Abstract
Background
Plants of the genus Taxus have attracted much attention owing to the natural product taxol, a successful anti-cancer drug. T. fuana and T. yunnanensis are two endangered Taxus species mainly distributed in the Himalayas. In our study, an untargeted metabolomics approach integrated with a targeted UPLC-MS/MS method was applied to examine the metabolic variations between these two Taxus species growing in different environments.
Results
The level of taxol in T. yunnanensis is much higher than that in T. fuana, indicating a higher economic value of T. yunnanensis for taxol production. A series of specific metabolites, including precursors, intermediates, competitors of taxol, were identified. All the identified intermediates are predominantly accumulated in T. yunnanensis than T. fuana, giving a reasonable explanation for the higher accumulation of taxol in T. yunnanensis. Taxusin and its analogues are highly accumulated in T. fuana, which may consume limited intermediates and block the metabolic flow towards taxol. The contents of total flavonoids and a majority of tested individual flavonoids are significantly accumulated in T. fuana than T. yunnanensis, indicating a stronger environmental adaptiveness of T. fuana.
Conclusions
Systemic metabolic profiling may provide valuable information for the comprehensive industrial utilization of the germplasm resources of these two endangered Taxus species growing in different environments.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1412-4) contains supplementary material, which is available to authorized users.
Keywords: Flavonoids, Germplasm resources, Metabolite, Metabolomics, Taxus, Taxol
Background
Plants of the genus Taxus, coniferous trees or shrubs known as yews, are major sources for various bioactive compounds, such as paclitaxel and its derivatives [1]. The generic paclitaxel, is marketed under the registered trade name Taxol, which is one of the most successful anti-cancer drugs utilized in the past years [2]. The market demand for taxol exceeds the supply, which is limited by several restrictive conditions, such as the slow growth of wild yews, the low taxoid contents and the complex synthesis routes [3].
The biosynthetic pathways of Taxol have been revealed, resulting in a large number of precursors, intermediates and derivatives of Taxol [2]. Firstly, the plastidial 2-C-methyl-D-erythritol phosphate pathway supplies 3 units of the C5 isoprenoid precursor isopentenyl diphosphate and one unit of dimethylallyl diphosphate to synthesize the precursor of the diterpenoid taxane core geranylgeranyl diphosphate [4, 5]. The formation of the taxane skeleton is initiated by the cyclization of geranylgeranyl diphosphate yielding taxa-4(5),11(12)-diene, which is mediated by the important enzyme taxadiene synthase [6, 7]. Then, several intermediate enzymatic reactions, such as hydroxylation, acetylation and N-benzoylation, occur in the pathway [2]. For example, the acetylation of 10-deacetylbaccatin III (10-DAB) is an essential step to produce baccatin III, which is an advanced precursor for Taxol biosynthesis [8, 9]. Thereafter, the C13-side chain is appended to baccatin III to form N-debenzoyl-2′-deoxytaxol [10, 11]. The final step of Taxol biosynthesis is conducted by the side chain N-benzoyl transferase using N-debenzoyltaxol as the actual substrate [12].
Over 500 taxane diterpenoid compounds, including cephalomannine, 7-epi 10-desacetyl paclitaxel and 7-epipaclitaxel, have been identified [13–15]. Recently, novel taxoids have been isolated from different Taxus species. For example, taxezopidine J and taxuspine D from the seeds of T. cuspidata, a new taxane glucoside from the bark of T. mairei, two new rearranged taxane diterpenoids from the barks of T. wallichiana, and three new taxane diterpenoids, namely baccatin VIII, IX and X, from the seeds of T. yunnanensis were identified over the last two decades [16–19]. Interestingly, intermediate oxygenation steps produce a broad range of taxusin-like metabolites, which are considered as dead-end metabolites and cannot be used for Taxol biosynthesis [20, 21].
An untargeted metabolome provides a shortcut to systematically analyze and compare the features of primary and secondary metabolites among species [22]. Studies of the metabolomes in the Taxus genus have been carried out since 2003, profiling the differential metabolites of T. media cultures with or without methyl jasmonate treatments [23]. In 2009, the defense mechanism of T. cuspidata cells to shear stress was uncovered using a metabolic profiling tool [24]. Fluctuations in the synthesis of taxoids in cultured seedlings of T. mairei have also been investigated using a metabolomics approach [25]. The crucial network that controls the biosynthesis of Taxol and its precursors under methyl jasmonate treatments was elucidated by metabolomics profiling data [26]. An integrated metabolomics and transcriptomics analysis of T. chinensis cells under different dissolved oxygen conditions suggests a relationship between the changes in taxane production caused by the shift in dissolved oxygen and the global variations in the cell’s central carbon metabolism [27]. Recently, an integrated proteomics/metabolomics analysis revealed an important role of a short-term high doses of ultraviolet (UV)-A radiation in promoting Taxol production in T. mairei [28].
To date, ~ 14 species of the Taxus genus have been identified; however, the levels of accumulated taxoids can vary significantly among different species and cultivars [29, 30]. T. fuana and T. yunnanensis are two slow-growing and endangered species mainly distributed in southwestern China [31]. Investigations into the metabolic features contributing to the interspecific differential accumulation of taxoids, as well as other secondary metabolites, between these two morphologically similar Taxus species will enhance the highest-yielding species breeding and reveal the mechanism underlying both of the species- and environment- dependent metabolic variations. Furthermore, the Himalayas is one of few paradises of rare vegetation in the world. Our study may provide valuable information for the conservation of these germplasm resources of these two endangered Taxus species.
Methods
Plant materials
Fresh twig samples were harvested from 15 independent 10-year-old wild Taxus trees of T. fuana in Jilong (29° 22’ N, 95° 26′ E, Tibet, China) and T. yunnanensis in Motuo (28° 28’ N, 85° 13′ E, Tibet, China), respectively (Fig. 1a). The average altitude of Jilong is 4,000 m and the average altitude of Motuo is 1,200 m.
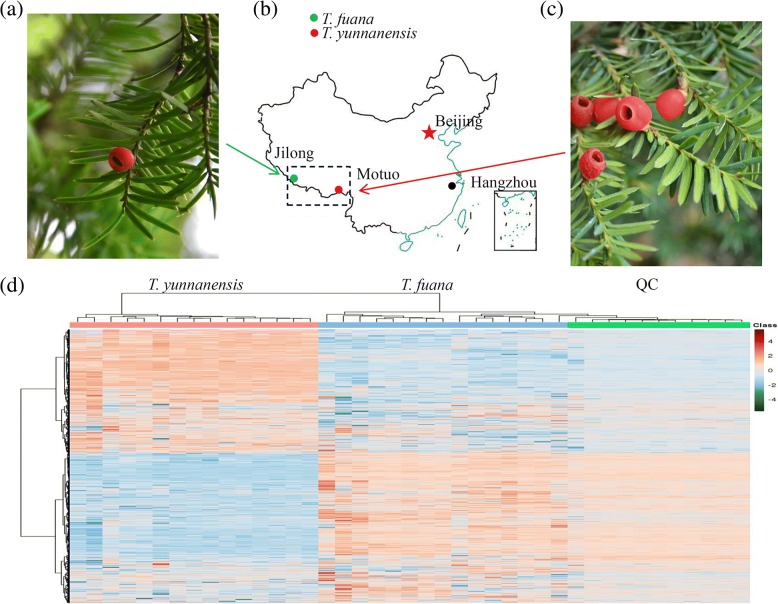
Fig. 1.
