This secondary analysis of tumor specimens collected for the LUX-Lung 8 randomized clinical trial assesses whether ERBB gene mutations of family members were associated with outcomes among patients with lung squamous cell carcinoma randomized to treatment with afatinib or erlotinib.
Key Points
Question
What is the association between ERBB family mutations and outcomes of patients with lung squamous cell carcinoma who received treatment with afatinib dimaleate or erlotinib hydrochloride?
Findings
In this secondary analysis of the LUX-Lung 8 trial, of 245 clinically selected patients, 21.6% had tumors with at least 1 ERBB mutation. Although progression-free survival and overall survival were improved with afatinib vs erlotinib treatment among patients with ERBB wild-type tumors, this was more pronounced among patients with tumors having at least 1 ERBB family mutation, with the largest benefits observed among those with HER2 mutations.
Meaning
Mutations of ERBB, particularly HER2 mutations, may be used as a biomarker to identify patients with lung squamous cell carcinoma who would derive additional benefit from afatinib.
Abstract
Importance
Treatment choice for lung squamous cell carcinoma could be aided by identifying predictive biomarkers.
Objective
To assess whether patient outcomes in the LUX-Lung 8 trial were associated with ERBB gene family member aberrations in tumor specimens.
Design, Setting, and Participants
Ad hoc secondary analysis of the LUX-Lung 8 trial conducted at 183 centers in 23 countries from March 30, 2012, to January 30, 2014. Eligible patients had stage IIIB or IV lung squamous cell carcinoma with progressive disease after 4 or more cycles of platinum-based chemotherapy. Tumor genetic analysis (TGA) was performed using next-generation sequencing in a cohort enriched for patients with progression-free survival (PFS) of more than 2 months. Epidermal growth factor receptor (EGFR) expression levels were assessed by immunohistochemistry in a separate cohort of patients from the LUX-Lung 8 population. Associations of PFS and overall survival (OS) with ERBB gene alterations and EGFR expression levels were assessed. This analysis was conducted from February 26, 2015, to June 12, 2017.
Interventions
Patients were randomized 1:1 to treatment with afatinib dimaleate (40 mg/d; n = 398) or erlotinib hydrochloride (150 mg/d; n = 397).
Main Outcomes and Measures
Overall survival, PFS, pooled and individual ERBB gene mutations, ERBB copy number alterations, and EGFR expression.
Results
Tumor specimens from 245 patients were eligible for next-generation sequencing (TGA subset: 132 patients treated with afatinib; 113 patients treated with erlotinib). In this population, outcomes were improved with afatinib vs erlotinib treatment (PFS: median, 3.5 vs 2.5 months; hazard ratio [HR], 0.69; 95% CI, 0.51-0.92; P = .01; OS: median, 8.4 vs 6.6 months; HR, 0.81; 95% CI, 0.62-1.05; P = .12). Of 245 patients in the TGA subset, 53 (21.6%) had tumors with 1 or more ERBB mutations. Among afatinib-treated patients, PFS (median, 4.9 vs 3.0 months; HR, 0.62; 95% CI, 0.37-1.02; P = .06) and OS (median, 10.6 vs 8.1 months; HR, 0.75; 95% CI, 0.47-1.17; P = .21) were longer among those with ERBB mutation–positive disease than among those without. The presence of HER2 mutations was associated with favorable PFS and OS following afatinib vs erlotinib treatment. There was no apparent association between copy number alteration or EGFR expression level and outcome.
Conclusions and Relevance
Next-generation sequencing may help identify patients with lung squamous cell carcinoma who would derive additional benefit from treatment with afatinib. The role of ERBB mutations, particularly HER2 mutations, as predictive biomarkers for afatinib treatment in this setting warrants further evaluation.
