Abstract
Calpain 9 (CAPN9) is expressed in the stomach and small intestine. CAPN9 has regulatory roles in hypertension, heart disease, gastric mucosal defense, and kidney disease. The involvement of CAPN9 has not been reported in the development of chickens. CAPN9 mRNA was found in adipose and muscle tissue in this study. Two linkage single nucleotide polymorphisms (SNP; G7518A and C7542G) in intron 4 were screened from 160 birds of the D2 chicken line. The 2 mutation sites were associated with carcass weight, evisceration weight, abdominal fat weight (AFW), abdominal fat percentage (AFP), and breast muscle percentage (all P < 0.05). Intramuscular fat (IMF) content was not significantly different in the 3 genotypes. But, the AA(7518)/GG(7542) genotype had the highest IMF content, highest breast muscle weight, and lower AFW and AFP. Moreover, the mRNA level of CAPN9 in abdominal fat tissue was significantly different (P < 0.05 or P < 0.01) between any 2 genotypes, consistent with AFW and AFP. In summary, the expression of CAPN9 in adipose and breast muscle tissue is reported for the first time. CAPN9 affected production performance of chickens. As a marker, the linkage G7518A and C7542G polymorphisms in intron 4 of CAPN9 could affect the production traits by regulating mRNA expression. The findings concerning the marker enrich the theoretical foundation for molecular breeding of high-quality broilers.
Keywords: Calpain 9 gene, expression, SNP, development, chicken
INTRODUCTION
The Calpain superfamily (CAPN) is a large group of calcium-activated cysteine proteases that are involved in a variety of functions, including cytoskeletal remodeling, cell signaling, and apoptosis (Shioda et al., 2006; Gafni et al., 2009; McClung et al., 2009). Among them, μ-calpain will play a major role in meat quality changes of chicken breast muscle (Huang et al., 2014; Zhao et al., 2018). In addition, Calpain 10 plays an important role in regulating obesity and diabetes in mice (Cheverud et al., 2010).
Calpain 9 (CAPN9, also known as nCL-4) is a typical calcium protease that is expressed mainly in the stomach and small intestine (Lee et al., 1998). CAPN 9 associates with the common 28 kDa regulatory subunit calpain-4 (Lee et al., 1999). Most studies of CAPN9 have focused on disease (Hata et al., 2010; Ookura et al., 2015), CAPN9 and CAPN8 have been functionally implicated in the gastrointestinal tract (Hata et al., 2010). But the involvement of CAPN9 on animal production performance is unclear.
In this study, the expression and function of CAPN9 gene were detected. It was found that CAPN9 gene was expressed in adipose and breast muscle tissue of chicken. Further, the polymorphism analysis of CAPN9 gene also was performed by direct PCR sequencing, and the linkage G7518A and C7542G polymorphisms in intron 4 of CAPN9 were found, and associated with the body weight, muscle development, and fat deposition of chicken. The findings provide a marker and enrich the theoretical foundation for molecular breeding of high-quality broilers.
MATERIALS AND METHODS
Animals and Ethics Statement
All of the animal experiments were conducted in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China). Animal experiments were approved by the Animal Management Committee (in charge of animal welfare issues) of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China). Ethical approval on animal survival was given by the animal ethics committee of IAS-CAAS.
D2 chickens (n = 160), a yellow feather broiler with dwarf consanguinity, were obtained from Institute of Animal Science, Chinese Academy of Agricultural Science (Beijing, China). All birds were slaughtered at 90 d of age. Blood samples were collected from the wing vein for DNA extraction. For the detection of gene expression, samples of abdominal fat (AF) and pectoralis major muscle from 6 random birds were snap-frozen in liquid nitrogen and stored at –80°C for reverse transcription polymerase chain reaction (RT-PCR). For every effective SNP, 6 AF samples of every genotypes were snap-frozen in liquid nitrogen and stored at –80°C for the real-time quantitative reverse transcription polymerase chain reaction (Q-PCR). Other pectoralis major muscle samples were stored at –20°C for intramuscular fat (IMF) assay.
