Abstract
Biomarkers for a more precise patient care are needed in metastatic prostate cancer (mPC). We have reported a Phase II trial (TOPARP-A) of the poly(ADP)-ribose polymerase (PARP) inhibitor olaparib in mPC, demonstrating antitumor activity associating with homologous recombination DNA repair defects. We now report targeted and whole exome sequencing of serial circulating-free DNA (cfDNA) samples collected during this trial. Decreases in cfDNA concentration independently associated with outcome in multivariable analyses (HR for overall survival at week 8: 0.19; 95%CI 0.06-0.56 p=0.003). All tumor tissue somatic DNA repair mutations were detectable in cfDNA; allele frequency of somatic mutations decreased selectively in responding patients (Chi-squared p<0.001). At disease progression, following response to olaparib, multiple sub-clonal aberrations reverting germline and somatic DNA repair mutations (BRCA2, PALB2) back in frame emerged as mechanisms of resistance. These data support the role of liquid biopsies as predictive, prognostic, response and resistance biomarker in mPC.
Keywords: prostate cancer, DNA repair, PARP inhibitor, circulating DNA, drug resistance
Introduction
Prostate cancer is a major cause of male cancer mortality globally. Studies indicate substantial inter- and intra-patient genomic heterogeneity although treatment to date has not incorporated molecular stratification. (1–7) Clinical qualification of predictive biomarkers is a major need for these invariably lethal tumors. We previously reported that 20-30% of lethal prostate cancers have deleterious aberrations in genes involved in DNA repair by homologous recombination (homologous recombination deficiency, HRD), including BRCA2, ATM, BRCA1, PALB2, FANCA, CHEK2 and CDK12. (4) Studies indicate that HRD-associated mutations associate with a worse prognosis, with these aberrations being inherited in approximately 10-12% of men with lethal prostate cancer. (8,9)
We also recently reported an investigator-initiated Phase II clinical trial (TOPARP-A) of the poly(ADP)ribose polymerase (PARP) inhibitor olaparib (Lynparza) in metastatic prostate cancer patients, describing antitumour activity associating with HRD. (10) These data led to olaparib being given ‘Breakthrough Designation’ by the Food and Drug Administration (FDA) for advanced prostate cancer associated to BRCA2/ATM defects, and registration trials of different PARP inhibitors being currently pursued.(11–13) A major regulatory approval challenge for these drug development efforts remains the lack of proven surrogates of survival benefit. We hypothesized that circulating biomarkers can enhance drug development and patient care, informing on treatment response and resistance. Response biomarkers are crucial to improving the care of advanced prostate cancer, which is often characterized by metastatic disease only to bone, which is not easily evaluable.(14) Circulating free DNA (cfDNA) in blood, acquired from plasma by a simple blood test, provides repeated serial access to tumor DNA as a minimally invasive ‘liquid biopsy’. (15–19)
We report here pre-planned cfDNA analyses from serial blood samples from the TOPARP-A trial. We had hypothesized that cfDNA analyses can enhance drug development and patient care and that: 1) Changes in cfDNA concentrations in response to treatment would be prognostic; 2) Next generation sequencing of cfDNA would detect the predictive HRD-associated mutations found in tumor biopsies; 3) Changes in cfDNA somatic mutation allele frequency on therapy would associate with response; and 4) Serial cfDNA analyses could detect the evolution of resistant sub-clones at disease progression.
Results
Patient Characteristics and Blood Sample Disposition
Fifty patients were treated on TOPARP-A; 49 were evaluable for response with 16 patients responding to treatment. Overall, 16 of these 49 patients had prostate cancers with a deleterious aberration in homologous recombination DNA repair, with 14 of these patients responding to treatment. Serial samples for cfDNA studies were available for 46/49 (94%) patients. We now report on these analyses utilizing updated outcome data based on a data snapshot by 24/05/2016. One patient with a BRCA2 homozygous deletion remained on therapy and free of progression at the data cut-off, after 22 months of therapy. Overall, the median cfDNA baseline concentration across the trial population was 31.6 ng/ml (IQR 19.4-57.1). Next-generation targeted sequencing of cfDNA was successful for 43/46 (93%) patients. (Supplementary Figure S1)
Prognostic Relevance of Changes in cfDNA Concentration Following PARP Inhibition
Changes in cfDNA concentration were evaluated in patients responding (n=16), or not responding (n=30), to olaparib. After 4-weeks of therapy, there was a median -51.4% change in responders (IQR -72.6, -29.5%) and a median -33.4% change in non-responders (IQR -52.3, +5.5%) (p=0.07). After 8-weeks of therapy, responders continued to experience sustained declines (median -49.6% change; IQR -76.5, -20.4%), differing significantly from non-responders (median +2.1% increase; IQR -43.6, +57.8%) (p=0.006). (Supplementary Figure S2)
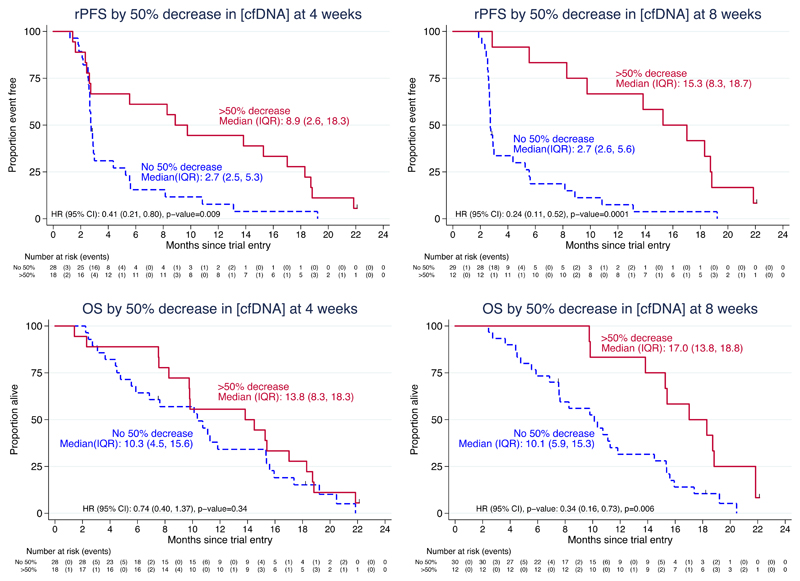
Next, we explored how declines in cfDNA concentrations correlated with patient outcome based on radiological progression-free survival (rPFS) and overall survival (OS). cfDNA concentration falls robustly correlated with rPFS as early as after 4-weeks of therapy (HR 1.70, 95%CI 1.13 to 2.55, p=0.01 for cfDNA log-fold change; HR 0.41, 95%CI 0.21 to 0.80, p=0.009 for absence/presence of ≥50% fall from baseline of cfDNA concentration). cfDNA concentration falls after 8-weeks of olaparib correlated with both prolonged rPFS and OS. (Figure 1)
Figure 1.
