Abstract
The ETS transcription factor ESE-1/Elf3 is overexpressed in breast cancer, controls transformation properties in mammary epithelial cells, and is most clinically relevant in HER2+ breast cancer. Here we show that ESE-1 knockdown, via shRNA, inhibits proliferation, clonogenicity, and anchorage-independent growth in HER2+, trastuzumab-resistant breast cancer cell lines HR20 (derived from HER2+ ER+ BT474) and Pool2 (derived from HER2+ ER- SKBR3 cells). ESE-1 knockdown in HR20 cells inhibited HER2/pHER2, pIGF-IR, pAkt and cyclin D1 levels; whereas ESE-1 knockdown in Pool 2 cells caused downregulation of IGF-IR/pIGF-IR, Src/pSrc, pAkt, mTOR/p-mTOR, and cyclin D1, but increased the levels of HER2 and pHER2. While certain signaling molecules (HER2/pHER2) responded in opposite fashion in HR20 and Pool 2 cells, inhibition of ESE-1 caused downregulation of pAkt and cyclin D1 in both trastuzumab-resistant cell lines, suggesting a common ESE-1-mediated downstream pathway leading to cell proliferation. In HER2+ parental lines, BT474 and SKBR3, knockdown of ESE-1 revealed a potent anti-proliferative effect that mimics the trastuzumab-mediated growth inhibition in these cells. However, ESE-1 knockdown did not enhance trastuzumab sensitivity in the resistant sublines. Collectively, these studies establish that ESE-1 controls cell proliferation in trastuzumab-resistant HER2+ breast cancer cells, and mechanistically does so by inhibiting pHER2, pAkt and downstream cyclin D1, which contribute to resistance of HER2 inhibitors. The significance of this study is that it provides a rationale for targeting ESE-1 and its downstream effectors as a novel means to treat HER2+ patients who show resistance to anti-HER2 therapy.
Keywords: ETS, Elf3, Her2 signaling, cyclin D1
1. Introduction
Trastuzumab, also known as Herceptin, is a monoclonal antibody that interrupts HER2-mediated downstream signaling by various mechanisms, such as disruption of HER2-HER3 dimerization, induction of antibody mediated cellular toxicity, and endocytic degradation of the HER2 receptor 1–3. In combination with chemotherapy, trastuzumab has been shown to increase overall survival in HER2+ breast cancer patients 4. However, only 30% of all HER2+ breast cancer patients respond to trastuzumab, and often duration of response to trastuzumab lasts only 5 to 9 months, indicating that both primary and acquired resistance to trastuzumab is common. As a consequence, several tyrosine kinase inhibitors, such as lapatinib and gefitinib, have been developed, which are selective inhibitors of the tyrosine kinase domains of HER2 and EGF-R 5. In an effort to achieve greater response rates, trastuzumab and lapatanib/gefitnib combination therapies have been attempted 5. Unfortunately, responses to such combination therapies have also been fraught with resistance arising from compensatory signaling molecules or pathways 1,6. There is, therefore, a need for studying other effectors or modulators that influence the durability of response and/or by-pass resistance to HER2-targeted therapies.
Several ETS transcription factors, such as ETS-1, ESE-1/Elf-3, ESE-2/Elf5 and PEA-3, appear to be important in human breast cancer. ESE-1 is particularly relevant in HER2+ breast cancer, because ESE-1 regulates the HER2+ promoter activity and protein levels 7, ESE-1 expression is required to maintain the transformed state in breast cancer8, and ESE-1 mRNA expression correlates with poor clinical outcomes (Kar and Gutierrez-Hartmann, unpublished data). In BT474 and SKBR3 HER2+ luminal breast cancer cell lines that are fully transformed, ESE-1 positively controls cell proliferation, clonogenicity, anchorage-independent growth, and xenograft tumor growth (Kar and Gutierrez-Hartmann, unpublished data). Moreover, neuregulin and other growth factor ligands induce ESE-1 expression 9, revealing an additional feed-forward level of control of ESE-1 and downstream effectors targeted by trastuzumab.
In this paper, we have investigated the role of ESE-1 in controlling transformation in trastuzumab-resistant HER2+ positive cell lines derived from parental HER2+, ER+ BT474 and HER2+, ER- SKBR3 breast cancer cell lines. These trastuzumab-resistant cell lines develop an interaction between HER2, HER3/ErbB3 and IGF-1R to form a hetero-trimeric receptor signaling complex as the mechanism mediating trastuzumab resistance 10. Investigating ESE-1’s role in the context of trastuzumab-resistance shows that knockdown of ESE-1 inhibits proliferation, clonogenicity and anchorage-independent growth in both BT474 and SKBR3 trastuzumab-resistant cell lines. Specifically, ESE-1 knockdown acts by modulating the activation and/or expression of HER3, mTOR, IGF-IR, Src and pAkt to control the transformation phenotype in these cell lines. Furthermore, the lack of a synergistic inhibitory response of trastuzumab plus ESE-1 knockdown in the parental lines reveals that HER2 and ESE-1 function in the same pathway. Taken together, these studies highlight ESE-1 as a by-pass effector in HER2 resistance and establish the utility of pursuing ESE-1 as a future therapeutic target to inhibit the counter–regulatory responses to trastuzumab-mediated tumor inhibition.
