Key Points
Question
Are adults with higher genetic risk for schizophrenia more likely to live in urbanized and populated areas than those with lower risk?
Findings
In this cross-sectional study of 4 community-based samples from Australia, the United Kingdom, and the Netherlands (N = 504 130), significantly higher genetic loading for schizophrenia was identified in participants living in more densely populated areas; mendelian randomization on a subsample suggests that schizophrenia may have a causal association with the tendency to live in urban areas.
Meaning
The higher rates of schizophrenia in cities may be accentuated by selective migration to cities of participants with higher genetic risks.
Abstract
Importance
Urban life has been proposed as an environmental risk factor accounting for the increased prevalence of schizophrenia in urban areas. An alternative hypothesis is that individuals with increased genetic risk tend to live in urban/dense areas.
Objective
To assess whether adults with higher genetic risk for schizophrenia have an increased probability to live in more populated areas than those with lower risk.
Design, Setting, and Participants
Four large, cross-sectional samples of genotyped individuals of European ancestry older than 18 years with known addresses in Australia, the United Kingdom, and the Netherlands were included in the analysis. Data were based on the postcode of residence at the time of last contact with the participants. Community-based samples who took part in studies conducted by the Queensland Institute for Medical Research Berghofer Medical Research Institute (QIMR), UK Biobank (UKB), Netherlands Twin Register (NTR), or QSkin Sun and Health Study (QSKIN) were included. Genome-wide association analysis and mendelian randomization (MR) were included. The study was conducted between 2016 and 2018.
Exposures
Polygenic risk scores for schizophrenia derived from genetic data (genetic risk is independently measured from the occurrence of the disease). Socioeconomic status of the area was included as a moderator in some of the models.
Main Outcomes and Measures
Population density of the place of residence of the participants determined from census data. Remoteness and socioeconomic status of the area were also tested.
Results
The QIMR participants (15 544; 10 197 [65.6%] women; mean [SD] age, 54.4 [13.2] years) living in more densely populated areas (people per square kilometer) had a higher genetic loading for schizophrenia (r2 = 0.12%; P = 5.69 × 10−5), a result that was replicated across all 3 other cohorts (UKB: 345 246; 187 469 [54.3%] women; age, 65.7 [8.0] years; NTR: 11 212; 6727 [60.0%] women; age, 48.6 [17.5] years; and QSKIN: 15 726; 8602 [54.7%] women; age, 57.0 [7.9] years). This genetic association could account for 1.7% (95% CI, 0.8%-3.2%) of the schizophrenia risk. Estimates from MR analyses performed in the UKB sample were significant (b = 0.049; P = 3.7 × 10−7 using GSMR), suggesting that the genetic liability to schizophrenia may have a causal association with the tendency to live in urbanized locations.
Conclusions and Relevance
The results of this study appear to support the hypothesis that individuals with increased genetic risk tend to live in urban/dense areas and suggest the need to refine the social stress model for schizophrenia by including genetics as well as possible gene-environment interactions.
This community-based study examines the association between population density in Australia, the United Kingdom, and the Netherlands and likelihood of residence in individuals of European ancestry with a genetic risk for schizophrenia.
Introduction
In 2011, Lederbogen and colleagues1 published a functional magnetic resonance imaging study that showed greater brain activation of the stress-processing pathways in participants living in urban vs rural areas and suggested there and in a later study2 that the greater social stress of urban living could explain the well-documented higher prevalence of schizophrenia observed in urban than rural environments (odds ratio, 1.72; 95% CI, 1.53-1.92).3 Herein, we investigate an alternative, but not incompatible, explanation: people with higher genetic risk for schizophrenia tend to live in more urbanized areas owing to selective migration4 in either past or current generations.
eAppendix 1 in the Supplement presents a short review of the literature in this area. In summary, living in an urban environment, which is itself partially heritable,5 is associated with increased risk of developing schizophrenia after controlling for potential confounders (age, sex, ethnicity, drug use, social class, family history, and season of birth) and using different measures of urbanicity (population size or density6,7), window of exposure (birth,4,8 upbringing,1,6 or illness onset7,9), and disease definition (narrow schizophrenia or broad psychosis7,10). Although the association is established, its putative (familial) environmental or genetic components are unclear. It has been suggested that approximately 30% of all schizophrenia cases could be potentially prevented if the exposure to urban environments was removed, assuming urban environment is a causal factor.10 However, it is not clear whether urban residence has a causal effect on mental health or whether urban residence is a consequence of the disease (eg, migration to the city of people in the prodromal stages of the disorder6).
In the present study, we sought to examine the nature of this association by testing whether adults older than 18 years with higher genetic risk for schizophrenia are more likely to live in urbanized and populated areas (measured as population density) than those with lower genetic risk. If so, this finding would suggest that the higher prevalence of schizophrenia in cities is not only a consequence of the urban environment. This determination has been made possible by the advances in the identification of common genetic variants associated with schizophrenia in large discovery samples11 and the development of polygenic risk scores (PRS) in independent samples.12 In addition, we checked that the association could not be explained by differences in socioeconomic status (SES) of the residential areas. We also investigated the direction of causation between schizophrenia and population density using multi-instrument mendelian randomization (MR).13,14 For completeness, we present the estimates of the twin heritability and genome-wide association analyses (GWAS) of our main phenotypes.
Methods
Cohorts and Variables
We performed the analyses using a discovery cohort of 15 544 participants genotyped as part of a series of studies of general health conditions conducted by the Genetic Epidemiology Unit at Queensland Institute for Medical Research Berghofer Medical Research Institute (QIMR), Australia15,16 (Table 1; eAppendix 2, eTable 1, and eFigure 1 in the Supplement provide cohort and variable descriptions). We used the UK Biobank (UKB) (n = 456 426),17,18 the Netherlands Twin Register (NTR) (n = 16 434),19 and the Australian QSkin Sun and Health Study (QSKIN) sample (n = 15 726)20 to replicate and extend our analyses (Table 1; eAppendixes 3-5, eTable 2, eTable 3, eFigures 2-5 in the Supplement provide cohort and variable descriptions).