Slaughter Indicators and IMF Content
After birds were slaughtered, carcass weight (CW), eviscerated weight (EW), breast muscle weight (BMW), and abdominal fat weight (AFW) were determined, and abdominal fat percentage (AFP, %, mass of AF as a percentage of BW) and breast muscle percentage (BMP, %, mass of breast muscle as a percentage of BW) were calculated.
IMF content of pectoralis major muscle, trimmed of visible fat surrounding and lying around muscles, was measured by Soxhlet extraction (AOAC, 1990) and is presented as the weight percentage of dry muscle tissue.
RNA Extraction
Total RNA was isolated from the frozen AF and pectoralis major muscle tissues using Trizol reagent according to the manufacturer's protocol (Takara Bio, Shiga, Japan). After removal of any genomic DNA using the DNA Removal Kit according to the manufacturer's protocol (Abnova, Wuhan, China), RNA was dissolved at 1 μg/μL and stored at –80°C for use in RT-PCR and Q-PCR. The RNA integrity and purity were assessed by electrophoresis on 1.5% agarose gels and ultraviolet spectrophotometry.
RT-PCR and Q-PCR
Total RNA (2.0 μg) from the frozen AF or pectoralis major muscle tissues was used to generate cDNA in a final volume of 20 μL using the GoScript™ reverse transcription system according to the manufacturer's instructions (Promega, Beijing, China). The specific primers of CAPN 9 gene (Table 1) was designed with Primer Premier 5.0 software. Based on the sequence of CAPN 9 gene (GenBank accession No. XM_419,585), the same specific primers (Q-1) were designed for RT-PCR and Q-PCR to detect the expression of CAPN 9 gene.
Table 1.
The specific primers of CAPN9 gene.
| Primer | Sequence | Length (bp) |
|---|---|---|
| Q-1 | 5΄-AGGGTCTGACACCACCTCC-3΄ | 158 |
| 5΄-GAGGTCTAGCAGAACAGGGC-3΄ | ||
| S-1 | 5΄-CTAGGGAGGGTCTGACAC-3΄ | 509 |
| 5΄-GGACAGGACCTCAAAAAGTA-3΄ | ||
| S-2 | 5΄-GATCACCATGCAAGTGTACT-3΄ | 537 |
| 5΄-TCTTCACAGAAACTGGCTAC-3΄ | ||
| S-3 | 5΄-ACCAACCTAGCATGACACAG-3΄ | 416 |
| 5΄-GGGCAGCAGCACATAATACT-3΄ |
Q-1: the primers used for Q-PCR;
S-1, -2, -3: the primers used for PCR of SNP.
RT-PCR was carried out in a 20-μL reaction volume containing 100 ng cDNA template of AF or pectoralis major muscle, 0.5 μL of each primer (10 pM), 10 μL of 2 × Taq PCR MasterMix (Tiangen, Beijing, China) and the remaining volume being distilled deionized water (ddH2O). The amplification conditions were 94°C for 3 min (preliminary denaturation), followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 40 s, and extension at 72°C for 45 s; and a final elongation step at 72°C for 10 min.
Q-PCR was carried out in a 20-μL reaction volume containing 10 μL of 2 × iQ™ SYBR Green Supermix, 0.5 μL (10 mM) of each primer, and 100 ng cDNA. Mixtures were incubated in an ABI 7500 Real-Time PCR Detection System (Applied Biosystems). A melting curve was constructed to verify that only a single PCR product was amplified. Samples were assayed in triplicate with standard deviations of CT values not exceeding 0.5 on a within-run basis.
DNA Extraction and PCR Amplification
Genomic DNA was isolated from non-coagulated blood by the phenol-chloroform method (Sambrook and Russell, 2001) for PCR. The DNA integrity and purity were assessed by electrophoresis on 1.0% agarose gels and UV spectrophotometry.
According to the reference sequences (Chromosome 3: 40,615,856 to 40,650,876, Ensembl version 421,544.1), 60 pairs of primers were designed to amplify the sequences of CAPN9 gene for screen the SNPs (Table 1, only listed 3 pairs primers, which had found the mutation in the according product). PCR was carried out in a 20-μL reaction volume containing 50 ng genomic DNA template, 0.5 μL of each primer (10 pM), 10 μL of 2 × Taq PCR MasterMix (Tiangen, Beijing, China), and the remaining volume being distilled deionized water (ddH2O). The amplification conditions were 94°C for 3 min (preliminary denaturation), followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 40 s, and extension at 72°C for 45 s, and a final elongation step at 72°C for 10 min.