Untargeted metabolite profiling reveals the variations in the abundance of metabolites between T. yunnanensis and T. fuana. a A picture of the twigs of T. fuana. b The collection sites of T. fuana (Jilong) and T. yunnanensis (Motuo) were indicated by green and red small dots, respectively. c A picture of the twigs of T. yunnanensis. d A heatmap of the metabolites identified in the metabolomes of the two Taxus species (n = 15). The heatmap scale ranges from − 4 to + 4 on a log2 scale
Standards and reagents
Paclitaxel (PTX; ≥99%), 10-deacetylbaccatin III (10-DAB; ≥98%), baccatin III (BAC; ≥99%), apigenin (API; ≥98%), quercetin (QUE; ≥98.5%), luteolin (LUT; ≥98%), kaempferol (KAE; ≥98%), (+)-catechin (CAT; ≥97%) and (−)-epicatechin (E-CAT; ≥98%) were obtained from Aladdin Biochemical Technology (Shanghai, China). 10-Deacetyl paclitaxel (DAP; 98%), 7-epipaclitaxel (7-E-PTX; 98%), amentoflavone (AMF; 98%) and ginkgetin (GKG; 98%) were purchased from J&K Scientific (Beijing, China). 7-Epi 10-desacetyl paclitaxel (7-E-DAP) was obtained from Toronto Research Chemicals (Toronto, Canada). Cephalomannine (CEP; ≥98.5%) was provided by MUST Bio-Technology (Chengdu, China). Methanol (HPLC grade) and formic acid (HPLC grade) were obtained from Merck (Kenilworth, NJ, USA).
Metabolite extraction for the untargeted metabolomic analysis
The twig samples from the two Taxus species (25 mg each, n = 15) were transferred to a microcentrifige tube. An aliquot of 800 μL pre-cooled 50% methanol was added to the tube. The mixture was ground at a rate of 1,900 strokes/min for 2 min using a 2010 Geno/Grinder (SPEX SamplePrep, Metuchen, NJ, USA). The samples were mixed with 500 μL of pre-cooled chloroform/methanol/water (v:v:v, 1:3:1), vortexed at 4 °C in the dark for 15 min, and subjected to ultrasonication for 5 min on ice. After centrifugation at 13,000 g for 15 min at 4 °C, the supernatant was transferred to a new tube. The extraction solution was vacuum-dried and resuspended in 50% methanol. Meanwhile, a quality control (QC) sample (a pool of all the samples) was prepared by mixing an equal volume of each extraction sample from one Taxus species. All the samples were stored at − 80 °C prior to Ultra-performance liquid chromatography (UPLC)-MS/MS analysis.
UPLC-MS/MS analysis for the untargeted metabolomics
The chromatographic separations of all the samples were performed using a SCIEX UPLC system (Applied Biosystems, Foster City, CA, USA). An ACQUITY BEH Amide column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, MA, USA) was used for the reversed-phase separation. The column oven was maintained at 35 °C and the flow rate was 0.4 mL/min. The mobile phase consisted of solvent A (aqueous solution with 25 mM ammonium acetate and 25 mM NH4H2O) and solvent B (IPA:ACN = 9:1 + 0.1% formic acid). The gradient elution procedures were set as follows: 0 ~ 0.5 min, 95% solvent A; 0.5 ~ 9.5 min, 95% to 65% solvent A; 9.5 ~ 10.5 min, 65% ~ 40% solvent A; 10.5 ~ 12.0 min, 40% solvent A; 12.0 ~ 12.2 min,40% ~ 95% solvent A; 12.2 ~ 15.0 min, 95% solvent A. The injection volume for each sample was 4 μL.
A high-resolution tandem mass spectrometer SCIEX Triple-TOF-5600 plus (Applied Biosystems) was used to detect metabolites eluted from the column. The Q-TOF was operated in both positive and negative ion modes. The curtain gas was set at a pressure of 30 psi, ion source gas 1 and ion source gas 2 were both set at 60 psi, and the interface heater temperature was set at 650 °C. The ion spray voltages were set at 5000 V and - 4500 V for the positive and negative ion modes, respectively. The mass spectrometry data were acquired in information-dependent acquisition (IDA) mode. The TOF mass range was from 60 to 1,200 Da. The survey scans were acquired over 150 ms and as many as 12 product ion scans were collected if exceeding a threshold of 100 counts per second and with a 1+ charge-state. The total cycle time was held constant at 0.56 s. Four time bins were summed for each scan at a pulser frequency value of 11 kHz by monitoring of the 40 GHz multichannel time-to-digital converter (TDC) detector with four-anode/channel detection. The dynamic exclusion was set to 4 s. During the acquisition, the mass accuracy was calibrated every 20 samples. Furthermore, in order to evaluate the system stability of UPLC-MS/MS analysis, a QC sample was analyzed right after every 10 samples.
Bioinformatic analysis of the untargeted metabolomic dataset
The acquired MS data pretreatments including peak picking, peak grouping, retention time correction, second peak grouping, and annotation of isotopes and adducts were performed using the XCMS software. LC−MS raw data files were converted into mz.XML format and then processed by the XCMS, CAMERA and metaX toolbox implemented with the R software. Each ion was identified by the retention time (RT) and m/z data. Intensities of each peak were recorded and a three dimensional matrix containing arbitrarily assigned peak indices (retention time-m/z pairs), sample names (observations) and ion intensity information (variables) was generated.
The online KEGG, HMDB databases were used to annotate the metabolites by matching the exact molecular mass data (m/z) of the samples with those from the databases. If a mass difference between the observed value and the database value was less than 10 ppm, the metabolite would be annotated and the molecular formula of the metabolite would further be identified and validated by the isotopic distribution measurement. In addition, an in-house fragment spectrum library of metabolites was also used to validate the metabolite identification.
The intensities of peak data were further preprocessed by an in-house software metaX. Those features that were detected in less than 50% of the QC samples or 80% of the biological samples were removed, and the remaining peaks with missing values were imputed with the k-nearest neighbor algorithm to further improve the data quality. Principal component analysis (PCA) was performed for outlier detection and batch effect evaluation using the pre-processed dataset. Quality control-based robust LOESS signal correction was fitted to the QC data with respect to the order of injection to minimize signal intensity drift over time. In addition, the relative standard deviations of the metabolic features were calculated across all the QC samples, and those > 30% were then removed.
Quantitative analysis of taxoids and flavonoids
Twigs were harvested from six trees of T. fuana and T. yunnanensis, respectively. The samples were thoroughly dried at 40 °C and then ground into powder. A modified version of a previously published method was used to prepare crude extracts [32]. In brief, the powder was passed through a filter (mesh size 0.42 mm) and 0.5 g of the fine powder was precisely weighed, and mixed with 15 mL of 100% methanol. The mixture was ultrasonicated at 150 W and 40 kHz for 30 min. After centrifugation at 5000 rpm for 5 min, the supernatant was collected. The residue was re-suspended in 15 mL methanol and subjected to the same extraction procedures. After centrifugation, the second supernatant was combined with the first extract. Before UPLC-MS/MS analysis, the crude Taxus extracts were diluted at a ratio of 1:10 and all the samples were passed through 0.22 μm membrane filters.
The quantification of seven taxoids and eight flavonoids was carried out using an LC-30 AD UPLC system (Shimadzu, Japan) coupled with a SCIEX QTRAP 6500 mass spectrometer (Applied Biosystems). The MultiQuant software (version 3.0) was used for data acquisition and processing.