Table 1. Description of the Cohorts Used for the Analysesa.
| Variable | Discovery Cohort | Replication Cohorts | ||
|---|---|---|---|---|
| QIMR15,16 (Australia) |
UKB17,18 (United Kingdom) |
NTR19 (the Netherlands) |
QSKIN20 (Australia [limited to Queensland]) |
|
| Sample size, No. | 15 544 | 456 426 (345 246 with PRS) |
16 434 (11 212 with PRS) |
15 726 |
| Inclusion criteriab | Adult genotyped participants of QIMR studies | Genotyped participants of the UKB | Adult genotyped participants of the NTR | Unrelated (GRM <0.1) adult genotyped participants of the QSKIN who had not previously participated in QIMR studies |
| Demographics | 10 197 (65.6%) Women | 187 469 (54.3%) Women | 6727 (60.0%) Women | 8602 (54.7%) Women |
| Age, mean (SD): 54.4 (13.2) | Age, mean (SD): 65.7 (8.0) | Age, mean (SD): 48.6 (17.5) | Age, mean (SD), 57.0 (7.9) | |
| From 7015 families | From 4456 families | |||
| Sample comprises 1119 complete MZ pairs of twins, 1104 complete DZ pairs, and 1448 singleton twins | Sample comprises 345 258 unrelated individuals (GRM <0.05) | Sample comprises 1740 complete MZ pairs, 1114 DZ complete twin pairs, and 812 singleton twins | ||
| Genetic data | Participants genotyped using commercial arrays | Participants genotyped using 2 closely related arrays (the UK BiLEVE and the UK Biobank Axiom Arrays) | Participants genotyped using commercial arrays | Same as QIMR |
| Genotype data cleaned (by batch) for call rate (≥95%) | We used HRC imputed data provided by the biobank | Genotype data cleaned for call rate (≥95%) | ||
| MAF (≥1%) | Variants with MAF <0.005%, missingness <0.05, pHWE <10−6, and MAC >5 excluded | MAF ≥0.5% | ||
| Hardy-Weinberg equilibrium (P ≥ 10−3), GenCall score (≥0.15 per genotype; mean, ≥0.7), standard Illumina filters | Data were checked for non-European ancestry | Hardy-Weinberg equilibrium (P ≥ 10−12), allele frequency difference with GONL <0.10 | ||
| Data checked for pedigree, sex, and mendelian errors and for non-European ancestry | Data checked for pedigree, sex, and mendelian errors and for non-European ancestry21 | |||
| Imputation to the 1000 genomes (phase 3, release 5) performed on the Michigan Imputation Server22 | ||||
| Measures of population density and SES | Population density, remoteness, and SES variables generated from the postcode provided by participants at the time of last contact (1990-2015) | From the Easting and Northing coordinates rounded to the kilometer, we performed reverse geocoding to identify the postcode district in which the participants likely lived | Population density and SES derived from the most recent participants’ postcodes | Population density and SES variables were generated from the postcode provided by the participants at the time of last contact (2010-2012) |
| We matched postcodes to the latest census data collected by the Australian Bureau of Statistics (2016 for population density, 2011 for remoteness and SES) | We crossed this information with the population density by postcode district calculated in the 2011 census | Numbers corresponded to the neighborhood data published in 2015-2016 by the Netherlands’s national statistical agency (CBS), which defines a neighborhood as the part of a municipality that is homogeneously demarcated from either a demographic or socioeconomic structure | We matched the postcodes as described in the QIMR sample | |
| Population density expressed in number of residents per squared kilometer | Population density expressed in number of residents per hectare | Population density expressed in number of residents per squared kilometer | Population density expressed in number of residents per squared kilometer | |
| SES based on the IRSAD,23 which can be used to measure socioeconomic well-being in a continuum, from the most disadvantaged areas (low values) to the most advantaged areas (high values) | We used the Townsend deprivation index17,18 as a measure of SES | SES measured using the average personal income per person and the average market value of residential properties | SES based on the IRSAD,23 as described in the QIMR sample | |
| Mean (SD) population density was 1169 (1350) people/km2 (range, 0.01-5506) | Mean (SD) population density was 24.10 (25.1) inhabitants per hectare (ie, 2410 per km2) (range, 0.1-222.5) | Mean (SD) population density in the total sample was 4669 (3793) (range, 1-17 797) | Mean (SD) population density was 748 people/km2 (945) (range, 0.01-3771) | |
Abbreviations: DZ, dizygotic; GONL, Genome of the Netherlands; GRM, genetic relationship matrices; HRC, Haplotype Reference Consortium; IRSAD, Index of Relative Socioeconomic Advantage and Disadvantage; MAC, minor allele count; MAF, minor allele frequency; MZ, monozygotic; NTR, Netherlands Twin Register; PHWE, P value for the Hardy-Weinberg test statistic; PRS, polygenic risk scores; QIMR, QIMR Berghofer Medical Research Institute; QSKIN, QSkin Sun and Health Study; SES, socioeconomic status; UKB, United Kingdom Biobank.
The Supplement contains documents on genetic and phenotypic data for all cohorts.
European ancestry was an inclusion criterion across all cohorts.
The QIMR Berghofer Medical Research Institute-Human Research Ethics Committee approved the study. The study was conducted between 2016 and 2018.
Statistical Analysis
Variance Component Analysis
We analyzed data from 5894 twins from the QIMR sample to estimate the contribution of additive genetic influences (narrow sense heritability), shared/familial environment, and unique environment to the interpersonal differences in population density, remoteness, and SES of the residential area (eAppendix 6 in the Supplement provides more information on twin and family studies). We used the OpenMx24 package in R25 to estimate the parameters of the mixed models. Significance of the variance components was tested using likelihood ratio tests on nested models.26 We used the same approach to replicate the results on population density in the NTR cohort.
In the QIMR data, we fit a gene-environment moderator effect model27 that allows the variance components (heritability, shared environment, and unique environment) to vary across age (eAppendix 7 in the Supplement). This approach reflected previous results from Whitfield et al5 that suggested that heritability of rural/urban living in Australia increases with age. Finally, we estimated the genetic and environmental correlations between population density, remoteness, and SES using bivariate twin models.26 All models included age, age,2 sex, age × sex, age2 × sex, GWAS array, and 4 genetic principal components as covariates.
PRS Analysis
The PRS in the QIMR sample were calculated from the imputed genotype dosage scores using GWAS summary statistics from the GWAS meta-analysis from the 2014 Psychiatric Genomics Consortium Schizophrenia Working Group (36 989 cases and 113 075 controls)11 following the method described by Wray et al.12 We excluded single-nucleotide polymorphisms (SNPs) with low imputation quality (r2<0.6) or minor allele frequency (MAF) below 1%. We selected the most significant independent SNPs using PLINK1.928 to correct for signal redundancy owing to linkage disequilibrium (LD) (criteria LD r2<0.1 within windows of 10 mb). We calculated 8 different PRS using different P value thresholding of the GWAS summary statistics (eTable 1 in the Supplement provides the number of SNPs included for each threshold, and eFigure 1 in the Supplement shows histograms of PRS for schizophrenia). We used mixed models to test the association between neighborhood variables (population density, remoteness, and SES) and PRS and set the significance threshold to 3.15 × 10−3 to account for multiple testing (eAppendix 8 in the Supplement).
