Abstract
Background:
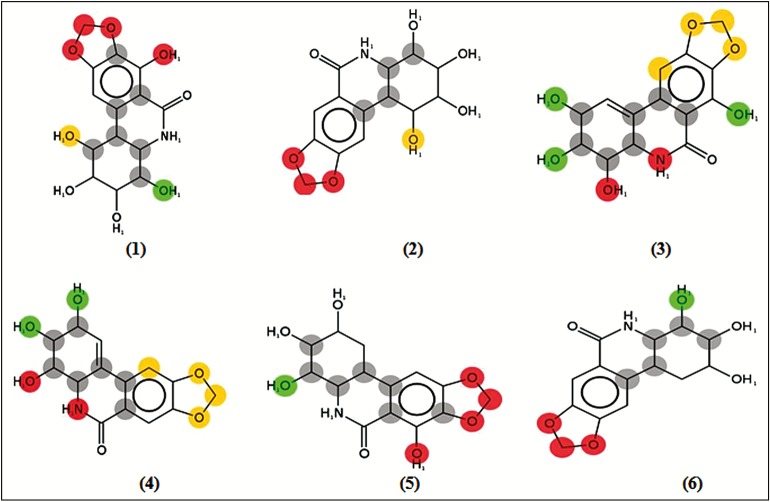
Plants of the Amaryllidaceae family have been under intense scrutiny for the presence of a couple of alkaloidal secondary metabolites with endued cytotoxic activity, such as pancratistatin (1), 7-deoxypancratistatin (2), narciclasine (3), 7-deoxynarciclasine (4), trans-dihydronarciclasine (5), and 7-deoxy-trans-dihydronarciclasine (6). Nevertheless, preclinical evaluation of these alkaloids has been put on hold because of the limited quantity of materials available from isolation.
Aim:
To explore the underlying cytotoxic molecular mechanisms of the Amaryllidaceae alkaloids (1–6) and to assess their absorption, distribution, metabolism, excretion, and toxicity (ADMET) profiles using chemoinformatic tools.
Materials And Methods:
AutoDock 4.0 software along with different in silico chemoinformatic tools, namely PharmMapper, Molinspiration, MetaPrint2D, and admetSAR servers, were used to assess the drugability of the Amaryllidaceae alkaloids (1–6).
Results:
Deoxycytidine kinase (dCK) (PDB: 1P60) was predicted as a potential target with fitting score of 5.574. In silico molecular docking of (1–6) into dCK revealed good interactions, where interesting hydrogen bonds were observed with the amino acid residues—Gly-28 and Ser-35—located in the highly conserved P-loop motif. This motif plays a special role in dCK function. Contrary to (1), in silico pharmacokinetic results have shown good absorption and permeation and thus good oral bioavailability for (2–6).
Conclusion:
The in silico docking data have proposed that the reported cytotoxic activity of the Amaryllidaceae alkaloids (1–6) could be mediated through dCK inhibition. In addition, the ADMET profile of these alkaloids is promising and thus (1–6) could be candidates for future drug development.
KEYWORDS: Amaryllidaceae alkaloids, cytotoxicity, deoxycytidine kinase, in silico
INTRODUCTION
Over the past several decades, the search for natural products in marine and terrestrial environments has led to the discovery of a number of biologically active alkaloids. Of those are the Amaryllidaceae alkaloids, which are structurally related compounds primarily isolated from the plants of the family Amaryllidaceae.[1] Among these, pancratistatin (1), 7-deoxypancratistatin (2), narciclasine (3), 7-deoxynarciclasine (4), trans-dihydronarciclasine (5), and 7-deoxy-trans-dihydronarciclasine (6) constitute an emblematic group. They are characterized by their highly oxygenated phenanthridinone core with contiguous chiral centers on one ring [Figure 1]. Amaryllidaceae alkaloids are known to have antitumor and antiviral activities along with other interesting biological activities.[2] Pancratistatin, which was isolated in 1984 by Pettit et al.[3] from the roots of the Hawaiian Pancratium littorale, has shown promising in vitro antineoplastic activity.[4,5] Furthermore, pancratistatin and other Amaryllidaceae alkaloids also proved to induce apoptosis against a large panel of cancer cell lines.[6] Interestingly, pancratistatin has an insignificant cytotoxic effect on noncancerous cell lines.[7,8] Nevertheless, the biochemical mechanism by which pancratistatin induces apoptosis in cancer cells is still unknown.[5,9] This can be attributed to the limited quantity of material available from either isolation or total syntheses.[10] Fascinatingly, the in silico prediction of leads’ molecular mechanisms is now widely accepted, where different computational tools are used to mimic the involved biological systems.[11,12,13] It was, therefore, hoped that the molecular mechanism of pancratistatin could be assessed without having to undergo the costly and tedious wet conventional experiments. Our preliminary in silico investigations revealed that deoxycytidine kinase (dCK) is a potential target for pancratistatin. This target is a crucial enzyme in deoxyribonucleoside salvage pathway and is involved in apoptosis inhibition. dCK is activated on deoxyribonucleic acid (DNA) damage, and the degradation products are recycled to aid in DNA repair and apoptosis inhibition.[14,15,16] On the other hand, inactivation of dCK causes DNA replication stress and subsequent cell cycle arrest, resulting in apoptosis induction.[17,18] Thus, dCK inhibition could be envisioned as a potential strategy for cancer therapy. We herein report the unprecedented use of an in silico approach to uncover the molecular mechanism of the anticancer properties of pancratistatin and its derivatives as a dCK inhibitor and, thus, as apoptosis inducers.