To separate seven taxoids, the UPLC was performed on a Kinetex C18 column (100 × 4.6 mm, 2.6 μm; Phenomenex, Torrance, CA, USA). The mobile phase consisted of 65% solvent A (methanol) and 35% solvent B (aqueous solution with 0.1% formic acid and 2 mM ammonium formate). The flow rate was set at 0.2 mL/min. The column oven was maintained at 30 °C and the injection volume was 5 μL. The capillary temperature was set at 270 °C and the ion spray voltage was set at 3.0 kV. Multiple reaction monitoring (MRM) was applied for the quantification in the positive ionization mode. The transitions of m/z 876.4 → 308.2, 567.4 → 445.3, 609.5 → 427.3, 834.4 → 308.2, 832.3 → 264.1, 876.4 → 591.4 and 834.4 → 308.2 were used for the quantification of PTX, 10-DAB, BAC, DAP, CEP, 7-E-PTX and 7-E-DAP, respectively.
The separation of eight flavonoids was achieved on an ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters). The gradient elution was performed with solvent A (methanol) and solvent B (aqueous solution with 0.1% formic acid and 2 mM ammonium formate) at a flow rate of 0.3 mL/min using the following programs: 0 ~ 2.0 min, 5% solvent A; 2.0 ~ 3.0 min, 5 ~ 35% solvent A; 3.0 ~ 10.0 min, 35 ~ 50% solvent A; 10.0 ~ 13.0 min, 50 ~ 65% solvent A; 13.0 ~ 14.5 min, 65% solvent A; 14.5 ~ 15.0 min, 65 ~ 90% solvent A; 15.0 ~ 17.0 min, 90% solvent A; 17.0 ~ 17.1 min, 90 ~ 5% solvent A; 17.1 ~ 20.0 min, 5% solvent A. The column oven was maintained at 40 °C and the injection volume was 5 μL. The determination of QUE, LUT, KAE, AMF and GKG was performed in the negative ion mode. The transitions of m/z 301.1 → 151.0, 285.0 → 133.0, 285.1 → 159.0, 537.0 → 374.9 and 565.2 → 533.1 were used for the quantification of QUE, LUT, KAE, AMF and GKG, respectively. The measurement of CAT, E-CAT and API was performed in the positive ion mode. The transitions of m/z 291.1 → 139.0, 291.0 → 139.0 and 271.0 → 153.0 were used for the quantification of CAT, E-CAT and API, respectively.
Statistical analysis
For the untargeted metabolomic analysis, Wilcoxon tests were carried out to examine metabolic differences between every two samples. The P values were adjusted for multiple testing correction by false discovery rate (FDR; Benjamini–Hochberg). The supervised partial least squares-discriminant analysis (PLS-DA) was conducted using the metaX to discriminate different variables between groups. The VIP value was calculated and a VIP cut-off value of 1.0 was used to select important features.
The quantification results of taxoids and flavonoids are presented as mean ± SD (n = 6). The statistical analysis was performed using SPSS software (version 19.0; SPSS Inc., Chicago, IL, USA). A one-way analysis of variance (ANOVA) was carried out to compare the content differences of taxoids and flavonoids between T. fuana and T. yunnanensis. A P value < 0.05 was considered statistically significant.
Results
Untargeted metabolite profiling of two endangered Taxus species
To explore global metabolic variations, an untargeted metabolomic approach was used (n = 15), which identified 4,986 annotated metabolites from 7,251 ion features (Additional file 1). To examine the quality of the acquired MS data, total ion chromatograms (TICs) were generated for all of the samples and revealed a high degree of overlap (Additional file 2). In addition, the m/z width and retention-time width analyses suggested that the sample preparation and instrument state met the required standards (Additional file 3). An overview of the metabolite profiling of T. fuana and T. yunnanensis is shown in Fig. 1d. Unlike the similarity in twig morphology, dramatic variations in the metabolomes between T. fuana and T. yunnanensis were observed.
Based on their annotations, a number of metabolites were assigned to at least one primary or secondary metabolic category. The top 20 largest metabolic categories, such as porphyrin and chlorophyll metabolism (134 metabolites), 2-oxocarboxylic acid metabolism (134 metabolites), biosynthesis of amino acids (127 metabolites), ABC transporters (126 metabolites), diterpenoid biosynthesis (103 metabolites), and other secondary metabolites (141 metabolites) are shown in Additional file 4 and Additional file 5.
Metabolite profiling reveals variations in the abundance levels of metabolites between T. fuana and T. yunnanensis
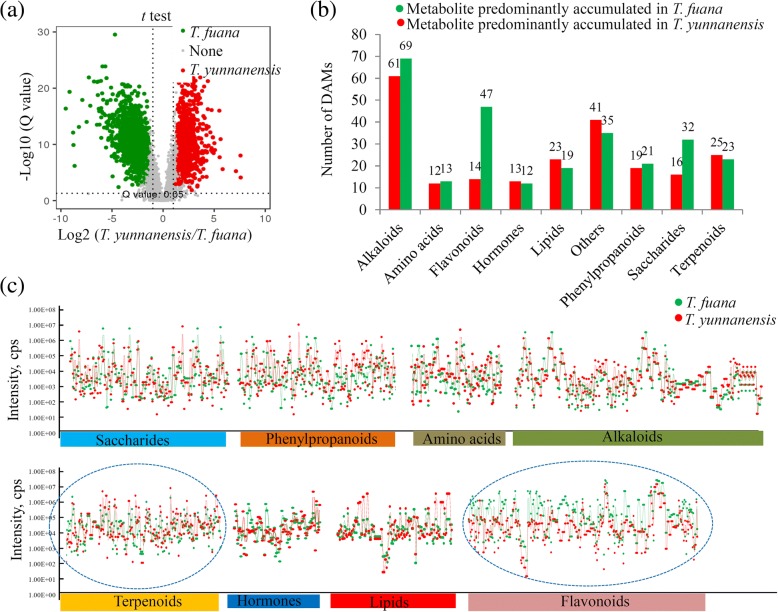
To provide a deep overview of the metabolic variations, several quality control parameters for the quantification, including coefficient of variation (CV), principal component (PC) and normalized intensity, were analyzed. The CV values were lower than 30%, suggesting a good repeatability (Additional file 6). The PC analysis showed that PC1 and PC2 were responsible for 59.75% and 5.43% of the variation, respectively, indicating a clear separation between these two Taxus species (Additional file 7). After filtering, 4,986 high-quality metabolites were used to screen the differentially accumulated metabolites (DAMs). The statistical analysis identified 1,972 significant DAMs, including 788 T. yunnanensis predominantly accumulated metabolites and 1,184 T. fuana predominantly accumulated metabolites (Additional file 8).
All of the DAMs were assigned to various major metabolic categories, including alkaloids, amino acids, flavonoids, hormones, lipids, terpenoids, phenylpropanoids, saccharides, and others. For most of these categories, the numbers of metabolites predominantly accumulated in T. fuana were similar to those in T. yunnanensis. For instance, the number of T. fuana-predominantly accumulated terpenoids was similar to that of T. yunnanensis-predominantly accumulated terpenoids (Fig. 2b). However, 47 flavonoids were predominantly accumulated in T. fuana, while only 14 flavonoids were predominantly accumulated in T. yunnanensis. The normalized ion intensity of each metabolite belonging to these categories was calculated and is shown in Fig. 2c.