SNPs Screen and Identification
For screening the SNPs of population, the pre-experiment was performed, and a genomic DNA pooling (100 ng/μL) with the equal amount DNA from every bird of 160 individuals was used for PCR as the template. Aliquots of the PCR products were analyzed by 1.5% agarose gel electrophoresis, and were then sequenced to identify the authenticity using an ABI3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Based on the results from an automated sequence shown in the form of a chromatogram, the sequence mutations would have been found, and the double peaks phenomenon in nucleotide base position was considered as the candidate SNP.
Based on these screened SNPs in pre-experiment, the fragments containing the SNPs were amplified again by the same PCR operation using the DNA from each bird. Every PCR product was directly sequenced to screen different genotypes. The sequence results with the chromatogram form were analyzed with the DNAMAN software (version 6.0, Lynnon Corporation, San Ramon, CA). As the evaluation criteria, the 2 homozygous types should have 1 peak for a single nucleotide base, and the heterozygous type has the double peaks for 2 different nucleotides. So the sequence mutations were identified between individuals, and the 3 different genotypes of every SNP were recorded.
Statistical Analysis
Data were analyzed using the general linear model procedure of statistical analysis systems software (SAS, Version 8.2; SAS Institute, Cary, NC). The general linear model Yi = μ + Gi + e was used to analyze the genotype effects of the CAPN9 gene and the phenotype trait, where Yi, μ, Gi, and e are the measured value of trait phenotype, population mean, genotype effect, and residual error, respectively.
Differences in allelic or genotypic frequencies and between least squares means for the measured parameters in each of the genotypes were assessed. Correlations between several of the measured variables were analyzed using the Pearson's 2 test and, where appropriate, partial correlations were estimated. All experimental data are presented as mean ± SEM. A P-value < 0.05 was considered statistically significant.
RESULTS
Expression of Calpain 9 gene in Pectoralis Major Muscle and AF Tissues of Chicken
To determine if CAPN9 was expressed in chicken muscle and adipose tissue, total RNA was isolated from AF and pectoralis major muscle tissues, respectively. CAPN9 mRNA expression was detected at the transcript level in AF and pectoralis major muscle tissues. After RT-PCR, the expected 158 bp product was verified by electrophoresis and confirmed by sequencing, and it was confirmed that CAPN9 mRNA was present in adipose and breast muscle tissues (Figure 1).
Figure 1.
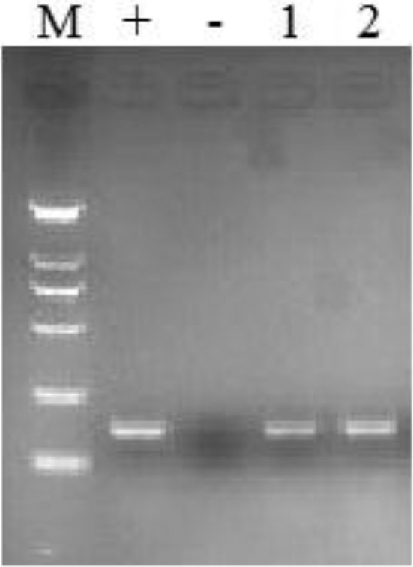
Expression of CAPN9 mRNA in chicken abdominal adipose and breast muscle tissues. CAPN9 mRNA expression was detected by RT-PCR. M: DL2000 Marker, +: positive control (stomach), –: negative control, 1: abdominal adipose tissue, 2: breast muscle tissue, respectively. The expected 158 bp product was verified by electrophoresis in abdominal adipose and breast muscle tissues.
Screening of Calpain 9 gene SNPs
After qualified genomic DNA was obtained, the pooled DNA with equal amounts from each sample was used to amplify the CAPN9 gene. Sequence analysis confirmed the correctness of the PCR products. Eight effective mutation sites were found in PCR products using 3 pair primers (S-1, S-2, S-3) (Table 1). Polymorphic loci were not found in other primers (not shown), so these 3 pair primers were chose for PCR amplification in the following experiment.