We replicated the PRS analysis for the neighborhood variables in the UKB, NTR, and QSKIN samples using the same GWAS summary statistics11 and mixed model approach (eAppendix 8 in the Supplement). eAppendixes 3-5 in the Supplement provide differences in phenotypic and genetic data and PRS calculation between these samples.
Genome-wide Association Analyses
We performed GWAS of population density or SES in our largest sample, the UKB, using BOLT-LMM29 including age, age,2 sex, age × sex, age2 × sex, and 4 genetic principal components 30 on 12 272 635 SNPs with MAF greater than 0.005%. We ran additional GWAS of population density controlling for SES and of SES controlling for population density.
We also conducted GWAS of population density in the QIMR, NTR, and QSKIN cohorts. In the QIMR sample, we used RAREMETALWORKER31 and controlled for age, age,2 sex, age × sex, age2 × sex, SES, GWAS array, and 4 genetic principal components as covariates. We explicitly corrected for relatedness using the kin pedigree option. Single-nucleotide polymorphisms with MAF less than 0.5% or imputation r2<0.6 were excluded, leaving 8 495 074 SNPs for analyses. In the NTR, the GWAS, corrected for age and sex, was performed using GCTA-MLMA32,33 to account for relatedness and population stratification using 2 genetic relationship matrices (one corrects for the related individuals; the second corrects for the population stratification in the distantly related individuals) and 5 genetic principal components. The number of markers included after quality control (r2>0.8 and MAF<0.01%) was 7 636 917. Finally, we performed the GWAS in QSKIN using PLINK, version 1.90b4.128 with age, sex, and 4 genetic principal components as covariates, using a total of 7 672 045 markers after selecting those with r2>0.6 and MAF less than 0.01%.
We used LD score regressions34 to confirm the SNP heritability and the genetic correlations between the measures of population density across samples. eAppendixes 2-5 in the Supplement provide details on genetic data of the 4 cohorts.
Mendelian Randomization
Mendelian randomization methods allow us to generate hypotheses concerning the direction of causation between 2 heritable variables.13 Herein, we relied on the 2014 GWAS meta-analysis summary results from the Psychiatric Genomics Consortium for schizophrenia and the GWAS results calculated from our samples for population density and SES. We used MR-Base35 (TwoSampleMR R package36) and GSMR37 to conduct MR using known schizophrenia SNPs as instruments, thus testing the selection hypothesis that having a higher propensity to schizophrenia (ie, higher PRS) may have a causal association with the tendency to live in a denser and less remote area (eAppendix 9 in the Supplement gives more details on MR and our analysis). We also investigated the reverse hypothesis (population density or SES inducing onset of schizophrenia) using GWAS results from our largest sample, the UKB.
Results
Variance Component Analysis
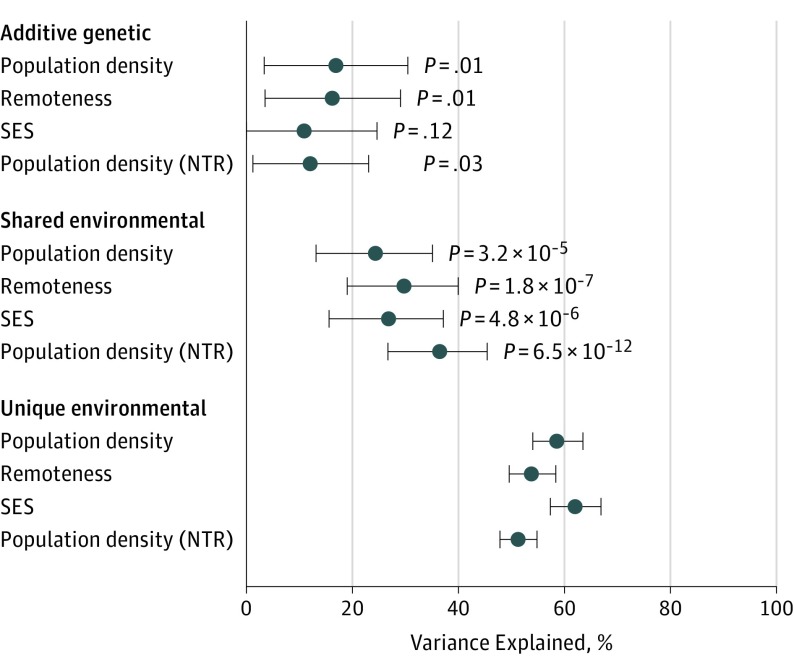
Population density, remoteness, and SES were all significantly correlated at a phenotypic level in all samples considered (eAppendix 10 and eTable 4 in the Supplement). Population density and remoteness were heritable (heritability [h2]) in the QIMR sample (Figure 1) (h2 for population density = 16.9%; 95% CI, 3.4-30.4; P = .01; h2 for remoteness = 16.3%; 95% CI, 3.5-29.0; P = .01); the heritability of SES was not significant (h2 = 11.0%; 95% CI, 0.00-24.6; P = .12). Shared environment (common environment [c2]) effects explained a more substantial and highly significant proportion of the trait variance (Figure 1) (c2 for population density = 24.3%; 95% CI, 13.1-35.1; c2 for remoteness = 29.1%; 95% CI, 19.0-40.0; and c2 for socioeconomic status = 26.8%; 95% CI, 15.6-37.1; all P < .001), which highlights that people tend to live with or close to their parents or other relatives. Population density was also heritable in the NTR (h2 = 12.1%; 95% CI, 1.3-23.2; P = .28) and showed shared environment sources of variance (c2 = 36.5; 95% CI, 26.8-45.7; P = 7 × 10−12).
Figure 1. Percentage of Variance.
Additive genetic, shared environmental, and unique environmental factors in population density, remoteness, and socioeconomic status (SES) in the Queensland Institute of Medical Research cohort (discovery). Results for population density in the Netherlands Twin Register (NTR) cohort are presented for replication. P values are not applicable for unique environmental factors.
In the QIMR cohort, population density was more heritable and less influenced by shared environmental sources as participants became older (eAppendix 11 in the Supplement), with the heritability increasing from 9.0% to 25.6% between ages 20 and 80 years. Over the same lifespan, c2 decreased from 44.5% to less than 10.0% (eAppendix 11 in the Supplement), but variance explained by unique environmental sources (including measurement error) remained constant (eAppendix 11 in the Supplement). Similar results were obtained using standardized estimates, suggesting constant phenotypic variance across age. In addition, population density, remoteness, and SES shared environmental influences as indicated by significant environmental correlations from the twin models (eTable 4 in the Supplement).