Fig. 2.
Identification of the DAMs between T. fuana and T. yunnanensis. a Significance analysis of the DAMs between the two Taxus species by Volcanoplot. b The numbers of T. yunnanensis predominantly accumulated metabolites and T. fuana predominantly accumulated metabolites in each metabolic category. c The abundances of the DAMs belonging to different metabolic categories
Untargeted metabolomics analysis reveals variations in the precursors, intermediates and competitors of Taxol
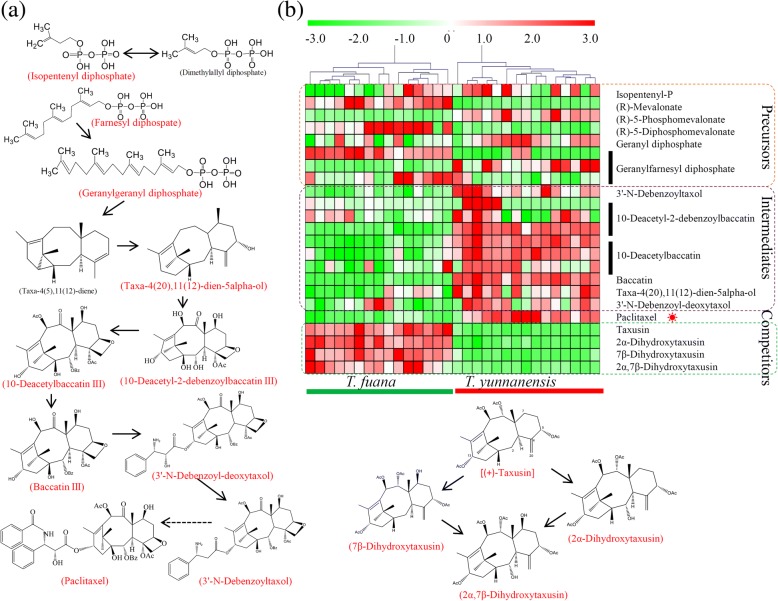
Taxol biosynthesis is a complicated metabolic pathway that involves a large number of secondary metabolites, including the precursors, intermediates and competitors of Taxol (Fig. 3a) [2]. After searching the metabolite pool, six important precursors from the 2-C-methyl-D-erythritol phosphate pathway, isopentenyl diphosphate, geranyl diphosphate and geranylfarnesyl diphosphate, six intermediates, 3’-N-debenzoyltaxol, 10-deacetyl-2-debenzoylbaccatin III, 10-deacetylbaccatin III, taxa-4(5),11(12)-dien-5α-ol, 3’-N-debenzoyl-deoxytaxol, and baccatin III, paclitaxel, and four Taxol competitors, taxusin, 2α-dihydroxytaxusin, 7β-dihydroxytaxusin, and 2α,7β-dihydroxytaxusin, were identified. There were no significant differences in the accumulations of the precursors between T. fuana and T. yunnanensis; however, paclitaxel was predominantly accumulated in T. yunnanensis. Intereastingly, most of the intermediates were highly accumulated in T. yunnanensis while all of the taxusin-like metabolites were significantly accumulated in T. fuana (Fig. 3b).
Fig. 3.
Variations in the abundances of the precursors, intermediates, competitors of taxol between T. fuana and T. yunnanensis. a An overview of the taxol biosynthesis pathway. b A heatmap of the relative amounts of the precursors, intermediates, competitors of taxol in the two Taxus species. The precursors from MEP pathway were included in a red box, the intermediates and derivatives were included in a purple box, and the competitors of taxol were included in a green box. The heatmap scale ranges from − 3 to + 3 on a log2 scale
Quantitative analysis confirms the variations in taxoids
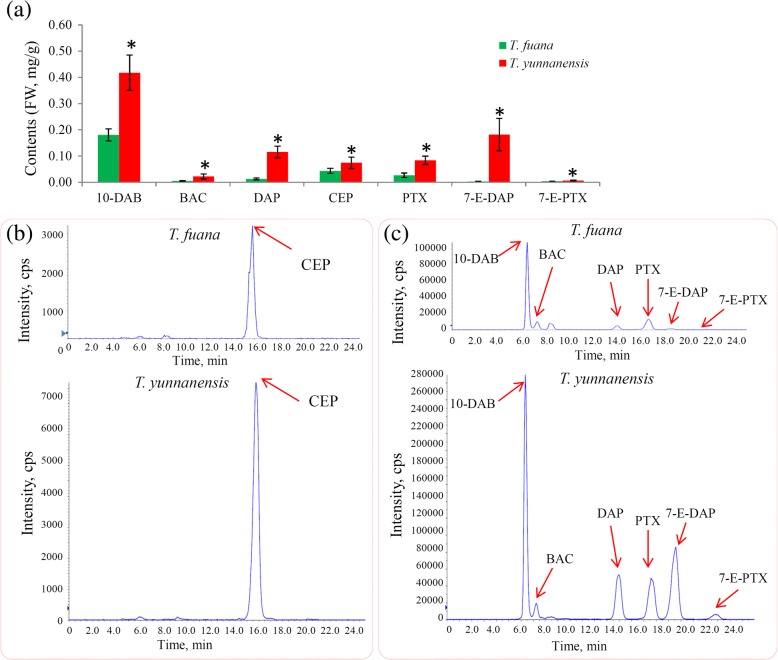
To precisely determine the content differences of Taxol and its derivatives between T. fuana and T. yunnanensis, seven taxoids, including Taxol, cephalomannine, baccatin III, 10-deacetylbaccatin III (10-DAB), 10-deacetylpaclitaxel (DAP), 7-epi 10-desacetyl paclitaxel (7-E-DAP) and 7-epipaciltaxel (7-E-PTX), were quantified using a UPLC-MS/MS method. The untargeted metabolomics analysis indicated that paclitaxel was predominantly accumulated in T. yunnanensis, and the direct quantification with an authentic standard of paclitaxel confirmed this phenomenon. The content of taxol in T. yunnanensis (0.084 mg/g) was 3.1-fold greater than that in T. fuana (0.027 mg/g). Furthermore, all the six other taxoids were significantly accumulated in T. yunnanensis compared with in T. fuana (Fig. 4a). The representative TIC chromatograms of these taxoids between T. fuana and T. yunnanensis are shown in Fig. 4b and c.
Fig. 4.