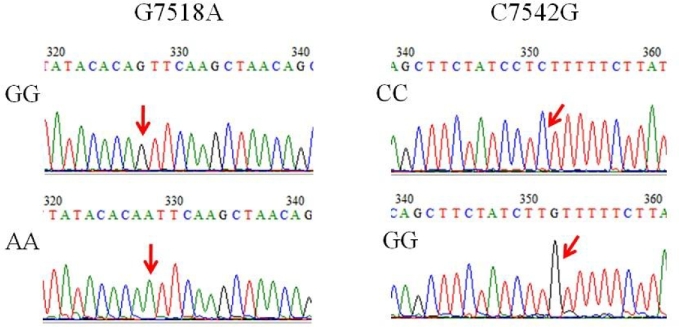
Among the 8 mutant sites (Table 2), 2 (G305A, A518G) were located in exon 1 and intron 1 for primer 1. They displayed only 2 genotypes (GG and AG, AA and AG). In addition, 3 sites were found in intron 2 (T6689C and C6769A) and exon 3 (T6820G) displayed 3 genotypes (TT, TC and CC; CC, AC and AA, TT, TG and GG) for primer 2. Similarly, 3 sites (G7518A, C7540T, and C7542G) were found in intron 4. These also displayed 3 genotypes (GG, AG and AA; CC, TC and TT; CC, GC and GG) for primer 3. Two sites (G7518A and C7542G) were identified as the linkage in intron 4. Their homozygous genotypes are provided in Figure 2.
Table 2.
The related information of 8 mutation sites of CAPN9 gene by 3 primers.
| Mutation site | Genotype | Location | Primer |
|---|---|---|---|
| 305 | GG and AG | Exon 1 | 1 |
| 518 | AA and AG | Intron 1 | 1 |
| 6689 | TT, TC and CC | Intron 2 | 2 |
| 6769 | CC, AC and AA | Intron 2 | 2 |
| 6820 | TT, TG and GG | Exon 3 | 2 |
| 7518 | GG, AG and AA | Entron 4 | 3 |
| 7540 | CC, TC and TT | Entron 4 | 3 |
| 7542 | CC, GC and GG | Entron 4 | 3 |
Figure 2.
The genotyping of the linkage SNPs (G7518A and C7542G) in CAPN9 gene. The mutation sites G7518A and C7542G were concatenated, and located in intron 4 of CAPN9 gene. The homozygous genotypes were GG and AA (7518 site), CC and GG (7542 site), respectively.
Effect of G7518A and C7542G Calpain SNPs on Carcass Traits
Analysis of genotypes and carcass traits by least square analysis revealed significantly different phenotypic values of CW, EW, AFW, AFP, and BMP traits (all P < 0.05), but no significant difference on BMW (P > 0.05) in birds with 2 linkage SNPs (G7518A and C7542G) among the 3 genotypes (Table 3). On the contrary, among the other SNPs, no significant differences were evident between genotypes and phenotypes (all P > 0.05).
Table 3.
Associations between genotypes with G7518A and C7542G and carcass traits and IMF content.
