Abstract
In both NOD mice and humans, the development of type 1 diabetes (T1D) is dependent in part on autoreactive CD8+ T-cells recognizing pancreatic ß-cell peptides presented by often quite common MHC class I variants. Studies in NOD mice previously revealed the common H2-Kd and/or H2-Db class I molecules expressed by this strain aberrantly lose the ability to mediate the thymic deletion of pathogenic CD8+ T-cell responses through interactions with T1D susceptibility (Idd) genes outside the MHC. A gene(s) mapping to proximal Chromosome 7 was previously shown to be an important contributor to the failure of the common class I molecules expressed by NOD mice to mediate the normal thymic negative selection of diabetogenic CD8+ T-cells. Using an inducible model of thymic negative selection and mRNA transcript analyses we initially identified an elevated Nfkbid expression variant as a likely NOD proximal Chromosome 7 region gene contributing to impaired thymic deletion of diabetogenic CD8+ T-cells. CRISPR/Cas9-mediated genetic attenuation of Nfkbid expression in NOD mice resulted in improved negative selection of autoreactive diabetogenic AI4 and NY8.3 CD8+ T-cells. These results indicated allelic variants of Nfkbid contribute to the efficiency of intrathymic deletion of diabetogenic CD8+ T-cells. However, while enhancing thymic deletion of pathogenic CD8+ T-cells, ablating Nfkbid expression surprisingly accelerated T1D onset in NOD mice that was associated with numeric decreases in both regulatory T- and B-lymphocytes (Tregs/Bregs).
Introduction:
In both the NOD mouse model and humans, type 1 diabetes (T1D3) results from the autoimmune destruction of insulin-producing pancreatic β-cells mediated by the combined activity of CD4+ and CD8+ T-cells, as well as B-lymphocytes (1). T1D is highly polygenic in nature, as more than 50 loci have been associated with disease susceptibility or resistance in both humans and NOD mice (2, 3). While its complete pathogenic etiology remains unsolved, there is a wide acceptance that disease develops as a consequence of interactions between T1D susceptibility (Idd) genes resulting in breakdowns in mechanisms controlling induction of immunological tolerance to self-proteins (4, 5). Some Idd genes appear to contribute to defects in central tolerance mechanisms that normally represent a first checkpoint engendering the thymic deletion of autoreactive CD4+ and CD8+ T-cells during early stages of their development (6–8). However, even in healthy individuals, some autoreactive T-cells escape central tolerance mechanisms (9). Such autoreactive effectors are normally prevented from mediating pathogenic effects by a second checkpoint, provided by multiple mechanisms of peripheral tolerance involving regulatory lymphocyte populations (Tregs/Bregs), with significant evidence for some of these processes also being disrupted by various Idd genes (5).
Immature CD4+CD8+ double positive (DP) thymocytes expressing TCR molecules allowing for high avidity interactions with APC displaying self-peptide-MHC complexes are normally deleted by apoptosis or diverted to a CD4+Foxp3+ Treg lineage (9). Idd gene-elicited deficiencies in this process contribute to the thymic survival and peripheral seeding of both autoreactive CD4+ and CD8+ T-cells that overwhelm the suppressive capacity of self-antigen-specific Tregs (4, 5). MHC class II-restricted autoreactive CD4+ T-cells clearly play a critical role in T1D development. However, they cannot do so by directly engaging insulin-producing pancreatic β-cells that only express MHC class I molecules. Hence, through an ability to directly engage and destroy pancreatic β cells, MHC class I restricted autoreactive CD8+ T-cells are likely the ultimate mediators of T1D development in both humans and NOD mice (10, 11). Indeed, epidemiological studies have shown that in addition to class II effects, particular HLA class I variants provide an independent T1D risk factor in humans (12, 13). Interestingly, this includes some quite common MHC class I variants that in both humans and NOD mice only acquire an aberrant ability to contribute to T1D when expressed in the context of other Idd genes.