Determination of the contents of taxoids in T. fuana and T. yunnanensis. a The contents of paclitaxel and six intermediates were quantified by a UPLC-MS/MS method. A P value < 0.001 was considered to be statistically significant and indicated by “*”. b Representative chromatograms of cephalomannine in T. yunnanensis and T. fuana in the positive ion mode. c Representative TIC chromatograms of six taxoids in T. yunnanensis and T. fuana in the positive ion mode. 10-DAB:10-deacetylbaccatin III; BAC: baccatin III; DAP: 10-deacetylpaclitaxel; CEP: cephalomannine; PTX: paclitaxel; 7-E-DAP: 7-epi 10-desacetyl paclitaxel; and 7-E-PTX: 7-epipaclitaxel
Untargeted metabolomics analysis reveals variations in the flavonoids
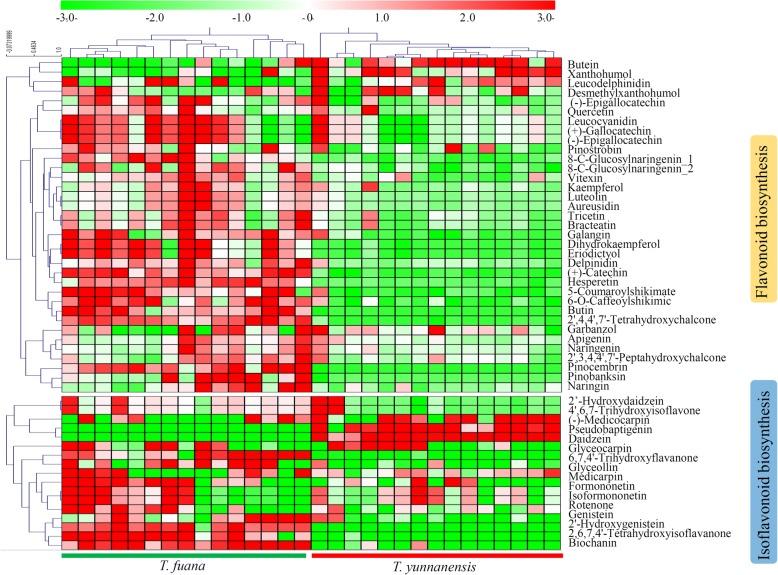
A large number of flavonoids have been isolated from various Taxus species to date. In our study, 35 metabolites involved in flavonoid biosynthesis and 17 metabolites involved in isoflavonoid biosynthesis were identified. For flavonoid biosynthesis, most of the metabolites, except for butein, xanthohumol, and leucodelphinidin, were significantly accumulated in T. fuana compared with in T. yunnanensis. For isoflavonoid biosynthesis, most of the metabolites, except for (−)-medicocarpin, pseudobaptigenin, and daidzein, were predominantly accumulated in T. fuana compared with in T. yunnanensis (Fig. 5).
Fig. 5.
Untargeted metabolomic analysis reveals the variations in the flavonoids between T. fuana and T. yunnanensis. A heatmap of the relative amounts of flavonoids in the two Taxus species. The heatmap scale ranges from − 3 to + 3 on a log2 scale
Quantitative analysis confirms variations in flavonoids
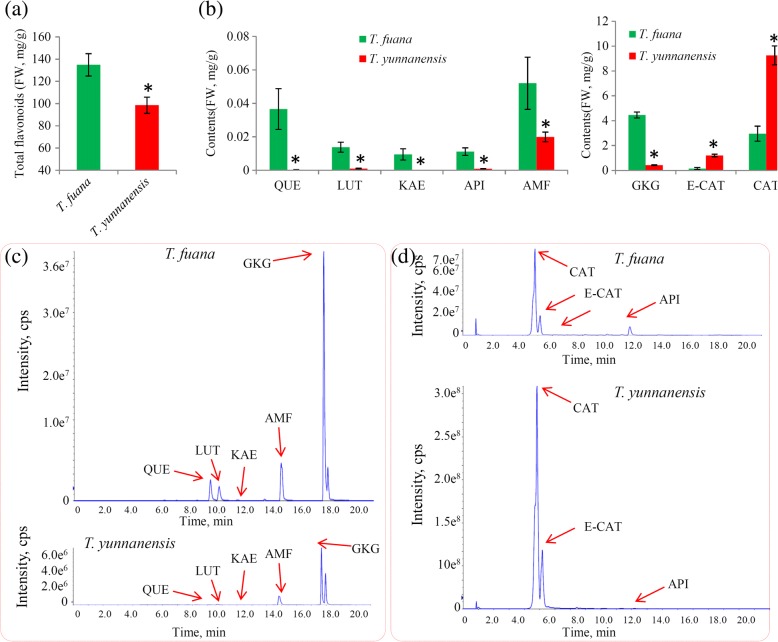
The levels of total flavonoids and eight individual flavonoids, including apigenin, luteolin, kaempferol, quercetin, (+)-catechin, (−)-epicatechin, amentoflavone and ginkgetin in T. fuana and T. yunnanensis were determined. The total flavonoid contents were significantly higher in T. fuana than in T. yunnanensis (Fig. 6a). Similarly, most of the individual flavonoids, including apigenin, luteolin, kaempferol, quercetin, amentoflavone and ginkgetin, were predominantly accumulated in T. fuana than T. yunnanensis. Only two flavonoids, (+)-catechin and (−)-epicatechin, were predominantly accumulated in T. yunnanensis rather than in T. fuana (Fig. 6b). The representative TIC chromatograms of these flavonoids in T. fuana and T. yunnanensis samples are shown in Fig. 6c and d.
Fig. 6.
Determination of the contents of total and individual flavonoids in T. fuana and T. yunnanensis. a The contents of total flavonoids in T. fuana and T. yunnanensis. b The contents of eight individual flavonoids in T. fuana and T. yunnanensis. A P value < 0.001 was considered to be statistically significant and indicated by “*”. c Chromatograms of individual flavonoids in negative mode. d Chromatograms of individual flavonoids in positive mode. CAT: (+)-Catechin; E-CAT: (−)-Epicatechin; API: Apigenin; QUE: Quercetin; LUT: Luteolin; KAE: Kaempferol; AMF: Amentoflavone; and GKG: Ginkgetin
Discussion
Trees from the Taxus genus are major natural resources for Taxol extraction [28]. To date, a large number of secondary metabolites, including taxane diterpenoids and flavonoids, have been isolated and identified in various Taxus species [13, 33]. T. fuana and T. yunnanensis are two endangered Taxus species sparsely distributed in southern Tibet and western Yunnan Province, China [34, 35]. In past decades, deforestation has driven these Taxus trees to the edge of extinction [36]. A metabolomics approach provides a good opportunity to systematically analyze the metabolic variations between T. fuana and T. yunnanensis, which grow in different environments. Our data will enhance the conservation of the germplasm resources of these endangered Taxus species [28].
A traditional metabolomics analysis could only identify a small number of metabolites. For example, only 65 intracellular metabolites were identified and quantified in T. cuspidata cells under laminar shear stress [24]. A metabolomics approach using LC-IT-TOF-MS identified 10 compounds in the cultured seedlings of T. mairei [25]. In 2014, a metabolomics dataset identified 12 metabolites in methyl jasmonate-induced T. chinensis cells [26]. In our study, a large number of metabolites, belonging to various metabolic pathways, were identified (4986 annotated metabolites), which is more than the previously published metabolomes. Embracing comprehensive metabolite profiling allowed us to explore new metabolites and potential interactions, directly or indirectly, among the metabolites in the Taxus genus.
The accumulation of metabolites is a complex and variable trait largely affected by genetic and environmental factors [37, 38]. T. fuana and T. yunnanensis are distributed in different regions of the Yalu Tsangpo river basin, and dramatic variations in the metabolomes between these Taxus species were observed (Fig. 1d) [39]. Our data showed that the Taxol level in T. yunnanensis was more than three times greater than that in T. fuana, indicating the greater economic value of T. yunnanensis for paclitaxel production. A series of specific metabolites, including precursors, intermediates, and competitors of Taxol, were identified in our study. No significant differences in the precursors between T. fuana and T. yunnanensis were observed, suggesting that the precursor supply is not a limiting factor in the differential accumulation of Taxol [2]. In the biosynthesis pathway of taxol, the oxygenation, hydroxylation, benzoylation and acetylation steps produce many intermediates [7, 40]. In our study, six important intermediates were identified (Fig. 3). P450-mediated hydroxylation of taxa-4(5),11(12)-diene to taxa-4(5),11(12)-dien-5α-ol is the first oxygenation step after the formation of the taxane skeleton [40, 41]. 10-DAB is a natural compound isolated from the needles of the Taxus species, and the conversion of taxane glycosides to 10-DAB improves the total yield of paclitaxel [42, 43]. The final acylation step is the conversion of 3’-N-debenzoyltaxol to mature Taxol by 3’-N-debenzoyltaxol N-benzoyltransferase [44]. Interestingly, all of these intermediates were predominantly accumulated in T. yunnanensis compared with in T. fuana, suggesting a reasonable explanation for the higher accumulation of Taxol in T. yunnanensis.