Figure 1.

Amaryllidaceae alkaloids
MATERIALS AND METHODS
Preparation of ligand and protein structures
ChemDraw Ultra 12 software (CambridgeSoft) was used for ligand preparation and optimization. The crystal structure of the predicted target protein was retrieved from the Protein Data Bank[19] and optimized using Swiss-PdbViewer 4.1.0 software (Swiss Institute of Bioinformatics).[20]
Biological activity
The potential protein targets for the tested compounds were predicted using PharmMapper server.[21] On the other hand, for each compound, Molinspiration server[22] was used to predict drug-likeness properties such as G-protein–coupled receptor (GPCR) ligands, ion channel modulators (ICM), kinase inhibitors (KI), nuclear receptor ligands (NRL), protease inhibitors (PI), and enzyme inhibitors (EI).
Molecular docking
Molecular docking was performed using AutoDock 4.0 software (Molecular Graphics Laboratory) based on Lamarckian Genetic Algorithm.[23,24] Polar hydrogen atoms were added to the protein target and Kollman united atomic charges were computed. All hydrogen atoms were added to the ligands before the Gasteiger partial charges were assigned. The cocrystal ligand was removed and the bond orders were checked. The target’s grid map was calculated and set to 60×60×60 points with grid spacing of 0.375 Ǻ. The grid box was then allocated properly in the target to include the active residue in the center. The default docking algorithms were set in accordance with the standard docking protocol. Docking results having less than 1.0 Å in positional root-mean-square deviation were clustered together, and the results were retrieved as binding energies. Poses that showed the lowest binding energies were visualized using molecular operating environment[25] and University of California San Francisco chimera.[26]
Physicochemical properties
LogP, topological polar surface area (TPSA), and the number of hydrogen bond donors (HBDs) and hydrogen bond acceptors (HBAs) for the tested leads were estimated using Molinspiration server.[22]
Pharmacokinetics and toxicity
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) were estimated using admetSAR[27] and MetaPrint2D[28] online servers.
