Abstract
The Pirarucu (Arapaima gigas) is one of the world’s largest freshwater fishes and member of the superorder Osteoglossomorpha (bonytongues), one of the oldest lineages of ray-finned fishes. This species is an obligate air-breather found in the basin of the Amazon River with an attractive potential for aquaculture. Its phylogenetic position among bony fishes makes the Pirarucu a relevant subject for evolutionary studies of early teleost diversification. Here, we present, for the first time, a draft genome version of the A. gigas genome, providing useful information for further functional and evolutionary studies. The A. gigas genome was assembled with 103-Gb raw reads sequenced in an Illumina platform. The final draft genome assembly was ∼661 Mb, with a contig N50 equal to 51.23 kb and scaffold N50 of 668 kb. Repeat sequences accounted for 21.69% of the whole genome, and a total of 24,655 protein-coding genes were predicted from the genome assembly, with an average of nine exons per gene. Phylogenomic analysis based on 24 fish species supported the postulation that Osteoglossomorpha and Elopomorpha (eels, tarpons, and bonefishes) are sister groups, both forming a sister lineage with respect to Clupeocephala (remaining teleosts). Divergence time estimations suggested that Osteoglossomorpha and Elopomorpha lineages emerged independently in a period of ∼30 Myr in the Jurassic. The draft genome of A. gigas provides a valuable genetic resource for further investigations of evolutionary studies and may also offer a valuable data for economic applications.
Keywords: Arapaima gigas, Pirarucu, Osteoglossomorpha, Teleostei, genome sequencing, phylogenomics
Introduction
Arapaima gigas, also known as Pirarucu or Paiche, is one of the world’s largest freshwater fishes (Wijnstekers 2011) whose body length and weight may attain 4.5 m (15 ft) and 200 Kg (440 lb), respectively (Nelson 1994; Froese and Pauly 2018). The genus Arapaima emerged in the Amazon floodplain basin and is presently distributed in Brazil, Colombia, Ecuador, and Peru (Hrbek et al. 2005, 2007; Froese and Pauly 2018), and also in Thailand and Malaysia where it has been introduced for commercial fishing (Froese and Pauly 2018). Arapaima gigas local name (Pirarucu) derives from the indigenous Tupi words “pira” and “urucum” for “fish” and “red,” respectively, presumably referring to its red tail scales flecks or to its reddish flesh (Marsden 1994; Godinho et al. 2005). The peculiarity of its breathing apparatus is characteristic of this Amazonian fish, comprising gills and a lung-like tissue devised for air-breathing derived from a modified and enlarged swim bladder (Burnie and Wilson 2001; Brauner et al. 2004). The Pirarucu has an attractive market value due to its low-fat and low bone content. Overfishing practices in the Amazonian region led to the banning of Pirarucu commercialization by the Brazilian government in 2001, although consumption by the native population is currently permitted under strict size and seasoning regulations (Bayley and Petrere 1989). Its main supply is provided by wild-caught fish and fish farming conducted by riverbank population of the Amazonas (Froese and Pauly 2018). Aquaculture production is attractive due to high carcass yields and rapid juvenile growth, with yearlings reaching up to 10 kg (22 lb) (Almeida et al. 2013).
Arapaima gigas belongs to the superorder Osteoglossomorpha of bony-tongued fishes whose tongue contains sharp bony teeth for disabling and shredding preys (Sanford and Lauder 1990; Burnie and Wilson 2001). Together with Elopomorpha (eels and tarpons) and Clupeocephala (most of extant fish species), the Osteoglossomorpha comprises one of the three main teleosts groups whose phylogenetic position has been controversial (Le et al. 1993; Inoue et al. 2003; Near et al. 2012; Betancur-R 2013; Faircloth et al. 2013; Chen et al. 2015; Hughes et al. 2018). Fossil records and some early molecular studies, including a recent comprehensive analysis of >300 Actinopterygii species (Hughes et al. 2018), placed Osteoglossomorpha as the oldest teleost group (Greenwood 1970; Inoue et al. 2003), while other studies placed Elopomorpha as the most ancestral one (Near et al. 2012; Betancur-R 2013; Faircloth et al. 2013). Recently, a phylogenetic study based on whole genome sequencing of the bony-tongued Asian arowana (Scleropages formosus) suggested that the branching of Elopomorpha and Osteoglossomorpha occurred almost simultaneously, placing them as sister lineages of Clupeocephala (Bian 2016). Within this context, the genome of the Pirarucu provides new insights to study the evolutionary history of teleosts as well as providing useful information for sustainable exploration of this giant Amazon fish. Here, we present the first whole genome assembly, gene annotation, and phylogenomic inference of the Pirarucu which should facilitate the molecular characterization and conservation of this economically important fish species.
Materials and Methods
Sample Collection and Sequencing
Genomic DNA was extracted from peripheral blood samples of four adult individuals (two males and two females) of Arapaima gigas: NCBI taxonomy ID 113544, FishBase ID: 2076. All samples were collected in accordance with the standards of the Federal University of Pará animal protocol. We applied a whole-genome shotgun sequencing strategy using two short-insert libraries (400 and 500 bp) in an Illumina HiSeq 2500 platform according to the manufacturer’s instructions (Illumina, San Diego, CA). HiSeq Rapid SBS Kits (FC-402-4021) and HiSeq Rapid Cluster Kits (PE-402-4002) were used to sequence paired-end read of 2 × 250 base pairs. Read quality was checked using FastQC, version 0.11.4 (Andrews 2010), and low-quality reads were trimmed with Sickle paired-end (pe), version 1.33 (Joshi and Fass 2011), under default parameters.
Genome Size Estimation and De Novo Assembly
Genome size was estimated based on the k-mer spectrum with the following formula: G= (N×(L−K + 1)−B)/D. Where N is the total read count, L is the read length, K is k-mer length (K = 31), B is the total low-frequency (frequency ≤1) k-mer count, D is the k-mer depth, and G is the genome size. Jellyfish 2.2.6 (Marçais and Kingsford 2011) was used to count k-mer frequencies of high-quality sequencing reads.
Genome assembly was performed using SOAPdenovo2 (version 2.04) (Luo et al. 2012) under default parameters (127mer version). Three assemblies were conducted: 1) using all reads; 2) with reads from male samples; and 3) with reads from female samples. Subsequently, gaps were filled using Redundants (Pryszcz and Gabaldón 2016) using three-run scaffolding steps: firstly with the default value of minimum read pairs to joining contigs (5 pairs), subsequently rerunning with previous data with a minimum value of four read pairs and, finally, using a minimum of three read pairs. Assembly quality and statistics were assessed with QUAST (version 4.4) (Gurevich et al. 2013).
Assessment of Genome Completeness
Assembly quality was measured by assessing gene completeness with Benchmarking Universal Single-Copy Orthologs (BUSCO) (Simão et al. 2015) based on 4,584 BUSCO groups derived from Actinopterygii orthologs.