Trial Registration
ClinicalTrials.gov Identifier: NCT01523587
Introduction
Squamous cell carcinoma (SqCC) of the lung is one of the most genetically complex and difficult-to-treat cancers.1 Until recently, platinum doublet chemotherapy was the first-line treatment of choice for most patients with lung SqCC, and second-line treatment options were also limited, with erlotinib hydrochloride (an epidermal growth factor receptor [EGFR] tyrosine kinase inhibitor) and docetaxel as the only approved options.2 Recently, several new agents have been approved for patients with lung SqCC, including the EGFR monoclonal antibody necitumumab (combined with standard first-line chemotherapy)3 and the immune checkpoint inhibitor pembrolizumab (in tumors with high expression levels of programmed cell death ligand 1 [PD-L1])4 as first-line treatments. The immune checkpoint inhibitors nivolumab,5 pembrolizumab,6 and atezolizumab7; the anti–vascular endothelial growth factor receptor 2 antibody ramucirumab (combined with docetaxel)8; and the ERBB family blocker afatinib have been approved as second-line treatments.9 This expansion of the armamentarium invites questions regarding optimal treatment of lung SqCC, which could be addressed by identifying biomarkers that may predict outcomes.
Afatinib dimaleate was approved as a second-line treatment of lung SqCC based on the phase 3 LUX-Lung 8 randomized clinical trial, which compared treatment with afatinib vs erlotinib following progression during and after chemotherapy.9 Compared with erlotinib therapy, treatment with afatinib significantly improved overall survival (OS; median, 7.9 vs 6.8 months; hazard ratio [HR]; 0.81: 95% CI, 0.69-0.95; P = .008) and progression-free survival (PFS; median, 2.6 vs 1.9 months; HR, 0.81; 95% CI, 0.69-0.96; P = .01). The tolerability profile was similar in both treatment arms and was consistent with prior experience.9
To investigate whether the efficacy of afatinib in lung SqCC varied depending on the molecular characteristics of tumors, we conducted a comprehensive genetic analysis using Foundation Medicine FoundationOne next-generation sequencing (NGS) in a cohort of 245 patients who were included in the LUX-Lung 8 trial. The aims were to (1) assess the genetic characteristics of SqCC tumors in this cohort; (2) identify individual genes that are commonly mutated in this cohort and assess outcomes with respect to these genes; and (3) characterize and calculate the cumulative mutation frequency in the ERBB family of genes, assessing outcomes among patients with tumors with or without such mutations. Previous studies have reported mutation frequencies in EGFR, HER2, HER3, and HER4 of approximately 1% to 3%, 4%, 1% to 2%, and 8%, respectively.10,11,12,13 Given that afatinib irreversibly inhibits signaling from all homodimers and heterodimers of the ERBB family and that these receptors cooperate via interconnected intracellular pathways to regulate cellular proliferation,14 it is possible that genetic aberrations within the family might identify a subgroup of patients who could particularly benefit from afatinib treatment. Therefore, analysis of ERBB/HER family aberrations was prespecified in the present analysis. Furthermore, given that up to 80% of lung SqCC tumors overexpress EGFR15 and that EGFR, HER2, and HER3 amplifications have been reported,10,13 we assessed outcomes based on EGFR expression levels and ERBB family copy number alterations (CNAs).
Methods
Study, Design, and Participants
The design of the LUX-Lung 8 trial has been previously described.9 In brief, patients had stage IIIB or IV non–small cell lung cancer (NSCLC) of squamous (including mixed) histology and had progressed during or after 4 or more cycles of platinum-based doublet chemotherapy as a first-line treatment. The included 795 patients were randomized (1:1) to treatment with afatinib (40 mg/d; 398 patients) or erlotinib (150 mg/d; 397 patients), and randomization was stratified by race/ethnicity (East Asian vs non–East Asian origin) to eliminate any potential bias in EGFR mutation frequency. The primary end point was PFS (independent review). The key secondary end point was OS. Tumor specimens and blood samples for exploratory biomarker analysis were required at study entry. The study was conducted in accordance with the Declaration of Helsinki,16 Good Clinical Practice guidelines, and applicable region-specific regulatory requirements and was approved by independent ethics committees at each center. All patients provided written informed consent. The present study is an ad hoc, retrospective secondary analysis of the LUX-Lung 8 trial. Patients included in LUX-Lung 8 were required to have archival tissue available for investigational assessment of tumor biomarkers. Therefore, collection of tumor samples was preplanned.