Polygenic Risk Scores Analysis
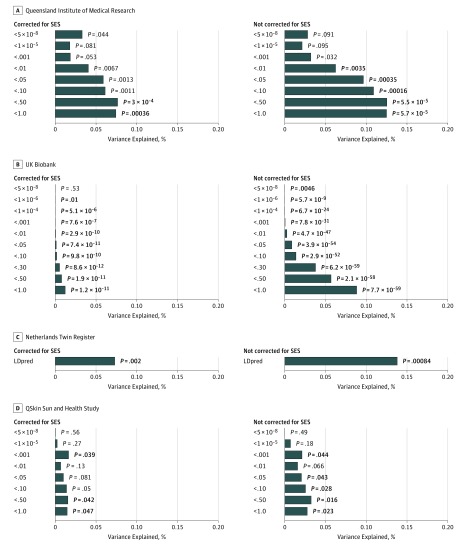
Figure 2 shows the percentage of variance of the population density of the place of residence explained by the PRS for schizophrenia, with and without controlling for SES. In the QIMR sample, PRS calculated from all semi-independent SNPs across the genome (Figure 2) explained the greatest amount of variance in population density (r2 = 0.12%; P = 5.69 × 10−5), and still explained (r2 = 0.074%; P < .001) when accounting for SES. Schizophrenia PRS also were significantly associated with remoteness (r2 = 0.06%; P = .003) when including all of the independent SNPs, although the association disappeared when correcting for SES. eFigure 6 and eAppendix 12 in the Supplement provide information on the association between genetic risk for schizophrenia, remoteness, and SES. We did not find evidence of interactions between sex or age and PRS for schizophrenia (P > .05) contributing to population density or remoteness in the QIMR sample.
Figure 2. Percentage of Variance of the Population Density of the Place of Residence Explained by the Genetic Risk for Schizophrenia, Both Without and With Socioeconomic Status (SES) as a Covariate.
A-D, Genetic risk for schizophrenia showed positive effects for population density in all significant associations. Results in bold highlight significant results after correction for multiple testing. In the discovery Queensland Institute of Medical Research sample, we accounted for the number of tests performed. In the Netherlands Twin Register, UK Biobank, and QSkin Sun and Health Study, we used a significance threshold of .05 as we aimed to replicate the results found with the polygenic risk scores (PRS) calculated over all independent genomic regions. However, when multiple PRS were available, we present all of the results for completeness. Python-based software (LDpred) was used to conduct the analyses in the NTR cohort.
The association between schizophrenia risk score and population density was replicated in the NTR (r2 = 0.14%; P = 8.3 × 10−4 and r2 = 0.073%; P = .002 when correcting for SES), in the UKB (r2 = 0.088%; P = 7.7 × 10−59; r2 = 0.012%; P = 1.2 × 10−11 when accounting for SES) and in QSKIN (r2 = 0.027%; P = .02 and r2 = 0.015%; P = .047 when correcting for SES) (Figure 2). All correlations were in the same direction, pointing to increased PRS for participants living in more densely populated areas. Results for remoteness and SES in QSKIN were consistent with those in QIMR and are presented in eFigure 7 in the Supplement.
In addition, we tested the association between SES and schizophrenia PRS. The association did not reach statistical significance in the QIMR or QSKIN sample when taking into account multiple testing (P > .01) (eFigures 6 and 7 in the Supplement) but was significant in the UKB (r2 = 0.084%; P = 8.6 × 10−64, correcting for population density) (eFigure 8 in the Supplement) likely because of the gain of power owing to its very large sample size.
Genome-wide Association Analyses
Six genomic regions reached genome-wide significance for population density in the UKB, and this number increased to 12 when correcting for SES. Similarly, we identified 13 loci associated with SES in the UKB when correcting for population density. We observed fewer significant associations with population density or remoteness in the smaller samples, but they did not correspond to the SNP associations found in the UKB. The SNP heritability ranged from 0.6% (QIMR: SE, 3.2%) to 9.3% (QSKIN: SE, 4.0%) as estimated by LD score regression (eTable 5 in the Supplement). The genetic correlation between population density across samples ranged from 0.30 (SE, 0.44; P = .49, QSKIN-NTR) to 0.61 (SE, 0.29; P = .04, UKB-NTR). eAppendix 13, eFigures 9-16, and eTable 5 in the Supplement provide all GWAS and LD score regression results (including GWAS meta-analysis).
Mendelian Randomization
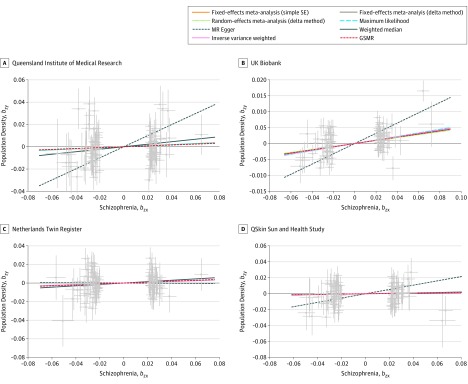
Finally, we selected between 88 and 94 genome-wide significant SNPs for schizophrenia as instruments to perform MR analyses with population density as the outcome variable after excluding SNPs showing evidence of pleiotropic effects by the heterogeneity in dependent instruments outlier analysis (implemented in the GSMR software).37 These numbers are consistent with the 108 independent associations reported by the Psychiatric Genomics Consortium11; the difference arose from SNPs not being present or not passing quality control in GWAS of population density and SES.
Estimates from MR analyses performed in the UKB sample were significant (b = 0.049; P = 3.7 × 10−7 using GSMR) (Table 2, Figure 3), suggesting that the genetic liability to schizophrenia has a causal association with the tendency to live in urbanized locations. We observed similar effect sizes in all other samples, although the MR results were not significant. We found no evidence of confounding heterogeneity of effect sizes (P > .30) or from pleiotropy (P > .05) using the tests implemented in MR-Base. Reverse MR testing (propensity to live in a more dense or low socioeconomic status area as a cause of schizophrenia) was only suggestive of a (larger) association with population density or SES on schizophrenia (b = 0.20; P = .01 using GSMR) as it did not survive multiple testing correction. This lack of evidence could have been the result of reduced statistical power, as both genetic instruments had a smaller number of SNPs (n = 12) (Table 2; eAppendix 14 and eTables 6-10 in the Supplement provide detailed MR results).
