080/03079457.2012.696311. [DOI] [PubMed] [Google Scholar]
- 67.Shabbir M.Z., Goraya M.U., Abbas M., Yaqub T., Shabbir M.A.B., Ahmad A., Anees M., Munir M. Complete genome sequencing of a velogenic viscerotropic avian paramyxovirus 1 isolated from pheasants (Pucrasia macrolopha) in Lahore, Pakistan. J. Virol. 2012;86(24):13828–13829. doi: 10.1128/JVI.02626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vijayarani K., Muthusamy S., Tirumurugaan K.G., Sakthivelan S.M., Kumanan K. Pathotyping of a Newcastle disease virus isolated from peacock (Pavo cristatus). Trop. Anim. Health Prod. 2010;42(3):415–419. doi: 10.1007/s11250-009-9436-2. [DOI] [PubMed] [Google Scholar]
- 69.Khulape S.A., Gaikwad S.S., Chellappa M.M., Mishra B.P., Dey S. Complete genome sequence of a Newcastle disease virus isolated from wild peacock (Pavo cristatus) in India. Genome Announc. 2014;2(3):e00495–e14. doi: 10.1128/genomeA.00495-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X., Wang J., Li S., Gong Z., Wang X., Zhong D., Hu Y., Zhang W. Pathohistological studies on ducks infected with NDV of duck origin. Chin. J. Prev. Vet. Med. 2010;32:27–31. [Google Scholar]
- 71.Abolnik C. Molecular epidemiology of Newcastle disease and avian influenza in South Africa. Ph.D. thesis, University of Pretoria, Pretoria, South Africa. 2007. [Google Scholar]
- 72.Kaleta E.F., Baldauf C. Newcastle disease in free-living and pet birds. In: Alexander DJ, ed. Newcastle Disease. Boston: Kluwer Academic Publishers 1988: 197-246. [DOI] [Google Scholar]
- 73.Zanetti F., Berinstein A., Pereda A., Taboga O., Carrillo E. Molecular characterization and phylogenetic analysis of Newcastle disease virus isolates from healthy wild birds. Avian Dis. 2005;49(4):546–550. doi: 10.1637/7381-051605R.1. [DOI] [PubMed] [Google Scholar]
- 74.Merino R., Villegas H., Quintana J.A., Calderon N. Characterization of Newcastle disease viruses isolated from chicken, gamefowl, pigeon and quail in Mexico. Vet. Res. Commun. 2009;33(8):1023–1030. doi: 10.1007/s11259-009-9321-5. [DOI] [PubMed] [Google Scholar]
- 75.Perozo F., Merino R., Afonso C.L., Villegas P., Calderon N. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 2008;52(3):472–479. doi: 10.1637/8276-022908-Reg.1. [DOI] [PubMed] [Google Scholar]
- 76.Senthuran S., Vijayarani K., Kumanan K., Nainar A.M. Pathotyping of Newcastle disease virus isolates from pet birds. Acta Virol. 2005;49(3):177–182. [PubMed] [Google Scholar]
- 77.Kim S.H., Wanasen N., Paldurai A., Xiao S., Collins P.L., Samal S.K. Newcastle disease virus fusion protein is the major contributor to protective immunity of genotype-matched vaccine. PLoS One. 2013;8(8):e74022. doi: 10.1371/journal.pone.0074022. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Bankowski R.A., Conrad R.D., Reynolde B. Avian influenza A and paramyxo viruses complicating respiratory disease diagnosis in poultry. Avian Dis. 1968;12(2):259–278. doi: 10.2307/1588226. [DOI] [PubMed] [Google Scholar]
- 79.Susta L., Cornax I., Diel D.G., Garcia S.C., Miller P.J., Liu X., Hu S., Brown C.C., Afonso C.L. Expression of interferon gamma by a highly virulent strain of Newcastle disease virus decreases its pathogenicity in chickens. Microb. Pathog. 2013;61-62:73–83. doi: 10.1016/j.micpath.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 80.Capua I., Marangon S. The use of vaccination as an option for the control of avian influenza. Avian Pathol. 2003;32(4):335–343. doi: 10.1080/0307945031000121077. [DOI] [PubMed] [Google Scholar]
