Abstract
Germinal heterozygous activating STAT3 mutations represent a novel monogenic defect associated with multi-organ autoimmune disease and, in some cases, severe growth retardation. By using whole-exome sequencing, we identified two novel STAT3 mutations, p.E616del and p.C426R, in two unrelated pediatric patients with IGF-I deficiency and immune dysregulation. The functional analyses showed that both variants were gain-of-function (GOF), although they were not constitutively phosphorylated. They presented differences in their dephosphorylation kinetics and transcriptional activities under interleukin-6 stimulation. Both variants increased their transcriptional activities in response to growth hormone (GH) treatment. Nonetheless, STAT5b transcriptional activity was diminished in the presence of STAT3 GOF variants, suggesting a disruptive role of STAT3 GOF variants in the GH signaling pathway. This study highlights the broad clinical spectrum of patients presenting activating STAT3 mutations and explores the underlying molecular pathway responsible for this condition, suggesting that different mutations may drive increased activity by slightly different mechanisms.
Keywords: STAT3, IGF-I deficiency, Growth hormone insensitivity, Activating mutations, Immune dysregulation
1. Introduction
Growth failure associated with severe primary insulin-like growth factor 1 (IGF-I) deficiency is characterized by an insufficient production of IGF-I, notwithstanding adequate secretion of growth hormone (GH). The classic form of severe primary IGF-I deficiency (IGFD) is Laron syndrome, where a homozygous or compound heterozygous mutation in the gene encoding the GH receptor (GHR) leads to low or undetectable IGF-I levels (Laron, 2015). Other well defined causes of IGFD are defects in genes encoding post-GHR signaling components, including the signal transducer and activator of transcription (STAT)-5b (Kofoed et al., 2003), the IGF-I (Woods et al., 1996) and the acid-labile subunit (ALS) (Domené et al., 2004). Recently, activating mutations in the STAT3 gene were described in children with severe growth failure associated with a spectrum of early-onset autoimmune disease, including type I diabetes, enteropathy, autoimmune cytopenia, hypothyroidism, arthritis, and interstitial lung disease (Flanagan et al., 2014; Haapaniemi et al., 2015; Milner et al., 2015). STAT3 is a cytosolic protein involved in intracellular signaling transduction from cytokines, including several interleukins (IL), interferons (IFN α/β and γ) and growth factors (Mogensen, 2013). It contains several domains, a coil-coiled, a DNA binding, a SH2 and a transactivation domain and it is involved in many biological processes, such as cell growth, apoptosis, organogenesis, inflammation, infection and oncogenesis (Akira, 2000). Response to cytokines and growth factors is mediated by JAK activation, which in turn, phosphorylates specific tyrosine residues of the receptor. The phosphotyrosine residues constitute the docking site for the SH2 domain of the STAT3 protein, leading to JAK-mediated STAT3 phosphorylation and dimer formation. STAT3 dimers translocate to the nucleus, bind to DNA and induce gene transcription (Groner, 2012). Activating STAT3 germline mutations are located in the DNA-binding, SH2, transactivation or coiled-coil domains.
We report two novel heterozygous de novo STAT3 mutations in two unrelated patients with severe growth failure and IGF-I deficiency. We have evaluated the effects of these variants on the endocrinological and the immunological system and characterized the in vitro functional activities in response to GH and IL-6 stimuli. In addition, we studied how these mutants affect STAT5b function in the GH-signaling pathway.
2. Subjects and methods
2.1. Case reports
2.1.1. Patient 1
Patient 1 was a female, the second daughter from healthy non consanguineous parents of normal height, born at term with normal weight (3.155 g, −0.18 SDS) and low birth length (44 cm, −2.76 SDS) (Table 1). Congenital autoimmune hypothyroidism was diagnosed with anti-thyroid antibodies persistently positive since birth. In the first two years of life she developed descamative eczema, chronic diarrhea, an episode of Citrobacter spp urinary tract infection, recurrent oral candidiasis, lymphocytic interstitial pneumonia with non-necrotizing granulomas associated with severe respiratory infections that required permanent oxygen supply and over 30 hospitalizations. She was referred to the pediatric endocrinologist at 2.4 years of age, when she presented a height of −6.4 SD. The immunological evaluation showed IgG levels between −1 and −2 SDS for age, with elevated IgA (>2 SDS), normal IgM and non-detectable IgE levels (Table 1). Lymphocyte subsets, including CD3+, CD4+, CD8+, CD19+, CD3−, CD56+ and regulatory T cells (CD4+ CD25++CD127lowFOXP3+ Tregs), were normal. Her cytokine profile revealed absence of Th17 with CD4+ T cells skewed to Th2. The patient also had elevated prolactin and GH serum levels, associated with non-detectable IGF-I and normal IGFBP-3 levels. She underwent an IGF generation test with recombinant human (rh) GH (33 mg/kg.day) for 7 days which showed a limited increase in IGF-I and normalization of IGFBP-3 levels (Table 1). She received levothyroxine treatment since 20 days of age and started rhGH treatment (0.43 mg/kg.wk) at 2.5 years, gaining 1.4 SDS in 2 years of treatment and increasing IGF-I levels to 240 ng/mL (Supplementary Fig. 1). She also received oral meprednisone, prophylactic trimethoprimsulfamethoxazole and intravenous γ globulin treatments. At the age of 3.2 years, oral sirolimus was added but was replaced by oral cyclosporine because of lymphedema complications. The patient died at 4 years of age, 18 days after a bone marrow transplantation with a matched unrelated donor due to a multiple organ failure.
Table 1.
Clinical characteristics of patients with de novo STAT3 mutations.
| Patient 1 (female) | Patient 2 (male) | ||
|---|---|---|---|
| Birth | Gestational Age (weeks) | 38 | 38 |
| Birth weight (g) | 3155 | 3586 | |
| Birth length (cm/SDS) | 44 (−2.76) | 50.8 (−0.75) | |
| First visit | Chronological Age (years) | 2.5 | 3.0 |
| Height (SDS) | −6.4 | −5.4 | |
| Weight (SDS) | −3.4 | −2.7 | |
| Clinical features | Congenital autoimmune hypothyroidism, descamative eczema, chronic diarrhea, recurrent oral candidiasis, severe respiratory infections | History of IPEX-like syndrome with dermatitis, chronic diarrhea, colitis, autoimmune hypothyroidism | |
| Immunological evaluation | IgG (RR: 760–1348mg/dL) | 637 | 760 |
| IgA(RR: 40–132 mg/dL) | 389 | 211 | |
| IgM (RR: 79–131 mg/dL) (mg/dL) | 103 | 134 | |
| IgE (RR: 8–32 UI/mL) | <5 | <1 | |
| CD3/CD4/CD8/CD19/CD3CD56(%) | 49/34/14/42/8 | 82/35/45/12/nd | |
| FOXP3/Treg CD127/Thl7 | N/N/low | N/N/N | |
| Endocrine evaluation | GH (ng/ml) | 20 | - |
| IGF-I (ng/ml) basal (RR: 35–160) | <12 | <25 | |
| post 1GF-GT (rhGH for 7d) | 20 | ||
| IGFBP-3 (ng/ml) basal (RR: 1.7–4.2) | 1.0 | 0.5 | |
| post 1GF-GT (rhGH for 7d) | 2.2 | ||
| Prolactin (ng/ml) (RR: 2–15) | 30,6 | ||
| TSH (mlU/ml) (RR: 0.5–6.5) | 238 | 364 | |
| FT4 (ng/dl) (RR: 0.8–2.0) | 0.4 | 0.2 | |
| TPO-Ab/TG-Ab (IU/ml) (RR:<20/<20) | 83/48 | >1000/165 | |
| rhGH treatment | Dose | 0.43 mg/kg.wk | 0.3 mg/kg.d |
| Height gain (SDS)/Period (years) | 1.4/2 | 0.42/1.4 | |
| IGF-I (ng/ml) | 240 | ||
| IGFBP-3 (ng/ml) | 4.4 | ||
| Molecular studies | WES: Heterozygous de novo STAT3 variants | c.l847_1849delAAG (p.Glu616del) | C.1276T>C (p.Cys426Arg) |
| SH2 domain | DNA binding domain |
RR: Reference range. N: Normal. nd: Not determined.