Repeat Analysis
Transposable elements (TEs) and other repetitive elements of the Pirarucu genome were identified by a combined, homology-based method and a de novo annotation approach. Initially, tandem repeats were identified with Tandem Repeats Finder 4.09 (Benson 1999) with the following parameters: “Match=2, Mismatch=7, Delta=7, PM=80, PI=10, Minscore=50, and MaxPerid=2,000.” Additionally, a de novo repeat library was built with RepeatModeler 1.0.9 and LTR_FINDER (Xu and Wang 2007), and filtered with LTR_retriever (Ou and Jiang 2017) under default parameters. Subsequently, known and novel transposable elements were identified by mapping the assembled sequences to the Repbase TE 22.05 (Bao et al. 2015) and de novo repeat libraries using RepeatMasker 4.0 (Tarailo-Graovac and Chen 2009). In addition, we annotated TE-related proteins using RepeatProteinMask 4.0 (Tarailo-Graovac and Chen 2009).
Gene Structure and Function Annotation
Genome annotation was carried out with the MAKER2 pipeline (Holt and Yandell 2011) in a two-pass iteration. First, homology annotation was performed with protein data from Homo sapiens (human), Danio rerio (zebrafish), Takifugu rubripes (Japanese fugu), Tetraodon nigroviridis (spotted green pufferfish), Gasterosteus aculeatus (three-spined stickleback), Oryzias latipes (Japanese medaka), Latimeria chalumnae (coelacanth) (Ensembl release 88), together with Scleropages formosus (Asian arowana) protein sequences from NCBI RefSeq annotation data. Subsequently, de novo annotations were performed using the homology-based results achieved in the first step. We also used the RepeatModeller 1.0.9 (Smit and Hubley 2008) to build a de novo repeat library with default parameters. The GFF output from the first step was used to train the SNAP 20131129 (Korf 2004) and AUGUSTUS 3.2.3 (Stanke et al. 2008) predictors. GeneMark-ES 4.32 (Lomsadze et al. 2005) was trained using the genome assembly itself. InterProScan 5.24-63.0 (Jones et al. 2014) was run on the protein output of MAKER, providing gene ontologies and classifying protein domains and families. Protein output was compared using BLAST against the NCBI NR database (available on May 29, 2017) for identifying putative gene names. Blast2GO v5 (Conesa et al. 2005) was subsequently used to obtain Gene Ontology mapping and annotation (supplementary file S2, Supplementary Material online).
Phylogenomic Analysis
Phylogenomics was based on protein data from 24 fish species. Transcriptome data from ENA database were used for species whose genome had not been sequenced (supplementary table S5, Supplementary Material online). Transcripts were assembled with Trinity (Haas et al. 2013) and protein sequences were deduced with Transdecoder. All redundant sequences (>99.5% of identity) were later removed with CD-HIT (Fu et al. 2012), and those with <200 residues were discarded, resulting in a data set with a total number of 651,482 protein sequences.
Subsequently, 17,031 orthogroups were built with OrthoFinder (Emms and Kelly 2015) using all-to-all BLASTP comparisons and MCL clustering (supplementary table S6, Supplementary Material online). Proteins in each group were aligned using MAFFT (Katoh and Standley 2013) and gene trees were estimated with FastTree (Price et al. 2010). Paralogous and spurious branches were removed by identifying clusters with monophyletic outgroups (“prune_ paralogs_MO.py”) as described by Yang and Smith (2014) and applying parameters suggested by Austin et al. (2015). Protein sequences in each cluster were realigned with MAFFT, trimmed with Gblocks (Talavera and Castresana 2007) and concatenated into a supermatrix of 282 loci and 188,505 aligned columns with an overall occupancy of 88.5%.
Phylogenomic analysis was conducted with ML and BI using the constructed supermatrix. ML analysis was conducted with RAxML (Stamatakis 2014) with 200 rapid bootstrap replicates considering each locus as a separate partition (278 loci, after merging partitions without occurrence of all amino acids). The final tree topology was selected as the tree with the best likelihood estimate. Bayesian inference was carried out using BEAST 2.4 (Bouckaert et al. 2014) under a LG substitution model. A Markov chain Monte Carlo (MCMC) was run for ten million generations and sampled every 5,000 generations. The consensus tree was determined after discarding (burn-in) 10% of initial trees.
Evolutionary rates were estimated by adding branch lengths from the tree tips to the teleost MRCA node (fig. 1, red star). Tajima’s relative rate tests (Tajima 1993) were performed with MEGA 7 (Kumar et al. 2016) with the same concatenated alignment used in phylogenomics analysis. Pirarucu rates were compared with rates of all other teleosts using the Spotted gar as outgroup; P < 0.05 was considered for rejecting the null hypothesis of equal rates between lineages.
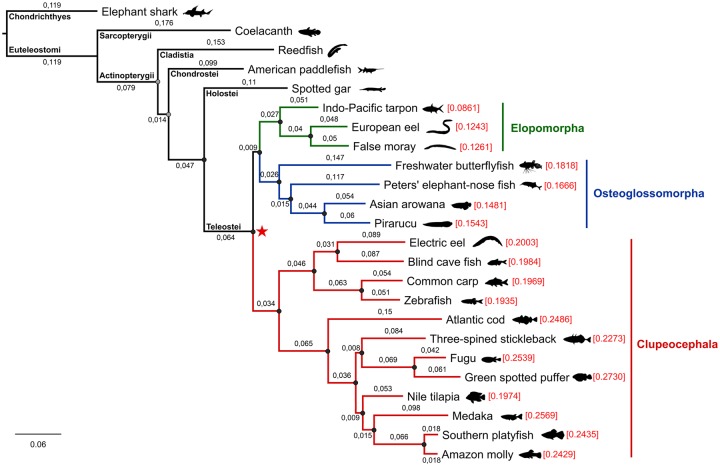
Fig. 1.
—Phylogenomics inference. Phylogenomic tree inferred by maximum likelihood (ML) based on a supermatrix of 278 orthologs loci (188,505 amino acid sites) from 24 species using Elephant shark as outgroup. Dark gray circles indicate coincident nodes with Bayesian inference (BI) and maximum support values in both approaches (bootstrap=100% and Bayesian posterior probability=1). Branch lengths represent number of substitutions/site. Rates of molecular evolution (i.e., number of amino acids substitutions per site) estimated from the teleost split (red star) to the tips of the topology are indicated in red font close to the name of each taxon.
Divergence Times Estimation
Estimations of divergence times were carried out with MCMCTree of PAML package (Yang 2007). Calibration times were obtained from TimeTree (Hedges et al. 2015) (supplementary table S8, Supplementary Material online), a public knowledge-base providing information of the evolutionary time of the tree of life (TTOL) based on >3,000 studies and comprising >97,000 species (at the time of this work). Time estimations were calculated using the amino acid supermatrix as input and the ML topology.
Whole Genome Analysis
Distributions of synonymous substitutions per synonymous site (Ks) were estimated with the wgd Python package (https://github.com/arzwa/wgd). Briefly, for each species, CDS sequences were first translated to protein sequences, compared all-versus-all using BLASTP and clustered using the MCL algorithm (Enright et al. 2002). Then, for each cluster, sequences were aligned using MUSCLE (Edgar 2004) and protein sequences were subsequently reverse translated to nucleotide sequences according to the input CDS. Finally, to estimate the Ks distributions, a maximum likelihood phylogenetic analysis was performed using the CODEML program from the PAML package (Yang 2007), and Ks values were corrected based on a phylogenetic tree constructed for each family using FastTree (Price et al. 2010).
Gene Family Evolution Analysis
To investigate gene gain and loss dynamics among ancestral lineages the previously inferred phylogenetic tree and the 17,031 orthogroups identified using OrthoFinder (see Materials and Methods—Phylogenomic analysis section) were used. Orthogroups consisting of species-specific single-genes or with >50 genes in one species were considered to be artefacts and were removed from downstream analysis, resulting in 16,968 orthogroups (each one representing a gene family). A Wagner parsimony approach, using the Count software (Csurös 2010) was used for identifying gene family gain and loss events, as well as family expansions and contractions. Gain to loss penalty ratio was set to 1, assuming expansions and contractions to be equally likely.
Maximum Likelihood Estimation of Gene Family Rates
Gene family expansion and contraction rates were estimated for gene families (see Materials and Methods—Gene family evolution analysis) with at least one gene present in the MRCA of teleosts (16,402 families on total). Estimations were carried out with CAFE v4.1 (Han et al. 2013) and the time-calibrated tree with MCMCTree (fig. 2). To account for potential errors in the data set, we used the “caferror.py” script for estimating global errors without a priori information.
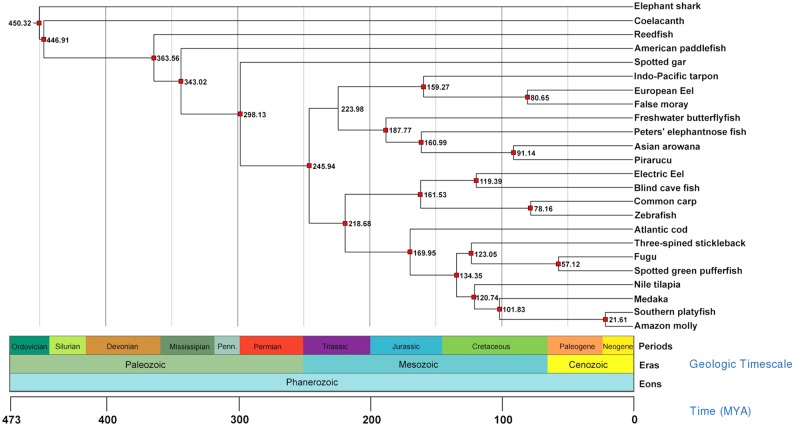
Fig. 2.
—Divergence time estimation between species. Numbers at nodes represent divergence time estimates in millions of years ago (Ma). Red squares indicate nodes calibrated by fossil records.
Rate estimations of gene family size evolution were assessed by equally considering rates of gains and losses (lambda) and different rates for each (lambdamu). We also performed estimations using a one-lambda(mu) model (i.e., average rate over all phylogenetic tree), and a two-lambda(mu) model (i.e., considering different rates for teleosts and nonteleosts). P threshold was set at 0.01.
Families Functional Annotation
Gene family functions were inferred by first selecting one representative sequence (the longest one) of each orthogroup. Subsequently, sequences were compared against the NR database with the DIAMOND tool (Buchfink et al. 2015) and against the InterProScan 5.24-63.0 (Jones et al. 2014) for domain annotation. Next, Gene Ontology terms were obtained with Blast2GO v5 (Conesa et al. 2005).
Sex Comparisons
To identify sex-specific genomic regions, we performed three different approaches:
Sex-specific trimmed reads were aligned against the main genome assembly (sex-mixed) using BWA (Li and Durbin 2009) and read mapping statistics was evaluated using SAMStat with default parameters (Lassmann et al. 2011). SAMtools package (Li et al. 2009) was used for creating and sorting binary (“.bam”) alignment files and, subsequently, for extracting only sequences mapped against the reference assembly, generating a file with genome length data. Statistics for assessing genome coverage were retrieved with BEDTools suit (Quinlan and Hall 2010) and shell command lines were used to calculate proportions of genomes/contigs coverage. Additionally, sex-specific coverages were summarized with Mosdepth (Pedersen and Quinlan 2018) considering read depths in window frames of 50 kb, and R for figure plotting.
In order to evaluate sex-specific genome assemblies, we used the Quality Assessment Tool for Genome Assemblies (QUAST) software (Gurevich et al. 2013). Comparisons were performed using the main genome assembly as reference and gene annotation data (“.gff”).
Lastly, a cross-read-assembly comparison was carried out for identifying sex-specific regions in each assembly. Firstly, reads of a given sex were aligned against the genome assembly of the opposite sex using BWA (Li and Durbin 2009). Unaligned reads considered to be sex-specific and were subsequently realigned against the genome assembly of the same sex for identifying regions mapped on the assembly. Mapped regions without mapped sex-specific reads were masked, and the remaining sequences were compared against the NR database using BLASTx to identify likely protein-coding genes associated with the sex-specific regions.
Results and Discussion
Genome Sequencing, Assembly, and Annotation
Whole genome sequencing of four adults (two males and two females) was performed with two paired-end short insert libraries using an Illumina HiSeq 2500 platform (2×250 bp), producing a total of 103.01-Gb raw sequences. After removing low-quality and redundant reads, we obtained ∼76.91 Gb of high-quality data for de novo assembling. Using a k-mer-based approach, genome size was estimated as 761 Mb (with ∼135× coverage) (supplementary table S1 and fig. S1, Supplementary Material online). Subsequently, de novo assembling generated a draft genome comprising 661,278,939 bp, 5,301 scaffolds, with scaffold N50 = 668 kb, and contig N50 = 51.23 kb. Assembly quality was measured with BUSCO (Simão et al. 2015) showing high-level completeness with 94.61% of complete BUSCOs groups (supplementary table S2, Supplementary Material online).
Repeat analysis showed a total of 143 Mb repetitive sequences, with DNA transposons representing the most predominant repeat, accounting for 46% of all transposable elements (TE) and 8.51% of the genome. Long repeat elements, like LTRs and LINEs, accounted for 3.07% and 4%, respectively (supplementary table S3, Supplementary Material online). In view that only short-insert libraries (400 and 500 bp long) were sequenced, complete LTR and LINE transposons were not expected to be fully identified.
Genome annotation predicted 24,655 protein-coding genes, covering 33.9% of the genome. Comparisons with the nonredundant (NR) NCBI database identified putative identities for 99% of the genes and Gene Ontology (GO) terms assigned to 12,460 proteins (50.5% of total) (supplementary table S4, Supplementary Material online). A summary of the sequencing data, genome assembly, and annotation is shown in table 1.
Table 1.
Summary Statistics of the Pirarucu Genome
| Sequencing Information | |
|---|---|
| Library insert size (bp) | 400–500 |
| Read length (bp) | 2×250 |
| Total raw bases sequenced (Gb) | 103.01 |
| Total filtered bases sequenced (Gb) | 76.91 |
| Genome Features | |
| Assembled genome size (Mb) | 661.28 |
| # scaffolds | 5,301 |
| Scaffold N50 (kb) | 668 |
| Contig N50 (kb) | 51.23 |
| Largest scaffold (bp) | 5,332,704 |
| GC (%) | 43.18 |
| Repeat content (% of genome) | 21.69 |
| Genome Annotation | |
| Protein-coding gene number | 24,655 |
| % of genome covered by genes | 33.9 |
| Mean transcript length (bp) | 9,150 |
| Mean exons per gene | 9 |
| Mean CDS length (bp) | 1,603 |
| Mean exon length (bp) | 174 |
| Mean intron length (bp) | 920 |
Compared with the Asian arowana genome, the genome of the Pirarucu is considerably smaller, with ∼60 Mb of difference in estimated genome size and 120 Mb in assembled genome size. However, the Pirarucu had more protein-coding genes identified (>2,000) and lower repeat content (21% against 27% in the Arowana) (Bian 2016).
Phylogenomic Analysis
Orthology inference was carried out with OrthoFinder (Emms and Kelly 2015) based on amino acid sequence data from 24 fish species (table 2). Due to the scarcity of genome data from Elopomorpha and Osteoglossomorpha species, available transcriptome data were also used for enriching these lineages and providing a better understanding of divergence between these taxa (supplementary table S5, Supplementary Material online). We identified 17,031 orthogroups, comprising a total of 630,993 genes, with 651 species-specific orthogroups and 1,436 orthogroups shared by all 24 fish species (supplementary table S6, Supplementary Material online). Following stringent procedures for identifying orthologs and excluding likely paralogs, 278 orthologous loci were found to be shared across species. Ortholog concatenation resulted in a supermatrix with 188,505 amino acid sites and overall occupancy of 88.5%. Tree topologies were inferred by maximum-likelihood (ML) and Bayesian inference (BI) with maximum values of bootstrap support and posterior probability for all nodes. Discordant arrangements were observed for the Cladistia and Chondrostei clades with ML supporting reedfishes as the sister lineage of all other Actinopterygii while BI placed reedfishes as the sister lineage of American paddlefishes (fig. 1). Evolutionary rates, based on number of amino acid substitutions per site, were found to be highly heterogeneous among teleost fishes (fig. 1; red numbers in brackets), indicating a rapid and divergent teleost evolution. The evolutionary rates of the Pirarucu were significantly different from all other teleost species, including the Asian arowana (P < 0.05; Tajima’s relative rate test, supplementary table S7, Supplementary Material online).
Table 2.
List of Species Included in Phylogenomic Analysis
| Organismsource | Scientific Name | Order | Reference |
|---|---|---|---|
| Ray-finned fish (Teleostei–Osteoglossomorpha) | |||
| Pirarucu* | Arapaima gigas | Osteoglossiformes | This study |
| Asian arowanaU | Scleropages formosus | Osteoglossiformes | Austin et al. (2015) |
| aFreshwater butterflyfishENA | Pantodon buchholzi | Osteoglossiformes | Pasquier et al. (2016) |
| aPeters’ elephantnose fishENA | Gnathonemus petersii | Osteoglossiformes | Pasquier et al. (2016) |
| Ray-finned fish (Teleostei–Elopomorpha) | |||
| European EelZ | Anguilla anguilla | Anguilliformes | Henkel et al. (2012) |
| aFalse morayENA | Kaupichthys hyoproroides | Anguilliformes | Gruber et al. (2015) |
| aIndo-Pacific tarponENA | Megalops cyprinoides | Elopiformes | Sun et al. (2016) |
| Ray-finned fish (Teleostei–Clupeocephala) | |||
| MedakaQFO | Oryzias latipes | Beloniformes | Kasahara et al. (2007) |
| Blind cave fishU | Astyanax mexicanus | Characiformes | McGaugh et al. (2014) |
| Nile tilapiaU | Oreochromis niloticus | Cichliformes | Brawand et al. (2014) |
| Common carpR | Cyprinus carpio | Cypriniformes | Xu et al. (2014) |
| ZebrafishQFO | Danio rerio | Cypriniformes | Howe et al. (2013) |
| Amazon mollyU | Poecilia formosa | Cyprinodontiformes | Unpublished |
| Southern platyfishU | Xiphophorus maculatus | Cyprinodontiformes | Schartl et al. (2013) |
| Atlantic codE | Gadus morhua | Gadiformes | Star et al. (2011) |
| Electric EelF | Electrophorus electricus | Gymnotiformes | Gallant et al. (2014) |
| Three-spined sticklebackU | Gasterosteus aculeatus | Perciformes | Jones et al. (2012) |
| Spotted green pufferfishU | Tetraodon nigroviridis | Tetraodontiformes | Jaillon et al. (2004) |
| FuguU | Takifugu rubripes | Tetraodontiformes | Kai et al. (2011) |
| Ray-finned fish (Holostei) | |||
| Spotted garQFO | Lepisosteus oculatus | Semionotiformes | Unpublished |
| Ray-finned fish (Chondrostei) | |||
| aAmerican paddlefishENA | Polyodon spathula | Acipenseriformes | Sun et al. (2016) |
| Ray-finned fish (Cladistia) | |||
| aReedfishENA | Erpetoichthys calabaricus | Polypteriformes | Sun et al. (2016) |
| Lobe-finned fish | |||
| CoelacanthU | Latimeria chalumnae | Coelacanthiformes | Unpublished |
| Cartilaginous fish | |||
| Elephant sharkR | Callorhinchus milii | Chimaeriformes | Venkatesh et al. (2014) |
Note.—Codes for source: Ensembl (E), efish genomics (F), Quest of Orthologs (QFO), RefSeq (R), EBI ENA (ENA), UniProt (U), ZF Genomics (Z), and this study (*).
Raw transcriptomics reads.
The branching order of the teleost superorders Osteoglossomorpha, Elopomorpha, and Clupeocephala has been controversial (Patterson and Rosen 1977; Le et al. 1993; Inoue et al. 2003; Near et al. 2012; Betancur-R 2013; Faircloth et al. 2013; Chen et al. 2015; Hughes et al. 2018). Our findings placed Osteoglossomorpha as a sister branch of Elopomorpha, both forming a monophyletic sister lineage with respect to Clupeocephala in a topology consistent with recent studies, suggesting a rapid, near-simultaneous emergence of teleost lineages (Bian 2016). This contradicts the current morphological view of basal teleost lineages, in which Osteoglossomorpha and Elopomorpha do not present identified synapomorphies, and the Elopomorpha is placed alone as the sister lineage to all other teleosts (Arratia 1997). Therefore, this might suggest a revaluation of morphological characters used to define these major teleost clades (Bian 2016), or a revaluation of phylogenetic assumptions by considering independent data sets or different hypothesis-testing procedures (Hughes et al. 2018).
The relationships within Osteoglossomorpha are also subject of controversy (Kumazawa and Nishida 2000; Hilton 2003; Lavoué and Sullivan 2004; Wilson and Murray 2008), and our findings showed the pantodontid freshwater butterflyfish (Pantodon buchholzi) as a sister lineage to all other Osteoglossiformes, while Mormyridae (represented by the Peters’ elephantnose fish) was placed as a sister branch of Osteoglossidae (Pirarucu and Asian arowana), in agreement with previous molecular studies (Lavoué and Sullivan 2004).
Estimation of Divergence Times
The divergence times of the ML topology were estimated with alignment data from 278 orthologous loci and calibration points (supplementary table S8, Supplementary Material online) obtained from the TimeTree database (Hedges et al. 2015) (fig. 2). Our findings were consistent with previous studies, including: 1) cartilaginous (chondrichthyes) and bony (Osteichthyes) fishes diverging at 450 Ma (Inoue et al. 2010; Dos Reis et al. 2015); 2) Actinopterygii and Sarcopterygii splitting ∼446 Ma (Patterson and Rosen 1977; Blair and Hedges 2005; Inoue et al. 2005; Azuma et al. 2008; Nakatani et al. 2011; Wei et al. 2014), and 3) teleosts emerging ∼245 Ma (Chen et al. 2013; Dornburg et al. 2014). Interestingly, in agreement with the proposed emergence of a monophyletic clade comprising Osteoglossomorpha and its sister Elopomorpha lineage, the divergence between these two superorders was estimated to have taken place 223 Ma, in the Late Triassic, with Osteoglossomorpha originating ∼187 Ma, in the Early Jurassic, and Elopomorpha almost 30 Myr after, in the Late Jurassic. These rapid cladogenic events occurring during this period (including the diversification of the Clupeocephala subgroups, Otomorpha and Euteleosteomorpha) might be attributed to the amelioration of restrictive environmental conditions ensuing periods of mass extinction (Broughton et al. 2013).
Whole Genome and Lineage-Specific Duplications among Teleosts
Events of whole genome duplication (WGD) are characterized by the occurrence of nondisjunction during meiosis that results in the duplication of the entire genome, including coding and noncoding regions like intronic and regulatory sequences. Along the history of vertebrates, at least two known WGD events occurred ∼500–600 Ma (Moriyama and Koshiba-Takeuchi 2018) while another event took place in the teleost lineage following divergence from land vertebrates, which is usually designated teleost-specific (TS) WGD or third round (3R) WGD (Glasauer and Neuhauss 2014). WGD events can be detected by estimating the number of synonymous substitutions per synonymous site (denoted as Ks, or ds). Since Ks is assumed to have remained constant throughout time, relict of WGDs are expected to be visualized by peaks in Ks distributions when comparing paralogous gene pairs (Lynch and Conery 2000; Zwaenepoel and Van de Peer 2017).
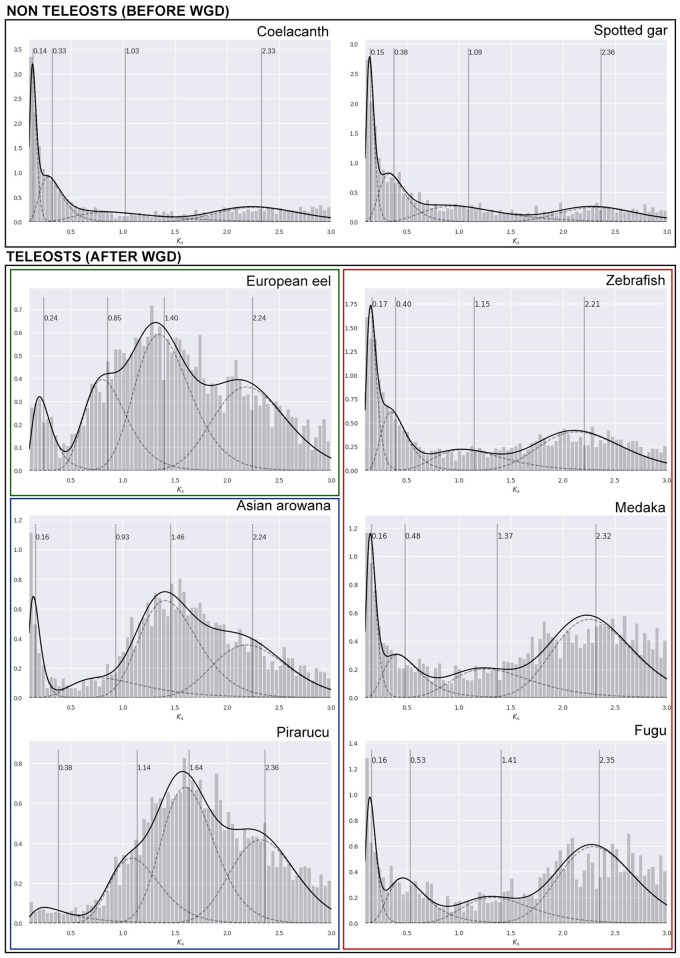
Analyses of Ks distributions of paralogous genes (paranome) of Pirarucu and other species in key branches of the phylogeny were analyzed before and after TS-WGD. Inferred paralogous families showed highly variable estimates across species, with fractions of outlying pairs (Ks > 5) ranging from 52% (Zebrafish) to 79% (Fugu) (supplementary table S9, Supplementary Material online). Peaks representing past duplication events were identified in teleost species, with evident differences in range between major teleost lineages (fig. 3). Osteoglossomorpha and Elopomorpha showed peaks with wide ranges around Ks=1.50, while Clupeocephala species showed higher peaks around Ks=2.30. Low Ks values indicate low mutational distances between duplicated genes, pointing to a recent evolutionary event (Lu et al. 2012). Previous studies suggested that peaks around Ks=2.30 and Ks=1.50 could be identified as remnants of the three major WGD events in teleost lineages (Vanneste et al. 2013). Differences in peak range across teleost groups may suggest a higher conservation of TS-WGD duplications in Osteoglossomorpha and Elopomorpha lineages than in Clupeocephala. Interestingly, an even more recent peak (Ks=0.85), with a wide range, was specifically observed in the European eel, which could support a recent hypothesis of a likely fourth WGD event in this species (Rozenfeld et al. 2017), however, we do not discard that such peak could be an artefact resulted from wrong fitting assumptions or due to sequencing or annotation faults. More research efforts should be directed at this topic to provide more reliable evidence.
Fig. 3.
—Empirical age distributions. Age distributions based on number of synonymous substitutions per synonymous site (Ks) estimated for paralogous gene families of each species. Distributions were modelled using a four component Gaussian mixture model (GMM). Solid black lines show mixture distributions, and dashed lines represent individual components. Vertical dashed lines correspond to the geometric mean of each component. Ks estimates (X axis) can be interpreted as age divergence between paralogous genes of a given species. The initial peak represents newly duplicated genes (usually derived from small-scale duplication events). Over time, duplications are eventually lost, and a decreasing slope is observed following the initial peak, outlining the steady decrease of retained duplicates. WGD events create distinct peaks to the distribution and can usually be observed as different components in a mixture distribution.
Evolution of Gene Families
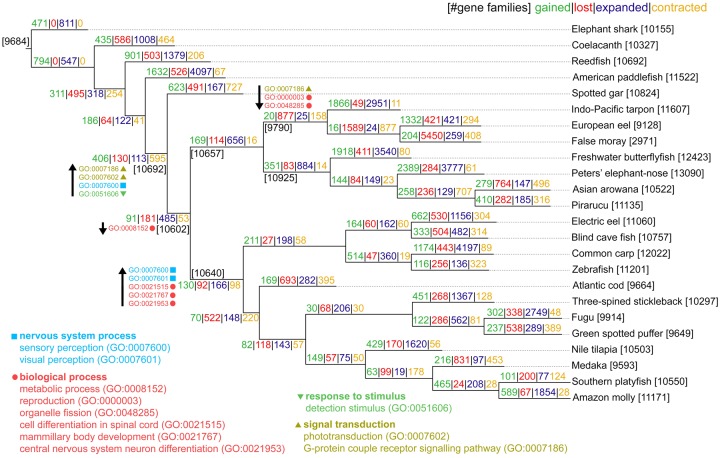
Gene gains and losses have been considered major sources of genomic variation and main drivers of phenotypic diversity (Ohno 1970; Zhang 2003). Based on homology data inferred with OrthoFinder, family gains and losses were mapped as discrete character-state changes. A parsimony approach was applied for inferring ancestral states and determining family gains, losses, expansions, and contraction events (fig. 4). In our inferred topology, 9,684 gene families were identified at the root of all sampled species, and 10,692 families were found in the Most Recent Common Ancestor (MRCA)—that is, the lowest common ancestor of two phylogenetic clades in an evolutionary tree—of Teleostei and Holostei. With respect to the MRCA of the three major teleost clades, Clupeocephala, Osteoglossomorpha, and Elopomorpha, 10,640, 10,925, and 9,790 gene families were respectively identified. Inference of ancestral gene duplication events revealed 485 gene family expansions in the MRCA of teleosts, followed by 656 expansions in the MRCA of Elopomorpha and Osteoglossomorpha, and 166 expansions in the MRCA of Clupeocephala. Osteoglossomorpha showed even further expansions (884 gene families) after diverging from Elopomorpha. Considerably losses were found in Elopomorpha, mainly in the Anguilliformes order (represented by the European eel and false moray) with 877 contractions, contrary to the Indo-Pacific tarpon (a representative of the Elopiformes order) with only 11.
Fig. 4.
—Reconstruction of gene family evolution. Events of gene family gains, losses, expansions, and contractions were inferred with Wagner parsimony. Number of families are indicated in black fonts near nodes. Gains (green numbers) indicate the number of families acquired along lineages leading to their respective MRCA node. Losses (red numbers) indicate lost families along lineages leading to their respective MRCA node. Expansions are indicated by numbers (in blue font) of expanded families (from size 1) and contractions by the number (in yellow font) of contracted families (to size 1) to their respective MRCA node. Gene Ontology (GO) terms associated with changes observed in key points of the phylogeny are shown near each node. GO enrichment was estimated based on Fisher’s exact test (FDR < 0.05) using, as background, population families present in each respective MRCA node. Arrows indicate terms associated to gains and/or expansions (upward) and losses and/or contractions (downward).
Analyses of the biological role of gene families that have gone through evolutionary change were carried out by assigning Gene Ontology (GO) terms at each major teleost group (fig. 4). Significant, enriched (FDR adjusted Fisher’s exact test; P < 0.05) GO terms of gained and expanded families in the MRCA of Teleostei and Holostei pointed to the G-protein couple receptor signalling pathway (GO: 0007186), phototransduction (GO: 0007602), sensory perception (GO: 0007600), and detection stimulus (GO: 0051606). At the teleost’s MRCA, enriched terms associated to gained or expanded families were not found, although terms related to metabolic processes (GO: 0008152) were enriched for lost families respective to the MRCA node. In Elopomorpha, considerably losses of functions related to G-protein couple receptor signalling pathway (GO: 0007186), reproduction (GO: 0000003), and organelle fission (GO: 0048285) were observed. In Clupeocephala, terms associated to sensory perception (GO: 0007600) and visual perception (GO: 0007601) were found to be enriched in gained and expanded families, while terms of cell differentiation in spinal cord (GO: 0021515), mammillary body development (GO: 0021767), and central nervous system neuron differentiation (GO: 0021953) were significant. With respect to Osteoglossomorpha, no significantly enriched terms were found in association with gained/expanded or lost/contracted families.
Likelihood Estimation of Gene Family Evolutionary Rates
The birth-and-death evolutionary model has been observed in several gene families, including sensory receptor and immune systems genes (Demuth and Hahn 2009; Innan and Kondrashov 2010). Using a ML framework of birth-and-death models, we carried out estimations of gain and loss rates across the tree. To account for potential erroneous gene number estimations, usually derived from low-quality genome assemblies or transcriptome-only data, we estimated the global error in the data set. We found that ∼14% (ɛ = 0.1417) of size groups estimates at the tree tips were prone to errors. The average rate of gene gain (λ) and loss (µ) was estimated for the 16,402 orthogroups with at least one representative teleost-species (supplementary table S10, Supplementary Material online). This was carried out for each group, namely teleosts and nonteleosts. The estimated rates of gene turnover of nonteleosts showed gains (λ0=0.0031), accounting for duplications/gene/Ma and losses (μ0=0.0016) for losses/gene/Ma. In teleosts, a more balanced gain/loss rates were observed, with λ1=0.0029 and µ1=0.0028 (supplementary table S10, Supplementary Material online; two-lambdamu model with global error correction).
Evidence of accelerated evolution was also inferred for some gene families based on the probability of any gene family of evolving under a birth-and-death process. Among the 16,402 gene families, 758 were found to be unlikely evolving under a random gain and loss process (P < 0.01; supplementary tables S11 and S12, Supplementary Material online). Of these, 714 were found at the tips of the topology, four shared among all teleosts, eight in at least two Osteoglossomorpha species, 13 shared by Anguilliformes, and a total of 19 rapidly evolving families in different branches of Clupeocephala (supplementary fig. S2, Supplementary Material online).
Sex-Specific Genomic Regions
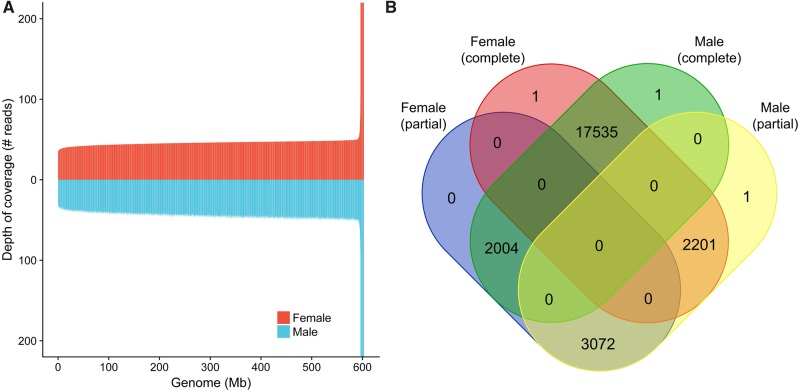
Studies on the mechanism of sex determination in fishes are important for preservation and commercial purposes. The Pirarucu does not show evident sexual dimorphism, and adults can only be reliably sexed during the reproductive phase (Chu-Koo et al. 2009). Previous karyotypic studies failed to identify chromosome differences between sexes, suggesting a nonchromosomal system of sex determination or recent loss of the sex-determining locus (SDL) in a carrier chromosome (Almeida et al. 2013). We herewith carried out computational analyses of genomic data from both sexes to identify potential genetic differences. Comparisons of sex-specific sequencing reads against the main genome assembly (built with data from both sexes) allowed for an initial evaluation of read depths across the genome. Female and male reads covered, with at least one read depth per base, 99.88% and 99.87% of the 661-Mb assembly, respectively. The average read depth per base over the whole genome equalled 55 female reads and 59 male reads without evident large regions of different depth coverage (fig. 5A).
Fig. 5.
—Comparison of sex-specific sequences and assemblies. Samples from each sex were compared against the main genome assembly containing data from both sexes. (A) Depth of coverage (i.e., average number of reads mapped to a specific region) of female (in red) and male (in blue) reads compared with the main assembly. Coverage was estimated for windows of 50 kb using Mosdepth (Pedersen and Quinlan 2018) and regions were ordered by female coverage estimates (ascending, left to right). Genome scaffolds with <50 kb were not included in the plot. Y axis was restricted to 200 for better visualization. (B) Number of complete and partial genes identified in each sex-specific assembly.
Comparative analysis of male and female genomes, carried out between sex-specific genome assemblies, showed minimal differences. Despite having higher contig N50 values and a slightly higher genome size, the female assembly was more fragmented, with 8,324 contigs and scaffold N50 of 295 kb than male reads, with 6,058 contigs and scaffold N50 of 471 kb (table 3). Comparisons of sex-specific assemblies against the main (mixed) assembly showed similar results, with both assemblies presenting 99.7% of aligned bases to the reference genome (genome fraction). The female assembly showed fewer misassemblies and longer continuous alignment with the reference genome than the male assembly despite being more fragmented. Considering the predicted gene regions using the main assembly as a reference, 19,737 complete genes and 5,076 partial ones were found in the female assembly, while 19,540 complete and 5,274 partial genes were found in the male assembly (fig. 5B). This suggested ∼2,000 specific genes for each sex, if only complete genes were considered. However, as a complete gene in one sex may be a partial gene in the opposite sex, these differences might be actually due to misassemblies. Functional analysis did not find significantly enriched GO terms in either set of sex-specific genes.
Table 3.
Comparison of Sex-Specific Genome Assemblies
| Genome Features | Arapaima gigas (female) | Arapaima gigas (male) |
|---|---|---|
| Assembled genome size (Mb) | 660.71 | 660.43 |
| Genome fraction (%) | 99.74 | 99.73 |
| # scaffolds | 8,324 | 6,058 |
| Scaffold N50 (kb) | 295 | 471 |
| Contig N50 (kb) | 40.75 | 35.19 |
| Largest scaffold (bp) | 2,179,931 | 2,199,363 |
| Largest alignment to the reference (bp) | 2,178,748 | 1,935,564 |
| GC (%) | 43.18 | 43.18 |
| # misassemblies | 2,041 | 2,342 |
| # complete genes | 19,737 | 19,540 |
| # partial genes | 5,076 | 5,274 |
| Number of sex-specific bases in assembly (bp) | 103,749 (0.0157%) | 64,323 (0.0097%) |
| Longest sex-specific sequence length (bp) | 2,881 | 1,658 |
In order to map sex-specific regions in each assembly, we mapped sex-specific reads to the opposite sex-specific assembly. We found that sex-specific regions were minimal, accounting for ∼0.01% of total base pairs and with continuous sequences as long as 2,881 and 1,658 bp in the female and the male, respectively (table 3). Comparison of sex-specific regions against the NR protein database showed inconclusive results, with few regions indicating coding potential, many of which with similarities to hypothetical or partial proteins (supplementary file S1, Supplementary Material online).
Conclusions
We report the first draft of the genome of Arapaima gigas, a fascinating Osteoglossomorpha species of bony-tongued fishes. The final draft assembly comprised ∼661.3 Mb, accounting for 86.9% of the estimated genome size (761.1 Mb). We also predicted 24,655 protein-coding genes from the generated assembly. Our phylogenomic analysis supported the postulation that Osteoglossomorpha and Elopomorpha are sister groups that diverged in a short period of time during the Jurassic. The data and resources produced in this study will be valuable to future analysis to understand the species evolutionary history, its breeding mechanism, and its large size. Additionally, these findings might contribute to the environmental protection of this ancient teleost species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Rede de Pesquisa em Genômica Populacional Humana (RPGPH)—3381/2013 CAPES-BioComputacional; FADESP/PROPESP/UFPA (Universidade Federal do Pará); and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). It was also supported by CNPq/Produtividade grant 304413/2015-1 to A.R.S. and CNPq/Produtividade grant 305258/2013-3 to S.S. The funders had no role in the study design, data collection and analysis, decision to publish or manuscript preparation. The authors declare that they have no competing interests.
Author Contributions
S.S., S.J.S., A.R.S., P.P.A., E.H.C.O., and I.G.H. designed the study. J.E.S.S. and P.A.A.S. performed sequencing quality control and assembled the genome. R.A.V. performed the genome annotation. R.A.V., K.P.L., A.M.R.S., and F.A.O.S. analyzed the data. R.A.V., T.S., D.G.T., and J.P.M.S.L. performed the evolutionary analysis. C.F. performed the DNA sequencing. R.A.V., H.N.S., K.P.L., and S.J.S. wrote the manuscript. All authors participated in discussions and provided advice. All authors read and approved the final manuscript.
Literature Cited
- Almeida IG, Ianella P, Faria MT, Paiva SR, Caetano AR.. 2013. Bulked segregant analysis of the pirarucu (Arapaima gigas) genome for identification of sex-specific molecular markers. Genet Mol Res. 124:6299–6308. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data [cited 2017 Dec 12]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Arratia G. 1997. Basal teleosts and teleostean phylogeny. Palaeo Ichthyol. 7:5–168. [Google Scholar]
- Austin CM, Tan MH, Croft LJ, Hammer MP, Gan HM.. 2015. Whole genome sequencing of the Asian arowana (Scleropages formosus) provides insights into the evolution of ray-finned fishes. Genome Biol Evol. 710:2885–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Kumazawa Y, Miya M, Mabuchi K, Nishida M.. 2008. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol Biol. 8:215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley PB, Petrere M.. 1989. Amazon fisheries: assessment methods, current status and management options. Can Spec Publ Fish Aquat Sci. 106:385–398. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 272:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb 165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C. 2016. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci Rep. 6:24501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Hedges SB.. 2005. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2211:2275–2284. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 104:e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner CJ, Matey V, Wilson JM, Bernier NJ, Val AL.. 2004. Transition in organ function during the evolution of air-breathing; insights from Arapaima gigas, an obligate air-breathing teleost from the Amazon. J Exp Biol. 2079:1433–1438. [DOI] [PubMed] [Google Scholar]
- Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 5137518:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton RE, Betancur-R R, Li C, Arratia G, Ortí G.. 2013. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLOS Curr. doi: 10.1371/currents.tol.2ca8041495ffafd0c9 2756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson D.. 2015. Fast and sensitive alignment using DIAMOND. Nat Methods 121:59–60. [DOI] [PubMed] [Google Scholar]
- Burnie D, Wilson D.. 2001. Animal: the definitive visual guide to the world’s wildlife. London & New York: DK Publishers. [Google Scholar]
- Chen MY, Liang D, Zhang P.. 2015. Selecting question-specific genes to reduce incongruence in phylogenomics: a case study of jawed vertebrate backbone phylogeny. Syst Biol. 646:1104–1120. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Lavoué S, Mayden RL.. 2013. Evolutionary origin and early biogeography of otophysan fishes (Ostariophysi: teleostei). Evolution 678:2218–2239. [DOI] [PubMed] [Google Scholar]
- Chu-Koo F, et al. 2009. Gender determination in the Paiche or Pirarucu (Arapaima gigas) using plasma vitellogenin, 17beta-estradiol, and 11-ketotestosterone levels. Fish Physiol Biochem. 351:125–136. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21(18):3674–3676. [DOI] [PubMed]
- Csurös M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 2615:1910–1912. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW.. 2009. The life and death of gene families. Bioessays 311:29–39. [DOI] [PubMed] [Google Scholar]
- Dornburg A, Townsend JP, Friedman M, Near TJ.. 2014. Phylogenetic informativeness reconciles ray-finned fish molecular divergence times. BMC Evol Biol. 14:169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, et al. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 2522:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 307:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC, Sorenson L, Santini F, Alfaro ME.. 2013. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS One 86:e65923.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese R, Pauly D, eds. 2018. FishBase. World Wide Web electronic publication [cited 06/2018]. Available from: http://www.fishbase.org/.
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2823:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JR, et al. 2014. Genomic basis for the convergent evolution of electric organs. Science 3446191:1522–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer SMK, Neuhauss SCF.. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 2896:1045.. [DOI] [PubMed] [Google Scholar]
- Godinho HP, Santos JE, Formagio PS, Guimarães-Cruz RJ.. 2005. Gonadal morphology and reproductive traits of the Amazonian fish Arapaima gigas (Schinz, 1822). Acta Zool. 864:289–294. [Google Scholar]
- Greenwood PH. 1970. On the genus Lycoptera and its relationship with the family Hiodontidae (Pisces, Osteoglossomorpha). Bull Br Museum Nat Hist Zool. 19:257–285. [Google Scholar]
- Gruber DF, et al. 2015. Adaptive evolution of eel fluorescent proteins from fatty acid binding proteins produces bright fluorescence in the marine environment. PLoS One 1011:e0140972.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 298:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 88:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFÉ 3. Mol Biol Evol. 308:1987–1997. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 324:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel CV, et al. 2012. Primitive duplicate Hox clusters in the European eel’s genome. PLoS One 72:e32231.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton EJ. 2003. Comparative osteology and phylogenetic systematics of fossil and living bony-tongue fishes (Actinopterygii, Teleostei, Osteoglossomorpha). Zool J Linn Soc. 1371:1–100. [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 4967446:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrbek T, Crossa M, Farias IP.. 2007. Conservation strategies for Arapaima gigas (Schinz, 1822) and the Amazonian várzea ecosystem. Braz J Biol. 67(4 Suppl):909–917. [DOI] [PubMed] [Google Scholar]
- Hrbek T, et al. 2005. Population genetic analysis of Arapaima gigas, one of the largest freshwater fishes of the Amazon basin: implications for its conservation. Anim Conserv. 83:297–308. [Google Scholar]
- Hughes LC, et al. 2018. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci U S A. 11524:6248–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 112:97–108. [DOI] [PubMed] [Google Scholar]
- Inoue JG, et al. 2010. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 2711:2576–2586. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Tsukamoto K, Nishida M.. 2003. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the “ancient fish”. Mol Phylogenet Evol. 261:110–120. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Venkatesh B, Nishida M.. 2005. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: coelacanthiformes) and divergence time estimation between the two coelacanths. Gene 349:227–235. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 4317011:946–957. [DOI] [PubMed] [Google Scholar]
- Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 4847392:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 309:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NA, Fass JN.. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files [cited 2017 Dec 12]. Available from: https://github.com/najoshi/sickle.
- Kai W, et al. 2011. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol Evol. 3:424–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 4477145:714–719. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa Y, Nishida M.. 2000. Molecular phylogeny of osteoglossoids: a new model for Gondwanian origin and plate tectonic transportation of the Asian arowana. Mol Biol Evol. 1712:1869–1878. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Hayashizaki Y, Daub CO.. 2011. SAMStat: monitoring biases in next generation sequencing data. Bioinformatics 271:130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoué S, Sullivan JP.. 2004. Simultaneous analysis of five molecular markers provides a well-supported phylogenetic hypothesis for the living bony-tongue fishes (Osteoglossomorpha: Teleostei). Mol Phylogenet Evol. 331:171–185. [DOI] [PubMed] [Google Scholar]
- Le HL, Lecointre G, Perasso R.. 1993. A 28S rRNA-based phylogeny of the gnathostomes: first steps in the analysis of conflict and congruence with morphologically based cladograms. Mol Phylogenet Evol. 21:31–51. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M.. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 3320:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Peatman E, Tang H, Lewis J, Liu Z.. 2012. Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genomics 13:246.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 11:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 2905494:1151–1155. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 276:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden PD. 1994. Letter from Brasilia: some primitive peoples of the tropics. BMJ 3086936:1095–1096. [Google Scholar]
- McGaugh SE, et al. 2014. The cavefish genome reveals candidate genes for eye loss. Nat Commun. 5:5307.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama Y, Koshiba-Takeuchi K.. 2018. Significance of whole-genome duplications on the emergence of evolutionary novelties. Briefings in Functional Genomics, https://doi.org/10.1093/bfgp/ely007. [DOI] [PubMed] [Google Scholar]
- Nakatani M, Miya M, Mabuchi K, Saitoh K, Nishida M.. 2011. Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: pangaean origin and Mesozoic radiation. BMC Evol Biol. 111:177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, et al. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. 10934:13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JS. 1994. Fishes of the world. New York: John Wiley & Sons. [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer. [Google Scholar]
- Ou S, Jiang N.. 2017. LTR_retriever: a highly accurate and sensitive program for identification of LTR retrotransposons. bioRxiv 137141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier J, et al. 2016. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17:368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C, Rosen DE.. 1977. Review of Ichthyodectiform and other Mesozoic teleost fishes and the theory and practice of classifying fossils. Bull Am Mus Nat Hist. 158:81–172. [Google Scholar]
- Pedersen BS, Quinlan AR.. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 345:867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 53:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Gabaldón T.. 2016. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 4412:e113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenfeld C, Blanca J, Gallego V.. 2017. Large scale gene duplication affected the European eel (Anguilla anguilla) after the 3R teleost duplication. bioRxiv. doi: http://dx.doi.org/10.1101/232918.
- Sanford CP, Lauder GV.. 1990. Kinematics of the tongue-bite apparatus in osteoglossomorph fishes. J Exp Biol. 154:137–162. [Google Scholar]
- Schartl M, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 455:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shujun Ou Ning Jiang. 2018. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiology 1762:1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R.. 2008. RepeatModeler Open-1.0 [cited 2017 Dec 12]. Available from: http://www.repeatmasker.org/RepeatModeler/.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 245:637–644. [DOI] [PubMed] [Google Scholar]
- Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 4777363:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, et al. 2016. Fish-T1K (Transcriptomes of 1,000 Fishes) Project: large-scale transcriptome data for fish evolution studies. Gigascience 5:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing molecular clock hypothesis. Genetics 1352:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 564:564–577. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 25:4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Van de Peer Y, Maere S.. 2013. Inference of genome duplications from age distributions revisited. Mol Biol Evol. 301:177–190. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 5057482:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Sun Y, Zhang B, Wang R, Xu T.. 2014. A mitogenomic perspective on the phylogenetic position of the Hapalogenys genus (Acanthopterygii: perciformes) and the evolutionary origin of Perciformes. PLoS One 97:e103011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnstekers W. 2011. The convention on international trade in endangered species of wild fauna and flora (CITES) – 35 years of global efforts to ensure that international trade in wild animals and plants is legal and sustainable. Forensic Sci Rev. 231:1–8. [PubMed] [Google Scholar]
- Wilson MVH, Murray AM.. 2008. Osteoglossomorpha: phylogeny, biogeography, and fossil record and the significance of key African and Chinese fossil taxa. Geol Soc Lond Spec Publ. 2951:185–219. [Google Scholar]
- Xu P, et al. 2014. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet. 4611:1212–1219. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H.. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server issue):W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Smith SA.. 2014. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol. 3111:3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang JZ. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 186:292–298. [Google Scholar]
- Zwaenepoel A, Van de Peer Y.. 2017. Cedalion: developing a highly efficient computational framework for the discovery of evolutionary and ecological adaptations in plants. Universiteit Gent. Faculteit Wetenschappen.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.