Taxusin and its analogues, the prominent metabolites of yew heartwood, are considered to be dead-end metabolites for Taxol biosynthesis [45]. Taxusin-like metabolites, such as 2α-hydroxytaxusin, 7β-hydroxytaxusin and 2α,7β-dihydroxytaxusin, have been used as surrogates in studies of the intermediate oxygenation steps of the biosynthesis pathway [20]. In our study, the accumulations of taxusin and its analogues were significantly higher in T. fuana than in T. yunnanensis. The high accumulation levels of taxusin and taxusin-like metabolites may consume limited intermediates and block the metabolic flow toward Taxol.
The taxoid and flavonoid contents were greatly affected by environmental factors, such as temperature, altitude and light [13, 28, 46]. In our study, the comprehensive analysis of the metabolites highlighted significant variations in the contents of various flavonoids (Fig. 5a). In general, flavonoids function as antioxidants in plants and can be induced by abiotic and biotic stresses [47, 48]. For example, the contents of flavonoids were elevated in Taxus trees under UV-B radiation [49]. Jilong (with an average altitude of 4,000 m) has a more extreme environment than Motuo (with an average altitude of 1,200 m) (www.chenwentan.com/). Consequently, the higher accumulation of flavonoids suggested a stronger environmental adaptiveness of T. fuana (collected from Jilong) than T. yunnanensis (collected from Motuo) [50]. Eastern Himalayan populations of T. wallichiana, located at higher altitudes, tend to accumulate more paclitaxel than populations from lower altitudes [51]. However, T. fuana from higher altitudes accumulated less paclitaxel than T. yunnanensis from lower altitudes, suggesting that paclitaxel accumulation is controlled by both hereditary and environmental factors.
Conclusions
In our study, a metabolomics approach was applied to examine the metabolic variations in the twigs between T. fuana and T. yunnanensis, which are two endanger species growing in different environments. In total, 4,986 annotated metabolites belonging to various metabolic categories were identified. The level of taxol in T. yunnanensis is significantly higher than that in T. fuana. All the identified intermediates for taxol biosynthesis are predominantly accumulated in T. yunnanensis while four competitors for taxol biosynthesis are predominantly accumulated in T. fuana. Furthermore, the contents of total and some individual flavonoids are significantly accumulated in T. fuana than T. yunnanensis. Our study may provide valuable information for the comprehensive utilization and conservation of the germplasm resources of these two endangered Taxus species growing in different environments.
Additional files
Table S1. Detail information of all identified metabolites. (XLSX 2024 kb)
Figure S1. The total ion chromatograms of all the samples. (DOCX 531 kb)
Figure S2. The m/z width (a) and retention time width (b) of our MS data. (DOCX 611 kb)
Figure S3. The top 20 largest metabolic categories of all identified metabolites. (DOCX 351 kb)
Table S2. Identification and classification of all metabolites. (XLSX 13 kb)
Figure S4. The coefficient of variation of the metabolites from T. yunnanensis and T. fuana. (DOCX 91 kb)
Figure S5. The principal component analysis of the data from these two Taxus species. (DOCX 64 kb)
Figure S6. The statistical analysis of the DAMs between T. fuana and T. yunnanensis. (DOCX 15 kb)
Acknowledgements
We thank Professor Deyou Qiu (The Research Institute of Forestry, Chinese Academy of Forestry) for providing plant materials and giving some comments. We also are grateful to Professor Kaidong Liu (Lingnan Normal University) for reading this manuscript.
Funding
This work was funded by the National Key R&D Program of China (2016YFC0503100). National Natural Science Foundation of China (31501810 and 31601343), and Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ14C060001.
Availability of data and materials
The datasets generated and analysed during the current study are available in the ‘Baidu Netdisk’ (https://pan.baidu.com/s/1TsjzE-VJR9_aWfSmFY9Vdg (password:rnid)).
Abbreviations
- 10-DAB
10-deacetylbaccatin III
- ANOVA
Analysis of variance
- CV
Coefficient of variation
- DAM
Differentially accumulated metabolite
- FDR
False discovery rate
- IDA
Information-dependent acquisition
- MRM
Multiple reaction monitoring
- PC
Principal component
- PCA
Principal component analysis
- QC
Quality control
- RT
Retention time
- TDC
Time-to-digital converter
- UPLC-MS/MS
Ultra Performance Liquid Chromatography-Coupled Mass Spectrometry
Authors’ contributions
All authors have read and approved the final version. CS and MD conceived and designed the study. CY, XL and XZ collected and taken care of the plant samples. CY, XL, JH, and YS performed the experiments. LZ analyzed the data. CS wrote the manuscript.
Ethics approval and consent to participate
This project uses plant materials and does not utilize transgenic technology. The authority responsible for the Taxus resources is the Mount Qomolangma National Nature Reserve, who provided permission to collect the samples for our scientific research.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chunna Yu, Email: yuchunna@hznu.edu.cn.
Xiujun Luo, Email: qw45128@163.com.
Xiaori Zhan, Email: cortex@163.com.
Juan Hao, Email: juanhao@hznu.edu.cn.
Lei Zhang, Email: lei.zhang@wsu.edu.
Yao-Bin L Song, Email: ybsong@hznu.edu.cn.
Chenjia Shen, Phone: +86-571-28865198, Email: shencj@hznu.edu.cn.
Ming Dong, Phone: +86-571-28865198, Email: dongming@hznu.edu.cn.
References
- 1.S J, R C. Taxol: biosynthesis, molecular genetics, and biotechnological applications. Appl Microbiol Biotechnol. 2001;57(1–2):13–19. doi: 10.1007/s002530100757. [DOI] [PubMed] [Google Scholar]
- 2.Croteau R, Ketchum RE, Long RM, Kaspera R, Wildung MR. Taxol biosynthesis and molecular genetics. Phytochem Rev. 2006;5(1):75–97. doi: 10.1007/s11101-005-3748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.John Goodman W. V: the story of Taxol: science and politics in the making of an anticancer drug. 2001. [Google Scholar]
- 4.Eisenreich W, Menhard B, Hylands PJ, Zenk MH, Bacher A. Studies on the biosynthesis of taxol: the taxane carbon skeleton is not of mevalonoid origin. Proc Natl Acad Sci U S A. 1996;93(13):6431–6436. doi: 10.1073/pnas.93.13.6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vranova E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 6.Harrison JW, Scrowston RM, Lythgoe B, Harrison JW, Scrowston RM, Lythgoe B. Taxine. Part IV. The constitution of taxine-I. J Chem Soc C. 1966:1933–45.