For Patient 1, height SDS was based on Argentinean growth references (Lejarraga et al., 2009) and for Patient 2, on 2000 CDC growth charts (Kuczmarski et al., 2002).
2.1.2. Patient 2
Patient 2 was a male, the second child from non consanguineous parents, born at term with normal weight and birth length (Table 1). His parents and older brother are of normal stature and healthy. At 2 weeks of life the patient was hospitalized for RSV bronchiolitis with hypoxia. In the first two years of life, he developed failure to thrive associated with chronic, intractable diarrhea, intermittent vomiting, abdominal distention, and severe eczema. Endoscopy at 2 years of age indicated marked lymphocytic gastritis and basal cell hyperplasia of the esophagus and stomach consistent with gastritis; mild lymphoid nodularity was observed in the duodenum. Acquired hypothyroidism was diagnosed at age 2, and levothyroxine (L-T4) therapy was started. At the age of 3, his height was −5.4 SD. Prior treatments included topical tacrolimus, sulfasalazine, inhaled corticosteroids, bronchodilators and proton pump inhibitors. He was also treated with a variety of immune-suppressive agents including prednisolone, Sirolimus, and Abatacept, all with minimal response. Growth improved but remained poor. The immunological phenotype was similar to that of Patient 1 (Table 1) with normal IgG and IgM levels, high IgA and non-detectable IgE levels. Lymphocyte subsets, including FOXP3+, Tregs and Th17 were all normal. Endocrine evaluation was remarkable for relatively high GH serum levels, normal prolactin, undetectable IGF-I and low IGFBP-3 levels. The patient’s hypothyroidism was not well-controlled. TSH values rarely normalized despite L-T4 supplementation of ~5 mcg/kg/day. At age 6, the patient passed a pseudo-malabsorption test suggesting non-adherence to prescribed L-T4 therapy rather than malabsorption of thyroxine compound. At age 7, he was started on rhGH (0.3 mg/kg/day) showing a partial response with gain of height SDS from −2.90 to −2.48 within 1.4 years of treatment. IGF-I has normalized to near 10–25thcentile for age. Previous gain in height SDS from −5.4 was attributed to thyroxine and immunologic treatments.
2.2. Molecular studies
Genomic DNA from patients and relatives was extracted from venous peripheral blood (Del Sal et al., 1989). Using a candidate gene approach, STAT5B and FOXP3 genes were analyzed for P1 and P2 respectively, by PCR amplification followed by Sanger sequencing. Whole Exome Sequencing for patient 1 was performed using Illumina HiSeq1500 at the Instituto de Agrobiotecnología de Rosario (INDEAR)-CONICET (Rosario, Argentina) in a quartet-design, including the index case, her parents, and her healthy sister, using Illumina Nextera Exome V1.2 kit (45 Mb - 214,405 exons) for exome capture. WES for Patient 2 and unaffected parents was performed through the DNA Sequencing and Genotyping Core at Cincinnati Children’s Hospital Medical Center (Cincinnati, Ohio, USA), employing Illumina HiSeq2500. Stringent filtering strategies for analyzing WES data include population frequency, pattern of inheritance, and specific filters for immune candidate genes. STAT3 likely pathogenic variants were identified in the index cases, and confirmed by Sanger sequencing in patients and family members (primers used are available upon request).
2.3. In silico bioinformatics analysis
STAT3 variants were analyzed in silico to predict their effects on protein function. For this purpose, different bioinformatics tools were used: PolyPhen-2 (Adzhubei et al., 2010), SIFT (Kumar et al., 2009), Mutation Taster (Schwarz et al., 2014), MutPred (Li et al., 2009), and SNAP2 (Hecht et al., 2015). The following NCBI reference sequences were used: NG_007370.1 (gene), NM_139,276.2 (mRNA) and NP_40,763.1 (protein). Different databases were used to evaluate the presence of the STAT3 variants found in the patients: dbSNP (the NCBI database for Short Genetic Variation, http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId 3483), COSMIC (the Catalogue Of Somatic Mutations In Cancer,http://cancer.sanger.ac.uk/cosmic) and ExAC (the Exome Aggregation Consortium, http://exac.broadinstitute.org). A multiple sequence alignment was done with PRALINE program (http://www.ibi.vu.nl/programs/pralinewww/) using NCBI reference sequences. In silico structural models for STAT3 variants were based on the crystal structure of the mouse STAT3B homodimer bound to DNA (PDB ID: 1BG1) (Becker et al., 1998). Protein structure was visualized using the molecular graphics program PyMOL (PyMOL Molecular Graphics System, Version 1.8.4.0, Schrödinger, LLC, http://www.pymol.org/) which was also used to generate in silico the studied mutations. Side chain conformation for p.C426R was selected from the PyMOL backbone-dependent rotamer library that resulted in less steric clashes with surrounding residues.
2.4. Site-directed mutagenesis
Gene variants were introduced into a commercial plasmid (pCMV6-Entry, RC215836, Origene, Rockville, MD, USA) containing the wild-type (WT) STAT3 cDNA (NM_139,276), using the Quick Change II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) and primers listed in Supplementary Table 1. All constructs were verified by sequencing.
2.5. Cell culture and transfections experiments
HEK293-T cells were routinely grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 units/mL), streptomycin (100 μg/mL) and L-gluta-mine (2 mM) at 37 °C in a humidified atmosphere with 5% CO2. For transfection experiments, HEK293-T cells were seeded at a density of 2 × 105 cells/well in a 24-multi well plate, grown to approximately 70–90% confluence, and transiently transfected with 500 ng Empty-pCMV6, or with a combination of 250 ng pcDNAI_Amp-GHRfl (Fang et al., 2008) and 250 ng vector carrying WT-STAT3 or variants using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). After 24 h transfection, cells were washed and serum starved for 6 h before a 15, 30 or 120-min treatment with 200 ng/mL recombinant human (rh) GH (Sandoz, Olivos, Argentina) or 20 ng/mL IL-6 (Gibco, Grand Island, NY, USA). Transfection experiments were performed in duplicates, at least three independent times.
2.6. Luciferase reporter assays
HEK293-T cells were seeded as described above and cotransfected with 125 ng of Cignal reporter assay constructs (Cignal STAT3 Reporter Assay kit (LUC), Qiagen, CA, USA), 187.5 ng of WT or mutant STAT3 containing plasmids and 187.5 ng of pcDNAI_Amp-GHRfl using the Lipofectamine 3000 transfection reagent. After 24 h transfection, cells were washed and serum starved for 6 h before 18-h treatment with 200 ng/mL rhGH or 20 ng/mL IL-6. STAT3 reporter activity was assessed using a dual luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The renilla plasmid was used to normalize transfection efficiency. Results represent the ratio of reporter (firefly) to control (renilla) luciferase or are normalized as fold-change in the ratio as compared with WT-STAT3 plasmid. Data are presented as the mean ± SEM of five independent experiments. For evaluating STAT5b reporter activity in presence of STAT3 GOF variants, HEK293-T cells expressing GHR were seeded as described above and transfected with a total input of 500 ng per well: 400 ng of the luciferase reporter construct carrying 8xGH response element (GHRE) from the rat Spi2.1 gene in pGL2 (pGHRE-LUC), 50 ng of WT or mutant STAT3 containing plasmids and 50 ng of pCMV6-STAT5b (Origene, Rockville, US). After treatment with GH (200 ng/mL) for 18e24 h, collected cell lysates were analyzed for reporter activity using the luciferase assay system (Promega Corp., Madison, WI) following the manufacturer’s protocol. The results are presented as relative fold induction ± SEM, compared with activities detected in 20 mg total protein of untreated, pcDNA3.1-transfected cell lysates, which was given an arbitrary unit value of 1. Calculated values were from five independent experiments, performed in duplicate.