| Traits (M±S) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | Genotype (n) | Genotype frequency (%) | CW (g) | EW (g) | BMW (g) | AFW (g) | AFP (%) | BMP (%) | IMF (%) |
| AA(64) | 40.00 | 1028.44±201.65a,b | 856.38±173.33a,b | 133.35±30.13 | 33.13±15.75a,b | 3.66±1.48a,b | 15.54±1.40b | 2.68±1.15 | |
| 7542 | AG(68) | 42.50 | 1096.41±157.41b | 910.19±131.22b | 128.89±35.46 | 37.77±14.88b | 3.98±1.46b | 14.16±3.51a | 2.56±1.00 |
| GG(28) | 17.50 | 975.0±154.72a | 810.76±132.12a | 124.96±17.96 | 25.71±15.11a | 2.98±1.47a | 15.57±1.90a,b | 2.15±0.67 | |
| GG(64) | 40.00 | 1028.44±201.65a,b | 856.38±173.33a,b | 133.35±30.13 | 33.13±15.75a,b | 3.66±1.48a,b | 15.54±1.40b | 2.68±1.15 | |
| 7518 | GC(68) | 42.50 | 1096.41±157.41b | 910.19±131.22b | 128.89±35.46 | 37.77±14.88b | 3.98±1.46b | 14.16±3.51a | 2.56±1.00 |
| CC(28) | 17.50 | 975.0±154.72a | 810.76±132.12a | 124.96±17.96 | 25.71±15.11a | 2.98±1.47a | 15.57±1.90a,b | 2.15±0.67 | |
AFP, abdominal fat percentage; AFW, abdominal fat weight; BMP, breast muscle percentage; BMW, breast muscle weight; CW, carcass weight; EW, eviscerated weight; IMF, intramuscular fat; different lowercase superscripts in each row indicate significant differences of phenotype.
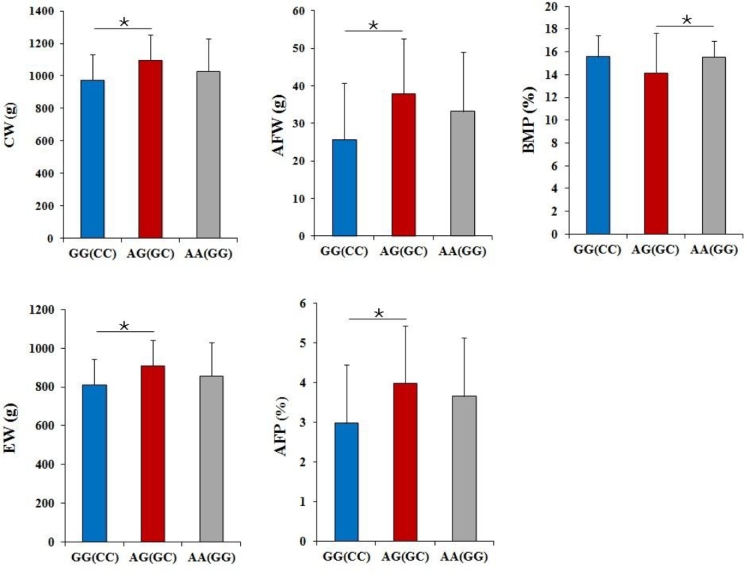
For the CW, EW, AFW, and AFP traits, the phenotype values in GG(7518)/CC(7542) homozygous genotype were significantly lower (all P < 0.05) than AG(7518)/GC(7542) heterozygous genotype. There were no significant differences (both P > 0.05) between GG(7518)/CC(7542) and AA(7518)/GG(7542), or AG(7518)/GC(7542) and AA(7518)/GG(7542) (Figure 3). However, for BMP, the phenotype values of AG(7518)/GC(7542) heterozygous genotype were significantly lower (P < 0.05) than of the AA(7518)/GG(7542) homozygous genotype. There were no significant differences (both P > 0.05) between GG(7518)/CC(7542) and AG(7518)/GC(7542), or GG(7518)/CC(7542) and AA(7518)/GG (7542) (Figure 3).
Figure 3.
Association between of genotype of G7518A (C7542G) and slaughter indicator. By least square analysis, the phenotypic values of CW, EW, AFW, AFP, and BMP traits in different genotypes with G7518A/C7542G polymorphism were significant different (all P < 0.05). Data are means ± SEM, n = 28(GG/CC), 64(AA/GG), and 64 (GA/CG).
Effect of Calpain 9 SNPs on IMF Content
Least square analysis for each of the 8 mutant sites concerning the association between genotypes and IMF trait showed that the phenotypic values of IMF trait were not significantly different (P > 0.05) among the 3 genotypes. However, for the 3 genotypes of SNPs G7518A and C7542G, the IMF content displayed a trend of gradual decrement of AA(7518)/GG(7542), AG(7518)/GC(7542), and GG(7518)/CC(7542) genotypes (Table 3). Similarly, BMW trait displayed the same trend with IMF content among 3 genotypes of SNPs G7518A and C7542G. In addition, the AA(7518)/GG (7542) genotype with the highest IMF content had lower AFW and AFP compared to the AG(7518)/GC(7542) genotype.