- 7.Williams David C, Wildung Mark R, Jin Alan Qingwu, Dalal Dolan, Oliver John S, Coates Robert M, Croteau Rodney. Heterologous Expression and Characterization of a “Pseudomature” Form of Taxadiene Synthase Involved in Paclitaxel (Taxol) Biosynthesis and Evaluation of a Potential Intermediate and Inhibitors of the Multistep Diterpene Cyclization Reaction. Archives of Biochemistry and Biophysics. 2000;379(1):137–146. doi: 10.1006/abbi.2000.1865. [DOI] [PubMed] [Google Scholar]
- 8.Thornburg CK, Walter T, Walker KD. Biocatalysis of a paclitaxel analogue: conversion of baccatin III to N-debenzoyl-N-(2-furoyl)paclitaxel and characterization of an amino phenylpropanoyl CoA transferase. Biochemistry. 2017;56(44):5920–5930. doi: 10.1021/acs.biochem.7b00912. [DOI] [PubMed] [Google Scholar]
- 9.Lee YH, Lee YR, Park CS, Im SA, Song S, Hong JT, et al. Baccatin III, a precursor for the semisynthesis of paclitaxel, inhibits the accumulation and suppressive activity of myeloid-derived suppressor cells in tumor-bearing mice. Int Immunopharmacol. 2014;21(2):487–493. doi: 10.1016/j.intimp.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez-Barbero J, Amat-Guerri F, Snyder JP. The solid state, solution and tubulin-bound conformations of agents that promote microtubule stabilization. Curr Med Chem Anticancer Agents. 2002;2(1):91–122. doi: 10.2174/1568011023354416. [DOI] [PubMed] [Google Scholar]
- 11.Guo B, Kai G, Gong Y, Jin H, Wang Y, Miao Z, et al. Molecular cloning and heterologous expression of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus × media. Mol Biol Rep. 2007;34(2):89–95. doi: 10.1007/s11033-006-9018-6. [DOI] [PubMed] [Google Scholar]
- 12.Long RM, Lagisetti C, Coates RM, Croteau RB. Specificity of the N-benzoyl transferase responsible for the last step of Taxol biosynthesis. Arch Biochem Biophys. 2008;477(2):384–389. doi: 10.1016/j.abb.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Li, Zheng Zan-Sheng, Cheng Fang, Ruan Xiao, Jiang De-An, Pan Cun-De, Wang Qiang. Seasonal Dynamics of Metabolites in Needles of Taxus wallichiana var. mairei. Molecules. 2016;21(10):1403. doi: 10.3390/molecules21101403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grobosch T, Schwarze B, Stoecklein D, Binscheck T. Fatal poisoning with Taxus baccata: quantification of paclitaxel (Taxol a), 10-deacetyltaxol, baccatin III, 10-deacetylbaccatin III, cephalomannine (Taxol B), and 3,5-dimethoxyphenol in body fluids by liquid chromatography-tandem mass spectrometry. J Anal Toxicol. 2012;36(1):36–43. doi: 10.1093/jat/bkr012. [DOI] [PubMed] [Google Scholar]
- 15.Shi Q, Oritani T, Sugiyama T, Murakami R, Wei H. Six new taxane diterpenoids from the seeds of Taxus chinensis var. mairei and Taxus yunnanensis. J Nat Prod. 1999;62(8):1114–1118. doi: 10.1021/np990106b. [DOI] [PubMed] [Google Scholar]
- 16.Wang LY, Ding LM, Huo SC, Sun L, Sun Z. Three new taxoids from the seed of Taxus cuspidata. J Nat Med. 2013;67(4):827–832. doi: 10.1007/s11418-012-0731-2. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary MI, Khan AM, Habib ur R, Atta ur R, Ashraf M. Two new rearranged taxoids from Taxus wallichiana ZUCC. Chem Pharm Bull (Tokyo) 2002;50(11):1488–1490. doi: 10.1248/cpb.50.1488. [DOI] [PubMed] [Google Scholar]
- 18.Sun ZH, Chen Y, Guo YQ, Qiu J, Zhu CG, Jin J, et al. Isolation and cytotoxicity evaluation of taxanes from the barks of Taxus wallichiana var. mairei. Bioorg Med Chem Lett. 2015;25(6):1240–1243. doi: 10.1016/j.bmcl.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 19.Hai Ping, Wen Shi-Zhen, Li Yan, Gao Yuan, Jiang Xian-Jun, Wang Fei. New Taxane Diterpenoids from Taxus yunnanensis. Natural Products and Bioprospecting. 2014;4(1):47–51. doi: 10.1007/s13659-014-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Horiguchi T, Croteau R, Williams RM. Studies on Taxol biosynthesis: preparation of taxadiene-diol- and triol-derivatives by deoxygenation of taxusin. Tetrahedron. 2008;64(27):6561–6567. doi: 10.1016/j.tet.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biondi-Zoccai GG, Agostoni P, Sangiorgi GM, Iakovou I, Antoniucci D, Grube E, et al. Comparison of ticlopidine vs. clopidogrel in addition to aspirin after paclitaxel-eluting stent implantation: insights from the TRUE (Taxusin real-life usage evaluation) study. Int J Cardiol. 2006;108(3):406–407. doi: 10.1016/j.ijcard.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Souard F, Delporte C, Stoffelen P, Thevenot EA, Noret N, Dauvergne B, et al. Metabolomics fingerprint of coffee species determined by untargeted-profiling study using LC-HRMS. Food Chem. 2018;245:603–612. doi: 10.1016/j.foodchem.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Ketchum RE, Rithner CD, Qiu D, Kim YS, Williams RM, Croteau RB. Taxus metabolomics: methyl jasmonate preferentially induces production of taxoids oxygenated at C-13 in Taxus × media cell cultures. Phytochemistry. 2003;62(6):901–909. doi: 10.1016/S0031-9422(02)00711-2. [DOI] [PubMed] [Google Scholar]
- 24.Han PP, Yuan YJ. Metabolic profiling as a tool for understanding defense response of Taxus cuspidata cells to shear stress. Biotechnol Prog. 2009;25(5):1244–1253. doi: 10.1002/btpr.209. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K, Li F, Morikawa K, Nobukawa T, Kadota S. Analysis of biosynthetic fluctuations of cultured Taxus seedlings using a metabolomic approach. Phytochemistry. 2011;72(14–15):1760–1766. doi: 10.1016/j.phytochem.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Song GH, Zhao CF, Zhang M, Fu CH, Zhang H, Yu LJ. Correlation analysis of the taxane core functional group modification, enzyme expression, and metabolite accumulation profiles under methyl jasmonate treatment. Biotechnol Prog. 2014;30(2):269–280. doi: 10.1002/btpr.1864. [DOI] [PubMed] [Google Scholar]
- 27.Zhao C, Song G, Fu C, Dong Y, Xu H, Zhang H, et al. A systematic approach to expound the variations in taxane production under different dissolved oxygen conditions in Taxus chinensis cells. Plant Cell Rep. 2016;35(3):541–559. doi: 10.1007/s00299-015-1902-x. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Wen, Komatsu Setsuko, Zhu Wei, Zhang Lin, Li Ximin, Cui Lei, Tian Jingkui. Response and Defense Mechanisms ofTaxus chinensisLeaves Under UV-A Radiation are Revealed Using Comparative Proteomics and Metabolomics Analyses. Plant and Cell Physiology. 2016;57(9):1839–1853. doi: 10.1093/pcp/pcw106. [DOI] [PubMed] [Google Scholar]