2.7. Western immunoblot (WIB)
The effects of GH and IL-6 on STAT3 phosphorylation status in HEK293-T cells expressing GHR were examined by WIB analysis. Cells were starved for 6 h in serum-free medium and then stimulated with 200 ng/mL rhGH or 20 ng/mL IL-6 at 37 °C for 15, 30 and 120 min. After treatment, cells were washed with PBS and lysed in RIPA lysis buffer (1 × phosphate-buffered saline, 1% v/v Nonidet P-40, 0.1% w/v SDS, 10 mg/mL phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and protease inhibitor mixture). In a separate experiment designed to study dephosphorylation kinetics, after 30 min-treatment with 200 ng/mL rhGH or 20 ng/mL IL-6 at 37 °C the stimuli were removed and the cells were washed with PBS and incubated for 15, 30 or 120 min at 37 °C in serum-free medium before solubilization. Extracts containing equal amounts of proteins, determined by the Bradford method (Bio-Rad, Michigan, USA), were separated by SDS-PAGE (12% acrylamide) and transferred to polyvinylidenedifluoride membranes (EMD Millipore, Billerica, MA, USA). Phosphorylation was detected using an anti-phospho-STAT3 (Tyr705) rabbit monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA) and protein abundance of STAT3, using a rabbit monoclonal antibody against STAT3 (Cell Signaling Technology, Danvers, MA, USA). The signal was developed with donkey anti-rabbit IgG-horseradish peroxidase (GE Health-care Life Sciences, Freiburg, Germany) by chemiluminescence using 20 × LumiGLO Reagent (Cell Signaling Technology, Danvers, MA, USA). The intensities of autoradiographic bands were estimated by densitometric scanning using Image Studio™ Lite software (LI-COR Corporation, Lincoln, Nebraska USA).
2.8. Immunohistochemistry
HEK293-T cells were seeded as described above and transiently transfected with 500 ng Empty-pCMV6, or with a combination of 250 ng pcDNAI_Amp-GHRfl (Fang et al., 2008) and 250 ng vector carrying WT-STAT3 or variants using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). After 24 h transfection, cells were washed and serum starved for 6 h before 120-min treatment with 200 ng/mL recombinant human (rh) GH (Sandoz, Olivos, Argentina). After GH treatment, cells were carefully washed with PBS and fixed with methanol:acetic acid (3:1) for 30 min followed by several washes in PBS. Immunostaining for phospho-Stat3 was performed using a rabbit monoclonal antibody (Phospho-Tyr705-Stat3; Cell Signaling Technology, Danvers, MA, USA) at a dilution 1:400, overnight at 4 °C. The immunohistochemical staining was performed manually at room temperature, using the avidin-biotinperoxidase complex method (Vectastatin Elite ABC kit; Vector Lab) following the manufacturer’s instructions. All the wells were lightly counterstained with hematoxylin for 30 s before dehydration and mounting. Digital images were acquired with a Cool Snap camera from Nikon and saved as .tif files. Images were taken randomly for counting the cells. Quantification of p-STAT3 (Tyr705) positive nuclei and total cells were manually performed from the images using Fiji Software (Schindelin et al., 2012). Minor adjustments in the captured images were performed identically and in parallel for the images presented using Corel Photo-Paint.
2.9. Real time quantitative reverse transcriptase-PCR
For real time PCR, total RNA was extracted using TRI Reagent (Sigma-Aldrich, Carlsbad, CA, USA) according to the manufacturer’s recommendations. The amount of RNA was estimated by spectrophotometry at 260 nm. Reverse transcription (RT) was performed on 2 μg RNA at 42 °C for 50 min using M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) containing 125 ng random primer and 0.5 mM dNTP Mix. The specificity of RT-PCR was evaluated by no template and no reverse-transcriptase controls. The cDNAs encoding SOCS3 were amplified from 1 μl of the cDNA reaction mixture using specific gene primers (Supplementary Table 2). qPCR was performed by a Step One Real Time PCR System (Applied Biosystems, Warrington, UK) using the FastStar Universal SYBR Green Master (ROX) (Merck KGaA, Darmstadt, Germany). Triplicate reactions were run for each sample and the data were normalized to human TBP (TATA-box binding Protein).
2.10. Statistical analysis
All experiments were repeated at least two times. Mean ± SEM of results from multiple experiments of the same study are reported. Results were analyzed with the t-test or Mann-Whitney test for non-parametric data, as appropriate using Prism 5 (GraphPad Software, San Diego, CA) and significance was set at P < 0.05.
3. Results
3.1. Whole exome sequencing: two novel variants identified in the STAT3 gene
In patients 1 and 2, the short stature, immunological and endocrinological evaluations, suggested possible defects in the STAT5B (Hwa et al., 2011; Kofoed et al., 2003) and FOXP3 (van der Vliet and Nieuwenhuis, 2007; Wildin et al., 2001), respectively, but candidate gene analyses were unrevealing. WES analysis was, therefore, undertaken.
In patient 1, WES performed in a quartet-design (patient, parents and healthy sister) revealed 42,639 variants in 13,110 genes. We applied different filtering strategies including mutation consequence (nonsense, missense, deletion or insertion in coding sequence), population frequency (<1%), inheritance pattern (auto-somal recessive or de novo) and a 250-candidate gene list considering the pathway involved. After a critical review of the literature and a final step of manual curation of the variants, a heterozygous in frame 3 bp deletion in STAT3 gene was the most likely candidate variant causing the observed phenotype of the patient. The variant c.1847_1849delAAG is predicted to result in deletion of glutamic acid 616 (p.Glu616del), located in the SH2 domain of the protein.
WES analysis of Patient 2 and unaffected parents led to the identification of a private, de novo, heterozygous STAT3 variant, c.1276T > C, in exon 14, as the top candidate variant in the patient. The c.1276T > C generates a predicted missense substitution, p.Cys426Arg, in the DNA binding domain of the protein.