RESULTS
Biological activity and molecular docking
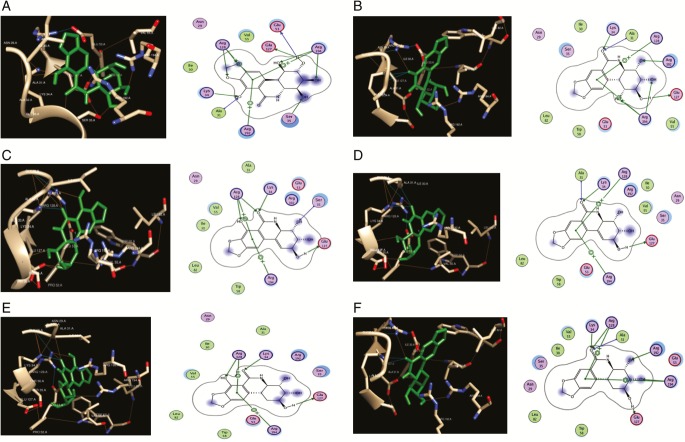
PharmMapper server predicted that dCK (PDB: 1P60)[29] is the best target in terms of fit score (5.574) for pancratistatin (1). Having nearly similar chemical scaffolds, we propose dCK as a potential target of the other Amaryllidaceae alkaloids (2–6). Docking results revealed that alkaloids (1–6) docked nicely into the active pocket and showed good biochemical interactions with essential amino acid residues [Figure 2]. Herein, (1) (E = –7.00 kcal/mol) docked into the highly conserved P-loop motif located at the amino acid residues—Gly-28 and Ser-35—in which this motif plays an essential role in dCK function. As for (2), a derivative of (1), a better binding affinity (E= –7.20 kcal/mol) and a superior interaction were observed. With a similar binding affinity to (1), (2) has showed an additional hydrogen bond with Arg-128. Interestingly, Arg-128 is noted to have a chief anchor function, as it interacts via its NH2 group with the 5ʹ-hydroxyl group of deoxycytidine and with Glu-35, which is strictly conserved within the dCK enzyme.[28] On the other hand, (3) and its derivatives (4, 5, and 6) have shown favorable interactions over the aforementioned compounds. The binding energies were –7.81, –7.79, –7.35, and –7.21 for (3), (4), (5), and (6), respectively. In addition, (3) and its derivatives (4, 5, and 6) have shown a hydrogen bonding with Arg-128 along with biochemical interactions with the residues of the conserved P-loop.
Figure 2.
Ligand–deoxycytidine kinase (dCK) interactions. (A) Pancratistatin–dCK interactions visualized by chimera (left) and molecular operating environment (MOE) (right). (B) 7-Deoxypancratistatin–dCK interactions visualized by chimera (left) and MOE (right). (C) Narciclasine–dCK interactions visualized by chimera (left) and MOE (right). (D) 7-Deoxynarciclasine–dCK interactions visualized by chimera (left) and MOE (right). (E) trans-Dihydronarciclasine–dCK interactions visualized by Chimera (left) and MOE (right). (F) 7-Deoxy-trans-dihydronarciclasine–dCK interactions visualized by Chimera (left) and MOE (right)
Molinspiration server was used to assess the physiochemical properties based on Lipinski’s rule of five.[30] With the exception of (1) (TPSA = 148 and HBD = 6), all the other compounds have shown good absorption and permeation and thus good oral bioavailability, as its predicted physicochemical properties were in agreement with Lipinski’s rule of five [Table 1].
Table 1.
Physicochemical parameters predicted by Molinspiration server
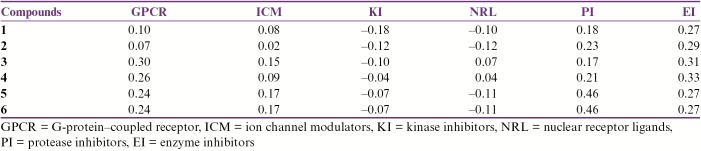
The compounds have also been predicted to be GPCR ligands, ICM, KI, NRL, PI, and EI, which were retrieved as bioactivity scores [Table 2]. Herein, scores more than 0.00 indicate high activity, between 0.00 and −0.5 indicate moderate activity, and less than −0.5 indicate inactivity.[31]
Table 2.
Drug-likeliness property estimations by Molinspiration server
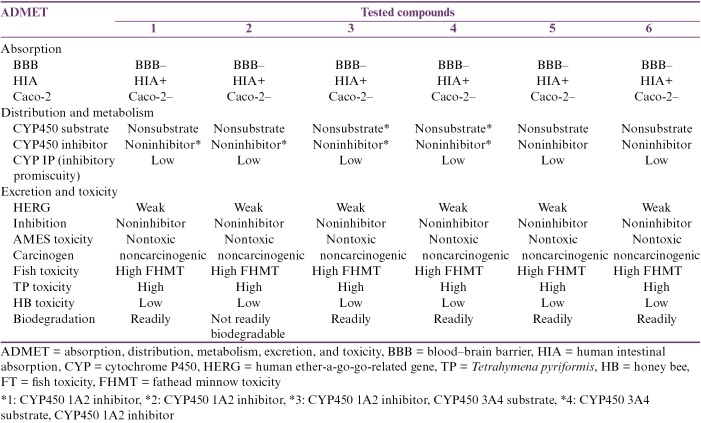
For ADMET profile prediction, we used Metaprint2D and admetSAR web-based servers. Having a glance at the Metaprint2D results [Figure 3], we could see that the tested compounds show a diverse pattern of metabolic transformations based on the normalized occurrence ratio (NOR). With the exception of (3) and (4), the metabolic transformation of the 1,3-dioxolane moieties in the four compounds (1, 2, 5, and 6) represented good sites for metabolism. Herein, the methylene group was predicted to undergo dealkylation and hydroxylation, whereas both oxygen atoms were predicted to undergo only dealkylation. As for (3) and (4), the –NH and –OH groups were predicted to undergo oxidative–deamination and sulfation, respectively. The compounds (1), (3), and (5) that bare phenol moieties were estimated to undergo a number of metabolic transformations. Both (1) and (5), which contain the highly metabolized –OH group, were predicted to undergo five transformations, namely glucuronidation, methylation, sulfation, glucosidation, and hydroxylation. Results of admetSAR revealed that all six compounds do not cross the blood–brain barrier (BBB). Further, these compounds were predicted not to penetrate through Caco-2 cell line. Obviously, all compounds did not show any acute toxicity and mutagenic effect with respect to the ADMET test data [Table 3].