Table 2. Summary of the MR-Based Analysis of Schizophrenia and Population Density Across the 4 Cohorts.
| Method | QIMR | UKB | NTR | QSKIN | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNPs | b (SE) | P Value | No. of SNPs | b (SE) | P Value | No. of SNPs | b (SE) | P Value | No. of SNPs | b (SE) | P Value | |
| Fixed-effects meta-analysis (simple SE) | 93 | 0.045 (0.050) |
.36 | 92a | 0.054 (0.009)a |
1.6 × 10−8a | 88 | 0.043 (0.050) |
.37 | 94 | 0.021 (0.049) |
.67 |
| Fixed-effects meta-analysis (delta method) | 93 | 0.047 (0.051) |
.35 | 92a | 0.049 (0.01)a |
3.4 × 10−7a | 88 | 0.042 (0.050) |
.40 | 94 | 0.019 (0.050) |
.69 |
| Random-effects meta-analysis (delta method) | 93 | 0.043 (0.054) |
.43 | 92a | 0.046 (0.013)a |
5.8 × 10−4a | 88 | 0.042 (0.050) |
.40 | 94 | 0.019 (0.050) |
.69 |
| Maximum likelihood | 93 | 0.045 (0.051) |
.37 | 92a | 0.056 (0.01)a |
9.6 × 10−9a | 88 | 0.044 (0.050) |
.37 | 94 | 0.021 (0.050) |
.66 |
| MR Egger | 93 | 0.49 (0.26) |
.06 | 92 | 0.16 (0.06) |
.013 | 88 | −0.006 (0.240) |
.97 | 94 | 0.27 (0.231) |
.24 |
| Weighted median | 93 | 0.11 (0.075) |
.13 | 92a | 0.053 (0.015)a |
7.7 × 10−4a | 88 | 0.074 (0.074) |
.31 | 94 | 0.026 (0.072) |
.71 |
| Inverse variance weighted | 93 | 0.045 (0.056) |
.41 | 92a | 0.053 (0.013)a |
1.0 × 10−4a | 88 | 0.043 (0.050) |
.37 | 94 | 0.021 (0.049) |
.67 |
| GSMR | 93 | 0.039 (0.051) |
.41 | 92a | 0.049 (0.01)a |
3.7 × 10−7 | 88 | 0.049 (0.051) |
.33 | 94 | 0.018 (0.050) |
.71 |
Abbreviations: GSMR, generalized summary data-based mendelian randomization; MR, mendelian randomization; NTR, Netherlands Twin Register; QIMR, QIMR Berghofer Medical Research Institute; QSKIN, QSkin Sun and Health Study; SNPs, single-nucleotide polymorphisms; UKB, United Kingdom Biobank.
Statistically significant results.
Figure 3. Mendelian Randomization Summary Results.
Hypothesis that schizophrenia influences the population density of the place or residence tested in the Queensland Institute of Medical Research (A), UK Biobank (B), Netherlands Twin Register (C), and QSkin Sun and Health Study (D) cohorts. Several methods yield exactly the same effect sizes (Table 2), and as a consequence, some lines may overlap. GSMR indicates generalized summary data-based mendelian randomization.
Discussion
The present study investigated the association between genetic risk for schizophrenia and characteristics of a person’s place of residence (population density, remoteness, and SES) to test the genetic nature of the association between schizophrenia and population density and infer the direction of causation. We used data on where people live collected as part of 4 studies from 3 countries (Australia, United Kingdom, and the Netherlands) for a total of 504 130 participants.
In all 4 nonclinical cohorts, genetic risk for schizophrenia was associated with greater population density of the postcode of residence beyond what could be explained by SES of the area (Figure 2). Results were consistent for remoteness. Our results show that the geographic distribution of the genetic risk for schizophrenia is not uniform and that participants with higher genetic risk levels live in areas with higher population density over what is expected by chance. We also found a significant association between genetic risk for schizophrenia and SES of the place of residence across the United Kingdom, although this association was not replicated in the 2 Australian samples.
Our MR results in the UKB suggest that schizophrenia risk could be a causal factor in the choice to live in more densely populated and low socioeconomic status areas, although more powered analyses would be required to confirm this result in the other cohorts (Figure 3). These results are consistent with the selective migration hypothesis that individuals with genetic liability for schizophrenia tend to move to or remain in urban areas.38 Larger GWAS for the environmental variables are required to confirm a reverse causation (eTable 9 and eTable 10 in the Supplement) and clarify how psychopathologic traits and residential location relate to each other. More data are needed to clarify the influence of comorbid psychiatric risks39,40,41 and associated traits (eg, educational attainment, creativity, or risk taking)11,40,41 on the reported association.
Our work builds on previous research that reported that the density of population of where one lives is significantly heritable5 and on nonmolecular studies showing evidence of a familial effect (ie, owing to genetics and/or family environment) in the association between schizophrenia and urban dwelling.4,42,43
Our results complement 2 recent publications on the interplay between schizophrenia risk and neighborhood.10,38 The first, from the Swedish registries (N = 759 536), reported an association between schizophrenia PRS and low socioeconomic status neighborhood,38 which we replicated in the UKB. In addition, we found that SES could not completely explain the association between schizophrenia PRS and population density.
We found a large environmental correlation between SES and population density in Australia (bivariate model including additive genetic, common environmental, and unique environmental factors, rC = 0.86, P < .0001; rE = 0.33, P < .0001) compared with the estimated genetic correlation (rG = 0.35, P = .34.) (eTable 4 in the Supplement). Thus, SES is a potential confounder of genetic analyses of population density, and conversely, population density is a confounder of SES genetic analyses. Composition of SES measures23 studied here differed in each country and also differ from the Swedish study.38
A study from the Danish registries found an association between schizophrenia PRS and urban living only for individuals aged 15 years, but not at birth,10 and did not study the neighborhood during adult age. Herein, we showed that the population density of where a person lives is mostly explained by shared and unique environment, with the heritability increasing with age (eAppendix 11 in the Supplement). Thus, we are uncertain whether shared environment confounded the results observed in the Danish population. However, we cannot rule out a genetic association between upbringing environment and the disease risk (ie, a passive gene-environment correlation) in which the association is driven by the genotype that a child inherits and the environment in which they are raised. Herein, we rather focused on an active gene-environment correlation, presumably driven by selective migration, by including only older participants who have a higher degree of independence in choosing where they live. More work is needed to confirm and examine these results over age groups, which will likely require large, longitudinal cohorts, such as national registries.
We highlighted the importance of age in our analysis by replicating and expanding previous results5: that place of residence is heritable and the heritability increases over time, while the influence of family environment declines (eAppendix 11 in the Supplement). This age effect, together with sex differences in prevalence and age of onset of schizophrenia,44,45 justified the study of interactions between PRS and age and sex that may contribute to the choice of neighborhood; however, these interactions were not significant in our analyses.
Limitations
A limitation of our study arises from the low power of the GWAS to detect all variants associated with schizophrenia.46 This lack of power results in a limited PRS instrument that explains only 11.6% of the total trait heritability in the population.11 Thus, the variance explained by the current PRS, which is based on common variants, may be only one-tenth of what one would observe with a PRS capturing the whole genetic signal. As a consequence, the small association with population density reported herein may account for 1.7% (95% CI, 0.8%-3.2%) of the schizophrenia risk (based on population density explaining 0.2% of schizophrenia PRS in the QIMR data and 0.002 of 0.116 = 0.017). Ancestry may also confound PRS analyses,47 especially for the variables studied herein (eAppendix 15 and eFigures 17-19 in the Supplement48). We tried to overcome this issue using mixed models that are equivalent to fitting all genetic principal components as covariates (eAppendixes 8 and 15 in the Supplement).