Previous studies found interactive contributions from some polymorphic genes outside of the MHC determine the extent to which particular class I molecules can allow the development and functional activation of diabetogenic CD8+ T-cells. This was initially demonstrated by congenically transferring a transgenic Vα8+Vβ2+ TCR from the H2g7 class I restricted AI4 diabetogenic CD8+ T-cell clone from a NOD genetic background strain (NOD-AI4) to a T1D resistant C57BL/6-stock (B6) congenic for the H2g7 haplotype (B6.H2g7). Autoreactive AI4 TCR transgenic T-cells were deleted to a significantly greater extent at the DP stage of thymic development in B6.H2g7 than NOD mice (7). This result indicated non-MHC genes allelically differing in NOD and B6 mice regulate the extent to which autoreactive diabetogenic CD8+ T-cells undergo thymic deletion. Such non-MHC genes controlling differing thymic negative selection efficiency in NOD and B6 background mice were found to function in a T-cell intrinsic manner (7). Subsequent linkage analyses mapped a gene(s) on the proximal region of Chromosome (Chr.) 7 strongly contributing to the differential levels of AI4 DP thymocytes in NOD and B6.H2g7 mice (8). This finding was confirmed by subsequent analyses of an NOD-AI4 TCR transgenic stock carrying a B6 origin proximal Chr. 7 congenic interval delineated by the flanking markers D7Mit267 (29.5 Mb) and D7Mit346 (58.7Mb) (14) (strain now designated NOD.Chr7B6FL-AI4). The Chr7B6FL congenic interval was found to contain a gene(s) intrinsically eliciting a significant decrease in numbers of AI4 DP thymocytes than in otherwise fully NOD genetic background mice (8).
A locus termed Idd7 locus had also been previously mapped to proximal Chr. 7 (15, 16). However, the Idd7 genetic locus has been enigmatic in that the NOD variant was originally identified in segregation studies as contributing to T1D resistance rather than susceptibility (15, 16). This original finding became a bit more perplexing by the above described finding that the Chr7B6FL congenic interval exerted an effect resulting in significantly fewer numbers of AI4 DP thymocytes than in otherwise fully NOD genetic background mice (8). Thus, it is unclear if the proximal Chr. 7 region gene(s) controlling the negative selection efficiency of diabetogenic AI4 CD8 T-cells is the same as that responsible for the Idd7 effect on disease development.
In this study, we identify a differential expression-level variants of Nfkbid (aka IκBNS) that can modulate NF-κB activity (17) as a likely proximal Chromosome 7 region gene regulating the efficiency of diabetogenic CD8+ T-cell thymic negative selection. Using TCR transgenic mouse models we show that a higher Nfkbid expression variant in NOD compared to B6 mice contributes to what is likely diminished negative selection in the former strain of both the AI4 and NY8.3 MHC class I restricted autoreactive diabetogenic CD8+ T-cell clonotypes. While genetic attenuation of Nfkbid expression to the lower levels observed in B6 background mice results in improved thymic deletion of pathogenic CD8+ T-cells, total ablation of this gene also numerically diminishes levels of peripheral Tregs and regulatory B-lymphocytes (Bregs) associated with a more rapid onset of T1D in the mutant stock than in standard NOD mice.
Materials and Methods
Mouse strains
NOD/ShiLtDvs (hereafter NOD), C57BL/6J (B6), and NOD.Cg-Prkdcscid Emv30b/Dvs (NOD.scid) mice are maintained in a specific pathogen free research colony at The Jackson Laboratory. NOD mice carrying a transgenic Vα8.3+Vβ2+ TCR derived from the AI4 diabetogenic CD8+ T-cell clone (NOD-AI4) have also been previously described (18). Previously generated NOD.LCMV mice transgenically express a Vα2+Vβ8+ TCR from a CD8+ T-cell clone recognizing the H2-Db restricted gp33 peptide (KAVYNFATM) derived from Lymphocytic Choriomeningitis Virus (LCMV) (19, 20). NOD.NY8.3 mice transgenically expressing the TCR derived from the H2-Kd restricted diabetogenic NY8.3 CD8+ T-cell clone (21) are maintained in an heterozygous state. A NOD stock congenic for a segment of B6-derived Chr. 7 delineated by the flanking markers D7Mit276 (29.5 Mb) and D7Mit346 (58.7 Mb) (14) (hereafter abbreviated NOD.Chr7B6FL) was used to introduce the congenic interval into NOD-AI4 mice (8).