Figure 3.
Metabolic predictions using Metaprint2D for each lead. The color highlighting an atom indicates its normalized occurrence ratio (NOR). Atoms are colored according to the likelihood of a metabolic site; high: red, medium: orange, low: green, very low: not colored, and no data: gray. A high NOR indicates a more frequently reported site of metabolism in the metabolite database for Amaryllidaceae alkaloids (1–6)
Table 3.
ADMET predictions using admetSAR
DISCUSSION
dCK is an interesting target to be considered for therapeutic targeting in cancer, being involved in DNA repair and apoptosis.[17,18] On the basis of the in silico pharmacophore mapping, we propose dCK as a possible target of pancratistatin. It is worth noting that dCK is highly expressed in thymus and bone marrow indicating its great role in hematopoiesis.[14] Furthermore, it has been reported that dCK has a crucial role in both DNA replication and repair process pertained with its constitutive expression throughout the cell cycle.[32]
Our in silico molecular docking results revealed that alkaloids (1–6) are possible dCK inhibitors. On the basis of the superlative knowledge of being cytotoxic to a panel of cancerous cell lines,[6] we attempted to explore their anticancer mechanisms at the molecular level. It is noteworthy that DNA damage induces rapid hyperphosphorylation and activation of p53.[33] This promotes DNA repair through dCK activation, leading to increase in the salvage pathway of deoxynucleoside triphosphate (dNTP) production.[34] Furthermore, dCK inactivates cyclin-dependent kinase-1 (Cdk1),[16] which induces G2/M transition and mitosis.[35] Therefore, inhibition of dCK by these alkaloids would hopefully lead to induction of apoptosis in cancer cells.
As most lead compounds fail to meet the pharmacokinetic criteria of a drug at the final stages of the drug discovery pipeline, it is advantageous to assess the physicochemical drug-likeness properties and ADMET profiles using advanced in silico tools. Molinspiration server was used to assess the physiochemical properties based on Lipinski’s rule of five.[30] Poor absorption or permeation is more likely when there are more than 5 HBDs, 10 HBAs, the molecular weight is greater than 500Da, and the calculated LogP (CLogP) is greater than 5 (or MlogP > 4.15). Moreover, good bioavailability is more likely for compounds with ≤10 rotatable bonds (nrotb) and TPSA of ≤140 Å.[36] Interestingly, all six compounds were predicted to be EI (score > 0.00) [Table 2]. In addition, (4), (5), and (6) have shown good activities such as GPCR and PI (score >0.00). From these results, we can confidently explain the predicted drugability of the tested compounds as dCK inhibitors. Nevertheless, other potential activities could be hopefully medicated via other pathways as PI or GPCR. Other ADMET properties such as BBB penetration, human intestinal absorption, human colon carcinoma cell (Caco-2) permeability, and Ames test were estimated via admetSAR. Having nearly similar chemical scaffolds, we expect that most of their ADMET patterns are quite similar. All six compounds are not expected to cross the BBB, as more polar compounds have lower lipid solubility, limiting their BBB penetration.[37] Furthermore, these compounds were predicted not to penetrate through Caco-2 cell line, which has also been proposed for the prediction of the BBB permeability of drugs. Herein, our tested compounds could be of low oral bioavailability and need further structural optimization. On the other hand, the metabolic performance of the compounds on cytochrome P450 (CYP) enzyme had been predicted on to 2C9, 2D6, 3A4, and 1A2 isoenzymes. It is worth noting that the drug that undergoes CYP-mediated oxidative biotransformation is responsible for the large number of clinically significant drug interactions during multiple drug therapy.[38]
CONCLUSION
To conclude, the cytotoxic mechanism of the studied Amaryllidaceae alkaloids (1–6) as dCK inhibitors and their ADMET profiles have been addressed using in silico approach. Wet experiments are needed to confirm these findings.