- 81.Cross T.A., Arsnoe D.M., Minnis R.B., King D.T., Swafford S., Pedersen K., Owen J.C. Prevalence of avian paramyxovirus 1 and avian influenza virus in double-crested Cormorants (Phalacrocorax auritus) in eastern North America. J. Wildl. Dis. 2013;49(4):965–977. doi: 10.7589/2012-06-164. [DOI] [PubMed] [Google Scholar]
- 82.Teske L., Ryll M., Rautenschlein S. Epidemiological investigations on the role of clinically healthy racing pigeons as a reservoir for avian paramyxovirus-1 and avian influenza virus. Avian Pathol. 2013;42(6):557–565. doi: 10.1080/03079457.2013.852157. [DOI] [PubMed] [Google Scholar]
- 83.Tolf C., Wille M., Haidar A.K., Avril A., Zohari S., Waldenström J. Prevalence of avian paramyxovirus type 1 in Mallards during autumn migration in the western Baltic Sea region. Virol. J. 2013;10(1):285. doi: 10.1186/1743-422X-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Durham P.J.K., Poole W.S.H., Gow A., Watters C.B. Characteristics of lentogenic strains of Newcastle disease virus isolated in New Zealand. N. Z. Vet. J. 1980;28(6):108–112. doi: 10.1080/00480169.1980.34713. [DOI] [PubMed] [Google Scholar]
- 85.Alexander D.J. Newcastle disease in countries of the European Union. Avian Pathol. 1995;24(1):3–10. doi: 10.1080/03079459508419045. [DOI] [PubMed] [Google Scholar]
- 86.Stanislawek W.L., Wilks C.R., Meers J., Horner G.W., Alexander D.J., Manvell R.J., Kattenbelt J.A., Gould A.R. Avian paramyxoviruses and influenza viruses isolated from mallard ducks (Anas platyrhynchos) in New Zealand. Arch. Virol. 2002;147(7):1287–1302. doi: 10.1007/s00705-002-0818-2. [DOI] [PubMed] [Google Scholar]
- 87.Liu H., Wang Z., Wang Y., Sun C., Zheng D., Wu Y. Characterization of Newcastle disease virus isolated from waterfowl in China. Avian Dis. 2008;52(1):150–155. doi: 10.1637/8030-061507-Reg. [DOI] [PubMed] [Google Scholar]
- 88.Lee E.K., Jeon W.J., Kwon J.H., Yang C.B., Choi K.S. Molecular epidemiological investigation of Newcastle disease virus from domestic ducks in Korea. Vet. Microbiol. 2009;134(3-4):241–248. doi: 10.1016/j.vetmic.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 89.Vidanović D., Sekler M., Asanin R., Milić N., Nisavić J., Petrović T., Savić V. Characterization of velogenic Newcastle disease viruses isolated from dead wild birds in Serbia during 2007. J. Wildl. Dis. 2011;47(2):433–441. doi: 10.7589/0090-3558-47.2.433. [DOI] [PubMed] [Google Scholar]
- 90.Bozorgmehri-Fard M.H., Keyvanfar H. Isolation of Newcastle disease virus from teals (Anas crecca) in Iran. J. Wildl. Dis. 1979;15(2):335–337. doi: 10.7589/0090-3558-15.2.335. [DOI] [PubMed] [Google Scholar]
- 91.Macpherson I., Watt R.G., Alexander D.J. Isolation of avian paramyxovirus other than Newcastle disease virus from commercial poultry in Great Britain. Vet. Rec. 1983;112(20):479–480. doi: 10.1136/vr.112.20.479. [DOI] [PubMed] [Google Scholar]