Kaplan-Meier plots showing differences in radiological progression-free survival (rPFS) and overall survival (OS) based on the presence or absence of a ≥50% fall in total cfDNA concentration after 4- and after 8-weeks of therapy with olaparib.
In multivariable analyses including established prognostic factors such as LDH and CTC count conversions, a ≥50% decline in cfDNA concentration after 4-weeks of olaparib was independently associated with longer rPFS. These changes at 4-weeks were not statistically significant for overall survival (HR 0.47; 95%CI 0.18-1.07; p=0.07). Nevertheless, a ≥50% fall in cfDNA concentration after 8-weeks of therapy was independently associated with longer overall survival (HR for ≥50% cfDNA decline and overall survival in multivariable analyses: 0.19; 95%CI 0.06-0.56; p=0.003). (Table 1)
Table 1.
Univariate and multivariable Cox regression analyses of the association cfDNA plasma concentrations (ng/mL) with patient outcome including established prognostic factors. (* indicates when proportional hazards assumption is not met)
| rPFS | Overall Survival | |||||
| Univariate analyses | N | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| 4 weeks | CTC conversion | 47 | 0.30 (0.14, 0.65) | 0.002 | 0.53 (0.27, 1.04) | 0.07 |
| ≥30% CTC decline | 47 | 0.33 (0.15, 0.74) | 0.007 | 0.86 (0.42, 1.74) | 0.67 | |
| cfDNA [c] log-fold change | 46 | 1.70 (1.13, 2.55) | 0.01 | 1.30 (0.87, 1.95) | 0.19 | |
| ≥30% cfDNA [c] decline | 46 | 0.61 (0.33, 1.14) | 0.12 | 0.70 (0.37, 1.32) | 0.27 | |
| ≥50% cfDNA [c] decline | 46 | 0.41 (0.21, 0.80) | 0.009 | 0.74 (0.40, 1.37) | 0.34 | |
| 8 weeks | CTC conversion | 41 | 0.38 (0.19, 0.74) | 0.005* | 0.74 (0.39, 1.42) | 0.37* |
| ≥30% CTC decline | 41 | 0.61 (0.31, 1.19) | 0.15 | 1.21 (0.62, 2.37) | 0.58 | |
| cfDNA [c] log-fold change | 42 | 1.83 (1.22, 2.74) | 0.003 | 1.43 (0.98, 2.09) | 0.06 | |
| ≥30% cfDNA [c] decline | 42 | 0.48 (0.25, 0.92) | 0.028 | 0.66 (0.34, 1.26) | 0.20 | |
| ≥50% cfDNA [c] decline | 42 | 0.24 (0.11, 0.52) | 0.0001 | 0.34 (0.16, 0.73) | 0.006 | |
| Multivariate analyses | HR (95% CI) | p-value | HR (95% CI) | p-value | ||
| 4 weeks | LDH | 1.00 (1.00, 1.00) | 0.058 | 1.00 (1.00, 1.00) | 0.012 | |
| ECOG status 2 (vs 0-1) | 1.48 (0.50, 4.38) | 0.48 | 1.74 (0.60, 5.06) | 0.31 | ||
| Radiological progression at trial entry (vs PSA progression only) | 1.01 (0.42, 2.43) | 0.98 | 0.20 (0.07, 0.52) | 0.001 | ||
| Measurable disease at trial entry | 0.32 (0.13, 0.79) | 0.014 | 0.50 (0.20, 1.29) | 0.154 | ||
| Baseline CTC | 1.00 (0.99, 1.00) | 0.20 | 1.00 (1.00, 1.00) | 0.34 | ||
| Baseline cfDNA | 1.00 (1.00, 1.01) | 0.54 | 1.00 (1.00, 1.01) | 0.23 | ||
| CTC conversion | 0.19 (0.07, 0.05) | 0.001 | 0.41 (0.17, 1.01) | 0.051 | ||
| ≥50% cfDNA [c] decline | 0.25 (0.09, 0.66) | 0.005 | 0.44 (0.18, 1.07) | 0.071 | ||