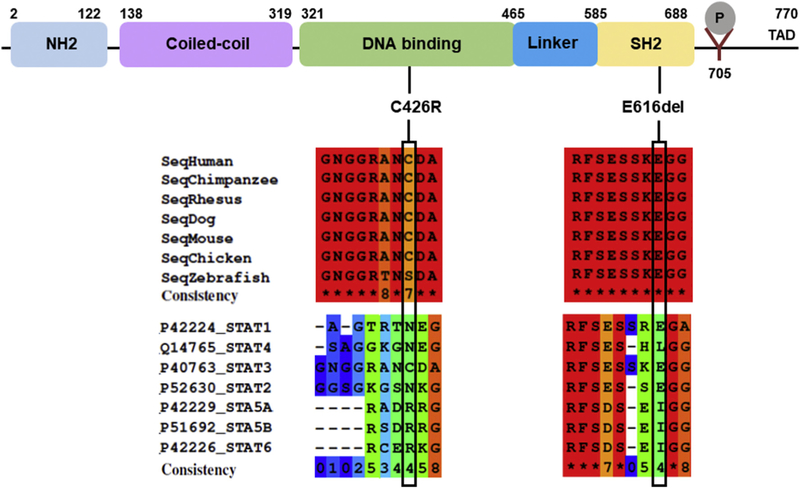
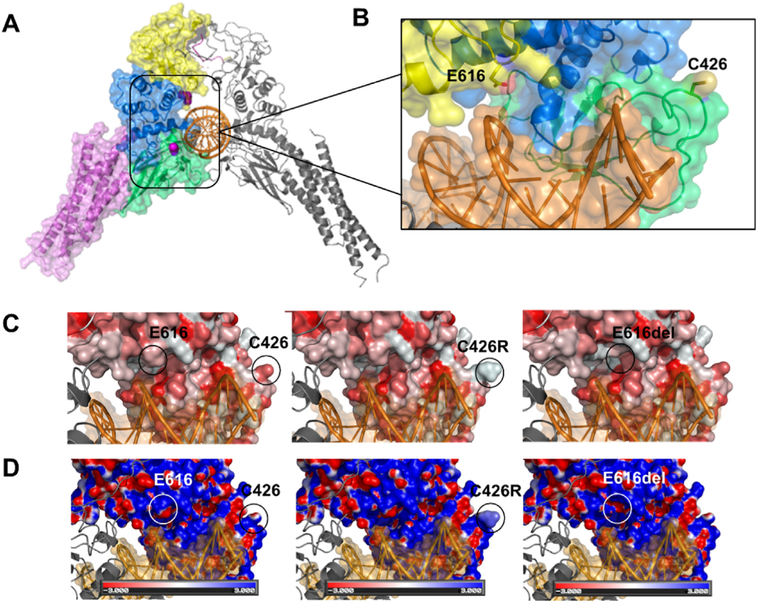
Analysis of parental DNA samples showed that both mutations had arisen de novo. According to bioinformatic algorithms and the ACMG recommendations (Richards et al., 2015), both variants were classified as likely pathogenic (Supplementary Table 3). The variant p.E616del has only been reported as a somatic mutation causing large granular lymphocytic (LGL) leukemia (COSMIC database) whereas variant p.C426R is absent from public databases of genetic variation (dbSNP, ExAC and COSMIC). The analysis of positional conservation of STAT3 residues by multiple sequence alignment from six species (human, chimpanzee, rhesus, mouse, chicken and zebrafish) showed that both residues C426 and E616 are highly conserved within the DNA binding and the SH2 domains, respectively, among species (Fig. 1). The E616 is also conserved among STAT1 and STAT2, in contrast to C426 which is not conserved among STATs proteins. Fig. 2A shows an illustration of the three-dimensional structure of the mouse Stat3b homodimer bound to DNA. The residues corresponding to the reported variants are shown in magenta. C426 is located in a region without known structure, within a stretch of 19 amino acids that is flanked by ß-sheets. Although E616 is located within the SH2 domain, it is close to the DNA binding surface and interactions between the STAT3 protein and the DNA duplex may involve this residue (Fig. 2A).
Fig. 1.
Schematics of human STAT3. The position of the two de novo mutations are shown below the STAT3 domains. Multiple sequence alignments among different species and among STAT proteins were done with PRALINE software. The color scheme indicates the least conserved alignment position (dark blue), to the most conserved alignment position (red). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
A) Structural model of the STAT3 dimer bound to DNA (PDB ID: 1BG1) (Becker et al., 1998) in cartoon representation. Individual domains are color coded as in Fig. 1 only on the left chain. Variant residues under study are shown as space-filling mode in magenta. B) The structure has been rotated and expanded to show residues E616 and C426 (sticks) close to the DNA duplex surface. C) Surface representation of the predicted hydrophobicity of WT-STAT3 (left) and variants p.C426R (center) and p.E616del (right). Coloring was achieved using the color_h.py python script in Pymol, and the color scale is based on the Eisenberg normalized hydrophobicity scale (Eisenberg et al., 1984) where hydrophilic residues are lighter/white, and hydrophobic residues are darker/red. Both variants show an increase in the hydrophobicity surface in regions where the residues were substituted or deleted. D) Electrostatic potentials are mapped onto the molecular surfaces of WT-STAT3 and mutants, with negative potentials colored red and positive potentials colored blue. The electrostatic potentials were calculated and visualized using APBS plugin -Adaptive PoissoneBoltzmann Solver (Baker et al., 2001)- in Pymol. The variants (center and right) show an increase in the positive electrostatic potential on the DNA binding surface compared to WT-STAT3 (left). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. STAT3 p.E616del and p.C426R are constitutively activated variants that further increase their transcriptional activities in response to GH
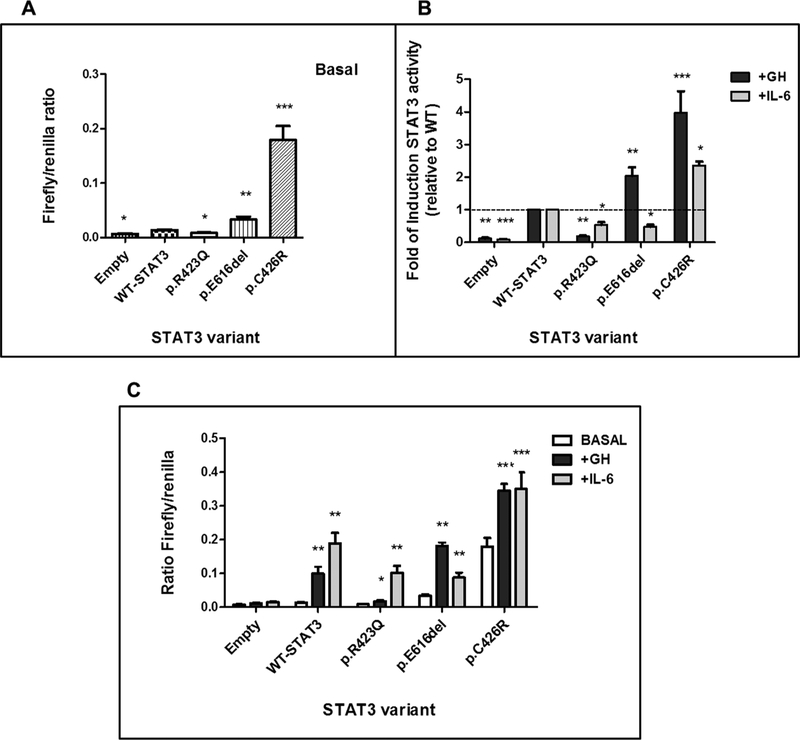
To investigate possible functional consequences of p.E616deland p.C426R-STAT3 variants, we evaluated the activity of each mutant using a STAT3-responsive dual-luciferase reporter assay. Constructs encoding a previously identified STAT3 mutation, a loss-of-function -LOF- mutation described in hyper IgE syndrome, p.R423Q (Hsu et al., 2007), and WT-STAT3 were generated and transiently transfected into cultured HEK293-T cells expressing GHR. Expression of p.E616del and p.C426R mutants resulted in a significant increase in reporter activity (P < 0.05) in comparison to WT-STAT3 or LOF-STAT3 under non-stimulated conditions (Fig. 3A), suggesting that these mutants are constitutively activated. The effect of GH on the transcriptional activity of p.E616del and p.C426R was also evaluated. In WT-STAT3 transfected cells, GH stimulation resulted in an 8-fold increase above basal levels. For constitutively activated p.E616del and p.C426R, GH treatment increased luciferase activities by 2- to 4-fold above the levels observed for WT-STAT3 (Fig. 3B). Although HEK293-T constitutively express GHR, in our system GH was not able to stimulate luciferase production in cells not overexpressing GHR (data not shown). These results suggest that GH stimulation is specifically mediated by the GH receptor. IL-6 stimulation of WT-STAT3 induced the luciferase reporter gene 15-to 20-fold (Fig. 3C). Both p.C426R and p.E616del variants showed ~2- to 2.5-fold increase in reporter activity under IL-6 stimulation (Fig. 3B). However, the fold change relative to WT-STAT3 was significantly higher only for p.C426R variant (Fig. 3C).
Fig. 3.
STAT3 transcriptional activity determined by luciferase reporter assay. A) STAT3 activity of WT and variants, p.E616del, p.C426R and LOF variant, p.R423Q, under non-stimulated conditions. Data represents the mean ratio of firefly/control luciferase activity for each construct ± SEM (n = 5). B) STAT3 activity of WT and mutants following 18 h activation with GH (200 ng/mL, black) or IL-6 (20 ng/mL, gray). Data are presented as average fold of change relative to WT ± SEM of at least 5 independent experiments. The dotted line represents a fold-change of 1 (no change from WT). C) The same experiment as B) but data are presented as the mean ratio of firefly/control for each construct ± SEM (n = 5) under GH (200 ng/mL, black) or IL-6 (20 ng/mL, gray) 18-h treatment in comparison with unstimulated conditions. *P < 0.05, **P < 0.01, ***P < 0.001, t-test.
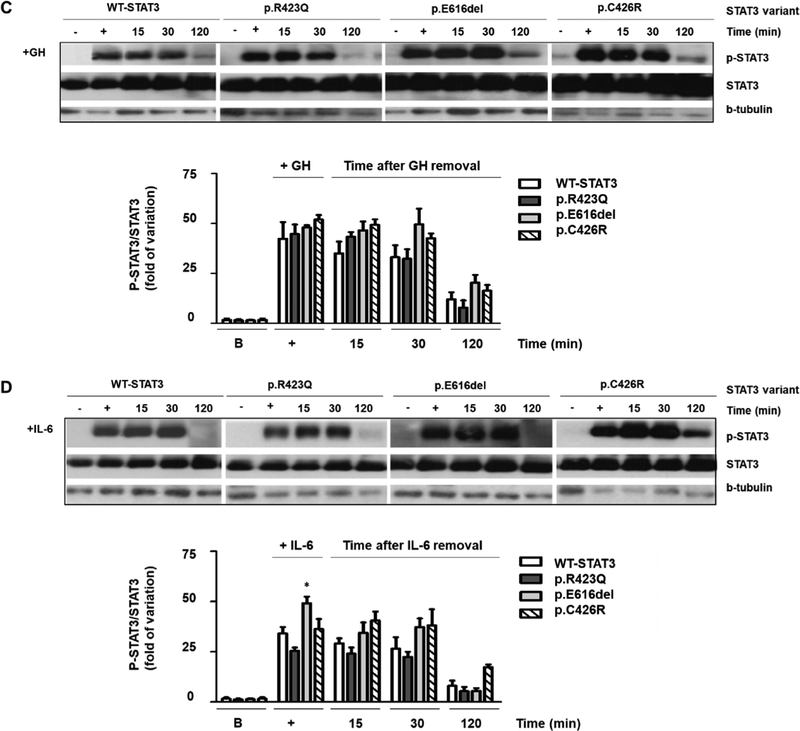
3.3. The STAT3 variants are not constitutively phosphorylated and demonstrate different dephosphorylation patterns under GH and IL-6 treatments
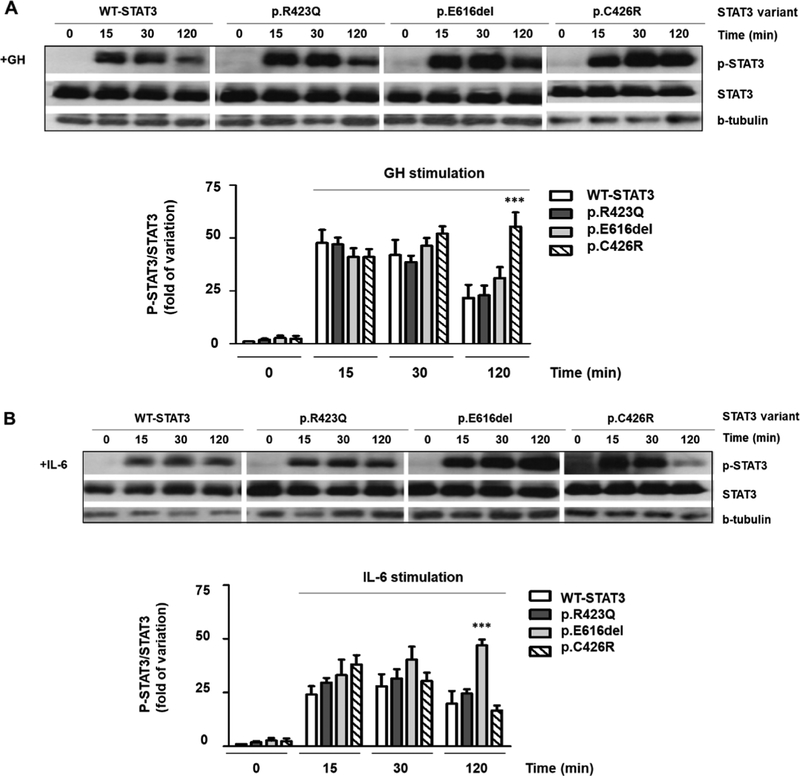
To further study the differences in the activation of p.E616del and p.C426R, we evaluated the effects of IL-6 and GH on Y705-STAT3 phosphorylation by WIB. Under basal conditions (unstimulated), STAT3 was not phosphorylated in either mutant (Fig. 4). WTSTAT3 and both variants were phosphorylated in response to IL-6 and GH and dephosphorylation was not observed within the first 30 min of stimulation (Fig. 4A and B). Phosphorylated 705Tyr-STAT3 was still detectable up to 120 min of treatment. However, at this time-point, phosphorylation was diminished for WT-STAT3 (Fig. 4A and B). A similar temporal pattern was observed for p.R423Q. This inactivating substitution is located in the DNA binding domain and the STAT3 loss of function is not a consequence of altered phosphorylation, as was previously reported (Minegishi et al., 2007).
Fig. 4.
Western Blot of STAT3 expression and phosphorylation. WT-STAT3 and mutants were transfected in HEK293-T cells expressing GHR and p-STAT3 and total STAT3 were determined under basal (B, 0 min) or stimulated conditions (15, 30, 120 min). GH (200 ng/mL) (A) and IL-6 (20 ng/mL) (B) induced phosphorylation of WT-STAT3 and mutants. p-STAT3 and total STAT3 were additionally evaluated at 15, 30 and 120 min after removing a 30-min treatment with GH (+, 200 ng/mL) (C) and IL-6 (+, 20 ng/mL) (D). β-tubulin was used as loading control. p-STAT3, phosphorylated STAT3. The upper panels show representative autoradiographies out of three. The lower panels show pooled data of three independent experiments indicating the fold variation in phosphorylation (ratio of p-STAT3 to total STAT3 in each sample) relative to WT-STAT3 for each time point. Results are expressed as means ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, t-test.
Both STAT3 activating mutants exhibited different dephosphorylation kinetics under GH and IL-6 treatments. While p.C426R exhibited delayed dephosphorylation only under GH treatment (Fig. 4A), p.E616del showed delayed dephosphorylation only when stimulated with IL-6 (Fig. 4B). To study differences in STAT3 dephosphorylation kinetics, we removed GH or IL-6 stimuli after 30 min of treatment and cell lysates were then collected 15, 30 and 120 min after removal of the stimuli. Cell lysates were analyzed by WIB (Fig. 4C and D). No differences regarding the temporal dephosphorylation pattern were observed for WT-STAT3 and p.R423Q when GH or IL-6 were depleted from the media. In contrast to the previous observed results when GH and IL-6 stimuli were maintained, no delayed dephosphorylation was observed for p.C426R and p.E616del once the stimuli were depleted from the media (Fig. 4C and D).
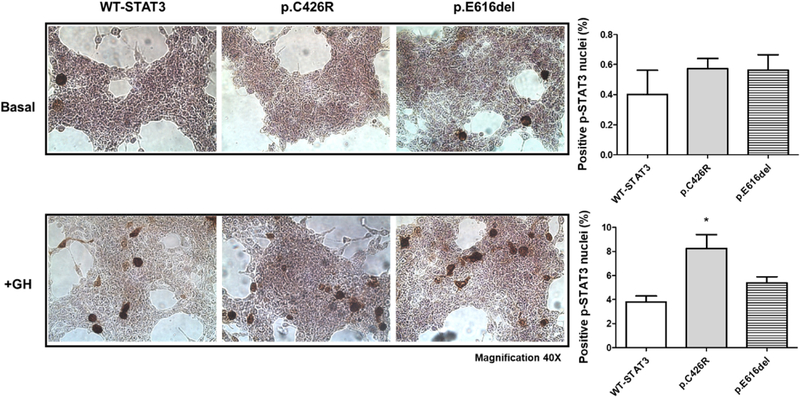
In agreement with WIB results, immunohistochemical (IHC) analysis showed a significantly increased presence of phospho-STAT3 positive nuclei after 2 h of GH-stimulation in cells transfected with p.C426R variant in comparison to those transfected with WT-STAT3 (Fig. 5).
Fig. 5.
HEK293-T cells overexpressing WT-STAT3 and variants were examined immunohistochemically for p-STAT3 (tyr705) detection under unstimulated conditions (upper panels) and after 2 h-GH treatment (200 ng/mL, lower panels). Number of positive p-STAT3 nuclei were normalized to total cells and expressed as Mean ± SEM from at least six random microscopic field images of two independent experiments. Mann-Whitney test (*P < 0.05) was used to compare positive p-STAT3 nuclei between cells expressing STAT3-GOF variants and WT-STAT3. Representative images are shown (Magnification 40 ×).
3.4. STAT5b function is partially impaired in presence of STAT3 activating variants
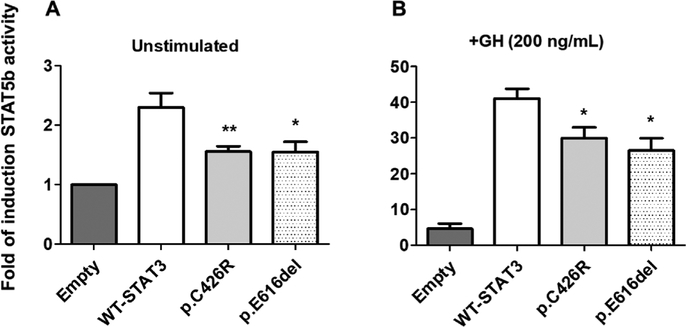
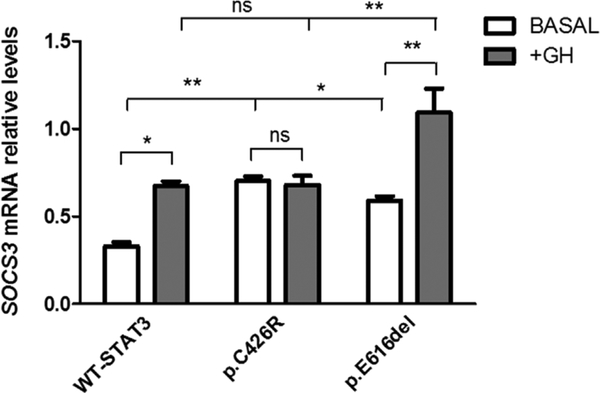
Since patients carrying these variants have growth retardation, we next decided to explore the impact of p.C426R and p.E616del STAT3 variants on STAT5b activity, a key intracellular mediator of the GH actions and inductor of IGF-1 transcription. HEK293-T cells overexpressing GHR were co-transfected with WT-STAT3 or variants and a plasmid encoding STAT5b in equimolar proportions. Transcriptional activity of STAT5b was evaluated by using a lucif-erase reporter assay. STAT5b transcriptional activity was diminished in the presence of STAT3 GOF variants under unstimulated conditions (p < 0.05) (Fig. 6A). The activity of STAT5b in cells expressing STAT3 activating variants was increased ~30-fold in response to GH, nonetheless this increase was significantly lower than in the presence of WT-STAT3 (Fig. 6B). Because activation of STAT5b can be negatively regulated by SOCS3, one of the major targets of STAT3, we evaluated SOCS3 mRNA levels in our system. We found that SOCS3 transcription levels were induced for both activating variants under unstimulated conditions (p < 0.05) but only p.E616del showed a >1.5-fold change in SOCS3 mRNA levels (p < 0.01) under GH treatment (Fig. 7).
Fig. 6.
STAT5b transcriptional activity in presence of WT-STAT3 or variants determined by luciferase reporter assay. (A) STAT5b activity in presence of WT-STAT3, p.C426R and p.E616del STAT3 variants under non-stimulated conditions. (B) Cells co-transfected with STAT5b, WT-STAT3 or variants, and pGHRE-LUC were treated with GH (200 ng/mL, 18 h) and cell lysates were analyzed for luciferase activities. Luciferase activities were normalized to total protein and expressed as Mean ± SEM from at least five independent experiments, each performed in duplicates. The normalized luciferase activity for Empty-pcDNA3.1 was set to an arbitrary value of 1. Relative induction of STAT5b transcriptional activity in presence of STAT3-GOF variants was compared to that in presence of WT-STAT3 using Mann-Whitney test (*P < 0.05 and **P < 0.01 vs. WT-STAT3).
Fig. 7.
SOCS3 mRNA levels were determined under unstimulated conditions or after 18 h-GH treatment (200 ng/mL) in HEK293-T cells expressing WT-STAT3 or variants. SOCS3 transcript levels were normalized to TBP. Data represent the mean ± SEM, n = 3. *P < 0.05, **P < 0.01, t-test.
4. Discussion
In this study, we presented two novel cases of IGF-I deficiency, severe short stature and immune dysregulation due to activating germline STAT3 mutations identified by WES. The two patients shared clinical features such as autoimmune hypothyroidism, dermatitis, chronic diarrhea and recurrent infections. They present a clinical phenotype overlapping with previously described patients with STAT3 GOF mutations (Flanagan et al., 2014; Haapaniemi et al., 2015; Milner et al., 2015), as well as some clinical similarities to other monogenic immune dysregulation disorders, including autoimmune lymphoproliferative syndrome (ALPS), immunodeficiency polyendocrinopathy enteropathy x-linked (IPEX), IPEX-like disorders, and STAT5b-deficiency. Of note, patients with STAT5B LOF mutations present severe postnatal growth failure and marked IGF-I deficiency as consequences of complete GH insensitivity, including poor responsiveness to GH treatment (Kofoed et al., 2003). In contrast, patient 1 had a good response to GH therapy. Good response to GH treatment was also reported in 2 other patients with activating STAT3 mutations (Milner et al., 2015).
P1 died after hematopoietic stem cell transplantation (SCT). Unfortunately, SCT was also unsuccessful in two out of three other patients, who succumbed shortly after the procedure (Haapaniemi et al., 2015; Milner et al., 2015; Sediva et al., 2017). This suggests that other therapeutic approaches, such as specific small-molecule STAT3 inhibitors, are perhaps more appropriate therapies for these patients.
To evaluate the impact of these mutations on STAT3 structure and activity we performed in silico and in vitro studies. Sequence alignments showed that C426 residue is located in a region containing basic amino acids in all STAT family proteins. STAT5a, 5b and 6 have an arginine residue in the equivalent position to STAT3 C426 (Fig. 1), suggesting that the change C426R in the STAT3 could be structurally tolerated. In silico mutagenesis of C426 by arginine using Pymol backbone-dependent rotamer library is predicted to cause an increase in the hydrophobicity in this region, compared to WT-STAT3 (Fig. 2B). Moreover, the calculation of electrostatic potential reveals an increase in the positively charged surface on the DNA-binding interface on p.C426R (Fig. 2C), as expected for the substitution of cysteine for the positively charged arginine residue. Likewise, in silico deletion of E616 predicts an increase in the positive electrostatic potential at the proximity of the DNA-binding surface (Fig. 2C). Therefore, both mutations would enhance the electrostatic interaction with the negatively charged DNA phosphate backbone, suggesting an increase in DNA binding affinity and, consequently, increased STAT3 transcriptional activities.
In vitro characterization of both variants indicated they were GOF variants since they activate STAT3 signaling pathway in absence of stimuli. As far as we know, the variant p.E616del is the only activating deletion in STAT3 described in patients with this syndrome. Although E616 is located within the SH2 domain, using molecular dynamics simulations Husby et al. predicted E616 as a key residue of monomer-B involved in STAT3 protein-DNA interaction (Husby et al., 2012), consistent with our molecular modelling. Therefore, enhanced activity of both variants, p.C426R and p.E616del, might be a consequence of electrostatic effects caused by the gain of positive charges at the DNA-binding surface, leading to increased DNA binding affinity and prolonged nuclear retention. However, each mutant responded differently to GH and IL-6 in vitro. When treated with IL-6, the transcriptional activity of p.E616del increased compared to unstimulated conditions. Nonetheless, this increase was lower than levels reached for WT-STAT3. This result is consistent with previously reported STAT3 mutations located within the same region, p.N646K (Flanagan et al., 2014) and p.T663I (Milner et al., 2015), where the significant increase in reporter activity under basal conditions was not above the levels reached by WT-STAT3 after stimulation with IL-6. In contrast, p.C426R had markedly elevated basal activity which was further increased with GH and IL-6 stimulation, as reported for other mutations in the DNA binding domain, as well as in the other protein domains (Flanagan et al., 2014; Haapaniemi et al., 2015; Milner et al., 2015). Therefore, increased basal STAT3 activity does not imply that response to a stimulus is also increased. The response will most likely depend on the type and location of the mutation and on the stimulus applied. In accordance with these findings, dephosphorylation patterns were different for each variant depending on the treatment. The p.C426R variant showed delayed dephosphorylation only under GH stimulus, in line with IHC results. Conversely, for p.E616del variant, delayed dephosphorylation was observed only when stimulated with IL-6. Delayed dephosphorylations were not observed once the stimuli were removed, which suggests that both variants are accessible to nuclear phosphatases that recycle STAT proteins back to the cytoplasm and mutations are not affecting phosphatases recognition processes.
The mechanism underlying these dephosphorylation patterns could probably involve the increased DNA affinity, as we already mentioned above, which induces a faster DNA binding (on-rate) and sensitizes STAT3 to cytokine stimulation, followed by a slow off-rate, which protects it from inactivation by phosphatases, as has been also suggested for the constitutively activated oncogenic Stat3 mutant (Stat3-C) (Li and Shaw, 2006). However, it is possible that the critical intracellular concentration of activated STAT3 needed to elicit the cellular response varies depending on the nature of each STAT3 mutation. Furthermore, other factors such as changes in partners involved in heterodimer formation and variations in co-activators and co-repressors recruitment, could also impact on the final transcriptional activity. Altogether, these aspects could also contribute to the broad spectrum of clinical manifestations among patients.
We also explored how these variants affect STAT5b in the GH signaling pathway. In most cell types, STAT5b and STAT3 play divergent and opposing effects on gene expression (Akira, 1999; Herrington et al., 2000). A previous study showed decreased STAT5 phosphorylation in patient-derived EBV-transformed cell lines carrying a STAT3 activating mutation (Milner et al., 2015). The authors suggest that STAT5 is negatively regulated by SOCS3, one of the major targets of STAT3. We found that SOCS3 transcription levels were induced for both GOF variants under unstimulated conditions but only p.E616del showed a significant increase in SOCS3 mRNA levels under GH treatment. This result is consistent with dephosphorylation patterns observed by WIB for both variants. Increased levels of SOCS3 for p.E616del in response to GH, could lead to a decrease in STAT3 phosphorylation via JAK2, as we detected by WIB. In contrast, p.C426R presented similar SOCS3 transcript levels under basal and stimulated conditions, in agreement with delayed dephosphorylation under GH treatment. Nonetheless, STAT5b transcriptional activity was partially impaired in presence of STAT3 GOF variants, both under unstimulated conditions and under GH treatment. These findings suggest that enhanced activity of STAT3 variants in response to GH decrease STAT5b function by slightly different mechanisms, leading to partial GH insensitivity. Further studies are necessary to elucidate the underlying mechanisms, that can include the formation of nonfunctional STAT5b/STAT3 heterodimers, competition for common receptor docking sites and regulation for posttranslational epigenetic modifications, as was recently described for GOF-STAT1 mutations and their effects on STAT3 function (Zheng et al., 2015).
Heterozygous STAT3 GOF mutations result in a broad spectrum of clinical phenotypes, involving infectious and autoimmune diseases, growth failure and IGF-I deficiency. No clear genotype/phenotype correlation has been observed. In some inherited reported cases, family members carrying the same STAT3 GOF mutation presented a milder phenotype, or were even asymptomatic, suggestive of incomplete penetrance (Milner et al., 2015). However, short stature is a key clinical finding in these patients. To our knowledge, 15 out of 22 reported patients with available growth data (Flanagan et al., 2014; Haapaniemi et al., 2015; Milner et al., 2015; Sediva et al., 2017; Velayos et al., 2017; Weinreich et al., 2017) (~68%, including our patients) had growth impairment (Supplementary Table 4 and Supplementary Fig. 2) and our in vitro studies suggest a disruptive role of STAT3 GOF variants in the GH signaling pathway. Accordingly, STAT3 activating mutations has already been considered an additional monogenic cause of primary IGF-I deficiency (Wit et al., 2015). Nonetheless, it is not entirely possible to rule out the potential secondary deleterious effects of the immune dysregulation (diabetes, hypothyroidism, recurrent infections) and the pharmacological treatments on the growth rate in these patients. As the effect of STAT3 GOF mutations on the clinical phenotype is difficult to accurately predict, our present report emphasizes the importance of complementing genetic analysis with functional studies, for a better understanding of potential therapeutic approaches.
Supplementary Material
Acknowledgements
We are grateful to the children, parents and relatives who agreed to take part in this study. We would also like to thank to Dr. Fernanda Riera for her outstanding advices in Real-Time qPCR.
Funding
This work was supported by PICT 2010 N° 1916 (ANPCYT), SANDOZ International GmbH Business Unit Biopharmaceuticals and the Fundación Alberto J. Roemmers and NIH NICHHD R01HD078592 to VH.
Footnotes
Declaration of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.mce.2018.01.016.