- 92.Rue C.A., Susta L., Brown C.C., Pasick J.M., Swafford S.R., Wolf P.C., Killian M.L., Pedersen J.C., Miller P.J., Afonso C.L. Evolutionary changes affecting rapid identification of 2008 Newcastle disease viruses isolated from double-crested cormorants. J. Clin. Microbiol. 2010;48(7):2440–2448. doi: 10.1128/JCM.02213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jindal N., Chander Y., Chockalingam A.K., de Abin M., Redig P.T., Goyal S.M. Phylogenetic analysis of Newcastle disease viruses isolated from waterfowl in the upper midwest region of the United States. Virol. J. 2009;6:191. doi: 10.1186/1743-422X-6-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim L.M., King D.J., Guzman H., Tesh R.B., Travassos da Rosa A.P., Bueno R., Jr, Dennett J.A., Afonso C.L. Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. J. Clin. Microbiol. 2008;46(10):3303–3310. doi: 10.1128/JCM.00644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schuler K.L., Green D.E., Justice-Allen A.E., Jaffe R., Cunningham M., Thomas N.J., Spalding M.G., Ip H.S. Expansion of an exotic species and concomitant disease outbreaks: Pigeon paramyxovirus in free-ranging Eurasian collared doves. EcoHealth. 2012;9(2):163–170. doi: 10.1007/s10393-012-0758-6. [DOI] [PubMed] [Google Scholar]
- 96.Alexander D.J., Pattison M., Macpherson I. Avian paramyxoviruses of PMV-3 serotype in British turkeys. Avian Pathol. 1983;12(4):469–482. doi: 10.1080/03079458308436192. [DOI] [PubMed] [Google Scholar]
- 97.Xie Z., Xie L., Chen A., Liu J., Pang Y., Deng X., Xie Z., Fan Q. Complete genome sequence analysis of a Newcastle disease virus isolated from a wild egret. J. Virol. 2012;86(24):13854–13855. doi: 10.1128/JVI.02669-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alexander D.J. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 2011;40(6):547–558. doi: 10.1080/03079457.2011.618823. [DOI] [PubMed] [Google Scholar]
- 99.Byarugaba D.K., Mugimba K.K., Omony J.B., Okitwi M., Wanyana A., Otim M.O., Kirunda H., Nakavuma J.L., Teillaud A., Paul M.C., Ducatez M.F. High pathogenicity and low genetic evolution of avian paramyxovirus type I (Newcastle disease virus) isolated from live bird markets in Uganda. Virol. J. 2014;11:173. doi: 10.1186/1743-422X-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulisa D.D., Alemu R.B., Keno M.S., Furaso A., Heidari A., Chibsa T.R., Chunde H.C. Characterization of Newcastle disease virus and poultry-handling practices in live poultry markets, Ethiopia. Springerplus. 2014;3:459. doi: 10.1186/2193-1801-3-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wakamatsu N., King D.J., Kapczynski D.R., Seal B.S., Brown C.C. Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002-2003. Vet. Pathol. 2006;43(6):925–933. doi: 10.1354/vp.43-6-925. [DOI] [PubMed] [Google Scholar]
- 102.Aldous E.W., Alexander D.J. Newcastle disease in pheasants (Phasianus colchicus): A review. Vet. J. 2008;175(2):181–185. doi: 10.1016/j.tvjl.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Higgins D.A. Nine disease outbreaks associated with myxoviruses among ducks in Hong Kong. Trop. Anim. Health Prod. 1971;3(4):232–240. doi: 10.1007/BF02359585. [DOI] [PubMed] [Google Scholar]
- 104.Jinding C., Ming L., Tao R., Chaoan X. A goose-sourced paramyxovirus isolated from southern China. Avian Dis. 2005;49(1):170–173. doi: 10.1637/7270-090101R1. [DOI] [PubMed] [Google Scholar]
- 105.Zou J., Shan S., Yao N., Gong Z. Complete genome sequence and biological characterizations of a novel goose paramyxovirus-SF02 isolated in China. Virus Genes. 2005;30(1):13–21. doi: 10.1007/s11262-004-4577-x. [DOI] [PubMed] [Google Scholar]
- 106.Wan H., Chen L., Wu L., Liu X. Newcastle disease in geese: Natural occurrence and experimental infection. Avian Pathol. 2004;33(2):216–221. doi: 10.1080/0307945042000195803. [DOI] [PubMed] [Google Scholar]
- 107.Alexander D.J., editor. Newcastle disease. Boston, MA: Kluwer Academic; 1988. Vindevogel H aJPD. Panzootic Newcastle disease virus in pigeon. pp. 184–196. [Google Scholar]
- 108.Zanetti F., Mattiello R., Garbino C., Kaloghlian A., Terrera M.V., Boviez J., Palma E., Carrillo E., Berinstein A. Biological and molecular characterization of a pigeon paramyxovirus type-1 isolate found in Argentina. Avian Dis. 2001;45(3):567–571. doi: 10.2307/1592896. [DOI] [PubMed] [Google Scholar]
- 109.Beck I., Gerlach H., Burkhardt E., Kaleta E.F. Investigation of several selected adjuvants regarding their efficacy and side effects for the production of a vaccine for parakeets to prevent a disease caused by a paramyxovirus type 3. Vaccine. 2003;21(9-10):1006–1022. doi: 10.1016/S0264-410X(02)00552-2. [DOI] [PubMed] [Google Scholar]
- 110.Panigrahy B., Senne D.A., Pearson J.E., Mixson M.A., Cassidy D.R. Occurrence of velogenic viscerotropic Newcastle disease in pet and exotic birds in 1991. Avian Dis. 1993;37(1):254–258. doi: 10.2307/1591484. [DOI] [PubMed] [Google Scholar]
- 111.Erickson G.A., Maré C.J., Gustafson G.A., Miller L.D., Proctor S.J., Carbrey E.A. Interactions between viscerotropic velogenic Newcastle diseases virus and pet birds of six species. I. Clinical and serologic responses, and viral excretion. Avian Dis. 1977;21(4):642–654. doi: 10.2307/1589424. [DOI] [PubMed] [Google Scholar]
- 112.Allison A.B., Gottdenker N.L., Stallknecht D.E. Wintering of neurotropic velogenic Newcastle disease virus and West Nile virus in double-crested cormorants (Phalacrocorax auritus) from the Florida Keys. Avian Dis. 2005;49(2):292–297. doi: 10.1637/7278-091304R. [DOI] [PubMed] [Google Scholar]
- 113.Kuiken T., Heckert R.A., Riva J., Leighton F.A., Wobeser G. Excretion of pathogenic Newcastle disease virus by double-crested cormorants (Phalacrocorax auritus) in absence of mortality or clinical signs of disease. Avian Pathol. 1998;27(6):541–546. doi: 10.1080/03079459808419381. [DOI] [PubMed] [Google Scholar]
- 114.Alexander D.J., Collins M.S. Pathogenicity of PMV-3/parakeet/Netherlands/449/75 for chickens. Avian Pathol. 1982;11(1):179–185. doi: 10.1080/03079458208436091. [DOI] [PubMed] [Google Scholar]
- 115.Tumova B., Robinson J.H., Easterday B.C. A hitherto unreported paramyxovirus of turkeys. Res. Vet. Sci. 1979;27(2):135–140. [PubMed] [Google Scholar]
- 116.Chen S., Hao H., Liu Q., Wang R., Zhang P., Wang X., Du E., Yang Z. Phylogenetic and pathogenic analyses of two virulent Newcastle disease viruses isolated from Crested Ibis (Nipponia nippon) in China. Virus Genes. 2013;46(3):447–453. doi: 10.1007/s11262-013-0881-7. [DOI] [PubMed] [Google Scholar]
- 117.Pchelkina I.P., Manin T.B., Kolosov S.N., Starov S.K., Andriyasov A.V., Chvala I.A., Drygin V.V., Yu Q., Miller P.J., Suarez D.L. Characteristics of Pigeon Paramyxovirus serotype-1 isolates (PPMV-1) from the Russian Federation from 2001 to 2009. Avian Dis. 2013;57(1):2–7. doi: 10.1637/10246-051112-Reg.1. [DOI] [PubMed] [Google Scholar]
- 118.Absalón A.E., Mariano-Matías A., Vásquez-Márquez A., Morales-Garzón A., Cortés-Espinosa D.V., Ortega-García R., Lucio-Decanini E. Complete genome sequence of a velogenic Newcastle disease virus isolated in Mexico. Virus Genes. 2012;45(2):304–310. doi: 10.1007/s11262-012-0782-1. [DOI] [PubMed] [Google Scholar]
- 119.Miller P.J., King D.J., Afonso C.L., Suarez D.L. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine. 2007;25(41):7238–7246. doi: 10.1016/j.vaccine.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 120.Bonfante F., Terregino C., Heidari A., Monne I., Salviato A., Taddei R., Raffini E., Capua I. Identification of APMV-1 associated with high mortality of collared doves (Streptoelia decaocto) in Italy. Vet. Rec. 2012;171(13):327. doi: 10.1136/vr.100448. [DOI] [PubMed] [Google Scholar]
- 121.Liu X., Wang X., Wu S., Hu S., Peng Y., Xue F., Liu X. Surveillance for avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated. Avian Pathol. 2009;38(5):377–391. doi: 10.1080/03079450903183637. [DOI] [PubMed] [Google Scholar]
- 122.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qiu X., Sun Q., Wu S., Dong L., Hu S., Meng C., Wu Y., Liu X. Entire genome sequence analysis of genotype IX Newcastle disease viruses reveals their early-genotype phylogenetic position and recent-genotype genome size. Virol. J. 2011;8:117. doi: 10.1186/1743-422X-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi S.H., Huang Y., Cui S.J., Cheng L.F., Fu G.H., Li X., Chen Z., Peng C.X., Lin F., Lin J.S., Su J.L. Genomic sequence of an avian paramyxovirus type 1 strain isolated from Muscovy duck (Cairina moschata) in China. Arch. Virol. 2011;156(3):405–412. doi: 10.1007/s00705-010-0866-y. [DOI] [PubMed] [Google Scholar]
- 125.Usachev E.V., Fediakina I.T., Shchelkanov M.Iu., L’vov D.N., Prilipov A.G., Iamnikova S.S. Molecular genetic characteristics of the Newcastle Sterna/Astrakhan/Z275/2001 virus isolated in Russia. Mol. Gen. Mikrobiol. Virusol. 2006;1(1):14–20. [PubMed] [Google Scholar]
- 126.Briand F.X., Henry A., Massin P., Jestin V. Complete genome sequence of a novel avian paramyxovirus. J. Virol. 2012;86(14):7710. doi: 10.1128/JVI.00946-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Desingu P.A., Singh S.D., Dhama K., Kumar O.R., Singh R., Singh R.K. A rapid method of accurate detection and differentiation of Newcastle disease virus pathotypes by demonstrating multiple bands in degenerate primer based nested RT-PCR. J. Virol. Methods. 2015;212:47–52. doi: 10.1016/j.jviromet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 128.Kumar S., Samal S.K. Pathogenicity of avian paramyxovirus serotype-3 in Chickens and Turkeys. In: Perez- Marin, C. C. (ed.), A Bird’s-Eye View of Veterinary Medicine, pp. 587-596. In Tech, Croatia 2012. [DOI] [Google Scholar]
- 129.Handel K.A., Pasick J. Newcastle Disease Virus NDV/Turkey/Ontario/CN36/2003 matrix protein (M) and fusion protein (F) genes partial cds: Direct submitted by Virology, CFIA-NCFAD, 1015 Arlington Street, Winnipeg, Manitoba R3E 3M4, Canada. https://www.ncbi.nlm.nih.gov/nuccore/ay289194. 2016.