| 8 weeks | LDH | 1.00 (1.00, 1.01) | 0.036 | 1.00 (1.00, 1.01) | <0.001 | |
| ECOG status 2 (vs 0-1) | 2.45 (0.73, 8.24) | 0.15 | 2.55 (0.81, 7.98) | 0.11 | ||
| Radiological progression at trial entry (vs PSA progression only) | 1.37 (0.47, 4.00) | 0.57 | 0.24 (0.08, 0.72) | 0.011 | ||
| Measurable disease at trial entry | 0.49 (0.18, 1.31) | 0.15 | 0.53 (0.19, 1.50) | 0.24 | ||
| Baseline CTC | 1.00 (1.00, 1.00) | 0.83 | 1.00 (0.99, 1.00) | 0.24 | ||
| Baseline cfDNA | 1.00 (0.99, 1.00) | 0.32 | 1.00 (0.99, 1.01) | 0.89 | ||
| CTC conversion | 0.10 (0.03, 0.28) | <0.001 | 0.35 (0.15, 0.85) | 0.020 | ||
| ≥50% cfDNA [c] decline | 0.09 (0.03, 0.30) | <0.001 | 0.19 (0.06, 0.56) | 0.003 | ||
Changes in Allele Frequency of Somatic Mutations
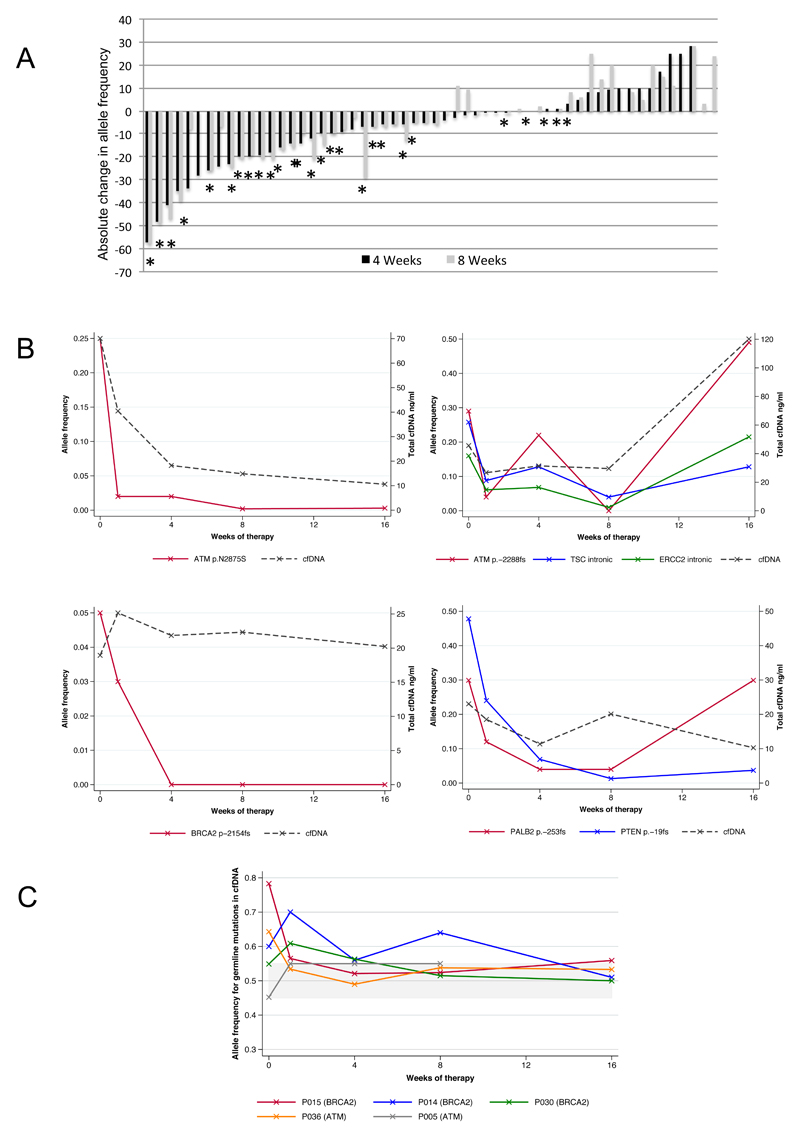
We then performed next-generation targeted sequencing of serial cfDNA samples to assess allele frequency of somatic mutations during PARP inhibitor therapy as an indirect estimate of tumor burden. A total of 254 plasma samples were analysed. We detected somatic mutations in cfDNA of 33/43 baseline samples. Olaparib treatment led to sustained decreases in cfDNA mutation allele frequencies in responding patients; sustained (≥8-weeks) falls in cfDNA somatic mutation allele frequencies were not observed in non-responding patients, although 3/29 detected somatic events in non-responding patients decreased transiently after 4-weeks of olaparib (Chi-squared p<0.001; Figure 2A)
Figure 2.
A) Waterfall plot summarizing change in allele frequency (AF) (defined as allele frequency on treatment subtracted by the allele frequency at baseline) after 4- and 8-weeks of therapy. Stars indicate patients considered responders to olaparib in the TOPARP-A trial in the predefined primary endpoint. An absolute decrease of ≥10% in allele frequency was observed in 18/27 somatic mutations detected in responding patients as compared with 3/29 somatic events monitored in non-responding patients (Chi-squared p<0.001). None of the three allele frequency falls in non-responding patients were maintained after 8-weeks of therapy. B) Four examples of how AF of somatic HRD-associated mutations decrease in response to therapy, in parallel with decreases in total cfDNA concentrations. A patient with an ATM p.-2288fs mutation had intermittent increases and decreases in AF in parallel to drug interruptions due to haematological toxicity. C) Changes in cfDNA mutation allele frequency over time in germline deleterious mutation carriers (BRCA2 and ATM) in five patients from the TOPARP-A trial. Those patients with LOH at baseline have the cfDNA mutation allele frequency trending towards 50% in response to therapy, probably due to elimination of the tumor clone. This is not seen in the serial cfDNA samples from the patient whose tumor did not have LOH.
Somatic HRD-Associated Mutations in cfDNA
Overall, 6 subjects in the TOPARP-A trial had tumors with somatic mutations likely to be associated with HRD (3 in ATM, 2 in BRCA2, 1 in PALB2; 5/6 responded to olaparib); all 6 mutations were detected in baseline cfDNA. In all 5 responding patients, these somatic mutation allele frequencies all decreased to <5% following olaparib treatment (Figure 2B). In the non-responding patient, the somatic mutation allele frequency remained unchanged at 4% throughout therapy.
Additionally, in one non-responding patient we detected two ATM frameshift mutations in cfDNA not previously detected in the tumor biopsy. Manual retrospective inspection of tumor data visualized both mutations in 3% of sequencing reads. We obtained a further metastatic biopsy from a different anatomic location after treatment; both of these ATM mutations were present in 18% and 29% of reads respectively. We also evaluated this patient’s primary prostate tumor biopsy taken at diagnosis; one of these mutations was not present and the other was visualized in 1% of reads. Overall, these data indicate that these were probably sub-clonal mutations.
Loss-of-Heterozygosity (LOH) in Germline Mutation Carriers
Five patients with pathogenic BRCA2 or ATM germline mutations responded to olaparib in the trial. In 4 of these, we had previously documented LOH for these pathogenic mutations in tumor biopsies. In these 4 patients, falls in the allele frequencies of these germline mutations towards 50% were observed as they responded to olaparib, suggesting elimination of the tumor clone with LOH. For the last patient with an ATM mutation, but no LOH in his tumor biopsy, allele frequencies remained between 45-55% in the cfDNA at all time points in concordance with the tumor tissue findings. (Figure 2C)
Mechanisms of PARP Inhibitor Resistance Detected in cfDNA
Ten of 16 patients having an initial tumor response to olaparib had cfDNA samples acquired at the time of resistance and disease progression. In addition to targeted sequencing, we successfully performed whole exome sequencing from paired plasma samples collected before olaparib treatment and at disease progression for 6 patients to study mechanisms of secondary resistance to PARP inhibition. In a seventh case we performed WES in the progression sample, and targeted sequencing in the baseline cfDNA sample, due to low DNA yield.
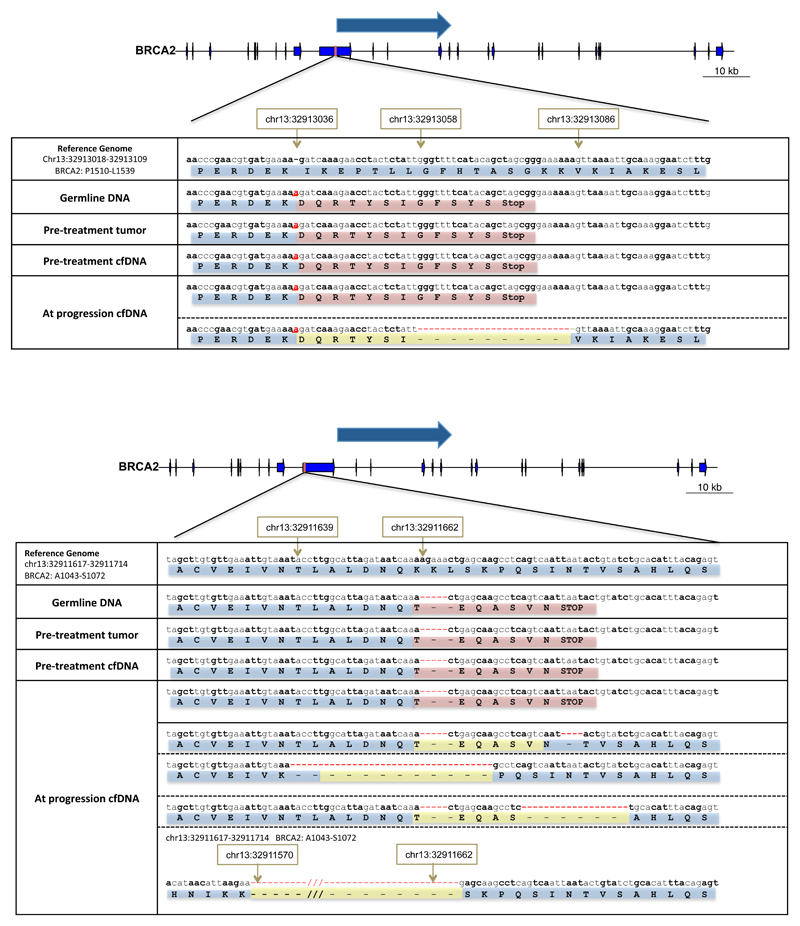
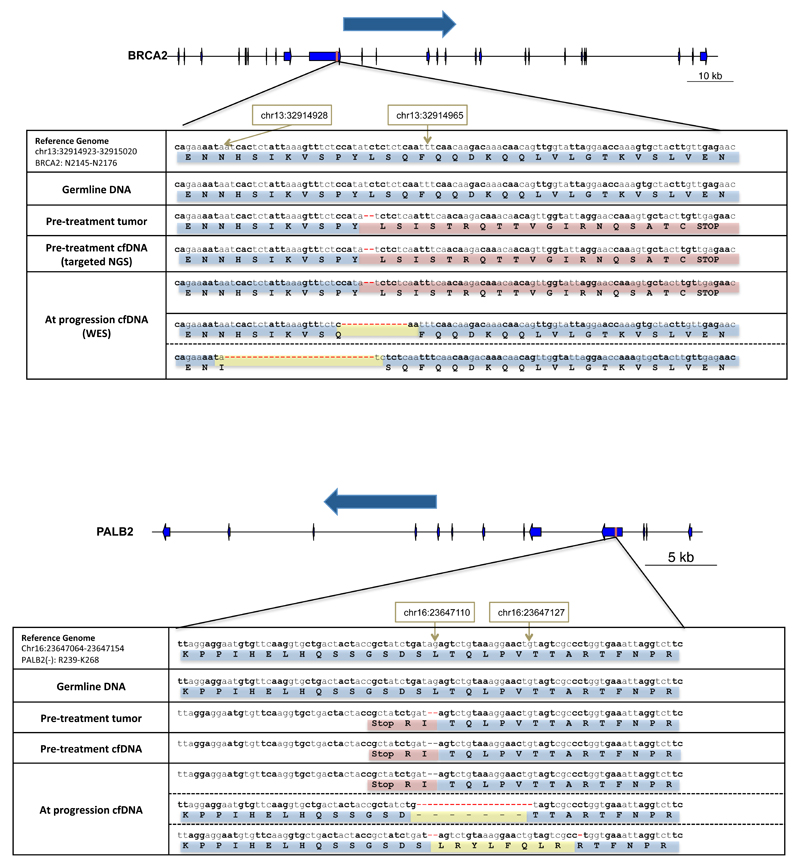
In both of the patients with a germline BRCA2 frameshift mutation, we identified at the time of tumor progression additional somatic BRCA2 mutations restoring the normal open reading frame (Figure 3).
Figure 3.
Visual representation of emerging de-novo mutations at progression that likely result in acquired drug resistance in two patients with germline BRCA2 mutations. In the top panel, a patient with a germline deleterious BRCA2 frameshift insertion that was present in both tumour and cfDNA at baseline presents at disease progression with a new frameshift deletion that restores the BRCA2 reading frame. In the lower panel, a second patient with a germline deleterious BRCA2 mutation is depicted; at progression cfDNA whole-exome sequencing identified multiple clones with different previously undetected mutations all resulting in reversion of the BRCA2 reading frame to normal.
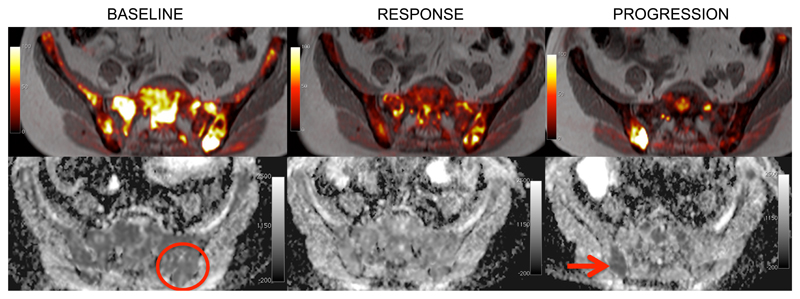
One of these patients had a germline BRCA2 p.-1056fs mutation and LOH in the pre-trial tumor biopsy. At the time of disease progression, a further somatic deletion of 4 base-pairs emerged, resulting in an overall in-frame change of 4 amino-acids (p.K1057_Q1063delinsTEQA). In parallel, 3 other events were detected in different positions in the same genomic region, all of them also restoring the BRCA2 normal open reading frame, and probably representing the coexistence of several resistant tumour sub-clones in cfDNA. This patient had bone metastases in the spine and pelvis; after an initial partial response to olaparib, he developed a heterogeneous pattern of progression after 9-months; a bone marrow biopsy of a relapse focus in the right hemipelvis was performed (Figure 4). In this tumor sample, only one of the three emerging somatic reversions (p.K1057_Q1063delinsTEQA) was detected; these data suggest these other emerging sub-clones may have originated in different metastases.
Figure 4.
Single site disease progression after 9-months of response to therapy in the right hemipelvis visualized by diffusion-weighted whole body MRI. The upper panels show fusion of the T1-weighted imaging and diffusion-weighted imaging. The bottom panels show apparent diffusion coefficient (ADC) maps. Areas of high signal on diffusion-weighted imaging and low ADC values indicate tumour bone marrow infiltration. The left panels depict the baseline MRI scan, showing diffuse tumour infiltration in the pelvic bone. The first trial biopsy was taken from the left iliac bone (red circle), and identified a deleterious BRCA2 mutation with LOH. After 12 weeks of therapy (middle panels) there was a major response to therapy reported. After 9-months (right panels) the MRI identified a focal area of tumour relapse in the right iliac bone, which was biopsied (red arrow). NGS of this biopsy confirmed a de-novo mutation in BRCA2 restoring the open reading frame.
A second germline BRCA2 mutation carrier (p.E1514fs*15) developed an additional deletion of 28-bp at progression reverting BRCA2 back in frame. We confirmed the emergence of this new event independently by targeted next generation sequencing (MiSeq). Additionally, we also detected the emergence at progression of a somatic ARID1A mutation (p.Q1145*). These aberrations again indicate possible divergent clonal evolutionary resistance mechanisms as a result of PARP inhibition generated selective pressures.
In non-germline mutation carriers, we also identified new emerging genomic events at progression leading to the reversion of a somatic BRCA2 mutation (Figure 5A). A somatic 2 base-pair frameshift deletion was detected in WES of the original tumor biopsy and in targeted sequencing of the baseline plasma sample (p.Y2154fs*21). Due to DNA yield, we could not obtain WES of the baseline cfDNA sample in this patient. At progression, WES revealed two alternative deletions resulting in an in-frame deletion but restoring the open reading frame of BRCA2 coexisting with the original clone containing the frameshift mutation.
Figure 5.
Visual representation of emerging de-novo mutations at progression that likely result in acquired drug resistance in two patients originally presenting somatic frameshift mutations in BRCA2 (upper panel) and PALB2 (bottom panel) respectively in the pre-treatment samples. In both cases, the sample at treatment progression showed 2 different new deletions resulting in in-frame deletions and restoring the reading frame for BRCA2 and PALB2 respectively. In both cases, these clones were coexisting with the original clone that was present prior to treatment.
Additionally, secondary genomic events causing reversion back to normal reading frame of a somatic PALB2 mutation were also detected for one patient. The pre-treatment sample contained a 2 base-pair frameshift deletion (p.-253fs*3) with LOH. At the time of progression, after 9-months of treatment, two different new mutations restoring the PALB2 reading frame were identified. (Figure 5B) Both events coexisted with the original clone, again indicating divergent resistant sub-clones.
We examined the nucleotide sequences flanking the BRCA2 and PALB2 original frameshift deletions in these cases with secondary gene mutations. In some of these multiple resistant subclones emerging in parallel we observed short regions of nucleotide sequence identity flanking the new deletions. This association with microhomology regions suggests that these might have evolved as a consequence of defective homologous recombination and the utilization of alternative error-prone DNA repair mechanisms.
Lastly, in one patient with HDAC2 biallelic somatic loss in the tumor biopsy, WES demonstrated subclonal divergent evolution and new mutations emerging at progression in TP53 and TSC2 (GAP domain), which have been associated with drug resistance (Supplementary Table S1).(20,21)
Among the remaining patients with plasma cfDNA evaluated at secondary resistance, we did not detect any other such events, although two of these patients discontinued drug due to tolerability issues prior to radiological progression. No emerging mutations or copy number changes in PARP1 or PARP2 were observed in any of these samples.
Discussion
In this study, we describe the clinical utility of cfDNA analyses as multi-purpose biomarkers for treatment with PARP inhibition in metastatic prostate cancer. Critically, our cfDNA analyses detected all somatic HRD-associated mutations identified in tumor biopsies as well as new mutations emerging at disease progression. These new mutations likely represent tumour sub-clones induced by therapeutic selective pressure driving drug resistance.
The emergence of secondary mutations in BRCA1/2 germline mutation carriers has been previously described in case reports and small retrospective series of breast or ovarian cancer patients after PARP inhibition and/or platinum chemotherapy. (22–24) Here, we report the first series of patients homogeneously treated within a prospective clinical trial; mutations reverting the reading frame of mutated homologous recombination genes were detected not only in germline BRCA2 mutation carriers but also, for the first time, in tumors harbouring somatic loss of BRCA2 and PALB2. These events were confirmed independently by orthogonal targeted sequencing of cfDNA and/or tumour biopsy DNA.
These multiple genomic events driving resistance emerged in parallel, indicating clonal divergence with functional convergence, restoring homologous recombination repair proficiency. A similar concept has been recently described after exposure to AR targeting agents in prostate cancer, with emergence in parallel of multiple AR aberrations. (25–30) Monitoring this sub-clonal equilibrium between the original clone and resistant clones merits further evaluation since PARP inhibitor discontinuation and administration of other treatments could potentially restore the dominance of the original clone sensitive to PARP inhibitor or platinum.
Interestingly, in two cases small tracts of DNA homology flanked all these multiple secondary mutations. These were reminiscent of similar deletions observed in preclinical models restoring the open reading frame of the gene and causing PARP inhibitor resistance, and probably arose from the use of error-prone DNA repair processes that predominate in the absence of functional BRCA2. (31–33)
In the case with the original somatic PALB2 mutation, the initial clinical response to olaparib, followed by PARPi resistance characterized by the emergence of secondary mutant PALB2 alleles with microhomology-associated intragenic deletions is strongly suggestive of the PALB2 mutation in this patient causing a homologous recombination defect which not only drives the initial PARPi sensitivity phenotype but also the mechanism of resistance that eventually emerges. Although pre-clinical studies have suggested that PALB2 defects are associated with defective homologous recombination, this is to the best of our knowledge the first evidence that the homologous recombination defect caused by a PALB2 mutation might not only drive drug sensitivity but also resistance.
These data are of major clinical relevance for subsequent treatment strategies for this subset of prostate cancers, and probably also for other cancer types. Platinum chemotherapy also has antitumor activity in BRCA2 deficient prostate cancers, (34) but our data indicate likely cross-resistance at least for some patients between PARP inhibitors and platinum. Clinical trials of PARP inhibitors and platinum-based chemotherapy in prostate cancer should therefore account for previous exposure to these drugs and the presence of these secondary reversion mutations.
Overall our data show the potential of serial cfDNA next generation sequencing to evaluate both temporal and spatial disease heterogeneity. In some cases, cfDNA could monitor the emergence of resistance mechanisms more comprehensively than single site biopsies. Such analyses may allow us to truly deliver more precise patient care by integrating real time, non-invasive, repeated assessment of disease biology. Our study also shows that simple, inexpensive, cfDNA quantification, and tumor mutation allele frequency analyses, have clinical utility as a response biomarker to guide early treatment switch decisions in the presence of ineffective therapies. While further validation studies are needed, these findings may be of huge importance since earlier discontinuation of therapy for futility can spare patients the toxicity of ineffective overtreatment, allowing these men to receive alternative therapy and decreasing treatment health economic costs to fund these cfDNA studies.
A limitation of our study is that we did not collect samples between week-16 of therapy and cancer progression, which in some cases meant over a year of olaparib therapy. Therefore, we were unable to determine precisely when these restoring resistance mutations emerged; this has clinical relevance since these may appear before disease progression detection by established methods, and allow earlier treatment changes. In the ongoing TOPARP-B trial, which is further evaluating responses to olaparib in metastatic prostate cancer patients, preselected based on DNA repair aberrations, monthly samples are being collected to address this. Furthermore, our assays were limited to exon sequencing, so it remains possible that some of these tumors had alternative undetected resistance mechanisms.
In conclusion, we provide strong evidence that serial cfDNA analyses are a powerful test for guiding prostate cancer care allowing disease molecular stratification, response assessment and the study of emerging resistant clones. Critically, we report previously undetectable divergent emerging sub-clones in cfDNA only at disease progression due to treatment-selective pressures, with multiple reversion genomic events restoring the DNA repair gene function, all causing acquired resistance through convergent mechanisms. Directing patient treatment through the early detection of emerging resistant tumor clones in cfDNA, either by adding other treatments or by switching therapy contingent on which clone is dominant, is envisioned. This could have a major impact on the treatment and outcome of not only prostate cancer but also other malignancies.
Methods
Study Population
All samples were collected prospectively as part of the TOPARP-A clinical trial (CRUK/11/029; NCT01682772). Patients provided written informed consent prior to trial participation. Full details of trial design, eligibility criteria, and response to treatment have been previously reported. (10) The study of cfDNA as response and resistance biomarker was included as an exploratory endpoint of the study.
cfDNA Extraction And Quantification
Thirty millilitres (ml) of blood were collected in CTP tubes from each patient at pre-specified time points: at baseline and after 1, 4, 8 and 16 weeks of therapy, and at disease progression when possible. Circulating-free DNA was extracted from 4-8 ml of plasma using the Qiasymphony™ (Qiagen) and the circulating DNA kit (Qiagen) and quantified by Quant-iT High Sensitivity Picogreen Kit (Invitrogen).
Targeted cfDNA Sequencing
Targeted next-generation sequencing was performed as previously described.(10) Libraries were constructed from 40ng of cfDNA using a customized Generead DNAseq Mix-n-Match v2 panel (Qiagen) and sequenced on the MiSeq Sequencer (Illumina). The somatic variant calls were manually inspected in IGV (Integrated Genome Viewer, Broad Institute).
Whole-Exome cfDNA Sequencing
Whole exome sequencing (WES) was perfomed using Kapa Hyper Plus library prep kits and the Agilent SureSelectXT V6 target enrichment system. Paired-end sequencing was performed using the NextSeq™ 500 (2x150 cycles; Illumina). FASTQ files were generated from the sequencer’s output using Illumina bcl2fastq2 software (v.2.17.1.14, Illumina) with the default chastity filter to select sequence reads for subsequent analysis. All sequencing reads were aligned to the human genome reference sequence (GRCh37) using the BWA (v. 0.7.12) MEM algorithm, with indels being realigned using the Stampy (v.1.0.28) package. Picard-tools (v.2.1.0) were used to remove PCR duplicates and to calculate sequencing metrics for QC check. The Genome Analysis Toolkit (GATK, v. 3.5-0) was then applied to realign local indels, recalibrate base scores, and identify point mutations and small insertions and deletions. Somatic point mutations and indels were called using MuTect2 by comparing tumour/plasma DNA to germline control and copy number estimation was obtained through modified ASCAT2 package.
Statistical Analyses
Radiological progression-free survival (rPFS) was defined as time from trial entry to progression (RECIST 1.1, bone scan-PCWG2)(35,36) or death. Overall survival (OS) was defined as time from trial entry to death. Response was defined in the trial protocol as either: response by RECIST 1.1, PSA decline of ≥50% and/or conversion of CTC from baseline ≥5 cells to <5 cells/7.5 ml on treatment, requiring a confirmatory assessment at least 4-weeks later. Mann-Whitney and Fisher’s exact tests were used to assess the association of continuous and categorical variables with response. Kaplan-Meier curves are presented for time-to-event endpoints. The prognostic significance of changes in cfDNA at week-4 and week-8 from baseline were explored using landmark analyses. Patients experiencing rPFS and/or OS events before the landmark time were excluded. Hazard-ratios (HR) were estimated utilizing Cox regression univariate and multivariable models; 95% confidence intervals are provided. In the absence of a validated cut off, log-fold change, 30% and 50% declines in cfDNA were evaluated. The proportional hazards assumption of the Cox model was tested using Schoenfeld residuals. Based on the exploratory nature of these analyses, Bonferroni-adjustment of p-values was not pursued but a p-value<0.01 was predefined as significant to account for multiple testing. Statistical analyses were conducted using STATA13 (StataCorp LP, College Station, TX, USA) on a data snapshot taken on 24/05/2016, when 49/50 patients had discontinued the trial.
Supplementary Material
Statement of Significance.
We report prospectively planned, serial, cell-free DNA (cfDNA) analyses from metastatic prostate cancer patients treated on an investigator-initiated Phase II of olaparib. These provide predictive, prognostic, response and resistance data with ‘second hit’ mutations first detectable at disease progression, suggesting clonal evolution from treatment selective pressure, and platinum resistance.
Funding support
We would like to acknowledge funding support from Movember (Movember/PCUK CEO13-2-002), Prostate Cancer Foundation (PCF grant 20131017), Prostate Cancer UK (PCUK PG12-49), Prostate Cancer Foundation/SU2C to the International Prostate Cancer Dream team (SU2C-AACR-DT0712, PCF grants 20131017 & 20131017-1), Cancer Research UK (Centre Programme grant), Experimental Cancer Medicine Centre grant funding from Cancer Research UK and the Department of Health, and Biomedical Research Centre funding to the Royal Marsden (ECMC CRM064X). TOPARP is an investigator-initiated study supported by Cancer Research UK (grants C12540/A12829, C12540/A13230, C1491/A9895, C1491/A15955) and conducted with support from the Investigator-Sponsored Study Collaboration between AstraZeneca and the National Institute for Health Research Cancer Research Network.
J Mateo was supported by a Prostate Cancer Foundation (PCF) Young Investigator Award (PCF-16-YOUN11) and a Prostate Cancer UK-Medical Research Council Fellowship (MR/M003272/1). G Seed was supported by a Prostate Cancer UK PhD Studentship (TLD-S15-006). A. Sharp was supported by a Medical Research Council Fellowship (MR/M018618/1).
Footnotes
Disclosures
JG, JM, WY, HM, NP, SM, RPL, DD, GF, BE, PF, GS, DNR, GB, CB, MA, MC, MC, IF, RR, SS, PR, ZZ, AS, NT, DV, AG, CJD, EH, SC and JSDB are employees of The Institute of Cancer Research, which is a joint applicant for the patent entitled ‘DNA damage repair inhibitors for treatment of cancer’ which includes the granted application US8143241. C. Lord reports holding patents related to the use of PARP inhibitors (WO2008020180 [A2] and WO2009027650 [A1]). J.S. de Bono has served as an advisor for AZ, Medivation, Pfizer, Merck, Tesaro and Biomarin. No other potential conflict of interest relevant to this article was reported.
Author’s contributions
Conception and design: Jane Goodall, Joaquin Mateo, Shahneen Sandhu, Suzanne Carreira, Emma Hall, Johann S. de Bono.
Development of methodology: J. Goodall, J. Mateo, H. Mossop, N. Porta, D. Robinson, D. Dolling. E. Hall, A. Chinnaiyian, S. Carreira, J.S. de Bono.
Acquisition of data: Jane Goodall, Joaquin Mateo, Wei Yuan, Helen Mossop, Nuria Porta, Susana Miranda, Raquel Perez-Lopez, David Dolling, Dan Robinson, Gemma Fowler, Berni Ebbs, Penny Flohr, George Seed, Daniel Nava-Rodrigues, G. Boysen, Claudia Bertan, Mark Atkin, Matthew Clarke, Mateus Crespo, Ines Figueiredo, Ruth Riisnaes, Semini Sumanasuriya, Pasquale Rescigno, Zafeiris Zafeiriou, Adam Sharp, Nina Tunariu, Diletta Bianchini, Alexa Gillman, Suzanne Carreira, Johann S. de Bono.
Analysis and interpretation of data: Jane Goodall, Joaquin Mateo, Wei Yuan, Helen Mossop, Nuria Porta, George Seed, Christopher J Lord, Emma Hall, Arul M. Chinnaiyan, Suzanne Carreira, Johann S. de Bono.
Manuscript writing: J. Mateo, J.S. de Bono
Manuscript revision: all authors
Study supervision: Emma Hall, Arul M. Chinnaiyan, Suzanne Carreira, Johann S. de Bono
References
- 1.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–6. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–45. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–78. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA Mutations Are Associated With Higher Risk of Nodal Involvement, Distant Metastasis, and Poor Survival Outcomes in Prostate Cancer. J Clin Oncol. 2013;31:1748–57. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. 2016;375:443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Eng J Med. 2015;373:1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono J, Ramanathan RK, Mina L, Chugh R, Glaspy J, Rafii S, et al. Phase I, Dose-Escalation, 2-Part Trial of Poly(ADP-Ribose) Polymerase Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;2045:1–11. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 13.Fong PCP, Boss DDS, Yap Ta, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Lopez R, Mateo J, Mossop H, Blackledge MD, Collins DJ, Rata M, et al. Diffusion-weighted Imaging as a Treatment Response Biomarker for Evaluating Bone Metastases in Prostate Cancer: A Pilot Study. Radiology. 2016 doi: 10.1148/radiol.2016160646. 160646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frenel J-S, Carreira S, Goodall J, Roda Perez D, Perez Lopez R, Tunariu N, et al. Serial Next Generation Sequencing of Circulating Cell Free DNA Evaluating Tumour Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res. 2015;15:4586–96. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. New Eng J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, et al. Circulating Tumor Cell Biomarker Panel As an Individual-Level Surrogate for Survival in Metastatic Castration-Resistant Prostate Cancer. J Clin Oncol. 2015;33:1348–55. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzenbach H, Alix-Panabières C, Müller I, Letang N, Vendrell J-P, Rebillard X, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res. 2009;15:1032–8. doi: 10.1158/1078-0432.CCR-08-1910. [DOI] [PubMed] [Google Scholar]
- 19.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–5. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 20.Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, et al. Activation of the PI3K/mTOR Pathway following PARP Inhibition in Small Cell Lung Cancer. PLoS One. 2016;11:e0152584. doi: 10.1371/journal.pone.0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aas T, Børresen A-L, Geisler S, Smith-Sørensen B, Johnsen H, Varhaug JE, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med. 1996;2:811–4. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- 22.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–15. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afghahi A, Timms KM, Vinayak S, Jensen KC, Kurian AW, Carlson RW, et al. Tumor BRCA1 Reversion Mutation Arising During Neoadjuvant Platinum-Based Chemotherapy in Triple-Negative Breast Cancer Is Associated with Therapy Resistance. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K, et al. Secondary Mutations in BRCA2 Associated with Clinical Resistance to a PARP inhibitor. J Pathol. 2013;229:422–9. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 25.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. New Eng J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6:254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanel A, Tandefelt DG, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10–312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016;2:1598–606. doi: 10.1001/jamaoncol.2016.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henzler C, Li Y, Yang R, McBride T, Ho Y, Sprenger C, et al. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat Commun. 2016;7 doi: 10.1038/ncomms13668. 13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Laere B, van Dam P-J, Whitington T, Mayrhofer M, Diaz EH, Van den Eynden G, et al. Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur Urol. 2017 doi: 10.1016/j.eururo.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 32.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–16. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 34.Cheng HH, Pritchard CC, Boyd T, Nelson PS, Montgomery B. Biallelic Inactivation of BRCA2 in Platinum-sensitive Metastatic Castration-resistant Prostate Cancer. Eur Urol. 2016;69:992–5. doi: 10.1016/j.eururo.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.