2. Materials and Methods
Cell lines and cell culture
Cell lines BT474 and SKBR3 were purchased from the tissue culture core at University of Colorado Anschutz Medical Campus and the trastuzumab resistant cell lines HR20 and Pool2 were provided by Dr. Bolin Liu. All cell lines were maintained in DMEM/F12 Ham in presence of 10% FBS. Trastuzumab resistant cell lines were cultured under the continuous presence of 20 ug/ml of trastuzumab.
shRNA transduction
The pLKO.1 shRNA construct targeting ESE-1 was purchased from Open Biosystems. Oligonucleotide shESE-1 (5’-gccatgaggtactactacaaac-3’) targets the ETS domain. The shScr is a scramble sequence control and was a gift from Dr. Bolin Liu 10. All shRNAs were packaged into lentivirus and cells were transduced at a ratio of 1:3 of viral supernatant to media 11 . The cells were selected with 0.75 ug/ml of puromycin for a period of three days post transduction and stable knockdown of ESE-1 protein was detected via immunoblotting after every selection. All experiments were plated eight days post transduction and long-term stable knockdown clones of ESE-1 were not selected to avoid a counter-selective response to cell growth inhibition mediated by ESE-1 knockdown.
Western blotting
Immunoblotting was performed using procedures described previously 8. Cells were lysed using RIPA buffer (150 mM sodium chloride, 1.0% NP-40 or Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS (sodium dodecyl sulphate), 50 mM Tris, pH 8.0), which was supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail from Roche. Protein level was quantified using the Bio Rad DC protein assay and 25–50 ug of protein were loaded on SDS PAGE gels for immunoblotting. Immunoblotting for ESE-1 was performed using anti-ESE-1 monoclonal antibody (mAb405) developed in our lab 12. Antibodies against IGFR (9750S), pIGF-IR (3021S), ERBB3 (12708S), pERBB3 (4791S), mTOR (2972S),pmTOR (2971S), Src (2109), pSrc(2101), HER2 (4290S), pHER2 (2243S),Akt, MAPK were purchased from cell signaling and were used following manufacturer’s protocol at a dilution of 1:1000.
MTS assay
MTS assay (Promega) was performed according to manufacturer’s protocol. Briefly, 5000 cells were plated, then at 3 days or 4 days post-plating 1X MTS assay buffer was added to the cells, and cells were allowed to incubate at room temperature for 10 min with 2 min of additional shaking to allow for cell lysis. Samples were then measured using the luminescence setting using an HT Synergy plate reader.
Proliferation, clonogenicity and soft agar assays
For cell proliferation assays, cells were transduced with shRNA and plated in duplicates in 10 cm plates and viable cells were counted by Vicell on days 1, 3, 5 and 7. For clonogenicity studies, 3000 cells per well were plated in quadruplicate in 6-well plates. Cell were then fixed with 4% PFA and stained with crystal violet on day 11. For soft agar assays, transduced Pool2 and HR20 cells were suspended at 30,000 cells/well and 20,000 cells/well, respectively, and plated in 6-well plates. A bottom layer of 0.6% and a top layer of 0.3% agar was used and cells were followed through day 17. At the end of day 17, cells were stained using nitroblue tetrazolium salt and colonies were counted and quantified using a NIH ImageJ software.
3. Results
ESE-1 knockdown inhibits proliferation, clonogenicity and anchorage-independent colony growth in trastuzumab-resistant HER2+ BT474 and SKBR3 breast cancer cell lines
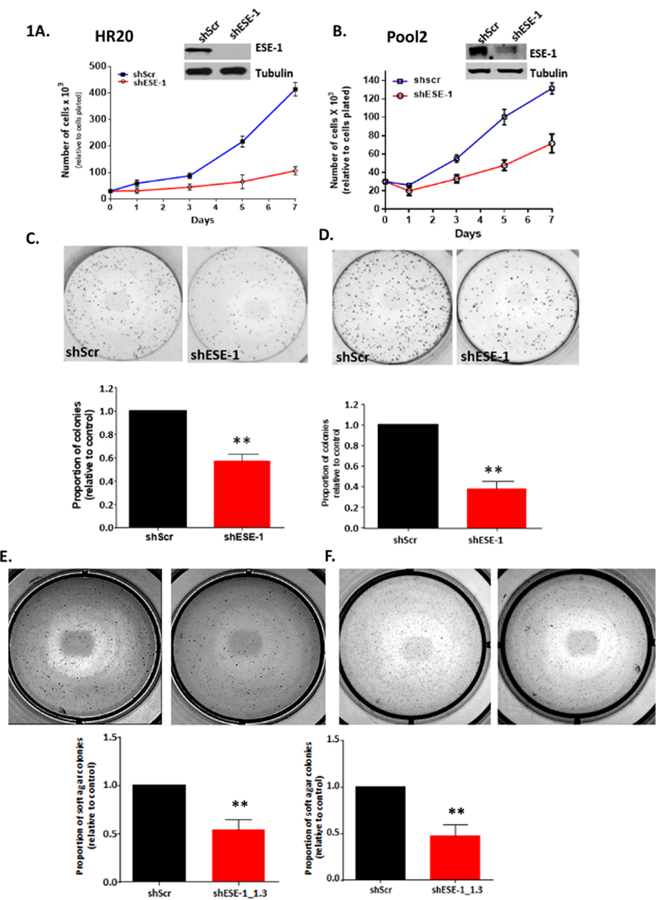
We first sought to determine whether ESE-1 is able to control cell proliferation, colony growth and anchorage-independent colony growth in trastuzumab-resistant cells HR20 and Pool2 cell lines, which were derived from parental HER2+ BT474 and SKBR3 cells, respectively, by continuous exposure to trastuzumab 13,14. These cells have been maintained under the continuous presence of 20 ug/ml of trastuzumab, a condition in which parental BT474 and SKBR3 cells do not survive. Knocking down ESE-1 in the trastuzumab-resistant lines resulted in a near complete inhibition of ESE-1 protein in HR20 (Figure 1A) and about a 60% decrease of ESE-1 in Pool2 (Figure 1B). Cell proliferation was measured in the scramble control and the ESE-1 knockdown groups, with viable cells counted from days 1 to day 7. As shown in Figures 1A and 1B, ESE-1 knockdown inhibited cell number at each of the 7 days in both HR20 and Pool2 cell lines, with the reduction in HR20 being more pronounced, with an 84% decrease in proliferation (Figure 1A) and Pool2 cells showing a 54% inhibition in cell number, each compared to the scramble control at day 7 (Figure 1B).
Figure 1.
ESE-1 knockdown inhibits transformation properties in trastuzumab resistant HER2+ cell lines. A, B. Stable ESE-1 knockdown inhibits proliferation in trastuzumab resistant HER2+ cell lines BT474-HR20 and SKBR3-Pool2 cell line. Proliferation was measured via trypan blue exclusion method by Vicell. C, D, E, F. Downregulation of ESE-1 inhibits clonogenicity, and soft agar colony formation in the two resistant sublines. Data represented is cumulative of three independent replicates. Error bars represent mean +/− SEM derived from three independent experiments.
To assess clonogenicity, an equal number of viable shScr and shESE-1 HR20 and Pool 2 cells were plated at low density in complete media. ESE-1 KD in HR20 cells resulted in a 50% reduction in 2D colony formation (p=0.0002) (Figure 1C) and a 70% reduction in colony number in Pool2 cells with shESE-1 compared to shScr control (p=0.002) (Figure 1D). Likewise, ESE-1 KD in HR20 cells resulted in a 62% decrease in soft agar colonies with shESE-1 (p=0.024) (Figure 1E) and ESE- KD in Pool2 cells resulted in 52% reduction (p=0.002) (Figure 1F).
Characterization of growth factor signaling in trastuzumab treated HER2+ BT474 and SKBR3 breast cancer cell lines, and their trastuzumab resistant counterpart
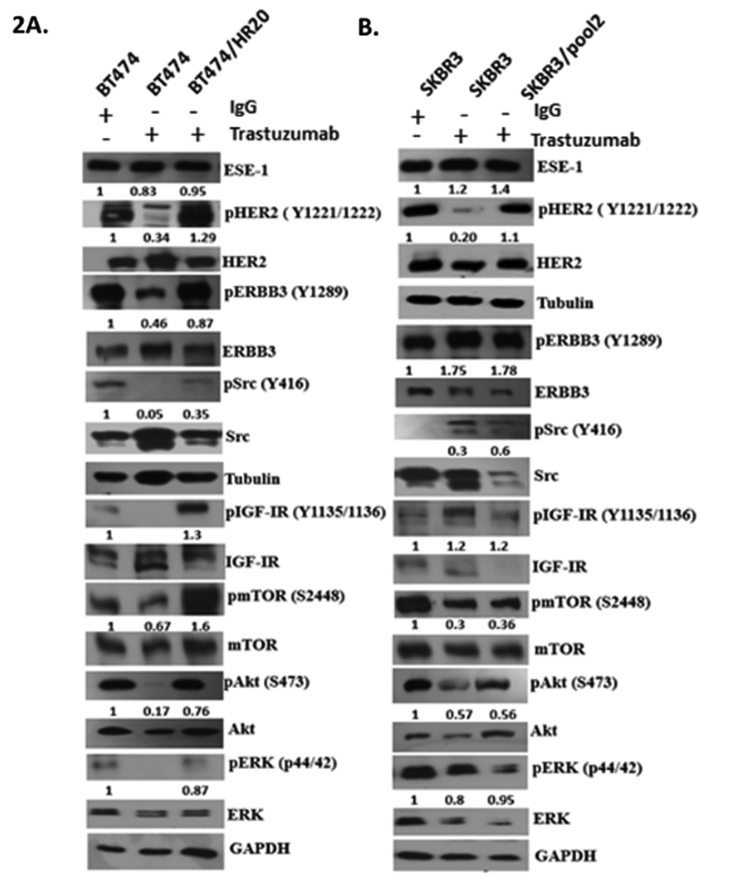
As a control for the effects of trastuzumab on HER2 signaling, we first added 20 μg/ml of trastuzumab to the parental BT474 and SKBR3 cells, and performed Western blot analysis on multiple downstream signaling effectors (Figures 2A & 2B). In both parental lines, trastuzumab treatment downregulated phosphorylation of HER2, Akt, as previously reported 14,15 (Figures 2A & 2B). However, the phosphorylation level of tyrosine kinase receptor HER3, which is considered to be the dimerizing partner of HER2, tyrosine kinase IGF-1R, and the non-receptor tyrosine kinase adaptor molecule Src, displayed cell type-specific responses to trastuzumab treatment (Figures 2A & 2B). Specifically, trastuzumab treatment inhibited HER3, IGF-IR and Src phosphorylation in BT474 cells, but induced their activation in the SKBR3 cells (Figure 2A). Trastuzumab treatment also caused a decrease in p-mTOR in both cell types (Figures 2A & 2B). Because ESE-1 and HER2 are involved in a positive feedback loop, we determined whether treatment of the parental cell lines with trastuzumab inhibited ESE-1 protein levels. While trastuzumab treatment of BT474 cells resulted in a 17 % inhibition of ESE-1 protein expression when normalized to GAPDH (Figure 2A), ESE-1 inhibition was not observed in the trastuzumab-treated SKBR3 cells, despite a detectable downregulation of pHER2 in these cells (Figure 2B).
Figure 2.
Immunoblot showing basal levels of signaling proteins in parental, trastuzumab treated, and trastuzumab resistant HER2+ cell lines. A, B. Immunoblotting was done with parental cell lines maintained in DMEM/F12 Ham, BT474 and SKBR3 cells treated with 20ug/ml trastuzumab for a period of 3 and 4 days respectively, and with trastuzumb resistant HER2+ lines BT474-HR20 and SKBR3-Pool2 stably maintained in 20ug/ml of trastuzumab. Quantification of the blot was done using the Image J software. All proteins were first normalized to the loading control. Phosphorylation levels were then estimated by normalizing the phosphorylation densities to their respective total.
We next investigated the basal activation or expression levels of HER2, HER3, Akt, ERK, and Src in the HR20 and Pool2 trastuzumab-resistant sublines compared to their parental lines and to trastuzumab-treated parental lines. Figures 2A and 2B are representative blots of three independent replicates. Both HR20 and Pool2 were grown in the constant presence of trastuzumab (20 μg/ml), and these cells exhibited either similar or slightly low levels of pHER2, HER2, pAKT, pERK expression compared to their basal expression in the parental counterparts (Figures 2A & 2B). In terms of HER3 activation, Pool2 cells showed high pHER3 compared to parental SKBR3, while HR20s displayed similar levels of pHER3 as their parental cells. Both cell lines, however, exhibited increased Src phosphorylation (Figures 2A & 2B). Src activation in Pool2 cells was higher than in parental SKBR3 cell lines, and Src phosphorylation levels in the HR20 cells, although lower than parental BT474 cells, was higher than the trastuzumab treated BT474 cells. These results suggest that sustained Src activation, albeit at a low level, contributes to their acquiring trastuzumab resistance. HR20 cells also evinced an induction in IGF-IR and mTOR phosphorylation, indicating additional pathways are active in these cells.
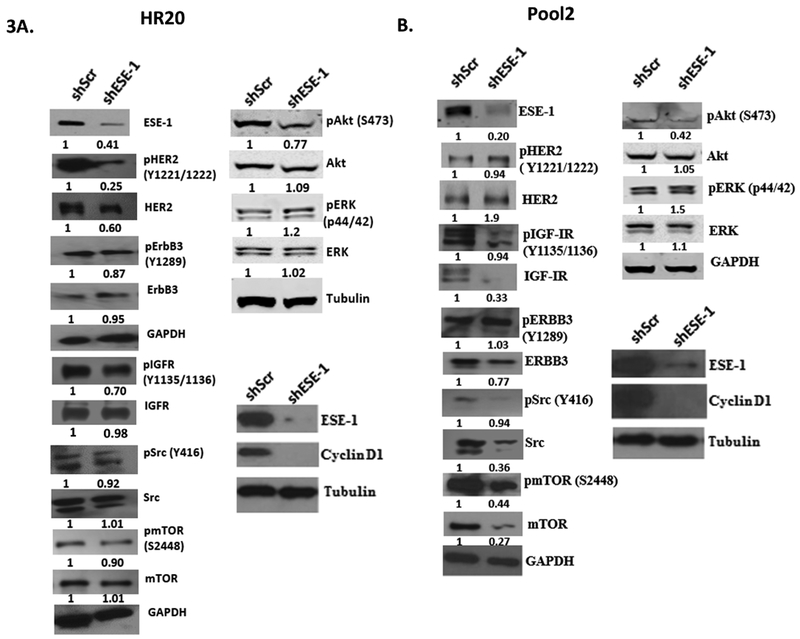
ESE-1 modulates ERBB3, IGF-IR, mTOR and downstream pAkt and cyclin D1 protein levels to control cell growth in trastuzumab-resistant HR20 and Pool2 sublines
We have previously established that ESE-1 knockdown does not induce apoptosis in transformed breast cancer cells, but rather inhibits cell proliferation 8. Knocking down ESE-1 in HR20 and Pool2 trastuzumab-resistant cells inhibited tyrosine kinases and their downstream effectors in a cell type-specific manner (Figure 3A-B). ESE-1 knockdown downregulated HER2 phosphorylation and HER2 protein expression in the HR20 cell line, although HER2 activation and protein expression in Pool2 cells were increased (Figure 3A-B). Since interaction between IGFR and HER2 or HER3 contributes to trastuzumab resistance for both HR20 and pool2 cell lines 10, we next determined if ESE-1 inhibition down-regulated either of these proteins. ESE-1 inhibition minimally downregulated phosphorylation of HER3 in HR20. In Pool2 resistant subline, there was about a 23% decrease in HER3 protein expression although HER3 activation remain unchanged (Figure 3A-B). With regards to IGF-IR, ESE-1 inhibition resulted in reduction in both pIGF-IR protein levels in both HR20 and Pool2 cells, with the latter being due to downregulation of IGF-IR protein (Figure 3A-B). The Pool2 cell line also exhibited a decrease in Src protein level, which largely accounted for the decrease in Src phosphorylation levels, while total Src levels in HR20 cells remain unchanged (Figure 3A-B). ESE-1 knockdown did not inhibit ERK protein levels or ERK phosphorylation in either of the cell lines, but downregulated Akt phosphorylation at Ser473 in both cell types (Figure 3A-B). We next investigated whether ESE-1 downregulation affects mTOR/p-mTOR, since the phosphorylation site at Ser473 is a bona fide substrate of mTORC2. ESE-1 knockdown downregulated phosphorylation of mTOR and mTOR protein levels in pool2 cells, indicating that regulation of mTOR, along with ERBB3, IGFR, and Src, upstream of Akt, contributes to ESE-1’s mode of cell transformation, specifically in these cell types. We surmise that the increase in HER2 and HER2 phosphorylation in Pool2 cells is also because of compensatory responses arising out of prolonged mTOR inhibition. HR20 are HER2+ and ER+ cells, and thus are considered luminal B breast cancer subtype, exhibited little or no changes in the level of Src or mTOR protein (Figures 3A & 3B) upstream of Akt. Both cell lines however evinced downregulation of cyclin D1, a known downstream target of Akt, which most likely contributed to the growth inhibitory phenotype of ESE-1 knockdown (Figure 3A-B).
Figure 3.
Immunoblot showing changes in signaling proteins in shESE-1 transduced trastuzumab resistant BT474-HR20 and SKBR3-Pool2 cell lines. A, B. Both resistant sublines were transduced with shESE-1, was puromycin selected and was harvested in RIPA buffer 8 days post transduction for subsequent immunoblotting. Blot shown is representative of three independent experiments. Quantification was done using the Image J software. All protein densities were first normalized to Tubulin or GAPDH. Levels of phosphorylation were estimated by normalizing the phosphorylation densities to their respective total.
ESE-1 knockdown alone exerts an anti-proliferative effect similar to trastuzumab in parental cell lines, but does not restore sensitivity to trastuzumab in trastuzumab resistant sublines
Because ESE-1 knockdown inhibited not only activation of Akt, but also modulated expression of key signaling molecules, such as ErbB3, mTOR, IGF-IR, and Src, we reasoned that ESE-1 inhibition, in combination with trastuzumab, could exert a more potent anti-proliferative effect in the parental cell lines especially in Pool2 than what might be achieved with either agent alone. Given the fact that ESE-1 also inhibited several signaling molecules that appear to function as counter-regulatory pathways to trastuzumab treatment, we tested if ESE-1 knockdown sensitizes the resistant cell lines to trastuzumab-mediated growth inhibition.
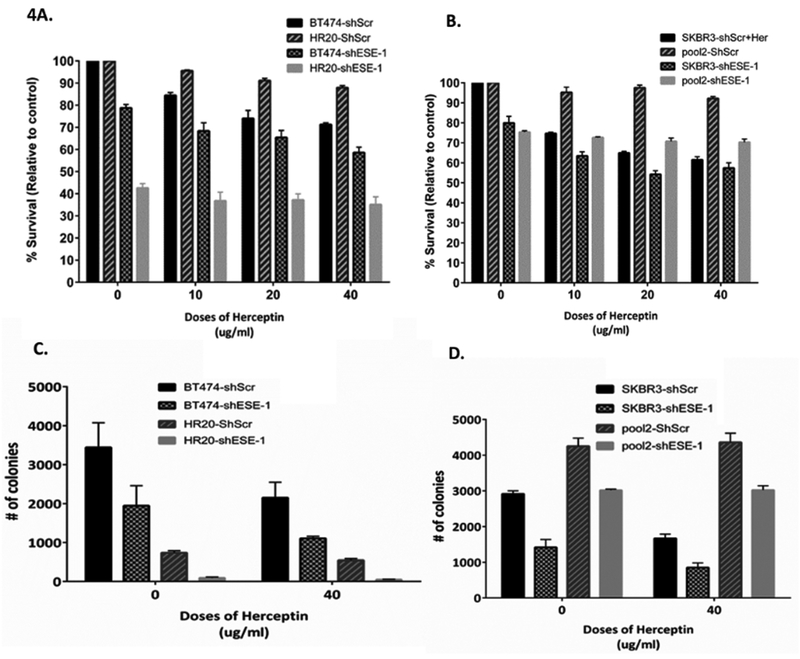
Since ESE-1 knockdown results in a fairly robust inhibition of cell proliferation, we designed an experiment to test whether synergy occurs with trastuzumab plus ESE-1 knockdown by achieving a lower level of ESE-1 knockdown resulting in persistent proliferation, so that synergy could be detected. To this end, we stably transduced and parental BT474 and SKBR3 cells, and their resistant sublines, HR-20 and pool2 cells, with shESE-1 and a shScr scramble control. To optimize for a lower level of ESE-1 knockdown, cells were subjected to only one round of lentiviral infection and selected with puromycin. Post-selection, cells were plated in 96 wells and treated with increasing doses of trastuzumab for a length of 3 days for BT474 cells and 4 days in SKBR3 cells (Figures 4A and 4B). In the parental BT474 cells, trastuzumab alone caused an 18%, 25% and 30% decrease in cell proliferation at 10, 20 and 40 μg/ml, respectively (Figure 4A, black bar). At the same doses of trastuzumab, the combination with ESE-1 knockdown resulted in 21%, 32%, 34% and 42% inhibition, at 0, 10, 20 and 40 μg/ml, respectively (Figure 4A, checkered bar). Thus, trastuzumab alone or ESE-1 knockdown alone achieved nearly the same level of growth inhibition (18% vs 21%), and the combination is better than either treatment alone, but is neither fully additive nor synergistic. In the parental SKBR3 cells, trastuzumab alone caused a 25%, 36% and 39% decrease in cell proliferation at 10, 20 and 40 μg/ml, respectively (Figure 4B, black bar). The same doses of trastuzumab in combination with ESE-1 knockdown resulted in 20%, 36%, 45% and 42% inhibition, at 0, 10, 20 and 40 μg/ml, respectively (Figure 4B, checkered bar). Again showing that trastuzumab alone or ESE-1 knockdown alone achieve nearly the same level of growth inhibition (25% vs 20%), and the combination is slightly better than either treatment alone, but is not synergistic.
Figure 4.
ESE-1 knockdown exerts trastuzumab like anti-proliferative effects. SKBR3, BT474, Pool2 and HR20 cells were transduced with shscr or shESE-1, and were treated with increasing doses of trastuzumab or IgG control. A,B. Cells were incubated for 3 days in case of BT474 or its resistant subline HR20 and 4 days for SKBR3 and its resistant subline Pool2. Cell survival was measured using MTS assay kit from Promega. Error bars are +/− SEM derived from three independent replicates. C,D. Cells were plated for clonogenicity and incubated for 11 days. Colonies were stained with crystal violet and quantified using Image J software. Error bars are +/− SEM derived from three replicates
We next investigated whether ESE-1 knockdown sensitizes trastuzumab resistant sublines to trastuzumab treatment. Compared to parental BT474 and SKBR3 cells, both HR20 and Pool2 scramble controls minimally responded to increasing doses of trastuzumab, as previously reported 10. By contrast, knocking down ESE-1 alone reduced % survival in HR20 and Pool2 cell lines by 58% and 25%, respectively, and trastuzumab treatment failed to enhance the inhibition beyond ESE-1 knockdown alone, achieving 58–60% inhibition in HR20 cells and 25–32% inhibition in Pool2 cells, at all doses of trastuzumab ± ESE-1 knockdown (Figures 4A and 4B). These results reveal that ESE-1 knockdown alone is fairly potent inhibitor of cell proliferation in these trastuzumab-resistant cell lines, and that ESE-1 knockdown does not rescue the inhibitory trastuzumab response.
To further test for any synergistic response, a clonogenicity assay was also used to measure colony numbers in ESE-1 knockdown parental and trastuzumab-resistant cell lines treated with trastuzumab. Treatment of scramble control BT474 parental cells with trastuzumab (40 μg/ml) alone inhibited colony numbers by 40% at 11 days post plating (Figure 4C, black bars). ESE-1 knockdown alone caused a 44% decrease in colony numbers, and a 68% decrease in combination with 40 μg/ml of trastuzumab. Trastuzumab-resistant HR20 cells formed many fewer colonies than the parental BT474 cells and showed a minimal response to trastuzumab treatment, whereas ESE-1 knockdown alone decreased HR20 colony numbers by 88% did not sensitize the cells to further trastuzumab treatment (Figure 4C, grey bars). In parental SKBR3 cells, trastuzumab treatment of 40 ug/ml inhibited colony numbers by 43% at 11 days post plating. ESE-1 knockdown alone caused a 51% decrease in colony numbers, and a 70 % decrease in colony numbers in combination with 40ug/ml of trastuzumab. Notably, trastuzumab resistant Pool2 cells formed 31% more colonies than their parental counterparts, and showed no sensitivity to trastuzumab. Knocking down ESE-1 caused a 30% decrease in Pool2 colony numbers with or without trastuzumab, and as before did not sensitize the cells to trastuzumab treatment (Figure 4D, grey bars).
4. Discussion
A major cause of trastuzumab resistance is an increase in expression of other tyrosine kinase receptors, resulting in by-pass activation of counter-regulatory signaling pathways. Increased signaling from EGFR and IGF-IR, and deregulation of the PI3K/PTEN/Akt pathway, are key contributors to trastuzumab resistance 1. It has been long established that prolonged trastuzumab treatment leads to a compensatory increase in PI3K/Akt signaling that arises because of a Akt-mediated feedback loop leading to HER3 upregulation and heregulin-mediated HER2 upregulation 16. Another key mediator of trastuzumab resistance is cyclin D1. In a doxycycline inducible MMTV-HER2 murine model, recurrent tumors had high expression of cyclin D1/CDK4 proteins resulting in cyclin D1/CDK4-mediated resistance to targeted therapy for HER2+. This was overcome using CDK4/6 inhibitors 17,18. These by-pass mechanisms contribute to clinical morbidity and mortality, due to relapse of trastuzumab-resistant breast cancer. Thus, a key gap is to identify targets that can overcome trastuzumab resistance. Here we show that knocking down ESE-1 in two distinct trastuzumab-resistant cell lines, HR20 and Pool2, result in a significant reduction in HR20 and Pool2 cell number, cell growth, colony formation and anchorage independent growth. Thus, ESE-1 and its key downstream effectors, serve as novel targets in the treatment of trastuzumab-resistant HER2 breast cancer.
Using cell culture models of trastuzumab resistance we show that ESE-1 knockdown in the resistant sublines have a cell type-specific effect on HER2 signaling and other protein kinases, such as HER3, IGF-1R, Src and mTOR. In the trastuzumab-resistant HR20 cells, which are ER+, HER2+ luminal B cells, ESE-1 inhibition decreases HER2 protein expression, HER2 phosphorylation, HER3 phosphorylation, IGF-IR phosphorylation when normalized to GAPDH . It appears that in trastuzumab-resistant SKBR3/pool2 cell lines, the anti-proliferative effects of ESE-1 knockdown are driven by inhibition in expression levels of mTOR, Src, and IGF-IR, and not by changes in phosphorylation of their respective proteins. In both resistant sublines, however, there is downstream inhibition in Akt activation, and a decrease in downstream cyclin D1. This reveals that ESE-1 is able to dictate cell growth responses by controlling key transformation nodes, such as Akt, which functions upstream of the cell cycle, and also by controlling cyclin D, which directly regulates the cell cycle.
While it is known that ESE-1 is a transcriptional activator of HER2 7,9, and EHF (an ETS transcription factor) transcriptionally regulates HER3 in thyroid cells 19, further studies are required to understand how ESE-1 controls expression of specific proteins, such as HER3, mTOR, IGFR, and Src in the trastuzumab-resistant sublines. It is unknown if ESE-1 transcriptionally controls each of the above genes in breast cancer cells. Whole genome analysis in ESE-1 knockdown parental SKBR3 cells does not show a decrease in the level of mTOR, Src, or IFG-1R mRNA (Kar and Gutierrez-Hartmann, unpublished data), suggesting that they are unlikely to be direct transcriptional targets of ESE-1 and are indirectly modulated by ESE-1 to control transformation properties in the resistant sublines.
The fact that ESE-1 knockdown inhibits mTOR and Src expression and activation has special implications in trastuzumab therapy. The baseline Src activation observed in the resistant sublines, especially in Pool2, corroborates with previous findings, which showed that Src activation following IGFR phosphorylation is a compensatory response in these particular cell types 10. An early increase in Src activation in response to trastuzumab treatment has far reaching implications in cases of resistance arising from trastuzumab and other tyrosine kinase inhibitors, such as lapatinib or gefitnib 20. Upregulation in Src or mTOR expression leading to aberrant Akt signaling is one of many mechanisms of lapatinib resistance 20,21. A therapeutic strategy for cancers that are refractory to anti-ERBB2 and/or anti-EGFR therapy is therefore to use mTOR or Src inhibitors, in combination with anti-HER inhibitors 1. Use of saracatinib in combination with lapatinib has been shown to synergistically inhibit proliferation, migration, and invasion in lapatinib resistant cell lines 6. In preclinical models of trastuzumab resistance, use of mTOR inhibitors has resulted in enhanced anti-proliferative effects, and there is some clinical evidence for the utility of mTOR inhibition in hormone receptor-positive and HER2-positive breast cancers 15,22,23.
We therefore tested if ESE-1 knockdown enhanced trastuzumab mediated growth inhibition in the parental cell lines or if it sensitized the resistant cells to trastuzumab treatment. Although ESE-1 knockdown did not enhance trastuzumab sensitivity in either case, it alone had a pronounced inhibitory effect on cell survival and colony growth in both parental BT474 and SKBR3 cell lines and trastuzumab-resistant derivative HR20 and pool2 cell lines. This was not surprising given that ESE-1 controls the transformation properties of the parental cells via PI3K/Akt activation (Kar and Gutierrez-Hartmann, unpublished data), a pathway that is commonly targeted by trastuzumab. One other mechanism by which trastuzumab controls growth response is via antibody-dependent cell cytotoxicity and apoptosis. ESE-1 knockdown had no effect on apoptosis and did not sensitize trastuzumab resistant cell for apoptosis. Nonetheless, collectively these studies established that ESE-1 modulates several signaling molecules, which commonly contribute to HER inhibitor resistance and therefore provides a rationale for investigating the signaling pathways of ESE-1 in HER2+ patients showing resistance to anti HER2 therapy.