- 29.Hao d C, Huang B, Yang L. Phylogenetic relationships of the genus Taxus inferred from chloroplast intergenic spacer and nuclear coding DNA. Biol Pharm Bull. 2008;31(2):260–265. doi: 10.1248/bpb.31.260. [DOI] [PubMed] [Google Scholar]
- 30.van Rozendaal EL, Lelyveld GP, van Beek TA. Screening of the needles of different yew species and cultivars for paclitaxel and related taxoids. Phytochemistry. 2000;53(3):383–389. doi: 10.1016/S0031-9422(99)00094-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Su J. Effects of light acclimation on shoot morphology, structure, and biomass allocation of two Taxus species in southwestern China. Sci Rep. 2016;6:35384. doi: 10.1038/srep35384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C, Guo H, Zhang Y, Song Y, Pi E, Yu C, et al. Identification of potential genes that contributed to the variation in the taxoid contents between two Taxus species (Taxus × media and Taxus mairei) Tree Physiol. 2017;37(12):1659–1671. doi: 10.1093/treephys/tpx091. [DOI] [PubMed] [Google Scholar]
- 33.Li Cunfang, Huo Changhong, Zhang Manli, Shi Qingwen. Chemistry of Chinese yew, Taxus chinensis var. mairei. Biochemical Systematics and Ecology. 2008;36(4):266–282. doi: 10.1016/j.bse.2007.08.002. [DOI] [Google Scholar]
- 34.Miao YC, Lang XD, Zhang ZZ, Su JR. Phylogeography and genetic effects of habitat fragmentation on endangered Taxus yunnanensis in Southwest China as revealed by microsatellite data. Plant Biol. 2014;16(2):365–374. doi: 10.1111/plb.12059. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Moller M, Gao LM, Zhang DQ, Li DZ. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol Ecol Resour. 2011;11(1):89–100. doi: 10.1111/j.1755-0998.2010.02907.x. [DOI] [PubMed] [Google Scholar]
- 36.Banskota AH, Usia T, Tezuka Y, Kouda K, Nguyen NT, Kadota S. Three new C-14 oxygenated taxanes from the wood of Taxus yunnanensis. J Nat Prod. 2002;65(11):1700–1702. doi: 10.1021/np020235j. [DOI] [PubMed] [Google Scholar]
- 37.Tang W, Hazebroek J, Zhong C, Harp T, Vlahakis C, Baumhover B, et al. Effect of genetics, environment, and phenotype on the metabolome of maize hybrids using GC/MS and LC/MS. J Agric Food Chem. 2017;65(25):5215–5225. doi: 10.1021/acs.jafc.7b00456. [DOI] [PubMed] [Google Scholar]
- 38.Aversano R, Contaldi F, Adelfi MG, D'Amelia V, Diretto G, De Tommasi N, et al. Comparative metabolite and genome analysis of tuber-bearing potato species. Phytochemistry. 2017;137:42–51. doi: 10.1016/j.phytochem.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Poudel RC, Gao LM, Moller M, Baral SR, Uprety Y, Liu J, et al. Yews (Taxus) along the Hindu Kush-Himalayan region: exploring the ethnopharmacological relevance among communities of Mongol and Caucasian origins. J Ethnopharmacol. 2013;147(1):190–203. doi: 10.1016/j.jep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 40.Jennewein S, Long RM, Williams RM, Croteau R. Cytochrome p450 taxadiene 5alpha-hydroxylase, a mechanistically unusual monooxygenase catalyzing the first oxygenation step of Taxol biosynthesis. Chem Biol. 2004;11(3):379–387. doi: 10.1016/j.chembiol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Vazquez A, Williams RM. Studies on the biosynthesis of taxol. Synthesis Of taxa-4(20), 11(12)-diene-2alpha,5alpha-diol. J Org Chem. 2000;65(23):7865–7869. doi: 10.1021/jo005537+. [DOI] [PubMed] [Google Scholar]
- 42.Yang T, Li M, Chen JM. Conversion of taxane glycosides to 10-deacetylbaccatin III. Nat Prod Res. 2006;20(2):119–124. doi: 10.1080/14786410500045218. [DOI] [PubMed] [Google Scholar]
- 43.Braga SF, Galvão DS. A semiempirical study on the electronic structure of 10-deacetylbaccatin-III. J Mol Graph Model. 2002;21(1):57–70. doi: 10.1016/S1093-3263(02)00121-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, Li ST, Fu CH, Zhou PP, Song FJ, Yu LJ. Overexpression of a 3’-N-Debenzoyltaxol-N-benzoyltransferase gene promotes Taxol yield in Taxus chinensis cells. Chin. J. Biochem. Mol. Biol. 2014;30(4):377–382. [Google Scholar]
- 45.Walker K, Croteau R. Taxol biosynthesis: Springer US; 1999.
- 46.Nasiri J, Naghavi MR, Alizadeh H, Moghadam MR. Seasonal-based temporal changes fluctuate expression patterns of TXS, DBAT, BAPT and DBTNBT genes alongside production of associated taxanes in Taxus baccata. Plant Cell Rep. 2016;35(5):1103–1119. doi: 10.1007/s00299-016-1941-y. [DOI] [PubMed] [Google Scholar]
- 47.Feng Hongjie, Tian Xinhui, Liu Yongchang, Li Yanjun, Zhang Xinyu, Jones Brian Joseph, Sun Yuqiang, Sun Jie. Analysis of Flavonoids and the Flavonoid Structural Genes in Brown Fiber of Upland Cotton. PLoS ONE. 2013;8(3):e58820. doi: 10.1371/journal.pone.0058820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M, Dong JF, Jin HH, Sun LN, Xu MJ. Ultraviolet-B-induced flavonoid accumulation in Betula pendula leaves is dependent upon nitrate reductase-mediated nitric oxide signaling. Tree Physiol. 2011;31(8):798–807. doi: 10.1093/treephys/tpr070. [DOI] [PubMed] [Google Scholar]
- 49.Zu YG, Pang HH, Yu JH, Li DW, Wei XX, Gao YX, et al. Responses in the morphology, physiology and biochemistry of Taxus chinensis var. mairei grown under supplementary UV-B radiation. J Photochem Photobiol B. 2010;98(2):152–158. doi: 10.1016/j.jphotobiol.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Mouradov A, Spangenberg G. Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front Plant Sci. 2014;5:620. doi: 10.3389/fpls.2014.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee S, Ghosh B, Jha TB, Jha S. Variation in content of taxol and related taxanes in eastern Himalayan populations of Taxus wallichiana. Planta Med. 2002;68(8):757–759. doi: 10.1055/s-2002-33808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detail information of all identified metabolites. (XLSX 2024 kb)
Figure S1. The total ion chromatograms of all the samples. (DOCX 531 kb)
Figure S2. The m/z width (a) and retention time width (b) of our MS data. (DOCX 611 kb)
Figure S3. The top 20 largest metabolic categories of all identified metabolites. (DOCX 351 kb)
Table S2. Identification and classification of all metabolites. (XLSX 13 kb)
Figure S4. The coefficient of variation of the metabolites from T. yunnanensis and T. fuana. (DOCX 91 kb)
Figure S5. The principal component analysis of the data from these two Taxus species. (DOCX 64 kb)
Figure S6. The statistical analysis of the DAMs between T. fuana and T. yunnanensis. (DOCX 15 kb)
Data Availability Statement
The datasets generated and analysed during the current study are available in the ‘Baidu Netdisk’ (https://pan.baidu.com/s/1TsjzE-VJR9_aWfSmFY9Vdg (password:rnid)).