Figure 5.
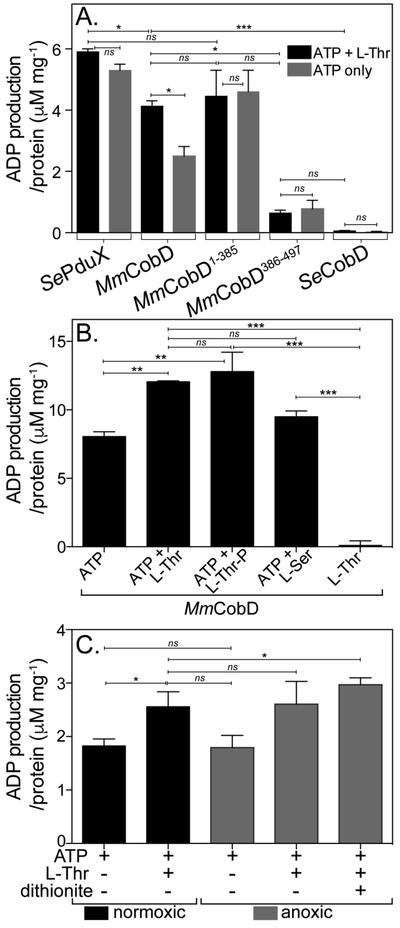
MmCobD has ATPase activity in vitro. ATPase activity assayed with ADP-Glo™ Kit (Promega). See Materials and Methods for a detailed description of the endpoint assay, which indirectly measures the enzymatic conversion of ATP to ADP via luminescence. The y-axis shows the conversion of ATP to ADP (μM) per mg of protein. The reaction mixture contained HEPES buffer (50 mM, pH 7.5 at 25°C), MgCl2 (1 mM), ATP (0.1 mM), L-Thr (0.3 mM), and normoxically purified protein (100 nM) incubated at 25°C for 1 h. (A) ATPase activity assayed in the presence and absence of L-Thr as co-substrate. Comparisons of SePduX, SeCobD, full-length MmCobD, and truncated enzymes in reactions with ATP + Thr (black bars) or ATP only (gray bars). Unpaired t test was used to calculate P values < 0.0003 (***), < 0.03 (*), or not significant (ns), with R2 = 0.96. (B) ATPase activity of MmCobD in the presence of cosubstrates or products. Reaction mixture containing HEPES buffer (50 mM, pH 7 at 25°C), MgCl2 (1 mM), MmCobD (72 nM) and ATP, L-Thr, L-Ser, or L-Thr-P (10 mM) where indicated; incubated at 25°C for 1 h. Unpaired t test was used to calculate P values < 0.0001 (***), <0.004 (**), and not significant (ns), with R2 = 0.99. (C) Reaction mixtures containing normoxically purified MmCobD enzyme were assayed under normoxic (black) and anoxic (gray) conditions. Reaction mixtures contained HEPES buffer (50 mM, pH 7.5 at 25°C), TCEP (2 μM MgCl2 (1 mM), ATP (10 mM), L-Thr (50 mM), protein (72 nM), and dithionite (2 mM) where indicated. Unpaired t test was used to calculate P values < 0.03 and 0.05 (*), and not significant (ns), R2 = 0.8. No-enzyme controls were subtracted to reduce background. Values were compared to a standard curve of luminescence vs ATP to ADP concentration, to generate a percent ATP conversion curve that was then transformed into units of ATP produced (μM) per mg of protein, with the standard error of the mean of triplicate reactions represented by the error bars. Calculations and graphs were generated with Prism v6 (GraphPad).