Another limitation is the possible sample overlap between the UKB sample and data used in the schizophrenia GWAS, which may inflate results from PRS and MR analyses (eAppendix 16 in the Supplement). However, we estimated the overlap to be negligible (64 participants or 0.01%, eAppendix 16 in the Supplement), and we did not observe larger effect sizes in the UKB compared with the 3 other cohorts.
Conclusions
Our findings support the notion that the increased schizophrenia prevalence in urbanized areas is not only owing to the environmental stressors of the city or other putative risk factors associated with urbanicity (eg, increased risk of infection, low vitamin D levels, and substance abuse)49 but also on the genetic risk for the disease. The associated PRS prediction was replicated across 3 different countries that likely differ in availability of space, social mobility/opportunities, associations between population density and SES of the area, and historical constraints on living environment. We showed that the distribution of the genetic risk for the disorder is not uniform and concentrates in more populated and urban areas, supporting the idea of an active gene-environment correlation because of selective migration. Previous evidence of an environmental association between city living and schizophrenia risk1,2 is compatible with our results and reflects that there are genetic as well as environmental risk factors for schizophrenia. Furthermore, we provide evidence that schizophrenia genetic risk may lead to individuals (or had led to their ancestors) seeking denser/urban and low socioeconomic status neighborhoods, which could in turn be risk factors for the disease.1,2,49
Future disease models will need to include both genetic selection and environmental factors of urban stress on schizophrenia to inform implications for intervention. In addition, there is a need to address the potential gene-by-environment interactions that would arise if genetic variants influencing schizophrenia also influence the choice of a stressful neighborhood, which would contribute to the interaction between urbanicity and family history of schizophrenia that has been reported in the Danish population.50 Such diathesis-stress interaction studies using PRS have been published for depression,51,52 but given the lower prevalence of schizophrenia, national registries will likely be required for investigation.
eAppendix 1. Short Review on the Gap in Schizophrenia Rates Between Urban and Rural Environments
eAppendix 2. Genetic and Phenotypic Data From the QIMR Sample
eTable 1. Number of SNPs Included in the Calculation of Each of the QIMR Polygenic Risk Scores (QIMR)
eFigure 1. Histograms of the Main Variables Used in the Analysis (QIMR)
eAppendix 3. Genetic and Phenotypic Data From the UK Biobank
eTable 2. UKB Sample Breakdown by Ancestry
eFigure 2. Histograms of the Main Variables Used in the Analysis (UKB)
eFigure 3. Scatter Plot of the Genetic Principal Component Projection That Was Used to Determine the Ancestry of Participants (UKB)
eAppendix 4. Genetic and Phenotypic Data From the NTR Sample
eFigure 4. Histograms of the Main Variables Used in the Analysis (NTR)
eAppendix 5. Genetic and Phenotypic Data From the QSKIN Sample
eTable 3. Number of SNPs Included in the Calculation of Each of the QSKIN Polygenic Risk Scores
eFigure 5. Histograms of the Variables Used in the Analysis (QSKIN)
eAppendix 6. Summary on Twin and Family Studies
eAppendix 7. GE Moderator Effect Model
eAppendix 8. PRS and Prediction Model Used
eAppendix 9. Summary on Mendelian Randomization (MR)
eAppendix 10. Phenotypic, Genetic and Environmental Correlations (95% CI and p-values) Between the Demographic Variables
eTable 4. Correlations (and 95% Confidence Intervals) in the QIMR Sample
eAppendix 11. Effect of Age on the Genetic (A) and Environmental (Common, C and Unique, E) Sources of Variances for Population Density
eAppendix 12. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia
eFigure 6. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia (QIMR)
eFigure 7. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia (QSKIN)
eFigure 8. Variance SES Explained by the Genetic Risk for Schizophrenia (UKB)
eAppendix 13. GWAS Results
eTable 5. SNP heritability (SNPh2) and standard error (SE) from the univariate analyses (left column) and genetic correlations (rg) and standard error (SE) from the bivariate analyses (right columns) performed in LD score for the GWAS results used in the present study.
eFigure 9. Manhattan Plot of Population Density of Place of Residence (UKB
eFigure 10. Manhattan Plot for Population Density of Residence, Correcting for SES (UKB)
eFigure 11. Manhattan Plot of SES of Residence (UKB)
eFigure 12. Manhattan Plot of SES, Correcting for Population Density (UKB)
eFigure 13. Manhattan Plot of Population Density of Residence in the (QIMR)
eFigure 14. Manhattan Plot of Population Density of Residence (NTR)
eFigure 15. Manhattan Plot of Population Density of Residence (QSKIN)
eFigure 16. Manhattan Plot of Population Density of Residence in All Cohorts (Meta-analysis of UKB+QIMR+NTR+QSKIN)
eAppendix 14. Detailed MR Results
eTable 6. MR Results Testing the Hypothesis That Schizophrenia Is Causal of Population Density of Residence (Correcting for SES in the GWAS of Population Density) in the QIMR (left panel) and UKB (right panel) Cohorts
eTable 7. MR Hypothesis That Schizophrenia Causes to Live in Deprived Neighbourhood (measured by SES of the area) in UKB
eTable 8. MR Hypothesis That Schizophrenia Causes to Live in Deprived Neighbourhood (measured by SES of the area) after adjusting for population density in UKB
eTable 9. Reverse MR Hypothesis That Population Density Corrected for SES Is Causal of Schizophrenia in UKB
eTable 10. Reverse MR Hypothesis That Deprived Neighbourhood (measured as SES) Adjusted for Population Density Is Causal for Schizophrenia in UKB
eAppendix 15. Sensitivity Analysis in the UKB
eFigure 17. Relationship Between PRS “p<1” for Schizophrenia and the First 20 Genetic PCs
eFigure 18. Relationship Between Population Density and the First 20 Genetic PCs
eFigure 19. Relationship Between PRS “p<0.05” for Schizophrenia and the First 20 Genetic PCs
eAppendix 16. Sample Overlap Between Schizophrenia GWAS and UKB
eReferences