Tumor Genetic Analysis
Formalin-fixed, paraffin-embedded tumor specimens were submitted to Foundation Medicine for genetic analysis. The DNA was extracted from sections (40-μm thick) of these specimens obtained from 245 patients. Next-generation sequencing was performed on hybridization-captured, adaptor ligation–based libraries for 4557 coding exons of 287 cancer-related genes, plus 47 introns from 19 genes frequently rearranged in cancer, and 3549 polymorphisms located throughout the genome to detect mutations (single-nucleotide variants), CNAs, and rearrangements. The NGS-based clinical cancer gene assay has been published previously, and the assay performance has been validated.17 The Foundation Medicine T7 assay used in this study is able to detect mutations in all 4 ERBB receptors (EGFR, HER2, HER3, and HER4), unlike more commonly used circulating tumor DNA platforms, such as Foundation ACT (which detects EGFR alterations only) or Guardant360 (which measures EGFR and HER2 alterations).
The specimens for tumor genetic analysis (TGA) were retrospectively selected and enriched for patients with PFS of more than 2 months (149 of 245 patients [60.8%] with specimens used for TGA had PFS of more than 2 months vs 341 of 795 patients [42.9%] in the overall LUX-Lung 8 population) to ensure that the data set reflected a range of responsiveness to EGFR-targeted tyrosine kinase inhibitors (eAppendix in the Supplement).
EGFR Immunohistochemistry
The expression of EGFR was assessed using immunohistochemistry (IHC) with an EGFR pharmDx kit (Dako). Two separate criteria for EGFR positivity, based on definitions from previous studies,15,18 were used. For the first criterion, a tumor was considered positive for EGFR if at least 10% of the tumor cells showed staining of any intensity. For the second criterion, a tumor was considered positive for EGFR if the H-score was 200 or greater when using the H-score method with magnification rule, which assigns a score from 0 to 300 to tumor specimens depending on the percentage of positive cells and the intensity of staining.15
Statistical Analysis
The OS and PFS were calculated using the Kaplan-Meier method. A Cox proportional hazards regression model was used to calculate HRs and 95% CIs. All analyses were conducted with SAS, version 9.2 (SAS Institute Inc). Two-sided P < .05 was considered statistically significant.
Results
Patients
The LUX-Lung 8 trial was conducted at 183 centers in 23 countries from March 30, 2012, to January 30, 2014. In total, 977 patients were enrolled and 795 patients were randomized, with 398 in the afatinib arm and 397 in the erlotinib arm (Figure 1). Among the 742 patients with archival tumor tissue specimens available, 440 specimens were selected for TGA (PFS >2 months, 320 specimens; PFS ≤2 months, 120 specimens). Of these specimens, 195 were ineligible for NGS; thus, the TGA subset consisted of 245 patients. Tumor specimens for IHC analysis were available for 345 patients from the LUX-Lung 8 population. Apart from 11 patients, the TGA subset and the IHC subset represented mutually exclusive populations. Blood samples from 675 patients were assessed using VeriStrat; the details for this analysis are published elsewhere.19 Tumor genetic analysis, IHC, and VeriStrat analysis were conducted from February 26, 2015, to June 12, 2017.
Figure 1. Patient Disposition.
EGFR indicates epidermal growth factor receptor; IHC, immunohistochemistry.
aSome specimens requested back by the sites; some patients randomized prior to discovery of inadequate tissue availability.
bSpecimens enriched for those with progression-free survival of 2 or more months.
cPathology laboratories unable to provide all tissue requested; specimens contained insufficient tumor content.
dEleven patients included in both the tumor genetic analysis and the EGFR IHC subsets.
eReceived at least 1 dose of study drug.
Baseline demographic and clinical characteristics were similar in the TGA and IHC cohorts and the overall LUX-Lung 8 population (eTable 1 in the Supplement). Current or ex-smokers made up 224 (91.4%) of the TGA cohort and 318 (92.2%) of the IHC cohort; 197 (80.4%) of the TGA cohort and 246 (71.3%) of the IHC cohort were of non–East Asian origin.
Tumor Genetic Analysis
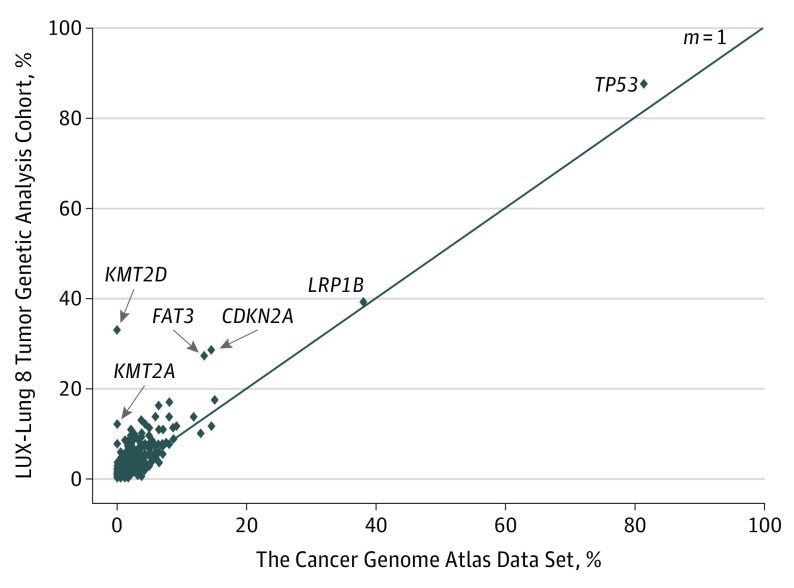
Figure 2 correlates the distribution of genetic aberrations detected in the TGA cohort with the findings previously reported by The Cancer Genome Atlas (TCGA) Research Network, which analyzed tumor specimens from 178 patients with lung SqCC.12 Overall, the genetic characteristics of SqCC tumors in the TGA cohort were similar to those in the TCGA analysis, although the mutation frequency of certain genes, including KMT2A, KMT2D, FAT3, and CDKN2A, was higher than that in the TCGA study. The most frequently observed genetic aberrations in the TGA cohort (245 patients) were somatic mutations in TP53 (214 patients [87.3%]), LRP1B (96 patients [39.2%]), KMT2D (81 patients [33.1%]), CDKN2A (70 patients [28.6%]), and FAT3 (67 patients [27.3%]) and CNAs (245 patients) in SOX2 (105 patients [42.9%]), KLHL6 (97 patients [39.6%]), PIK3CA (89 patients [36.3%]), MAP3K13 (79 patients [32.2%]), BCL6 (75 patients [30.6%]), and FGF12 (69 patients [28.2%]). All aberrations detected are given in eTable 2 in the Supplement.
Figure 2. Comparison of Mutation Allele Frequencies in the LUX-Lung 8 Tumor Genetic Analysis Cohort and in The Cancer Genome Atlas Data Set.
The m = 1 line shows the theoretical line on which all points would lie if the frequencies in both studies were identical.
In the TGA cohort, both PFS (median, 3.5 vs 2.5 months; HR, 0.69; 95% CI, 0.51-0.92; P = .01) and OS (median, 8.4 vs 6.6 months; HR, 0.81; 95% CI, 0.62-1.05; P = .12) favored treatment with afatinib over erlotinib, with systematically lower HRs and longer median PFS and OS than those observed in the overall LUX-Lung 8 population (eFigure 1 in the Supplement). This finding reflects the selection of a high proportion of specimens from patients with PFS of more than 2 months. Therefore, HRs in subgroups should be interpreted relative to the HR in the TGA cohort and not the overall LUX-Lung 8 population (eAppendix in the Supplement). None of the most frequently observed aberrations identified by TGA predicted PFS or OS benefit with afatinib over erlotinib treatment (eFigure 2 in the Supplement).
Outcomes Among Patients With ERBB Mutation–Positive Tumors
Overall, 53 of 245 patients (21.6%) had tumors with at least 1 ERBB family mutation (Table). Of 245 patients, 1 patient (0.4%) had tumors with EGFR, HER2, and HER3 mutations, and 2 patients (0.8%) had tumors with HER3 and HER4 mutations. Sixteen patients (6.5%) had tumors with at least 1 EGFR mutation; only 7 of the 17 EGFR mutations detected had previously been described (L861Q, E114K, Q1021, E746, A750del, V843I, and L858R). No tumor tested positive for EGFR variant III mutations, although such mutations are detectable using the Foundation Medicine NGS assay. The HER2 mutations were detected in tumors from 12 patients (4.9%), HER3 mutations in 15 patients (6.1%), and HER4 mutations in 14 patients (5.7%). Two patients (0.8%) had both a HER3 and a HER4 mutation. One patient (0.4%) had an EGFR, a HER3, and a HER4 mutation. Four of the 16 HER3 mutations (P1212S, R103H, and V104L in 2 patients) and 1 of the HER4 mutations (D931Y) have been previously described. Details of the ERBB mutation–positive tumors and the location of the HER2 mutations (detected in both the intracellular and extracellular domains) are shown in eFigure 3 in the Supplement.
Table. Frequency of ERBB Family Mutations in the Overall TGA Cohort and in Patients Who Were LTRs to Treatment With Afatiniba.
| Gene | TGA subset, % (n = 245) | Afatinib LTRs, % (n = 10) |
|---|---|---|
| ERBB wild type | 78.4 | 50.0 |
| ERBB mutation | 21.6 | 50.0 |
| EGFR | 6.5 | 20.0 |
| HER2 | 4.9 | 20.0 |
| HER3 | 6.1 | 0 |
| HER4 | 5.7 | 10.0 |
Abbreviations: EGFR, epidermal growth factor receptor; LTR, long-term responder; TGA, tumor genetic analysis.
Three patients were long-term responders to erlotinib: 1 patient had a tumor with an EGFR mutation, and 2 patients had tumors that were ERBB wild type.
In the overall LUX-Lung 8 data set of 398 patients, 21 patients (5.3%) were long-term responders to treatment with afatinib (received ≥12 months of treatment). Ten of these patients were in the TGA cohort, among whom 5 (50.0%) had ERBB mutation–positive tumors (Table). Therefore, the frequency of ERBB mutation–positive tumors was nominally higher among long-term responders than among the overall afatinib-treated population.
For patients receiving afatinib, both PFS and OS were longer among patients with ERBB mutation–positive tumors than among those without (PFS: median, 4.9 vs 3.0 months; HR, 0.62; 95% CI, 0.37-1.02; P = .06; OS: median, 10.6 vs 8.1 months; HR, 0.75; 95% CI, 0.47-1.17; P = .21) (Figure 3). By contrast, in the erlotinib arm, PFS and OS were similar among patients with tumors with or without ERBB mutations (PFS: median, 2.7 vs 2.4 months; HR, 0.76; 95% CI, 0.46-1.26; P = .29; OS: median, 7.2 vs 6.4 months; HR, 0.84; 95% CI, 0.54-1.32; P = .46).
Figure 3. Comparison of Progression-Free Survival (PFS) and Overall Survival (OS) Among Patients With Tumors Treated With Afatinib or Erlotinib in the Presence or Absence of ERBB Gene Family Mutations.
Outcomes assessed with respect to individual ERBB gene family members (Figure 4) indicated that EGFR mutations, in isolation, did not predict PFS or OS benefit of treatment with afatinib over erlotinib; indeed, the accentuated benefit of treatment with afatinib over erlotinib among patients with ERBB mutation–positive tumors appeared to be driven by HER3, HER4, and, in particular, HER2. Among the 12 patients with tumors having a HER2 mutation, PFS (HR, 0.06; 95% CI, 0.01-0.59; P = .02) and OS (HR, 0.06; 95% CI, 0.01-0.57; P = .02) strongly favored treatment with afatinib vs erlotinib. This observation was further underpinned by standardized effect plots (eFigure 4 in the Supplement). Despite the small sample size, the interaction P values were significant for PFS (P = .006) and OS (P = .003), indicating that the presence of a HER2 mutation may predict better outcomes with afatinib vs erlotinib treatment.
Figure 4. Comparison of Progression-Free Survival (PFS) and Overall Survival (OS) Among Patients in the Presence or Absence of EGFR, HER2, HER3, or HER4 Mutations.
EGFR indicates epidermal growth factor receptor; HR, hazard ratio; LL8, LUX-Lung 8; and TGA, tumor genetic analysis.
Outcomes According to ERBB CNAs
Seventeen (6.9%) and 9 (3.7%) of 245 patients had tumors with CNAs in EGFR and HER2, respectively. No CNA was detected in HER3 or HER4. There was no apparent correlation between CNAs and outcomes among patients treated with afatinib or erlotinib (data not shown).
Outcomes According to EGFR Overexpression
Outcomes in the IHC cohort (n = 345) were similar to those in the overall LUX-Lung 8 data set (PFS: median, 2.0 vs 1.9 months; HR, 0.74; 95% CI, 0.58-0.95; P = .14; OS: median, 7.4 vs 6.6 months; HR, 0.79; 95% CI, 0.63-0.99; P = .04). EGFR overexpression (by either method) did not predict PFS or OS benefit with afatinib vs erlotinib treatment (eTable 3 in the Supplement).
Discussion
To our knowledge, this secondary analysis of the LUX-Lung 8 trial represents the largest and most detailed evaluation of genetic mutations among patients with lung SqCC. Tumors were selected for analysis with Foundation Medicine NGS17 from 245 patients whose baseline demographic and clinical characteristics were similar to those in the overall LUX-Lung 8 population. Because afatinib is an ERBB family blocker and because other ERBB family members have been implicated in the pathogenesis of lung SqCC,12,20 the focus of this study was to assess outcomes among patients with ERBB mutation–positive tumors vs those with ERBB wild-type tumors. Although aberrations among individual ERBB family members were rare, cumulatively, 53 of the 245 patients (21.6%) had tumors with mutations in at least 1 ERBB family member. Although PFS and OS benefit with afatinib over erlotinib treatment was apparent among patients with ERBB wild-type tumors, the effects were more pronounced among patients with tumors that had at least 1 ERBB family mutation. Accentuated benefit with afatinib treatment did not appear to be driven by EGFR mutations; indeed, the largest benefits were observed among patients with tumors having HER2 or HER4 mutations.
Given the molecular heterogeneity of lung SqCC and the expanding armamentarium of available agents in this setting along with the ability of afatinib to irreversibly inhibit signaling via all ERBB heterodimers and homodimers, the present hypothesis-generating results suggest that identification of mutations in any ERBB family member might identify a subgroup of patients who may particularly benefit from treatment with afatinib following the failure of chemotherapy. Therefore, the role of HER2 as a potential biomarker warrants further investigation. Although HER2 mutations are rare, afatinib has previously demonstrated activity in heavily pretreated patients with adenocarcinoma NSCLC whose tumors had such a mutation.21 Notably, the pattern of HER2 mutations in the present study was different from that observed in adenocarcinoma NSCLC, in which the most frequently occurring mutations are in-frame insertion alleles in exon 20.22 In squamous disease, HER2 mutations are individually rarer and more heterogeneously distributed. In the present study, only 2 of the 9 mutations (Q57R and P489L) identified in tumors derived from afatinib-treated patients occurred in more than 1 patient, with 7 of the mutations being detected in the extracellular domain of HER2 and only 2 mutations occurring in the intracellular domain (G815R and P1037L). Overall, HER2 mutations were associated with better outcomes, suggesting that these mutations are important for tumor growth or survival, although more research is needed to characterize the individual aberrations and their likely variable role in receptor activation and response to afatinib. Mutations occurring outside the HER2 kinase domain are reported to be transforming in several cancers. Rare mutations occurring in the HER2 transmembrane domain23 and in the HER2 extracellular domain24 have been shown to be oncogenic in adenocarcinoma NSCLC. The HER2 extracellular domain mutations are also activating in colorectal25 and breast cancer26 and have been associated with response to afatinib treatment in urothelial cancer.27
The present study assessed the broad genetic characteristics of SqCC tumors in a large number of patients, potentially providing insights into the molecular pathogenesis of the disease. We found that the prevalence of the molecular aberrations in the LUX-Lung 8 population was consistent with data reported by the TCGA.12 The higher mutation frequencies observed in certain genes in the LUX-Lung 8 population (KMT2A, KMT2D, FAT3, and CDKN2A) may be due to the high read coverage obtained, which enabled the detection of mutations with lower variant allele frequency than in the TCGA analysis. In the cases of KMT2A and KMT2D, the mutation frequencies were reported as zero by the TCGA but were found to be 12.2% and 33.1%, respectively, in this study. The KMT2A and KMT2D mutations have been reported in squamous NSCLC in the COSMIC database,28 albeit at a lower frequency, suggesting that mutation detection in these genes is challenging and should be interpreted cautiously. None of the commonly mutated genes predicted superior OS or PFS outcomes with afatinib over erlotinib treatment.
In line with other studies,13,15 we identified high levels of EGFR overexpression in the IHC cohort (n = 345) of the LUX-Lung 8 population. Previous analyses have indicated that EGFR overexpression can predict outcomes with first-generation EGFR-targeted tyrosine kinase inhibitors in EGFR wild-type NSCLC.18 However, we found that the benefits of treatment with afatinib over erlotinib in the LUX-Lung 8 population were apparent regardless of EGFR expression levels or EGFR gene copy number. It seems, therefore, that the efficacy of afatinib in the LUX-Lung 8 population is not driven by EGFR overexpression.
In addition to the emergence of afatinib as a second-line treatment option for lung SqCC, the immune checkpoint inhibitors nivolumab, pembrolizumab, and atezolizumab have been approved on the basis of the phase 3 studies CheckMate 017 (in lung SqCC), Keynote 010, and OAK (the latter 2 in NSCLC of any histology), respectively.5,6,7 There is some evidence for PD-L1 as a predictive biomarker for these agents. For example, pembrolizumab is more active in patients with NSCLC tumors expressing high PD-L1 levels,6 whereas nivolumab efficacy in CheckMate 017 appears to be independent of PD-L1 expression.5 In other settings, nivolumab has demonstrated differential activity according to PD-L1 expression levels across different tumor types.29 Based on available data, checkpoint inhibitors are a second-line treatment option for lung SqCC.30 However, treatment with afatinib may be a viable alternative among those patients for whom checkpoint inhibitors are unsuitable. Furthermore, given our findings, it will be interesting to assess checkpoint inhibitors in tumors with ERBB family mutations. It may be that afatinib represents a preferable treatment option for these patients.
Limitations
Although the biomarker data presented herein are extensive, the present study has a number of limitations. First, because of the retrospective nature of the analysis and the lack of statistically significant findings (with the exception of outcomes among patients with HER2 mutations), the results should be considered hypothesis generating rather than hypothesis testing. Second, TGA is limited in the ability to detect functional aberrations (eg, hypermethylation and phosphorylation), which may be key to differential efficacy. Third, only a minority of patients from the overall LUX-Lung 8 population could be included in tumor genetic and IHC analyses. As such, it is difficult to extrapolate the results to the overall LUX-Lung 8 population given that the subgroups were relatively small and subject to selection bias. Indeed, the TGA cohort was enriched for patients with PFS of 2 or more months; consequently, the OS and PFS benefit with afatinib over erlotinib treatment was greater than that observed in the overall LUX-Lung 8 data set. Finally, we do not present independent evidence for the activating role of the individual ERBB mutations or for their sensitization of the tumors to afatinib.
Conclusions
Treatment with afatinib showed better outcomes than that with erlotinib across patient subgroups in this secondary analysis of the LUX-Lung 8 trial. The PFS and OS benefit with afatinib over erlotinib treatment was more pronounced among patients with ERBB mutation–positive tumors than among those without, especially among patients with tumors having HER2 or HER4 mutations. We hypothesize that the additional benefit with afatinib is attributable to its broad, irreversible inhibition of the entire ERBB signaling network. Finally, afatinib is a second-line treatment option for lung SqCC and may be particularly suitable for patients whose tumors carry at least 1 ERBB mutation.