Expression of Calpain 9 with Linkage SNPs (G7518A and C7542G) in AF Tissue of 3 Genotypes
Considering the important intron-mediated regulation of gene expression, the mRNA levels of CAPN9 with linkage SNPs (G7518A and C7542G) in AF tissue of 3 genotypes were detected to explain the molecular regulation of SNP. As shown in Figure 4, the level of CAPN9 mRNA in any 2 of 3 genotypes was significantly different (P < 0.05 or P < 0.01), and was significantly upregulated in the AG(7518)/GC(7542) genotype compared to the GG(7518)/CC(7542) genotype. The findings were consistent with the phenotypic values of AFW and AFP.
Figure 4.
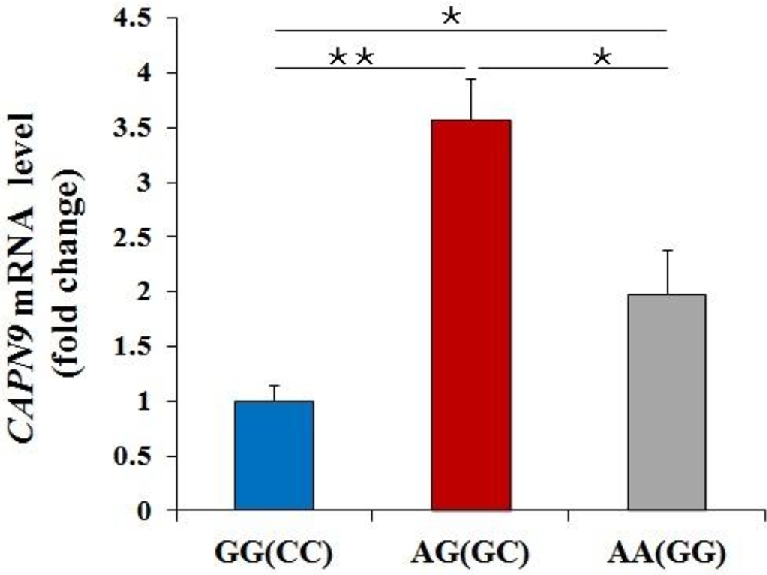
Expression level of CAPN9 gene in AF tissue. The mRNA levels of CAPN9 gene in AF tissue from different genotypes with G7518A and C7542G polymorphism were detected by Q-PCR. The level of CAPN9 mRNA in any 2 of 3 genotypes had the significant difference (P < 0.05 or P < 0.01). Data are means ± SEM, n = 6.
DISCUSSION
The Calpain superfamily is a large group of calcium-activated cysteine proteases involved in a variety of functions, including cytoskeletal remodeling, cell signaling, apoptosis (Shioda et al., 2006; Gafni et al., 2009; McClung et al., 2009), and adipocyte differentiation (Patel and Lane, 1999; Yajima et al., 2006). CAPN9, also known as nCL-4, a member of calpains (Lee et al., 1998), is expressed in the stomach and small intestine. In our current research, the expression of CAPN9 gene in AF tissue and breast muscle tissue of chicken was detected, and the result shows for the first time that CAPN9 is expressed in adipose and muscle tissue of chicken, and it was considered that CAPN9 might regulate the development of muscle or adipose tissue. Further, the polymorphism analysis of CAPN9 by direct sequencing was performed to identify if CAPN9 is functionally important in the development of muscle or adipose tissue of chicken.
DNA pooling was done to preliminarily screen the mutation site for CAPN9 nucleotide sequences. After PCR amplification and sequencing, 8 mutation sites were found in different exons and introns by 3 pair primers. The corresponding nucleotide sequences with mutation sites in each bird in the detected population were sequenced to differentiate the genotypes. Least square analysis was done to explore the association between genotype and carcass traits. Two linkage SNPs (G7518A and C7542G) were found. The phenotype values of CW, EW, AFW, AFP, and BMP were significantly different among the 3 genotypes.
The phenotype values of CW and EW in GG(7518)/CC(7542) homozygous genotype were significantly lower than the AG(7518)/GC(7542) heterozygous genotype. Similarly, the phenotype values of AFW and AFP for the G7518A and C7542G linkage SNPs had the same result. Previous studies revealed the involvement of CAPN9 in the regulation of gastric mucosal defense, hypertension, kidney disease, heart disease, and cancer (Markmann et al., 2005; Hata et al., 2010; Davis et al., 2014). However, the association of CAPN9 with body weight, muscle development, and fat deposition has been unknown. The present findings support the view that CAPN9 affects the development of body weight and fat deposition in chickens. The AG(7518)/GC(7542) heterozygote and GG(7518)/CC(7542) homozygote were the effective genotypes for body weight and fat deposition, respectively. However, different from other slaughter traits, the phenotype values of BMP for the G7518A and C7542G linkage SNPs in the GG(7518)/CC(7542) genotype were significantly lower than of the AG(7518)/GC(7542) genotype, although BMW was not significantly different. So, we believe that CAPN9 could regulate the development muscle in chicken, but possibly in a different way than in adipose tissue.
IMF is deposited in muscle tissue. It is an important factor of meat quality (Farmer, 1999; Fernandez et al., 1999; Cui et al., 2017). The performance of high IMF and low AF is pursued in production of yellow-feathered broilers, and this goal was focal in this study. The AA(7518)/GG(7542) genotype had the highest IMF content, with the highest BMW and lower AFW and AFP. Although the phenotypic values of IMF were not significantly different among the 3 genotypes, the AA(7518)/GG(7542) genotype remains valid in production by long-term population selection.
Finally, the expression of CAPN9 gene in AF tissue from 3 genotypes with linkage G7518A and C7542G polymorphisms was explored to uncover the expression regulation of CAPN9 gene for this mutation site. An intron-mediated regulation on gene expression has been described (Krizek, 2015; Visser et al., 2015; Cavalli et al., 2016). The mRNA levels of CAPN9 in any 2 of 3 genotypes had a significant difference and consistency with the phenotypic value of AFW and AFP. So, it was confirmed that the G7518A and C7542G polymorphisms in intron 4 regulate CAPN9 expression.
CONCLUSION
In summary, we report for the first time that CAPN9 is expressed in chicken adipose and breast muscle tissue, and affects production performance. Linkage G7518A and C7542G polymorphisms in intron 4 of CAPN9 were found. Both affected body weight, muscle development, and fat deposition by regulating mRNA expression. The findings provide a marker and add to the knowledge of the molecular breeding of high-quality broilers. It is necessary to further clarify the molecular regulation mechanism of CAPN9 on the development of chickens.
Acknowledgements
The research was supported by grants from National Natural Science Foundation of China (31372305), the Agricultural Science and Technology Innovation Program (ASTIP-IAS-TS-11 and ASTIP-IAS04).
REFERENCES
- AOAC Official Methods of Analysis. 1990. Arlington, 15th edn. Association of Official Analytical Chemists, Virginia. [Google Scholar]
- Cavalli M., Pan G., Nord H., Wadelius C.. 2016. Looking beyond GWAS: allele-specific transcription factor binding drives the association of GALNT2 to HDL-C plasma levels. Lipids Health Dis. 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., Fawcett G. L., Jarvis J. P., Norgard E. A., Pavlicev M., Pletscher L. S., Polonsky K. S., Ye H., Bell G. I., Semenkovich C. F.. 2010. Calpain-10 is a component of the obesity-related quantitative trait locus Adip1. J. Lipid Res. 51:907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X. Y., Liu R. R., Cui H. X., Zhao G. P., Zheng M. Q., Li Q. H., Liu J., Liu Z., Wen J.. 2017. Effects of caponization and ovariectomy on objective indices related to meat quality in chickens. Poult. Sci. 96:770–777. [DOI] [PubMed] [Google Scholar]
- Davis J., Martin S. G., Patel P. M., Green A. R., Rakha E. A., Ellis I. O., Storr S. J.. 2014. Low calpain-9 is associated with adverse disease-specific survival following endocrine therapy in breast cancer. BMC Cancer. 14:995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer L. J. 1999. Poultry meat flavor [A]. Richardson Rl et al. Poultry meat science [C] Pages 127–158CAB International, New York. [Google Scholar]
- Fernandez X., Monin G., Talmant A., Mourot J., Lebret B.. 1999. Influence of intramuscular fat content on the quality of pig meat-1. Composition of the lipid fraction and sensory characteristics of m.longissmus lumborum. Meat Sci. 53:59–65. [DOI] [PubMed] [Google Scholar]
- Gafni J., Cong X., Chen S. F., Gibson B. W., Ellerby L. M.. 2009. Calpain-1 cleaves and activates caspase-7. J. Biol. Chem. 284:25441–25449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S., Abe M., Suzuki H., Kitamura F., Toyama-Sorimachi N., Abe K., Sakimura K., Sorimachi H.. 2010. Calpain 8/nCL-2 and calpain 9/ nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense. PLoS Genet. 6:e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. C., Huang M., Wang P., Zhao L., Xu X. L., Zhou G. H., Sun J. X.. 2014. Effects of physical restraint and electrical stunning on plasma corticosterone, postmortem metabolism, and quality of broiler breast muscle. J. Anim. Sci. 92:5749–5756. [DOI] [PubMed] [Google Scholar]
- Krizek B. A. 2015. Intronic sequences are required for AINTEGUMENTA-LIKE6 expression in Arabidopsis flowers. BMC Res. Notes 8:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Sorimachi H., Jeong S. Y., Ishiura S., Suzuki K.. 1998. Molecular cloning and characterization of a novel tissue-specific calpain predominantly expressed in the digestive tract. Biol. Chem. 379:175–183. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Tomioka S., Kinbara K., Masumoto H., Jeong S. Y., Sorimachi H., Ishiura S., Suzuki K.. 1999. Characterization of a human digestive tract-specific calpain, nCL-4, expressed in the baculovirus system. Arch. Biochem. Biophys. 362:22–31. [DOI] [PubMed] [Google Scholar]
- Markmann A., Schäfer S., Linz W., Löhn M., Busch A. E., Wohlfart P.. 2005. Down-regulation of calpain 9 is linked to hypertensive heart and kidney disease. Cell Physiol. Biochem. 15:109–116. [DOI] [PubMed] [Google Scholar]
- McClung J. M., Judge A. R., Talbert E. E., Powers S. K.. 2009. Calpain-1 is required for hydrogen peroxide-induced myotube atrophy. Am. J. Physiol. Cell Physiol. 296:C363–C371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookura T., Koyama E., Hansen A., Teeter J. H., Kawamura Y., Brand J. G.. 2015. Phylogenetic analysis and taste cell expression of calpain 9 in catfish. Nat. Sci. 7:143–150. [Google Scholar]
- Patel Y. M., Lane M. D.. 1999. Role of calpain in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 96:1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W.. 2001. Molecular Cloning: A Laboratory Manual, 1st edn Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Shioda N., Moriguchi S., Shirasaki Y., Fukunaga K.. 2006. Generation of constitutively active calcineurin by calpain contributes to delayed neuronal death following mouse brain ischemia. J. Neurochem. 98:310–320. [DOI] [PubMed] [Google Scholar]
- Visser M., Palstra R. J., Kayser M.. 2015. Allele-specific transcriptional regulation of IRF4 in melanocytes is mediated by chromatin looping of the intronic rs12203592 enhancer to the IRF4 promoter. Hum. Mol. Genet. 24:2649–2661. [DOI] [PubMed] [Google Scholar]
- Yajima Y., Sato M., Sorimachi H., Inomata M., Maki M., Kawashima S.. 2006. Calpain system regulates the differentiation of adult primitive mesenchymal ST-13 adipocytes. Endocrinology 147:4811–4819. [DOI] [PubMed] [Google Scholar]
- Zhao L., Xing T., Huang J., Qiao Y., Chen Y., Huang M.. 2018. Involvement of μ/m- calpain in the proteolysis and meat quality changes during postmortem storage of chicken breast muscle. Anim. Sci. J. 89:423–431. [DOI] [PubMed] [Google Scholar]