References
- 1.Lederbogen F, Kirsch P, Haddad L, et al. City living and urban upbringing affect neural social stress processing in humans. Nature. 2011;474(7352):498-501. doi: 10.1038/nature10190 [DOI] [PubMed] [Google Scholar]
- 2.Lederbogen F, Haddad L, Meyer-Lindenberg A. Urban social stress—risk factor for mental disorders: the case of schizophrenia. Environ Pollut. 2013;183:2-6. doi: 10.1016/j.envpol.2013.05.046 [DOI] [PubMed] [Google Scholar]
- 3.Krabbendam L, van Os J. Schizophrenia and urbanicity: a major environmental influence—conditional on genetic risk. Schizophr Bull. 2005;31(4):795-799. doi: 10.1093/schbul/sbi060 [DOI] [PubMed] [Google Scholar]
- 4.Mortensen PB, Pedersen CB, Westergaard T, et al. Effects of family history and place and season of birth on the risk of schizophrenia. N Engl J Med. 1999;340(8):603-608. doi: 10.1056/NEJM199902253400803 [DOI] [PubMed] [Google Scholar]
- 5.Whitfield JB, Zhu G, Heath AC, Martin NG. Choice of residential location: chance, family influences, or genes? Twin Res Hum Genet. 2005;8(1):22-26. doi: 10.1375/twin.8.1.22 [DOI] [PubMed] [Google Scholar]
- 6.Pedersen CB, Mortensen PB. Evidence of a dose-response relationship between urbanicity during upbringing and schizophrenia risk. Arch Gen Psychiatry. 2001;58(11):1039-1046. doi: 10.1001/archpsyc.58.11.1039 [DOI] [PubMed] [Google Scholar]
- 7.Sundquist K, Frank G, Sundquist J. Urbanisation and incidence of psychosis and depression: follow-up study of 4.4 million women and men in Sweden. Br J Psychiatry. 2004;184:293-298. doi: 10.1192/bjp.184.4.293 [DOI] [PubMed] [Google Scholar]
- 8.Pedersen CB, Mortensen PB. Family history, place and season of birth as risk factors for schizophrenia in Denmark: a replication and reanalysis. Br J Psychiatry. 2001;179:46-52. doi: 10.1192/bjp.179.1.46 [DOI] [PubMed] [Google Scholar]
- 9.Marcelis M, Takei N, van Os J. Urbanization and risk for schizophrenia: does the effect operate before or around the time of illness onset? Psychol Med. 1999;29(5):1197-1203. doi: 10.1017/S0033291799008983 [DOI] [PubMed] [Google Scholar]
- 10.Paksarian D, Trabjerg BB, Merikangas KR, et al. The role of genetic liability in the association of urbanicity at birth and during upbringing with schizophrenia in Denmark. [published online June 29, 2017]. Psychol Med. 2018;48(2):305-314. doi: 10.1017/S0033291717001696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripke S, Neale BM, Corvin A, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421-427. doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55(10):1068-1087. doi: 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- 13.Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015;16:327-350. doi: 10.1146/annurev-genom-090314-050016 [DOI] [PubMed] [Google Scholar]
- 14.Katikireddi SV, Cezard G, Bhopal RS, et al. Assessment of health care, hospital admissions, and mortality by ethnicity: population-based cohort study of health-system performance in Scotland. Lancet Public Health. 2018;3(5):e226-e236. doi: 10.1016/S2468-2667(18)30068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath AC, Bucholz KK, Madden PA, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27(6):1381-1396. doi: 10.1017/S0033291797005643 [DOI] [PubMed] [Google Scholar]
- 16.Knopik VS, Heath AC, Madden PA, et al. Genetic effects on alcohol dependence risk: re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34(8):1519-1530. doi: 10.1017/S0033291704002922 [DOI] [PubMed] [Google Scholar]
- 17.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on ~500,000 UK Biobank participants [published online July 20, 2017]. bioRxiv. doi: 10.1101/166298 [DOI] [Google Scholar]
- 19.Willemsen G, Vink JM, Abdellaoui A, et al. The Adult Netherlands Twin Register: twenty-five years of survey and biological data collection. Twin Res Hum Genet. 2013;16(1):271-281. doi: 10.1017/thg.2012.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen CM, Green AC, Neale RE, et al. ; QSkin Study . Cohort profile: the QSkin Sun and Health Study. Int J Epidemiol. 2012;41(4):929-929i. doi: 10.1093/ije/dys107 [DOI] [PubMed] [Google Scholar]
- 21.Abdellaoui A, Hottenga JJ, de Knijff P, et al. Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet. 2013;21(11):1277-1285. doi: 10.1038/ejhg.2013.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitcher B, Schmid V, Thornton A. Working with the DICOM and NIfTI data standards in R. J Stat Softw. 2011;44(6):1-28. doi: 10.18637/jss.v044.i06 [DOI] [Google Scholar]
- 23.Australian Bureau of Statistics. Measures of Socioeconomic Status: ABS catalogue no. 1244.0.55.001. Commonwealth of Australia. http://www.ausstats.abs.gov.au/Ausstats/subscriber.nsf/0/367D3800605DB064CA2578B60013445C/$File/1244055001_2011.pdf. Published 2011. Accessed May 24, 2018.
- 24.Boker S, Neale M, Maes H, et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika. 2011;76(2):306-317. doi: 10.1007/s11336-010-9200-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing; 2012.
- 26.Neale MC, Cardon LR. Methodology for Genetic Studies of Twins and Families. Kluwer Academic Publishers; 1992. doi: 10.1007/978-94-015-8018-2 [DOI] [Google Scholar]
- 27.Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Res. 2002;5(6):554-571. doi: 10.1375/136905202762342026 [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh PR, Tucker G, Bulik-Sullivan BK, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284-290. doi: 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed model association for biobank-scale data sets [published online September 27, 2017]. bioRxiv. doi: 10.1101/194944 [DOI] [Google Scholar]
- 31.Feng S, Liu D, Zhan X, Wing MK, Abecasis GR. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30(19):2828-2829. doi: 10.1093/bioinformatics/btu367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76-82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46(2):100-106. doi: 10.1038/ng.2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemani G, Zheng J, Wade KH, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations [published online December 16, 2016]. bioRxiv. doi: 10.1101/078972 [DOI] [Google Scholar]
- 36.Hemani G, Haycock P, Zheng J TwoSampleMR: Two Sample MR functions and interface to MR Base database. https://mrcieu.github.io/TwoSampleMR/. Published 2017. Accessed May 24, 2018.
- 37.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9(1):224. doi: 10.1038/s41467-017-02317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sariaslan A, Fazel S, D’Onofrio BM, et al. Schizophrenia and subsequent neighborhood deprivation: revisiting the social drift hypothesis using population, twin and molecular genetic data. Transl Psychiatry. 2016;6:e796. doi: 10.1038/tp.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 . An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236-1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain [published online September 6, 2017]. bioRxiv. doi: 10.1101/048991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Power RA, Steinberg S, Bjornsdottir G, et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nat Neurosci. 2015;18(7):953-955. doi: 10.1038/nn.4040 [DOI] [PubMed] [Google Scholar]
- 42.Cantor-Graae E, Selten JP. Schizophrenia and migration: a meta-analysis and review. Am J Psychiatry. 2005;162(1):12-24. doi: 10.1176/appi.ajp.162.1.12 [DOI] [PubMed] [Google Scholar]
- 43.Pedersen CB, Mortensen PB. Are the cause(s) responsible for urban-rural differences in schizophrenia risk rooted in families or in individuals? Am J Epidemiol. 2006;163(11):971-978. doi: 10.1093/aje/kwj169 [DOI] [PubMed] [Google Scholar]
- 44.Abel KM, Drake R, Goldstein JM. Sex differences in schizophrenia. Int Rev Psychiatry. 2010;22(5):417-428. doi: 10.3109/09540261.2010.515205 [DOI] [PubMed] [Google Scholar]
- 45.McGrath J, Saha S, Welham J, El Saadi O, MacCauley C, Chant D. A systematic review of the incidence of schizophrenia: the distribution of rates and the influence of sex, urbanicity, migrant status and methodology. BMC Med. 2004;2:13. doi: 10.1186/1741-7015-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. doi: 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Curtis D. Polygenic risk score for schizophrenia is more strongly associated with ancestry than with schizophrenia [published online March 23, 2018]. bioRxiv. doi: 10.1101/287136 [DOI] [PubMed] [Google Scholar]
- 48.Haworth S, Mitchell R, Corbin L, et al. Common genetic variants and health outcomes appear geographically structured in the UK Biobank sample: old concerns returning and their implications [published online April 11, 2018]. bioRxiv. doi: 10.1101/294876 [DOI] [Google Scholar]
- 49.Plana-Ripoll O, Bocker Pedersen C, McGrath JJ. Urbanicity and risk of schizophrenia—new studies and old hypotheses [published online May 16, 2018]. JAMA Psychiatry. doi:10.1001/jamapsychiatry.2018.0551 [DOI] [PubMed] [Google Scholar]
- 50.van Os J, Pedersen CB, Mortensen PB. Confirmation of synergy between urbanicity and familial liability in the causation of psychosis. Am J Psychiatry. 2004;161(12):2312-2314. doi: 10.1176/appi.ajp.161.12.2312 [DOI] [PubMed] [Google Scholar]
- 51.Mullins N, Power RA, Fisher HL, et al. Polygenic interactions with environmental adversity in the aetiology of major depressive disorder. Psychol Med. 2016;46(4):759-770. doi: 10.1017/S0033291715002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colodro-Conde L, Couvy-Duchesne B, Zhu G, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . A direct test of the diathesis-stress model for depression. [published online July 11, 2017]. Mol Psychiatry. 2017. doi: 10.1038/mp.2017.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Short Review on the Gap in Schizophrenia Rates Between Urban and Rural Environments
eAppendix 2. Genetic and Phenotypic Data From the QIMR Sample
eTable 1. Number of SNPs Included in the Calculation of Each of the QIMR Polygenic Risk Scores (QIMR)
eFigure 1. Histograms of the Main Variables Used in the Analysis (QIMR)
eAppendix 3. Genetic and Phenotypic Data From the UK Biobank
eTable 2. UKB Sample Breakdown by Ancestry
eFigure 2. Histograms of the Main Variables Used in the Analysis (UKB)
eFigure 3. Scatter Plot of the Genetic Principal Component Projection That Was Used to Determine the Ancestry of Participants (UKB)
eAppendix 4. Genetic and Phenotypic Data From the NTR Sample
eFigure 4. Histograms of the Main Variables Used in the Analysis (NTR)
eAppendix 5. Genetic and Phenotypic Data From the QSKIN Sample
eTable 3. Number of SNPs Included in the Calculation of Each of the QSKIN Polygenic Risk Scores
eFigure 5. Histograms of the Variables Used in the Analysis (QSKIN)
eAppendix 6. Summary on Twin and Family Studies
eAppendix 7. GE Moderator Effect Model
eAppendix 8. PRS and Prediction Model Used
eAppendix 9. Summary on Mendelian Randomization (MR)
eAppendix 10. Phenotypic, Genetic and Environmental Correlations (95% CI and p-values) Between the Demographic Variables
eTable 4. Correlations (and 95% Confidence Intervals) in the QIMR Sample
eAppendix 11. Effect of Age on the Genetic (A) and Environmental (Common, C and Unique, E) Sources of Variances for Population Density
eAppendix 12. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia
eFigure 6. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia (QIMR)
eFigure 7. Variance of Remoteness and SES Explained by the Genetic Risk for Schizophrenia (QSKIN)
eFigure 8. Variance SES Explained by the Genetic Risk for Schizophrenia (UKB)
eAppendix 13. GWAS Results
eTable 5. SNP heritability (SNPh2) and standard error (SE) from the univariate analyses (left column) and genetic correlations (rg) and standard error (SE) from the bivariate analyses (right columns) performed in LD score for the GWAS results used in the present study.
eFigure 9. Manhattan Plot of Population Density of Place of Residence (UKB
eFigure 10. Manhattan Plot for Population Density of Residence, Correcting for SES (UKB)
eFigure 11. Manhattan Plot of SES of Residence (UKB)
eFigure 12. Manhattan Plot of SES, Correcting for Population Density (UKB)
eFigure 13. Manhattan Plot of Population Density of Residence in the (QIMR)
eFigure 14. Manhattan Plot of Population Density of Residence (NTR)
eFigure 15. Manhattan Plot of Population Density of Residence (QSKIN)
eFigure 16. Manhattan Plot of Population Density of Residence in All Cohorts (Meta-analysis of UKB+QIMR+NTR+QSKIN)
eAppendix 14. Detailed MR Results
eTable 6. MR Results Testing the Hypothesis That Schizophrenia Is Causal of Population Density of Residence (Correcting for SES in the GWAS of Population Density) in the QIMR (left panel) and UKB (right panel) Cohorts
eTable 7. MR Hypothesis That Schizophrenia Causes to Live in Deprived Neighbourhood (measured by SES of the area) in UKB
eTable 8. MR Hypothesis That Schizophrenia Causes to Live in Deprived Neighbourhood (measured by SES of the area) after adjusting for population density in UKB
eTable 9. Reverse MR Hypothesis That Population Density Corrected for SES Is Causal of Schizophrenia in UKB
eTable 10. Reverse MR Hypothesis That Deprived Neighbourhood (measured as SES) Adjusted for Population Density Is Causal for Schizophrenia in UKB
eAppendix 15. Sensitivity Analysis in the UKB
eFigure 17. Relationship Between PRS “p<1” for Schizophrenia and the First 20 Genetic PCs
eFigure 18. Relationship Between Population Density and the First 20 Genetic PCs
eFigure 19. Relationship Between PRS “p<0.05” for Schizophrenia and the First 20 Genetic PCs
eAppendix 16. Sample Overlap Between Schizophrenia GWAS and UKB
eReferences