- 130.Liu H., Zhang P., Wu P., Chen S., Mu G., Duan X., Hao H., Du E., Wang X., Yang Z. Phylogenetic characterization and virulence of two Newcastle disease viruses isolated from wild birds in China. Infect. Genet. Evol. 2013;20:215–224. doi: 10.1016/j.meegid.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 131.Duan X., Zhang P., Ma J., Chen S., Hao H., Liu H., Fu X., Wu P., Zhang D., Zhang W., Du E., Yang Z. Characterization of genotype IX Newcastle disease virus strains isolated from wild birds in the northern Qinling Mountains, China. Virus Genes. 2014;48(1):48–55. doi: 10.1007/s11262-013-0987-y. [DOI] [PubMed] [Google Scholar]
- 132.Hines N.L., Killian M.L., Pedersen J.C., Reising M.M., Mosos N.A., Mathieu-Benson C., Miller C.L. An rRT-PCR assay to detect the matrix gene of a broad range of avian paramyxovirus serotype-1 strains. Avian Dis. 2012;56(2):387–395. doi: 10.1637/10035-120811-Reg.1. [DOI] [PubMed] [Google Scholar]
- 133.Ramey A.M., Reeves A.B., Ogawa H., Ip H.S., Imai K., Bui V.N., Yamaguchi E., Silko N.Y., Afonso C.L. Genetic diversity and mutation of avian paramyxovirus serotype 1 (Newcastle disease virus) in wild birds and evidence for intercontinental spread. Arch. Virol. 2013;158(12):2495–2503. doi: 10.1007/s00705-013-1761-0. [DOI] [PubMed] [Google Scholar]
- 134.Wise M.G., Suarez D.L., Seal B.S., Pedersen J.C., Senne D.A., King D.J., Kapczynski D.R., Spackman E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004;42(1):329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bhuvaneswari S., Tirumurugaan K.G., Jones J.C., Kumanan K. Complete genome sequence of a Newcastle disease virus from a Coturnix coturnix japonica (Japanese Quail) covey in India. Genome Announc. 2014;2(3):e00374-14. doi: 10.1128/genomeA.00374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shi S.H., Huang Y., Cui S.J., Cheng L.F., Fu G.H., Li X., Chen Z., Peng C.X., Lin F., Lin J.S., Su J.L. Genomic sequence of an avian paramyxovirus type 1 strain isolated from Muscovy duck (Cairina moschata) in China. Arch. Virol. 2011;156(3):405–412. doi: 10.1007/s00705-010-0866-y. [DOI] [PubMed] [Google Scholar]
- 137.Aldous E.W., Fuller C.M., Mynn J.K., Alexander D.J. A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathol. 2004;33(2):258–269. doi: 10.1080/0307945042000195768. [DOI] [PubMed] [Google Scholar]
- 138.Cai S., Li J., Wong M.T., Jiao P., Fan H., Liu D., Liao M., Jiang J., Shi M., Lam T.T., Ren T., Leung F.C. Genetic characterization and evolutionary analysis of 4 Newcastle disease virus isolate full genomes from waterbirds in South China during 2003-2007. Vet. Microbiol. 2011;152(1-2):46–54. doi: 10.1016/j.vetmic.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 139.Alexander D.J. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 2011;40(6):547–558. doi: 10.1080/03079457.2011.618823. [DOI] [PubMed] [Google Scholar]
- 140.Zhu W., Dong J., Xie Z., Liu Q., Khan M.I. Phylogenetic and pathogenic analysis of Newcastle disease virus isolated from house sparrow (Passer domesticus) living around poultry farm in southern China. Virus Genes. 2010;40(2):231–235. doi: 10.1007/s11262-009-0436-0. [DOI] [PubMed] [Google Scholar]
- 141.Ghalyanchi-Langeroudi A., Hosseini H., Madadgar O., Karimi V., Shahraeini A., Ghafari M.M. Sequence Analysis of Fusion Gene of Newcastle Disease Viruses Isolated from Ostrich (Struthio camelus) in Iran. Indian J. Virol. 2011;5(3):12–17. [Google Scholar]