Financial support and sponsorship
Financial support and assistance was provided by the Faculty of Pharmacy, University of Khartoum, Sudan, and also by the Faculty of Pharmacy, Sudan International University, Sudan.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Bastida J, Berkov S, Torras L, Pigni NB, de Andrade JP, Martinez V, et al. Chemical and biological aspects of Amaryllidaceae alkaloids. In: Munoz-Torrero D, editor. Recent advances in pharmaceutical sciences. Kerala, India: Transworld Research Network; 2011. pp. 65–100. [Google Scholar]
- 2.Cordell GA, Quinn-Beattie ML, Farnsworth NR. The potential of alkaloids in drug discovery. Phytother Res. 2001;15:183–205. doi: 10.1002/ptr.890. [DOI] [PubMed] [Google Scholar]
- 3.Pettit GR, Gaddamidi V, Cragg GM, Herald DL, Sagawa Y. Isolation and structure of pancratistatin. J Chem Soc Chem Commun. 1984;24:1693–4. [Google Scholar]
- 4.Pettit GR, Gaddamidi V, Herald DL, Singh SB, Cragg GM, Schmidt JM, et al. Antineoplastic agents, 120. Pancratium littorale. J Nat Prod. 1986;49:995–1002. doi: 10.1021/np50048a005. [DOI] [PubMed] [Google Scholar]
- 5.McLachlan A, Kekre N, McNulty J, Pandey S. Pancratistatin: a natural anti-cancer compound that targets mitochondria specifically in cancer cells to induce apoptosis. Apoptosis. 2005;10:619–30. doi: 10.1007/s10495-005-1896-x. [DOI] [PubMed] [Google Scholar]
- 6.Evidente A, Kireev AS, Jenkins AR, Romero AE, Steelant WF, Van Slambrouck S, et al. Biological evaluation of structurally diverse Amaryllidaceae alkaloids and their synthetic derivatives: discovery of novel leads for anticancer drug design. Planta Med. 2009;75:501–7. doi: 10.1055/s-0029-1185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin C, Karnik A, McNulty J, Pandey S. Pancratistatin selectively targets cancer cell mitochondria and reduces growth of human colon tumor xenografts. Mol Cancer Ther. 2011;10:57–68. doi: 10.1158/1535-7163.MCT-10-0735. [DOI] [PubMed] [Google Scholar]
- 8.Pandey S, Kekre N, Naderi J, McNulty J. Induction of apoptotic cell death specifically in rat and human cancer cells by pancratistatin. Artif Cells Blood Substit Immobil Biotechnol. 2005;33:279–95. doi: 10.1081/bio-200066621. [DOI] [PubMed] [Google Scholar]
- 9.Havelek R, Seifrtova M, Kralovec K, Bruckova L, Cahlikova L, Dalecka M, et al. The effect of Amaryllidaceae alkaloids haemanthamine and haemanthidine on cell cycle progression and apoptosis in p53-negative human leukemic jurkat cells. Phytomedicine. 2014;21:479–90. doi: 10.1016/j.phymed.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Ghavre M, Froese J, Pour M, Hudlicky T. Synthesis of Amaryllidaceae constituents and unnatural derivatives. Angewandte Chemie (International Ed. in English) 2016;55:5642–91. doi: 10.1002/anie.201508227. [DOI] [PubMed] [Google Scholar]
- 11.Barlow DJ, Buriani A, Ehrman T, Bosisio E, Eberini I, Hylands PJ. In silico studies in Chinese herbal medicines’ research: evaluation of in silico methodologies and phytochemical data sources, and a review of research to date. J Ethnopharmacol. 2012;140:526–34. doi: 10.1016/j.jep.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirar AI, Waddad AY, Mohamed MA, Mohamed MS, Osman WJ, Mohammed MS, et al. In silico pharmacokinetics and molecular docking of three leads isolated from Tarconanthus camphoratus L. Int J Pharm Sci. 2016;8:71–7. [Google Scholar]
- 13.Mohamed MA, Dirar AI, Hamdoun S. Discovery of two diacetylene glycosides as human uridine-cytidine kinase 2 inhibitors: an in silico approach. J Appl Pharm Sci. 2016;6:34–9. [Google Scholar]