eAppendix. Methods
eTable 1. Baseline Clinical Characteristics
eTable 2. All Genetic Aberrations Detected in the TGA Cohort
eTable 3. Relationship Between EGFR Expression Status and PFS and OS
eFigure 1. Comparison of Clinical Outcomes in the TGA Cohort and Overall LUX-Lung 8 (Intent-to-Treat) Population
eFigure 2. Relationship Between the Most Frequent Tumor Genetic Aberrations and (A) PFS and (B) OS in the LUX-Lung 8 TGA Cohort
eFigure 3. (A) Details of ErbB Family Mutations; (B) Location of HER2 Mutations
eFigure 4. Standardized Effect Plot for PFS (Left) and OS (Right)
References
- 1.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reck M, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S; ESMO Guidelines Working Group . Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iii27-iii39. [DOI] [PubMed] [Google Scholar]
- 3.Thatcher N, Hirsch FR, Luft AV, et al. ; SQUIRE Investigators . Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): an open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2015;16(7):763-774. [DOI] [PubMed] [Google Scholar]
- 4.Reck M, Rodríguez-Abreu D, Robinson AG, et al. ; KEYNOTE-024 Investigators . Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823-1833. [DOI] [PubMed] [Google Scholar]
- 5.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540-1550. [DOI] [PubMed] [Google Scholar]
- 7.Rittmeyer A, Barlesi F, Waterkamp D, et al. ; OAK Study Group . Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon EB, Ciuleanu TE, Arrieta O, et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): a multicentre, double-blind, randomised phase 3 trial. Lancet. 2014;384(9944):665-673. [DOI] [PubMed] [Google Scholar]
- 9.Soria JC, Felip E, Cobo M, et al. ; LUX-Lung 8 Investigators . Afatinib versus erlotinib as second-line treatment of patients with advanced squamous cell carcinoma of the lung (LUX-Lung 8): an open-label randomised controlled phase 3 trial. Lancet Oncol. 2015;16(8):897-907. [DOI] [PubMed] [Google Scholar]
- 10.Jaiswal BS, Kljavin NM, Stawiski EW, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23(5):603-617. [DOI] [PubMed] [Google Scholar]
- 11.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869-873. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers [published correction appears in Nature. 2012;491(7423):288]. Nature. 2012;489(7417):519-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Malpartida AV, Ludeña MD, Varela G, García Pichel J. Differential ErbB receptor expression and intracellular signaling activity in lung adenocarcinomas and squamous cell carcinomas. Lung Cancer. 2009;65(1):25-33. [DOI] [PubMed] [Google Scholar]
- 14.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12(8):553-563. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch FR, Varella-Garcia M, Bunn PA Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798-3807. [DOI] [PubMed] [Google Scholar]
- 16.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 17.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133-144. [DOI] [PubMed] [Google Scholar]
- 19.Gadgeel S, Goss G, Soria JC, et al. Evaluation of the VeriStrat®serum protein test in patients with advanced squamous cell carcinoma of the lung treated with second-line afatinib or erlotinib in the phase III LUX-Lung 8 study. Lung Cancer. 2017;109:101-108. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch FR, Franklin WA, Veve R, Varella-Garcia M, Bunn PA Jr. HER2/neu expression in malignant lung tumors. Semin Oncol. 2002;29(1)(suppl 4):51-58. [DOI] [PubMed] [Google Scholar]
- 21.De Grève J, Moran T, Graas MP, et al. Phase II study of afatinib, an irreversible ErbB family blocker, in demographically and genotypically defined lung adenocarcinoma. Lung Cancer. 2015;88(1):63-69. [DOI] [PubMed] [Google Scholar]
- 22.Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31(16):1997-2003. [DOI] [PubMed] [Google Scholar]
- 23.Ou SI, Schrock AB, Bocharov EV, et al. HER2 transmembrane domain (TMD) mutations (V659/G660) that stabilize homo- and heterodimerization are rare oncogenic drivers in lung adenocarcinoma that respond to afatinib. J Thorac Oncol. 2017;12(3):446-457. [DOI] [PubMed] [Google Scholar]
- 24.Greulich H, Kaplan B, Mertins P, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109(36):14476-14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5(8):832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhury NJ, Campanile A, Antic T, et al. Afatinib activity in platinum-refractory metastatic urothelial carcinoma in patients with ERBB alterations. J Clin Oncol. 2016;34(18):2165-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805-D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbognin L, Pilotto S, Milella M, et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS One. 2015;10(6):e0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novello S, Barlesi F, Califano R, et al. ; ESMO Guidelines Committee . Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5):v1-v27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Methods
eTable 1. Baseline Clinical Characteristics
eTable 2. All Genetic Aberrations Detected in the TGA Cohort
eTable 3. Relationship Between EGFR Expression Status and PFS and OS
eFigure 1. Comparison of Clinical Outcomes in the TGA Cohort and Overall LUX-Lung 8 (Intent-to-Treat) Population
eFigure 2. Relationship Between the Most Frequent Tumor Genetic Aberrations and (A) PFS and (B) OS in the LUX-Lung 8 TGA Cohort
eFigure 3. (A) Details of ErbB Family Mutations; (B) Location of HER2 Mutations
eFigure 4. Standardized Effect Plot for PFS (Left) and OS (Right)