Congenic truncation analysis
The NOD.Chr7B6FL stock was intercrossed with NOD-AI4. Resultant F1 progeny were intercrossed and all F2 offspring were analyzed for recombination events within the original Chr. 7 FL congenic region. Genomic tail DNA samples were screened for the microsatellite markers D7Mit267, D7Mit117, D7Mit155, Gpi1b, D7Mit79, D7Mit225, D7Mit78, D7Mit247, D7Mit270, D7Mit230, and D7Mit346 by PCR. Identified recombinant mice were backcrossed to NOD-AI4 and then intercrossed to generate progeny homozygous for the sub-congenic regions. The homozygous state for each sub-congenic line was verified by PCR analyses of the markers listed above. Two lines resulted from this process, Ln82 with a sub-congenic region D7Mit117–D7Mit247 and Ln16, with a sub-congenic region D7Mit79–D7Mit247.
Generation of NOD Nfkbid-deficient mice
CRISPR/Cas9 technology was utilized to directly ablate the Nfkbid gene in NOD mice. NOD/ShiLtDvs embryos were microinjected with 3pl of a solution containing Cas9 mRNA and single guide RNA (sgRNA) at respective concentrations of 100ng/μl and 50ng/μl. The sgRNA sequence (5’-AGGCCCATTTCCCCTGGTGA-3’) was designed to delete the transcriptional start site of Nfkbid in exon 3. Genomic tail DNA was screened by targeted Sanger sequencing: the genomic region around exon 3 was amplified by PCR with primers Nfkbid-KO-F1 (5’-TGCTTGAGATCCAGTAG-3’) and Nfkbid-KO-R1 (5’-CCCTGACATCTCAGAATA-3’). The resulting PCR product was purified and sequenced using an ABI 3730 DNA analyzer (Applied Biosystems-Thermo Fisher Scientific, Waltham, MA, USA). Analyses of Sanger sequencing data was done using the software Poly Peak Parser (22) which allows identification of the different alleles present in heterozygous mutant mice. During the screening of N1 mutants, we identified an allele characterized by an 8-nucleotide deletion and an insertion of 154-nucleotides originated by copy and inversion of a DNA segment from Nfkbid exon 2. This produced a major disruption of the Nfkbid gene and deletion of the reference translation start site (Supplementary Fig. 1). N1F1 NOD-Nfkbid+/− mutant mice were intercrossed and the disrupted allele fixed to homozygosity. The new stock, formally designated NOD/ShiLtDvs-Nfkbid<em3Dvs>/Dvs (hereafter abbreviated NOD-Nfkbid−/−), was maintained by brother-sister mating.
Nfkbid gene expression analysis
Total RNA from whole thymus or sorted DP cell lysates was extracted using the RNeasy Mini kit (Qiagen, Germantown, MD, USA). For whole thymus tissue RNA, the complete organ was put in 2ml of RNAlater stabilization solution (Thermo Fisher Scientific), incubated 24 h at room temperature and then stored at −20˚C until processing. In other experiments 2×105 DP cells were sorted in 100% FBS, washed 2 times in cold PBS and resuspended in RLT lysis buffer (Qiagen) and stored at −20˚C until processing. Extracted total RNA was quantified by NanoDrop (Thermo Fisher Scientific) and quality assessed in Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with a RNA integrity index (RIN index) of 7 or higher were used for gene expression analyses. For cDNA synthesis 500ng of total RNA was diluted to 5μl in DEPC treated water and mixed with 5μl of SuperScript IV VILO Master Mix (Thermo Fisher Scientific) following vendor instructions.
Expression of Nfkbid was assessed by a predesigned and validated TaqMan qPCR assay using Mm.PT.58.12759232 (IDT Integrated Technologies, Coralville, Iowa, USA) targeting the exon 3–4 region and able to identify all four possible transcripts from wild type alleles. The assay consists of a FAM-labeled and double-quenched probe (5’-/56-FAM-CCTGGTGAT-/ZEN/-GGAGGACTCTCTGGAT-/3IABkFQ/−3’) that spans the 8-nucleotide deletion site and the primers p1 (5’-GACAGGGAAGGCTCAGGATA-3’) and p2 (5’-GCTTCCTGACTCCTGATTTCTAC-3’). The assay was used at a final concentration of 500nM primers and 250nM probe. As an endogenous control, we used a predesigned and validated TaqMan assay for Gapdh (ID: Mm99999915_g1) (VIC-labeled) (Applied Biosystems – Thermo Fisher Scientifc). The qPCR analyses were done using a ViiA-7 real time PCR system (Applied Biosystems - Thermo Fisher Scientific). Relative gene expression was determined by the ddCt method using the application RQ in Thermo Fisher Cloud Software, Version 1.0 (Thermo Fisher Scientific).