Acknowledgments
Grant support: NIH/NCI R01CA141201, Cancer League of Colorado
Footnotes
Disclosure: Authors have nothing to disclose
5. References
- 1.Luque-Cabal M, Garcia-Teijido P, Fernandez-Perez Y, Sanchez-Lorenzo L, Palacio-Vazquez I. Mechanisms Behind the Resistance to Trastuzumab in HER2-Amplified Breast Cancer and Strategies to Overcome It. Clin Med Insights Oncol 2016;10(Suppl 1):21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res June 15 2001;61(12):4744–4749. [PubMed] [Google Scholar]
- 3.Maximiano S, Magalhaes P, Guerreiro MP, Morgado M. Trastuzumab in the Treatment of Breast Cancer. BioDrugs April 2016;30(2):75–86. [DOI] [PubMed] [Google Scholar]
- 4.Dahabreh IJ, Linardou H, Siannis F, Fountzilas G, Murray S. Trastuzumab in the adjuvant treatment of early-stage breast cancer: a systematic review and meta-analysis of randomized controlled trials. Oncologist June 2008;13(6):620–630. [DOI] [PubMed] [Google Scholar]
- 5.Segovia-Mendoza M, Gonzalez-Gonzalez ME, Barrera D, Diaz L, Garcia-Becerra R. Efficacy and mechanism of action of the tyrosine kinase inhibitors gefitinib, lapatinib and neratinib in the treatment of HER2-positive breast cancer: preclinical and clinical evidence. Am J Cancer Res 2015;5(9):2531–2561. [PMC free article] [PubMed] [Google Scholar]
- 6.Madrid-Paredes A, Canadas-Garre M, Sanchez-Pozo A, Calleja-Hernandez MA. Non-HER2 signaling pathways activated in resistance to anti-HER2 therapy in breast cancer. Breast Cancer Res Treat October 2015;153(3):493–505. [DOI] [PubMed] [Google Scholar]
- 7.Eckel KL, Tentler JJ, Cappetta GJ, Diamond SE, Gutierrez-Hartmann A. The epithelial-specific ETS transcription factor ESX/ESE-1/Elf-3 modulates breast cancer-associated gene expression. DNA Cell Biol February 2003;22(2):79–94. [DOI] [PubMed] [Google Scholar]
- 8.Darius M Walker JMP, Melissa S. Gonzales, Henrick Horita and, Gutierrez-Hartmann A. ESE-1 is Required to Maintain the Transformed Phenotype of MCF-7 and ZR-75–1 Human Breast Cancer Cells. The Open cancer Journal. 2010;3:77–88. [Google Scholar]
- 9.Neve RM, Ylstra B, Chang CH, Albertson DG, Benz CC. ErbB2 activation of ESX gene expression. Oncogene May 30 2002;21(24):3934–3938. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Gao L, Wang S, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-i receptor in breast cancer cells resistant to herceptin. Cancer Res February 1 2010;70(3):1204–1214. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Wang D, Mo J, Li B. [Research progress of cell sheet technology in oral tissue engineering]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi September 2014;28(9):1168–1172. [PubMed] [Google Scholar]
- 12.Prescott JD, Poczobutt JM, Tentler JJ, Walker DM, Gutierrez-Hartmann A. Mapping of ESE-1 subdomains required to initiate mammary epithelial cell transformation via a cytoplasmic mechanism. Mol Cancer 2011;10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nahta R, Takahashi T, Ueno NT, Hung MC, Esteva FJ. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res June 1 2004;64(11):3981–3986. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Wang S, Lyu H, et al. The anti-erbB3 antibody MM-121/SAR256212 in combination with trastuzumab exerts potent antitumor activity against trastuzumab-resistant breast cancer cells. Mol Cancer 2013;12(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurvitz SA, Andre F, Jiang Z, et al. Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): a phase 3, randomised, double-blind, multicentre trial. Lancet Oncol July 2015;16(7):816–829. [DOI] [PubMed] [Google Scholar]
- 16.Gijsen M, King P, Perera T, et al. HER2 phosphorylation is maintained by a PKB negative feedback loop in response to anti-HER2 herceptin in breast cancer. PLoS Biol 2010;8(12):e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goel S, Wang Q, Watt AC, et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell March 14 2016;29(3):255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witkiewicz AK, Cox D, Knudsen ES. CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes Cancer July 2014;5(7–8):261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv Y, Sui F, Ma J, et al. Increased expression of EHF contributes to thyroid tumorigenesis through transcriptionally regulating HER2 and HER3. Oncotarget September 06 2016;7(36):57978–57990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Formisano L, Nappi L, Rosa R, et al. Epidermal growth factor-receptor activation modulates Src-dependent resistance to lapatinib in breast cancer models. Breast Cancer Res 2014;16(3):R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady SW, Zhang J, Tsai MH, Yu D. PI3K-independent mTOR activation promotes lapatinib resistance and IAP expression that can be effectively reversed by mTOR and Hsp90 inhibition. Cancer Biol Ther 2015;16(3):402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien NA, McDonald K, Tong L, et al. Targeting PI3K/mTOR overcomes resistance to HER2-targeted therapy independent of feedback activation of AKT. Clin Cancer Res July 1 2014;20(13):3507–3520. [DOI] [PubMed] [Google Scholar]
- 23.Andre F, O’Regan R, Ozguroglu M, et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol May 2014;15(6):580–591. [DOI] [PubMed] [Google Scholar]