- 14.Toy G, Austin WR, Liao HI, Cheng D, Singh A, Campbell DO, et al. Requirement for deoxycytidine kinase in T and B lymphocyte development. Proc Natl Acad Sci u s a. 2010;107:5551–6. doi: 10.1073/pnas.0913900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunimovich YL, Nair-Gill E, Riedinger M, McCracken MN, Cheng D, McLaughlin J, et al. Deoxycytidine kinase augments ATM-mediated DNA repair and contributes to radiation resistance. PLoS One. 2014;9:e104–25. doi: 10.1371/journal.pone.0104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Lee M, Hao J, Cui X, Guo X, Smal C, et al. Deoxycytidine kinase regulates the G2/M checkpoint through interaction with cyclin-dependent kinase 1 in response to DNA damage. Nucleic Acids Res. 2012;40:9621–32. doi: 10.1093/nar/gks707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy JM, Armijo AL, Nomme J, Lee CH, Smith QA, Li Z, et al. Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography. J Med Chem. 2013;56:6696–708. doi: 10.1021/jm400457y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomme J, Li Z, Gipson RM, Wang J, Armijo AL, Le T, et al. Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability. J Med Chem. 2014;57:9480–94. doi: 10.1021/jm501124j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Protein Data Bank [homepage on the internet]. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, et al. The protein data bank. [Last accessed on 2017 April];Nucleic Acids Res. 2000 28:235–42. doi: 10.1093/nar/28.1.235. Available from: http://www.rcsb.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-pdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–60. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molinspiration Cheminformatics [homepage on the internet] [Last accessed on 2017 January]. Available from: http://www.molinspiration.com/
- 23.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–62. [Google Scholar]
- 24.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. Autodock4 and autodocktools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molecular Operating Environment (MOE) [homepage on the internet], Chemical Computing Group ULC. [Last accessed on 2017 January]. Available from: http://www.chemcomp.com/
- 26.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. [Last accessed on 2017 January];J Chem Inf Model. 2012 52:3099–105. doi: 10.1021/ci300367a. Available from: http://lmmd.ecust.edu.cn/admetsar1/ [DOI] [PubMed] [Google Scholar]
- 28.Carlsson L, Spjuth O, Adams S, Glen R, Boyer S. Use of historic metabolic biotransformation data as a means of anticipating metabolic sites using MetaPrint2D and Bioclipse. [Last accessed on 2017 January];BMC bioinformatics. 2010 11:362. doi: 10.1186/1471-2105-11-362. Available from: http://wwwmetaprint2d.ch.cam.ac.uk/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10:513–9. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 30.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 31.Paramashivam SK, Elayaperumal K, Natarajan BB, Ramamoorthy MD, Balasubramanian S, Dhiraviam KN. In silico pharmacokinetic and molecular docking studies of small molecules derived from Indigofera aspalathoides Vahl targeting receptor tyrosine kinases. Bioinformation. 2015;11:73–84. doi: 10.6026/97320630011073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staub M, Eriksson S. The role of deoxycytidine kinase in DNA synthesis and nucleoside analog activation. Deoxynucleoside analogs in cancer therapy. Springer. 2006:29–52. [Google Scholar]
- 33.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis: the p53 network. J Cell Sci. 2003;116:4077–85. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 34.Keszler G, Virga S, Spasokoukotskaja T, Bauer PI, Sasvari-Szekely M, Staub M. Activation of deoxycytidine kinase by deoxyadenosine: implications in deoxyadenosine-mediated cytotoxicity. Arch Biochem Biophys. 2005;436:69–77. doi: 10.1016/j.abb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 36.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–23. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 37.Waterhouse RN. Determination of lipophilicity and its use as a predictor of blood–brain barrier penetration of molecular imaging agents. Mol Imaging Biol. 2003;5:376–89. doi: 10.1016/j.mibio.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Palleria C, Di Paolo A, Giofrè C, Caglioti C, Leuzzi G, Siniscalchi A, et al. Pharmacokinetic drug–drug interaction and their implication in clinical management. J Res Med Sci. 2013;18:601–10. [PMC free article] [PubMed] [Google Scholar]