References
- Adzhubei I.a, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR, 2010. A method and server for predicting damaging missense mutations. Nat. Meth 7, 248–249. 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19, 2607–2611. 10.1038/sj.onc.1203478. [DOI] [PubMed] [Google Scholar]
- Akira S, 1999. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cell. 17, 138–146. 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A 98, 10037–10041. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Groner B, Muller CW, 1998. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 394, 145–151. 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Del Sal G, Manfioletti G, Schneider C, 1989. The CTAB-DNA Precipitation Method: a simple common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7, 514–518. [PubMed] [Google Scholar]
- Domené HM, Bengolea SV, Martínez AS, Ropelato MG, Pennisi P, Scaglia P, Heinrich JJ, Jasper HG, 2004. Deficiency of the circulating insulin-like growth factor system associated with inactivation of the acid-labile subunit gene. N. Engl. J. Med 350, 570–577. 10.1056/NEJMoa013100. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC, 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. U. S. A 81, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Girgis R, Little BM, Pratt KL, Guevara-Aguirre J, Hwa V, Rosenfeld RG, 2008. Growth hormone (GH) insensitivity and insulin-like growth factor-I deficiency in Inuit subjects and an Ecuadorian cohort: functional studies of two codon 180 GH receptor gene mutations. J. Clin. Endocrinol. Metab 93, 1030–1037. 10.1210/jc.2007-2022. [DOI] [PubMed] [Google Scholar]
- Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Lango Allen H, De Franco E, McDonald TJ, Rajala H, Ramelius A, Barton J, Heiskanen K, Heiskanen-Kosma T, Kajosaari M, Murphy NP, Milenkovic T, Seppanen M, Lernmark A, Mustjoki S, Otonkoski T, Kere J, Morgan NG, Ellard S, Hattersley AT, 2014. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat. Genet 46, 812–814. 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner B, 2012. Determinants of the extent and duration of STAT3 signaling. JAKSTAT 1, 211–215. 10.4161/jkst.21469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapaniemi EM, Kaustio M, Rajala HLM, van Adrichem AJ, Kainulainen L, Glumoff V, Doffinger R, Kuusanmaki H, Heiskanen-Kosma T, Trotta L, Chiang S, Kulmala P, Eldfors S, Katainen R, Siitonen S, Karjalainen-Lindsberg M-L, Kovanen PE, Otonkoski T, Porkka K, Heiskanen K, Hanninen A, Bryceson YT, Uusitalo-Seppala R, Saarela J, Seppanen M, Mustjoki S, Kere J, 2015. Autoimmunity, hypogammaglobulinemia, lympho-proliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 125, 639–648. 10.1182/blood-2014-04-570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B, 2015. Better prediction of functional effects for sequence variants. BMC Genom. 16 (S1) 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J, Smit LS, Schwartz J, Carter-Su C, 2000. The role of STAT proteins in growth hormone signaling. Oncogene 19, 2585–2597. 10.1038/sj.onc.1203526. [DOI] [PubMed] [Google Scholar]
- Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns DB, Ph D, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Ph D, Whitney AR, Voyich JM, Ph D, Musser JM, Ph D, Woellner C, Sc M, Schäffer AA, Ph D, Puck JM, Grimbacher B, 2007. Mutations in the Hyper-IgE Syndrome, pp. 1608–1619. 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- Husby J, Todd AK, Haider SM, Zinzalla G, Thurston DE, Neidle S, 2012. Molecular dynamics studies of the STAT3 homodimer:DNA complex: relationships between STAT3 mutations and protein-DNA recognition. J. Chem. Inf. Model 52, 1179–1192. 10.1021/ci200625q. [DOI] [PubMed] [Google Scholar]
- Hwa V, Nadeau K, Wit JM, Rosenfeld RG, 2011. STAT5b deficiency: lessons from STAT5b gene mutations. Best Pract. Res. Clin. Endocrinol. Metabol 25, 61–75. 10.1016/j.beem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Kofoed E, Hwa V, Little B, Woods K, Buckway C, Tsubaki J, Pratt K, Bezrodnik L, Jasper H, Tepper A, Heinrich J, Rosenfeld R, 2003. Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med 1139–1147. 10.1056/NEJMoa022926. [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z,Wei R, Curtin LR, Roche AF, Johnson CL, 2002. 2000 CDC Growth Charts for the United States: methods and development. Vital Health Stat 11, 1–190. [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC, 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc 4, 1073–1081. 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Laron Z, 2015. Lessons from 50 years of study of Laron syndrome. Endocr. Pract 21, 1395–1402. 10.4158/EP15939.RA. [DOI] [PubMed] [Google Scholar]
- Lejarraga H, del Pino M, Fano V, Caino S, Cole TJ, 2009. Growth references for weight and height for Argentinian girls and boys from birth to maturity: incorporation of data from the World Health Organisation from birth to 2 years and calculation of new percentiles and LMS values. Arch. Argent. Pediatr 107, 126–133. 10.1590/S0325-00752009000200006. [DOI] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P, 2009. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750. 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shaw PE, 2006. Elevated activity of STAT3C due to higher DNA binding af-finity of phosphotyrosine dimer rather than covalent dimer formation. J. Biol. Chem 281, 33172–33181. 10.1074/jbc.M606940200. [DOI] [PubMed] [Google Scholar]
- Milner JD, Vogel TP, Forbes L, Ma CA, Stray-Pedersen A, Niemela JE, Lyons JJ, Engelhardt KR, Zhang Y, Topcagic N, Roberson EDO, Matthews H, Verbsky JW, Dasu T, Vargas-Hernandez A, Varghese N, McClain KL, Karam LB, Nahmod K, Makedonas G, Mace EM, Sorte HS, Perminow G, Rao VK, O’Connell MP, Price S, Su HC, Butrick M, McElwee J, Hughes JD, Willet J, Swan D, Xu Y, Santibanez-Koref M, Slowik V, Dinwiddie DL, Ciaccio CE, Saunders CJ, Septer S, Kingsmore SF, White AJ, Cant AJ, Hambleton S, Cooper MA, 2015. Early-onset lympho-proliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 125, 591–599. 10.1182/blood-2014-09-602763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H, 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–1062. 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- Mogensen TH, 2013. STAT3 and the Hyper-IgE syndrome: clinical presentation, genetic origin, pathogenesis, novel findings and remaining uncertainties. JAKSTAT 2, e23435 10.4161/jkst.23435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee O, behalf of the ALQA, 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med 17, 405–424. 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-YJ-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Liceiri K, Tomancak PAC, 2012. Fiji: an open source platform for biological image analysis. Nat. Meth 9, 676–682. 10.1038/nmeth.2019.Fiji. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D, 2014. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Meth 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Sediva H, Dusatkova P, Kanderova V, Obermannova B, Kayserova J, Sramkova L, Zemkova D, Elblova L, Svaton M, Zachova R, Kolouskova S, Fronkova E, Sumnik Z, Sediva A, Lebl J, Pruhova S, 2017. Short stature in a boy with multiple early-onset autoimmune conditions due to a STAT3 activating mutation: could intracellular growth hormone signalling Be compromised? Horm. Res. Paediatr 10.1159/000456544. [DOI] [PubMed] [Google Scholar]
- van der Vliet HJJ, Nieuwenhuis EE, 2007. IPEX as a result of mutations in FOXP3. Clin. Dev. Immunol 2007, 89017 10.1155/2007/89017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos T, Martinez R, Alonso M, Garcia-Etxebarria K, Aguayo A, Camarero C, Urrutia I, Martinez de LaPiscina I, Barrio R, Santin I, Castano L, 2017. An activating mutation in STAT3 results in neonatal diabetes through reduced insulin synthesis. Diabetes 66, 1022–1029. 10.2337/db16-0867. [DOI] [PubMed] [Google Scholar]
- Weinreich MA, Vogel TP, Rao VK, Milner JD, 2017. Up, down, and all around: diagnosis and treatment of novel STAT3 variant. Front. Pediatr 5 (49). 10.3389/fped.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, Bricarelli FD, Byrne G, McEuen M, Proll S, Appleby M, Brunkow ME, 2001. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet 27, 18–20. 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wit JM, Oostdijk W, Losekoot M, van Duyvenvoorde HA, Ruivenkamp CAL, Kant SG, 2015. Mechanisms in endocrinology: novel genetic causes of short stature. Eur. J. Endocrinol 145–173. 10.1530/EJE-15-0937. [DOI] [PubMed] [Google Scholar]
- Woods KA, Camacho-Hubner C, Savage MO, Clark AJ, 1996. Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N. Engl. J. Med 335, 1363–1367. 10.1056/NEJM199610313351805. [DOI] [PubMed] [Google Scholar]
- Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, Abinun M, Gennery AR, Mann J, Lendrem DW, Netea MG, Rowan AD, Lilic D, 2015. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur. J. Immunol 45, 2834–2846. 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.