Microarray analysis4
Five-week-old NOD.LCMV and NOD.Ln82-LCMV female mice were i.v. injected with 0.5μg of gp33 or Db binding control peptide (ASNENMETM) (n=3–5 biological replicates per strain and treatment type). Two hours post-injection, whole thymus tissue was processed as described above for total RNA extraction. RNA samples were hybridized to Affymetrix Mouse Gene 1.0 ST Arrays (Affymetrix – Thermo Fisher Scientific). Average signal intensities for each probe set within arrays were calculated by and exported from Affymetrix’s Expression Console (Version 1.1) software using the RMA method. Two pairwise comparisons were used to statistically resolve gene expression differences between experimental groups using the R/maanova analysis package (23). Specifically, differentially expressed genes were detected by using F1, the classical F-statistic that only uses data from individual genes. Statistical significance levels of the pairwise comparisons were calculated by permutation analysis (1000 permutations) and adjusted for multiple testing using the false discovery rate (FDR), q-value, method (24). Differentially expressed genes are declared at an FDR q-value threshold of 0.05. Two contrast groups were established: control Db peptide (NOD.LCMV vs NOD.Ln82-LCMV), and gp33 peptide (NOD.LCMV vs NOD.Ln82-LCMV). Data were filtered for an expression fold change (EFC) > 2 and an expression FDR (q-value) < 0.05. Genes meeting these criteria are identified in Supplementary Table 1, and NF-κB target genes are further annotated for function as documented in Dr. Thomas Gilmore’s curated database (http://www.bu.edu/nf-kb/gene-resources/target-genes/, accessed 3/26/2018).
Gene Expression analysis of NF-κB family members and NF-κB regulated genes
Live DP thymocytes were sorted from 5–6-week-old NOD-AI4 and NOD-AI4-Nfkbid−/− mice directly into fetal bovine serum using a FACSAria II sorter (BD Biosciences San Jose, CA, USA). Cells were pelleted, resuspended in TRIzol Reagent (Thermo Fisher Scientific) and frozen until use. Following phenol-chloroform phase separation, RNA was purified from the aqueous phase using a Quick-RNA MiniPrep Plus column (Zymo Research, Irvine, CA). Following RNA quantitation and reverse transcription as described above, qPCR was conducted using Power SYBR Green PCR master mix (Applied Biosystems – Thermo Fisher Scientific) with 250nM primers and 6ng of cDNA template. Primers are documented in Supplementary Table 2. Standard curves for each gene were performed using pooled cDNA, gene expression was normalized to Gapdh and expressed as fold change compared to NOD-AI4 control samples.
Flow cytometry analysis
Single cell suspensions of thymus (Thy), spleen (SPL) and pancreatic lymph nodes (PLN) from 5–11-week-old mice were prepared. Red blood cells in splenocyte samples were lysed with Gey’s buffer (25). Aliquots of 2–4×106 cells were stained with the following monoclonal antibodies: CD4-BV650, -BV785, or FITC (GK1.5), CD8-PE-Cy7, -BV480 or -APC (53–6.72), TCRβ-BV711 (H57–597), TCRVα8.3-FITC or -PE (B21.14), TCRVβ2-A647 (B20.6), TCRVβ8.1,2,3-FITC (F23.1) CD19-A700 or -RG710 (ID3), B220-BUV496 (RA3–6B2), CD11b-PB (M1/70), CD11c-PB (N418), CD21-FITC (7G6), CD23-PE (B3B4), CD45.1-BV421 (A201), CD5-PE (53–7.313), IL-10-APC (JES5–16E3), Foxp3-AF488 (FJK-16s) acquired from BD Bioscience, Invitrogen - Thermo Fisher Scientific, or BioLegend (San Diego, CA, USA), MHC class I tetramers Tet-AI4-PE (H2-Db/MimA2, sequence YAIENYLEL) (26) and Tet-NRPV7-PE (H2-Kd/NRPV7, sequence KYNKANVFL) acquired from the National Institutes of Health tetramer core facility (Atlanta, GA, USA). Dead cells were excluded by DAPI or propidium iodide (PI) staining for unfixed samples, or by Fixable Viability Dye eFluor 780 (eBioscience - Thermo Fisher Scientific) staining for samples fixed with Mouse Foxp3 Buffer Set (BD Bioscience). Stained cells were acquired using a BD LSRII (BD Bioscience), BD FACSymphony A5 (BD Bioscience), or Attune 1st Generation Flow Cytometer (Thermo Fisher Scientific). All flow cytometric data were analyzed with FlowJo (FlowJo LLC, Ashland, OR, USA).
T-cell Proliferation assay
Total splenic T-cells were purified from 5-week-old NOD and NOD-Nfkbid−/− female mice by negative depletion of CD11c+, CD11b+, B220+, Ter19+, CD49b+ cells with biotin-conjugated antibodies and streptavidin magnetic beads over MACS columns (Miltenyi Biotech, Cologne, Germany). Purified T-cells were washed with cold PBS, counted and adjusted to a concentration of 2×107/ml for labeling with 10μM cell tracer eFluor670 (eBioscience - Thermo Fisher Scientific). Labeled cells were adjusted to 4×106/ml. Whole collagenase digested splenocytes from NOD.scid mice were used as APCs. Labeled T-cells were seeded in triplicate into a 96 well plate at a density of 2×105/well with 4×105 APC/well. Anti-CD3 monoclonal antibody (145–2C11) was added at final concentrations ranging from 4–0.125μg/ml. The cells were incubated at 37˚C, 5% CO2 for 48h. T-cell proliferation was assessed by flow cytometric analysis of cell tracer eFluor670 dilution. Proliferation parameters were obtained using the proliferation platform analysis in FlowJo (FlowJo LLC).
Cytokine secretion analysis
IL-2 and IFN-γ secretion was assessed in supernatants of T-cell proliferation cultures by ELISA using BD OptEIA kits (BD Bioscience).
Western blot
Whole thymus lysates were prepared in RIPA buffer (Cell Signaling Technologies, Danvers, MA, USA) containing Halt™ Protease Inhibitor Cocktail (Thermo Fisher Scientific). Total protein content was quantified by Bradford assay (Thermo Fisher Scientific) and adjusted to 4mg/ml in RIPA buffer. Protein lysates were prepared for automated western blot using the Simple Wes system (ProteinSimple, San Jose, CA, USA). Briefly, samples were mixed at a 4:1 ratio with 5x fluorescent master mix and heated at 95°C for 5 min. Samples plus biotinylated molecular weight standards (ProteinSimple) were loaded along with blocking solution, biotin labeling reagent, wash buffers, primary antibodies, horseradish-peroxidase conjugated secondary antibodies and chemiluminescent substrate into a plate prefilled with stacking and separation matrices. In order to perform total protein normalization, each sample was loaded in duplicate; one capillary was used for immunodetection of Nfkbid and the second for total protein quantification. A 1:100 dilution of rabbit polyclonal anti-Nfkbid (27) with a 60 min incubation time was used for immunodetection. Chemiluminescent signal was captured and the resulting image was analyzed by the Compass for Simple Western software package (ProteinSimple). Nfkbid peak area was normalized to total protein area following ProteinSimple recommendations and standard guidelines (28, 29). Specifically, the Nfkbid area for each sample was multiplied by a normalization factor (calculated as the total protein for the B6 sample divided by the total protein for each individual sample).
Diabetes incidence
Diabetes was monitored once weekly using urine glucose strips (Diastix, Bayer, Leverkusen, Germany). Mice with two consecutive readings >250mg/dl (corresponding to a blood glucose of >300mg/dl) were considered diabetic.
Statistical analysis
Data analysis and graphs were made using Prism 6 software (GraphPad, San Diego, CA, USA). Statistical analyses are detailed in the corresponding Figure Legends.
Results
Mapping an Idd7 region gene(s) regulating thymic numbers of diabetogenic AI4 CD8+ T-cells to a 5.4 Mb region on Chr. 7.
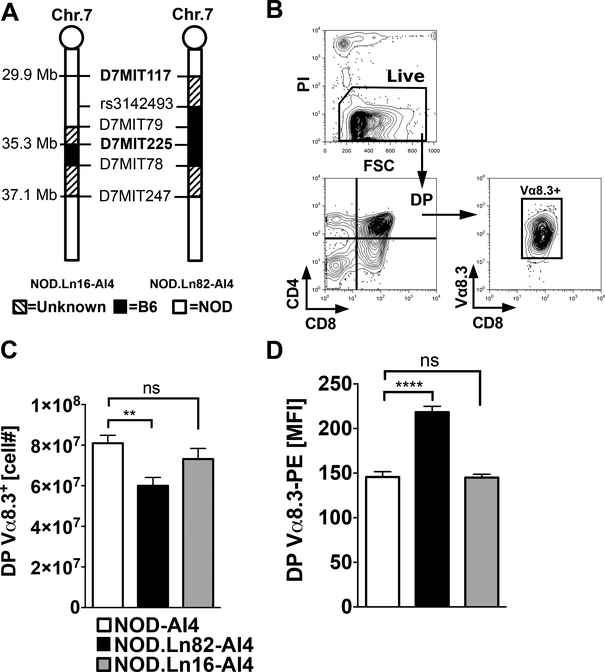
Among the multiple epitopes recognized by the promiscuous AI4 TCR is an insulin derived peptide (30). Insulin is thymically expressed in NOD mice (30). Thus, AI4 T-cells developing in the thymus are antigenically exposed. Such thymic antigenic availability indicates a gene(s) allelically varying between NOD and B6 mice on proximal Chr. 7 likely plays a strong role in controlling the extent of diabetogenic AI4 CD8+ T-cell negative selection. We subsequently derived from the original NOD.Chr7B6FL-AI4 stock two sub-congenic lines designated NOD.Ln82-AI4 (maximum length defined by the markers D7Mit117–D7Mit247) and NOD.Ln16-AI4 (maximum length defined by the markers D7Mit79–D7Mit247) (Fig. 1A). T1D development in the original NOD.Chr7B6FL-AI4 congenic stock did not differ from standard NOD controls (8), indicating shared loci between the two strains likely contributed to autoreactive T-cell activation, expansion, and pathogenicity separate from the observed thymic selection effects of the congenic region. Therefore, thymic selection, and not overt T1D development, was evaluated in the Ln82 and Ln16 sub-congenic stocks. Flow cytometric analyses found that compared to NOD-AI4 controls, the yield of DP Vα8.3+ AI4 thymocytes was significantly decreased in the NOD.Ln82-AI4, but not the NOD.Ln16-AI4 sub-strain (Fig. 1B, C). These differences indicated a polymorphic gene(s) influencing numbers of DP AI4 thymocytes resides within a 5.4 Mb region on Chr.7 maximally delineated by the markers D7Mit117-D7Mit225. Expression levels of the clonotypic Vα8.3 TCR element was higher in the NOD.Ln82-AI4 stock compared to NOD-AI4 controls (Fig. 1D), a trait previously observed in NOD.Chr7B6FL-AI4 mice (8). These results indicated a proximal Chr. 7 region gene(s) modulating TCR expression levels maps to the 5.4 Mb region between D7Mit117 and D7Mit225.
Figure 1: Mapping a proximal Chr. 7 region gene(s) controlling the thymic negative selection efficiency of diabetogenic AI4 CD8+ T-cells to a 5.4 Mb region.
(A) Schematic diagram of B6 derived Chr.7 congenic regions in NOD-AI4 mouse strains. Marker positions are indicated in Mb based on genome assembly release GRCm38.p5 (33) (B) Flow cytometric gating strategy to enumerate AI4 DP thymocytes. Thymocytes from 5-week-old female NOD-AI4 (n=32), NOD.Ln82-AI4 (n=20) and NOD.Ln16-AI4 (n=14) were stained with CD4, CD8 and Vα8.3 specific antibodies. (C) Numbers of live (PI negative) CD4+CD8+ (DP) AI4 (Vα8.3+) thymocytes in the indicated strains. (D) Mean fluorescence intensity (MFI) of staining of DP thymocytes from the indicated strains by the Vα8.3-PE monoclonal antibody B21.14. All values represent mean±SEM of more than 3 independent experiments, statistical significance was determined by Log-rank (Mantel-Cox) t-test (ns p>0.05, ** p<0.001, **** p<0.00001).
Nfkbid is the only differentially expressed proximal Chr. 7 region gene in thymocytes undergoing low versus high levels of deletion.
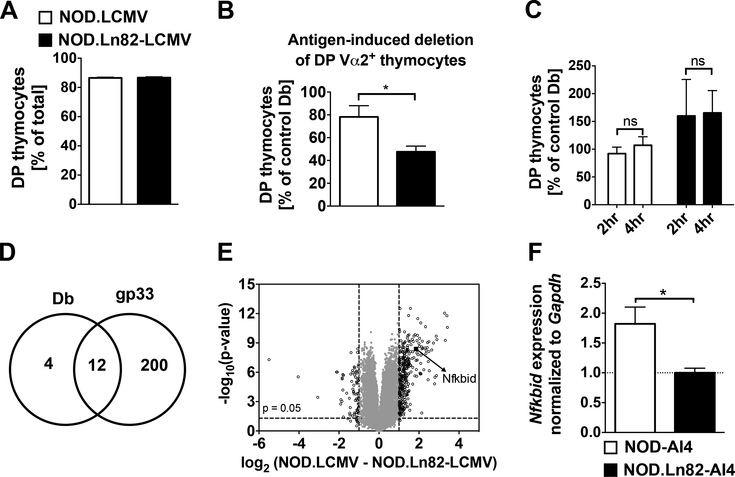
Due to differing cellular composition, we did not feel it appropriate to utilize thymii from NOD-AI4 and NOD.Ln82-AI4 mice to carry out initial mRNA transcript analyses to identify genes possibly contributing to their potentially differing negative selection of diabetogenic CD8+ T-cells. Instead, we reasoned a more appropriate platform to carry out such analyses would be provided by a previously produced NOD background stock transgenically expressing a Vα2+Vβ8+ TCR recognizing the H2-Db class I restricted gp33 peptide (KAVYNFATM) derived from Lymphocytic Choriomeningitis Virus (LCMV) (19, 20). The basis for choosing this NOD.LCMV stock was that LCMV reactive CD8+ T-cells developing in the thymus would not be under any negative selection pressure until such time their cognate antigen is exogenously introduced. We generated an additional NOD.LCMV stock homozygous for the Ln82 congenic interval. At baseline, proportions of DP thymocytes did not differ in NOD.LCMV and NOD.Ln82-LCMV mice (Figure 2A). This indicated a proximal Chr. 7 region gene(s) does not control the positive selection or expansion of DP thymocytes. NOD.LCMV and NOD.Ln82-LCMV mice were i.v. injected with 0.5μg of the gp33 or an H2-Db binding control peptide (ASNENMETM). At 24 hours post-injection, we assessed the proportion of Vα2+ TCR LCMV-specific DP thymocytes present in NOD.LCMV and NOD.Ln82-LCMV mice that had received the gp33 peptide relative to control cohorts. Compared to those receiving control peptide, treatment with gp33 elicited a greater reduction of DP thymocytes in NOD.Ln82-LCMV than NOD.LCMV mice (Fig. 2B). The finding that the greater diminution of DP thymocytes in NOD.Ln82-LCMV than NOD.LCMV mice was only observed under antigen stimulated conditions indicates this phenotype likely represents a proximal Chr. 7 region gene(s) controlled strain difference in the induction of negative rather than positive selection.
Figure 2: Nfkbid is the only differentially expressed proximal Chr. 7 region gene in diabetogenic CD8+ T-cells undergoing low versus high levels of thymic deletion.
(A). Similar proportions of DP thymocytes in untreated NOD.LCMV and NOD.Ln82-LCMV mice. (B) Either 0.5 μg of gp33 (cognate antigen) or Db binding control peptide were i.v. injected into five-week-old female NOD.LCMV (n=18 for controls and n=6 for gp33 treated) or NOD.Ln82-LCMV (n=22 for controls and n=6 for gp33 treated) mice. Remaining DP Vα2+ thymocytes were quantified at 24 hours post-injection in each group. The efficiency of deletion was calculated as a ratio of the number DP Vα2+ thymocytes remaining in gp33 treated mice to the average numbers in controls and expressed as a percentage (). Statistical significance was determined by Log-rank (Mantel-cox) t-test (ns p>0.05, * p<0.05). (C) At 2–4 hours post-i.v. injection with gp33 or Db binding control peptide numbers of DP Vα2+ thymocytes do not differ in five-week-old NOD.LCMV (n=18 for controls and n=7 for gp33 treated) or NOD.Ln82-LCMV (n=22 for controls and n=6 for gp33 treated) female mice. Calculation of deletion efficiencies and statistical comparisons as in panel B. (D) As assessed by microarray analyses number of genes deferentially expressed declared at a false discovery rate (q-value) <0.05 and a fold change ≥2 in thymocytes from NOD.LCMV and NOD.Ln82-LCMV mice 2 hours after i.v. injection with Db control or gp33 peptide (n=4–5 biological replicates per strain and treatment type). (E) Volcano plot showing microarray assessed relationship between fold change and level of significance (F1 permutation p-value) in thymic gene expression profiles in NOD.LCMV (n=5) and NOD.Ln82-LCMV (n=3) mice 2 hours after i.v. injection with gp33. Data points for the 212 probe sets with a fold-change > 2 (log2=1) are shown left or right of the dashed vertical lines. The arrow depicts differential Nfkbid expression. The other 211 genes are identified in Supplementary Table 1. (F) Total RNA was extracted from 2×105 DP Vß8+ thymocytes sorted from 6-week-old female NOD-AI4 and NOD.Ln82-AI4 mice. Nfkbid mRNA relative quantification was done using the ddCt method using Gapdh as an endogenous control and NOD.Ln82-AI4 mice as reference, mean relative fold change (RFC) ± SEM is indicated. * p<0.05 unpaired t-test.
The LCMV TCR transgenic system provided an opportunity to compare the influence of the proximal Chr. 7 region over the transcriptional profile of thymocytes undergoing different levels of antigen-induced negative selection. Time-course experiments indicated that at 2 and 4 hours post-injection with the cognate antigen or peptide control there was no significant reduction in the number of Vα2+ DP thymocytes (Fig. 2C). However, we hypothesized that at 2-hours post-antigen injection, a differential gene expression profile may have already been induced in NOD.LCMV and NOD.Ln82-LCMV thymocytes contributing to their subsequent varying levels of deletion seen at 24 hours. Therefore, we conducted microarray analysis of whole thymus comparing gp33 or control peptide treated NOD.LCMV and NOD.Ln82-LCMV mice at this time point. Based on cutoff criteria of a q-value <0.05 and minimum two-fold strain variation, 16 and 212 genes were found to be differentially expressed in thymocytes from NOD.LCMV and NOD.Ln82-LCMV mice injected with the Db binding control or gp33 peptide respectively (Fig. 2D, Supplementary Table 1). This included 12 genes that were commonly differentially expressed in thymocytes from NOD.LCMV and NOD.Ln82-LCMV mice treated with either the control or gp33 peptide (Fig. 2D, Supplementary Table 1). None of the 16 genes differentially expressed in thymocytes from NOD.LCMV and NOD.Ln82-LCMV mice treated with the control peptide mapped to the earlier defined 5.4 Mb proximal Chr. 7 support interval (genomic coordinates 7:29,926,306–35,345,930). Intriguingly, of the 212 genes matching our dual-cutoff differential expression criteria under gp33 stimulated conditions, Nfkbid was the sole one mapping within the above defined 5.4 Mb proximal Chr. 7 support interval (Fig. 2E). Nfkbid was expressed at a 3.6-fold higher level in thymocytes from gp33 treated NOD.LCMV than NOD.Ln82-LCMV mice (Fig. 2E). