Abstract
Purpose of review
Diarrhoea is a major global health problem, and recent studies have confirmed Shigella as a major contributor to this burden. Here, we review recent advances in Shigella research; focusing on their epidemiology, pathogenesis, antimicrobial resistance, and the role of the gut microbiome during infection.
Recent findings
Enhanced epidemiological data, combined with new generation diagnostics, has highlighted a greater burden of Shigella disease than was previously estimated, which is not restricted to vulnerable populations in low-middle income countries. As we gain an ever more detailed insight into the orchestrated mechanisms that Shigella exploit to trigger infection, we can also begin to appreciate the complex role of the gut microbiome in preventing and inducing such infections. The use of genomics, in combination with epidemiological data and laboratory investigations, has unravelled the evolution and spread of various species. Such measures have identified resistance to antimicrobials as a key contributor to the success of specific clones.
Summary
We need to apply novel findings towards sustainable approaches for treating and preventing Shigella infections. Vaccines and alternative treatments are under development and may offer an opportunity to reduce the burden of Shigella disease and restrict the mobility of antimicrobial resistant clones.
Keywords: diarrhoeal disease, epidemiology, genomics pathogenesis, Shigella
INTRODUCTION
Diarrhoea is a major global health issue. It accounts for approximately 1.3 million deaths each year, of which 500,000 are young children worldwide [1,2]. Despite the impressive reduction in diarrhoea-associated mortality over the past decade, there are still ∼950 million diarrhoea cases occurring in children less than 5 years annually [1]. This burden is mainly felt by low and middle-income countries in Asia and Africa, where accessibility to clean water, good nutrition, sustained sanitation, and healthcare is restricted. Tackling diarrhoea is complicated as the disease is caused by an array of bacterial, viral, and parasitic pathogens. Although improved sanitation has a major impact on lowering the incidence of all aetiologies, other public health measures, including appropriate treatment, education, and immunization remain crucial in furthering this success. Vaccines against rotavirus, the most common childhood diarrhoeal pathogen, are effectively alleviating diarrhoeal burden [3]. However, this global reduction in rotavirus disease is raising the profile, as well as the proportional burden, of other pathogens. This is particularly pronounced for bacterial agents such as Shigella, for which there is no licensed vaccine and treatment options become dwindling due to increasing resistance to key antimicrobials [4].
Recent estimates attribute Shigella to cause ∼125 million diarrhoeal episodes annually [5], leading to around 160 000 deaths, with a third of these associated with young children [1]. Shigella, along with enterotoxigenic Escherichia coli (E. coli), were identified as the predominant bacterial diarrhoeal pathogens in paediatric populations of South Asia and sub-Saharan Africa [6,7]. This research, the Global Enteric Multicentre Study (GEMS), also revealed that Shigella was the most prevalent aetiology in children aged 2 to 5 years who experienced diarrhoea. Reanalysis of the GEMS samples using quantitative molecular diagnostics suggested that Shigella-induced burden may actually be twice as high as previously estimated, ranking it as the most common detected pathogen [8▪▪]. Therefore, Shigella are a major contributor to the global diarrhoea burden and are arguably, given the associated disease severity and increasing antimicrobial resistance, the principal bacterial cause of sustained endemic diarrhoea. In the scope of this review, we highlight recent insights into the biology of Shigella and the disease that it causes, focusing on its pathogenesis, interaction with the microbiome, and the epidemiology of shigellosis.
Box 1.
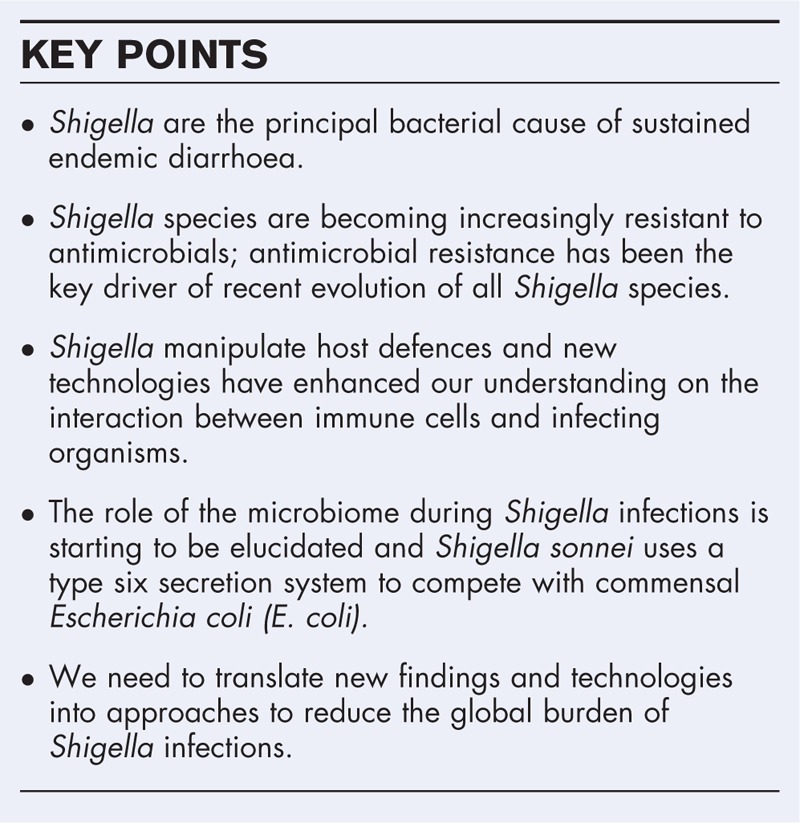
no caption available
SHIGELLA PATHOGENESIS: THE BALANCE BETWEEN VIRULENCE AND PROTECTION
Shigella is a member of the Gram-negative Enterobacteriaceae family, and current classification divides the genus into four species based on serological typing: S. dysenteriae, S. boydii, S. flexneri and S. sonnei. Ingestion of Shigella, which typically has a low infectious dose, commonly results in an aggressive watery or mucoid/bloody diarrhoea. This clinical presentation is a direct consequence of Shigella invasion and destruction of the large intestinal epithelium. Briefly, the bacterium crosses the epithelium via M cells, and induces phagocytosis by macrophages in the submucosa. Shigella quickly activate macrophage death and interact with the epithelium's basolateral surface, triggering its uptake through the reorganization of host cell cytoskeleton [9–11]. Once inside the epithelial cell, they again lyse the surrounding phagosome and replicate, before disseminating intracellularly to adjacent cells using actin polymerization [12,13]. Central to this well-choreographed pathogenesis is the large virulence plasmid (more than 200 kbp), which encodes the syringe-like type three secretion system (T3SS) and an arsenal of effector proteins, including several invasion plasmid antigens (Ipas) [14–16]. Various reviews have been dedicated to detail Shigella's pathogenesis [17,18] as well as to elucidate the role of each virulence factor [19▪].
Survival within host cells poses monumental challenges unmet by the free-living E. coli cousins, namely the detection and elimination by the host immune system. Decades of extensive research has portrayed Shigella as a master of survival, maintaining the subtle balance between virulence and immune protection. A classic example is the modulation of O-antigen (OAg) chain length in S. flexneri. Chromosomal wzz produces short-chain OAg to maximize the T3SS machinery's exposure to host cells whereas the pHS2 counterpart promotes long-chain OAg to mask S. flexneri from serum complement killing [20,21]. S. sonnei utilizes a different strategy to attain this same effect. It possesses a unique capsule made of OAg polysaccharides, decreasing its invasiveness in return for increased protection [22]. In addition, Shigella is particularly adept in subversion of the host immune response, targeting both the innate and adaptive systems [18,23]. Specifically, it is known to invade T lymphocytes via T3SS and arrest their migration in lymph nodes [24], and B lymphocytes are targeted for apoptosis via interaction with the T3SS effector IpaD [25]. These potentially deprive the human host to mount an effective and prolonged adaptive immune response. The initial process upon Shigella infection is the induction of macrophage pyroptosis, allowing the release of invading bacteria but compromising its survival by igniting a proinflammatory state [17]. Recently, IpaD was shown to mediate a noninflammatory macrophage apoptosis, thus trapping the pathogen within apoptotic bodies [26▪]. It is proposed that these parallel pathways are complementary to balance the trade-off between infectiousness and immune evasion. This same theme also underlies the functions of the IpaH family, a bacterial E3 ubiquitin ligase of research interest in recent years [27]. This enzyme catalyses the ligation of ubiquitin to target eukaryotic host proteins, usually designating them for degradation via proteasomes. Shigella carries numerous IpaH genes (situated both on chromosome and the virulence plasmid), which potentially affect ubiquitination in differing protein substrates [28]. Indeed, IpaH7.8 targets glomulin for proteolysis, thus indirectly activating inflammasomes and leading to macrophage pyroptosis [29]. In contrast, IpaH1.4 and IpaH2.5 were shown to suppress the NF-κB immune signalling by interfering with the linear ubiquitin chain assembly complex (LUBAC) machinery [30▪▪]. Immune suppression is also achieved through the IpaH9.8-mediated destruction of interferon-induced guanylate-binding proteins (GBPs), and this circumvents the host's cell-autonomous defence against intracellular microbes [31▪▪]. In addition, this same degradation is also essential in promoting cell-to-cell dissemination in Shigella infection [32▪]. The maintenance of the large virulence plasmid comes with a significant metabolic cost, which could be detrimental to Shigella's survival in resource-limited environments outside the host. It may counteract this expenditure by integrating pINV into the chromosome, thus downregulating the expression of virulence genes. This phenomenon has been observed in vitro during S. flexneri's growth at environmental temperatures, and reversible pINV excision restores its virulence at 37oC [33▪].
NEW FRONTIERS: SHIGELLA'S INTERACTION WITH THE GUT MICROBIOME
Until recently, the focus of Shigella pathogenesis research has been on its interaction with the human host, and this overlooks the roles of the heterogenous colonic landscape and its coinhabiting microbial communities. Use of innovative 3D fluorescent imaging and analyses help track S. flexneri journey in vivo, revealing that the pathogen targets colonic crypts during the early phase of infection [34]. These crypts house the intestinal stem cells at their base and harbour their own crypt-specific core microbiota (CSCM) [35]. Though Shigella's invasive zone rarely reaches the crypt base to disrupt stem cells progeniture, its interaction with the CSCM and indirect consequences on gut health remain unexplored. Successful invasion requires Shigella to overcome two gut-specific barriers: the microbiota and the mucus layer [36]. Colonic commensals could prevent pathogen proliferation by either direct competition for space and nutrient, secretion of antimicrobials, or modulation of immune response. Additionally, S. sonnei, but not S. flexneri, harbours an active type VI secretion system (T6SS), which kills co-inhabiting E. coli at infecting tissues [37▪▪]. A defective T6SS phenotype leads to reduced persistence in the colon, indicating that this apparatus is crucial for S. sonnei to overcome E. coli-established colonization resistance.
Two interesting questions remained insufficiently answered regarding Shigella's relationship with the gut microbiome: Which microbial communities are protective of Shigella infection in humans? And how does the human gut microbiome respond to a Shigella infection? Breakthroughs in sequencing, commonly employed as 16S rRNA profiling and shotgun metagenomics, have allowed an interrogation of microbial communities at the molecular level. In order to investigate the first question, it is important to evaluate the subjects clinically and microbiologically, pre and postinfection. However, data of such resolution is realistic from human challenge and longitudinal cohort studies, which are scarce. Previous immunization trials in macaques showed that Prevotella-rich microbiota was associated with asymptomatic infections upon challenge with wildtype S. dysenteriae[38]. Nonetheless, this effect is only apparent in one macaque genotype, prompting the contribution of other host factors. Prevotella are considered biomarkers for plant-based diets rich in fibre [39], and low fibre uptake prompts the gut microbiota to digest host's mucus glycoprotein [40]. This may result in rapid degradation of the mucus barrier, ultimately leading to increased susceptibility to invasion by bacterial pathogens, such as Shigella. Besides, the abundance of Prevotella species was shown to be negatively correlated with the copy number of Shigella/EIEC specific IpaH in diarrhoeal stools [41]. These studies suggest that Prevotella-rich microbiota is potentially protective for Shigella infections, but this will require further investigations. Regarding the second question, an examination on the diarrhoeal microbiome in Vietnamese young children indicated that the gut microbiota's response to Shigella infections is varied and nonspecific to the pathogen [42▪]. Instead, factors such as age, nutritional status, breastfeeding practice, and type of infection (virus/bacteria) are more indicative of the initial gut microbiota structures upon diarrhoea.
CHANGING EPIDEMIOLOGY AND THE CHALLENGE OF MULTIDRUG RESISTANCE
The four Shigella species and their various serotypes have differing geographical distribution and epidemiological significance. S. boydii infections are uncommon outside the Indian subcontinent, and there is currently limited epidemiological data regarding this species. S. dysenteriae, specifically S. dysenteriae 1, was the causative agent of multiple fatal dysentery epidemics since its first isolation in 1897 [43]. However, this species is rarely being isolated in current surveillance, and its decline is likely due to improvements in sanitation and antimicrobial access [5,44]. The current global epidemiological burden for shigellosis is attributed to two species, S. flexneri and S. sonnei, which were conventionally associated with developing and developed regions, respectively. Nevertheless, recent evidence points to the emergence of S. sonnei in economically transitional states, effectively replacing S. flexneri to become the predominant shigellosis aetiology [45]. This species replacement phenomenon is repeatedly documented in many countries in Asia, such as Vietnam [46], Thailand [47], and Bangladesh [48]. This shifting epidemiology is again reflected in the Shigella collection from GEMS, in which the authors argued that a quadrivalent vaccine targeting S. sonnei, S. flexneri 2a, S. flexneri 3a, and S. flexneri six is desired to provide sufficient coverage and protection against shigellosis in endemic regions [49].
Studies combining epidemiological and high-resolution pathogen's genomic data are increasingly common. This approach has untangled the evolutionary history and ecological dynamic of various Shigella species. Specifically, phylogenomic analyses of more than 300 temporally and spatially diverse S. dysenteriae one sequences proposed its existence as early as since the 18th century [50▪▪]. Intercontinental transmissions heightened quickly since the late 19th and throughout the 20th century, and recent waves of introductions from South Asia to Africa were responsible for multiple epidemics. Similarly, existing S. sonnei have been shown to likely descend from a common ancestor in the 17th century in Europe, and the expansions of the two main lineages (II and III) have led to their global dissemination since the 20th century [51,52▪]. These studies emphasize a pattern recognized between many Shigella species, whereby organisms are mobilized globally and then form localized endemic transmission. This is exemplified at the genomic scale by S. sonnei's introduction and subsequent establishment in Vietnam [53] and Latin America [52▪]. Alternatively, due to its low infectious dose and human-restricted nature, Shigella is able to induce sustained transmissions in close contact communities. Shigella causing several outbreaks in Orthodox Jewish communities in the United Kingdom, mainland Europe and North America are genetically closely related and clustered with those sampled in Israel, forming a single population diverging since the late 1980s [54▪]. In the United Kingdom, domestic Shigella transmissions have been exclusively noted in MSM communities, resulting in at least four discrete S. sonnei transmission chains with low genetic diversity [55].
Shigellosis is a self-limiting disease, with patients usually fully recovered within 7–10 days. However, the infection is known to cause potential complications, most severely encephalopathy [56]. Therefore, antimicrobial treatment is recommended to prevent further complications, reduce diarrhoeal output, and limit postsymptomatic faecal shedding [57,58]. However, the appropriate choice of antimicrobials is subject to debate, and no agent emerges to be superior clinically [59▪]. Unfortunately, resistance to antimicrobials appears to arise comparatively effortlessly in Shigella and may be a consequence of an unrestricted barrier for horizontal gene transfer between Shigella and other Enterobacteriaceae. Sulphonamide, tetracycline, streptomycin, and chloramphenicol were initially deployed to treat Shigella infections, but organisms that were nonsusceptible to all four antimicrobials emerged during the late 1950s. This phenotype was later determined to be conferred by small plasmids, such as spA in S. sonnei. Ampicillin, and later co-trimoxazole were used as alternatives, but these soon again met resistance in the 1980s [60]. Resistance to these agents could have facilitated the expansion and global spread of fit clones, exemplified by the integration of Tn7 transposon (encoding dfrA1 for trimethoprim resistance) in successful S. sonnei lineage III and S. dysenteriae one lineage IV [50▪▪,51]. Subsequent use of a quinolone, nalidixic acid, led to rapid and independent developments of resistance in endemic areas by 2000. The current recommended first-line treatment for shigellosis is fluoroquinolones, such as ciprofloxacin, and these quickly become the mainstay prescription for shigellosis as well as acute diarrhoea in endemic regions [58]. Mainly due to its common use, resistance to ciprofloxacin is widespread among Shigella retrieved globally since the turn of this century, and Asia serves as a likely reservoir for the rise and spread of resistant organisms [61]. Specifically, ciprofloxacin-resistant S. sonnei has evolved as a single clone, most likely in South Asia, before spreading internationally to Southeast Asia and Europe [62▪▪]. Such resistance relies on gradual accumulation of the triple mutations in chromosomal gyrA and parC. Additionally, horizontally transferred elements could help shape and establish emerging resistant clones. Recent years have witnessed a stark increase in azithromycin resistant S. flexneri 3a in MSM communities worldwide, which is caused by the propagation of a single sublineage of this species since 1998 [63]. Resistance to azithromycin is induced by the mobile plasmid pKSR100, which was recently shown to be acquired in separate S. sonnei and S. flexneri 2a populations [64▪▪]. This greatly facilitated new transmission chains, creating multiple co-circulating resistant Shigella epidemics in the United Kingdom's MSM community.
The evolutionary pressure created by antimicrobial usage fuels new resistances among Shigella, and globalization has enhanced an unprecedented mobility of this human restricted pathogen. In the present and coming age when multidrug resistance (MDR) is becoming the norm, much remains unanswered on how Shigella's state-of-resistance translates to clinical care. A recent study on paediatric diarrhoea in Vietnam found that hospitalization length for Shigella infected patients is similar regardless of the ciprofloxacin susceptibility profile of the associated organism [65▪]. Therefore, MDR likely poses a more significant threat to certain high-risk cohorts, including the malnourished, the elderly, and the immunocompromised. The latter is of increasing concern for MDR Shigella is surging in HIV-positive MSM, who present more severe clinical symptoms and require effective antimicrobial therapy [66].
CONCLUSION
The combination of larger epidemiological studies, more sophisticated in-vitro technologies, and genomics have provided unprecedented insights into the success of the genus Shigella. These could be invaluable to the development of future vaccines and alternative therapies. Namely, Shigella vaccines should account for the pathogen's numerous tricks to manipulate the immune response as well as the rapidly changing epidemiology. Novel therapies could benefit from the detailed portrayal of Shigella's pathogenesis and interactions with the gut microbiota. These tools need to be accelerated to stem the tide of increasingly antimicrobial resistant Shigella clones. Shigella research has reached a pivotal state, and we now need to apply our knowledge, technologies and experience to reduce the disease burden of this bacterial pathogen.
Acknowledgements
None.
Financial support and sponsorship
S.B. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (100087/Z/12/Z).
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 2017; 17:909–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO:Global Health Estimates 2015: disease burden by cause, age, sex, by country and by region, 2000-2015. 2016. [Google Scholar]
- 3.Lamberti LM, Ashraf S, Walker CLF, Black RE. A systematic review of the effect of rotavirus vaccination on diarrhoea outcomes among children younger than 5 years. Pediatr Infect Dis J 2016; 35:992–998. [DOI] [PubMed] [Google Scholar]
- 4.Kotloff KL, Riddle MS, Platts-Mills JA, et al. Shigellosis. Lancet 2018; 391:801–812. [DOI] [PubMed] [Google Scholar]
- 5.Bardhan P, Faruque a SG, Naheed A, Sack D a. Decrease in shigellosis-related deaths without Shigella spp.-specific interventions, Asia. Emerg Infect Dis 2010; 16:1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotloff KL, Blackwelder WC, Nasrin D, et al. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis 2012; 55 Suppl 4:S232–S245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–222. [DOI] [PubMed] [Google Scholar]
- 8▪▪.Liu J, Platts-Mills JA, Juma J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016; 388:1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identified a major increase in Shigella cases through molecular diagnostics
- 9.Zychlinsky A, Prevost M, Sansonetti P. Shigella flexneri induces apoptosis in infected macrophages. Nature 1992; 358:167–169. [DOI] [PubMed] [Google Scholar]
- 10.Hilbi H, Moss JE, Hersh D, et al. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB∗. J Biol Chem 1998; 273:32895–32900. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida S, Katayama E. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J 2002; 21:2923–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-prada CM, Hoover DL, Tall BEND, et al. Shigella flexneri IpaH7.8 facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect Immun 2000; 68:3608–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egile C, Loisel T, Laurent V. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol 1999; 146:1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sansonetti PJ, Kopecko DJ, Formal SB. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 1982; 35:852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchrieser C, Glaser P, Rusniok C, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 2000; 38:760–771. [DOI] [PubMed] [Google Scholar]
- 16.Venkatesan MM, Goldberg MB, Rose DJ, et al. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun 2001; 69:3271–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 2008; 21:134–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carayol N, Tran Van Nhieu G. Tips and tricks about Shigella invasion of epithelial cells. Curr Opin Microbiol 2013; 16:32–37. [DOI] [PubMed] [Google Scholar]
- 19▪.Mattock E, Blocker AJ. How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol 2017; 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thorough review on Shigella pathogenesis.
- 20.Hong M, Payne SM. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol 1997; 24:779–791. [DOI] [PubMed] [Google Scholar]
- 21.Morona R, Daniels C, Van Den Bosch L. Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology 2003; 149:925–939. [DOI] [PubMed] [Google Scholar]
- 22.Caboni M, Pédron T, Rossi O, et al. An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei. PLOS Pathog 2015; 11:e1004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phalipon A, Sansonetti PJ. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol Cell Biol 2007; 85:119–129. [DOI] [PubMed] [Google Scholar]
- 24.Salgado-Pabón W, Celli S, Arena ET, et al. Shigella impairs T lymphocyte dynamics in vivo. Proc Natl Acad Sci USA 2013; 110:4458–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nothelfer K, Arena ET, Pinaud L, et al. B lymphocytes undergo TLR2-dependent apoptosis upon Shigella infection. J Exp Med 2014; 211:1215–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪.Arizmendi O, Picking WD, Picking WL. Macrophage apoptosis triggered by IpaD from Shigella flexneri. Infect Immun 2016; 84:1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]; Important insights into the role of ipaD during infection.
- 27.Rohde JR, Breitkreutz A, Chenal A, et al. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 2007; 1:77–83. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, Yang J, Zhang X, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res 2005; 33:6445–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki S, Mimuro H, Kim M, et al. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proc Natl Acad Sci USA 2014; 111:E4254–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪▪.de Jong MF, Liu Z, Chen D, Alto NM. Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol 2016; 8:16084. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an important paper describing how Shigella flexneri supresses innate immune responses.
- 31▪▪.Li P, Jiang W, Yu Q, et al. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 2017; 551:378–383. [DOI] [PubMed] [Google Scholar]; Defining the mechanism of how ipaH interact with interferon-inducible guanylate-binding proteins.
- 32▪.Wandel MP, Pathe C, Werner EI, et al. GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 2017; 22:507–518e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Further insights into the interaction of ipaH and interferon-inducible guanylate-binding proteins.
- 33▪.Pilla G, McVicker G, Tang CM. Genetic plasticity of the Shigella virulence plasmid is mediated by intra- and inter-molecular events between insertion sequences. PLoS Genet 2017; 13:e1007014. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes function of Shigella virulence plasmid plasticity.
- 34.Arena ET, Campbell-Valois F-X, Tinevez J-Y, et al. Bioimage analysis of Shigella infection reveals targeting of colonic crypts. Proc Natl Acad Sci USA 2015; 112:E3282–E3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pédron T, Mulet C, Dauga C, et al. A crypt-specific core microbiota resides in the mouse colon. MBio 2012; 3:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson M, Sansonetti PJ, Marteyn BS. Shigella diversity and changing landscape: insights for the twenty-first century. Front Cell Infect Microbiol 2016; 6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37▪▪.Anderson MC, Vonaesch P, Saffarian A, et al. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 2017; 21:769–776e3. [DOI] [PubMed] [Google Scholar]; This paper describing a type six secretion system in Shigella sonnei, which aids competition with gut organisms
- 38.Seekatz AM, Panda A, Rasko D a, et al. Differential response of the cynomolgus macaque gut microbiota to Shigella infection. PLoS One 2013; 8:e64212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorvitovskaia A, Holmes SP, Huse SM. Interpreting prevotella and bacteroides as biomarkers of diet and lifestyle. Microbiome 2016; 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai MS, Seekatz AM, Koropatkin NM, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016; 167:1339–1353e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindsay B, Oundo J, Hossain MA, et al. Microbiota that affect risk for shigellosis in children in low-income countries. Emerg Infect Dis 2015; 21:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42▪.Chung The H, Sessions PF de, Jie S, et al. Assessing gut microbiota perturbations during the early phase of infectious diarrhea in Vietnamese children. Gut Microbes 2018; 9:38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes changes in the gut microbiome during infection with a range of pathogens.
- 43.Shiga K. Ueber den erreger der dysenterie in Japan (vorläufige mitteilung). Zentralbl Bakteriol Mikrobiol 1898; 23:599–600. [Google Scholar]
- 44.Gu B, Cao Y, Pan S, et al. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents 2012; 40:9–17. [DOI] [PubMed] [Google Scholar]
- 45.Thompson CN, Duy PT, Baker S. The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis 2015; 9:e0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinh H, Nhu NT, Nga TV, et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis 2009; 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bangtrakulnonth A, Vieira AR, Lo DMA, et al. Shigella from humans in Thailand during 1993 to 2006: spatial-time trends in species and serotype distribution. Foodborne Pathog Dis 2008; 5:773–784. [DOI] [PubMed] [Google Scholar]
- 48.Ud-Din AIMS, Wahid SUH, Latif H a, et al. Changing trends in the prevalence of Shigella species: emergence of multidrug resistant Shigella sonnei biotype g in Bangladesh. PLoS One 2013; 8:e82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livio S, Strockbine N a, Panchalingam S, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis 2014; 59:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50▪▪.Njamkepo E, Fawal N, Tran-Dien A, et al. Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat Microbiol 2016; 1:16027.doi:10.1038/nmicrobiol.2016.27. [DOI] [PubMed] [Google Scholar]; This paper defines the evolution and international spread of Shigella dysenteriae type 1.
- 51.Holt KE, Baker S, Weill FX, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet 2012; 44:1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52▪.Baker KS, Campos J, Pichel M, et al. Whole genome sequencing of Shigella sonnei through PulseNet Latin America and Caribbean: advancing global surveillance of foodborne illnesses. Clin Microbiol Infect 2017; 23:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genomic study of Shigella sonnei in Latin America.
- 53.Holt KE, Vu T, Nga T, et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc Natl Acad Sci USA 2013; 110:17522–17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54▪.Baker KS, Dallman TJ, Behar A, et al. Travel- and community-based transmission of multidrug-resistant Shigella sonnei lineage among international orthodox Jewish communities. Emerg Infect Dis 2016; 22:1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes transmission networks of MDR Shigella sonnei in orthodox Jewish communities.
- 55.Baker KS, Dallman TJ, Field N, et al. Genomic epidemiology of Shigella in the United Kingdom shows transmission of pathogen sublineages and determinants of antimicrobial resistance. Sci Rep 2018; 8:7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afroze F, Ahmed T, Sarmin M, et al. Risk factors and outcome of Shigella encephalopathy in Bangladeshi children. PLoS Negl Trop Dis 2017; 11:e0005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinh H, Main J, Chinh M, et al. Treatment of bacillary dysentery in Vietnamese children: two doses of ofloxacin versus 5-days nalidixic acid. Trans R Soc Trop Med Hyg 2000; 94:323–326. [DOI] [PubMed] [Google Scholar]
- 58.Legros D. WHO. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. XXX 2005. [Google Scholar]
- 59▪.Tickell KD, Brander RL, Atlas HE, et al. Identification and management of Shigella infection in children with diarrhoea: a systematic review and meta-analysis. Lancet Glob Heal 2017; 5:e1235–e1248. [DOI] [PMC free article] [PubMed] [Google Scholar]; Meta-analysis of Shigella treatment in children.
- 60.Niyogi SK. Increasing antimicrobial resistance--an emerging problem in the treatment of shigellosis. Clin Microbiol Infect 2007; 13:1141–1143. [DOI] [PubMed] [Google Scholar]
- 61.Chung The H, Baker S. Out of Asia: the independent rise and global spread of fluoroquinolone-resistant Shigella. Microb Genom 2018; doi: 10.1099/mgen.0.000171. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪▪.Chung The H, Rabaa MA, Pham Thanh D, et al. South Asia as a reservoir for the global spread of ciprofloxacin resistant Shigella sonnei: a cross-sectional study. PLoS Med 2016; 13:e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper highlights the role of Asia in the emergence and spread of drug resistant Shigella.
- 63.Baker KS, Dallman TJ, Ashton PM, et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis 2015; 15:913–921. [DOI] [PubMed] [Google Scholar]
- 64▪▪.Baker KS, Dallman TJ, Field N, et al. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat Commun 2018; 9:1462. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper again highlights the role of antimicrobial resistance in inducing epidemics of Shigella.
- 65▪.Duong VT, Tuyen HT, Minh P Van, et al. No clinical benefit of empirical antimicrobial therapy for pediatric diarrhea in a high-usage, high-resistance setting. Clin Infect Dis 2018; 66:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that ciprofloxacin therapy has a limited clinical impact during diarrhea
- 66.Mohan K, Hibbert M, Rooney G, et al. What is the overlap between HIV and shigellosis epidemics in England: further evidence of MSM transmission? Sex Transm Infect 2017; 94:67–71. [DOI] [PubMed] [Google Scholar]


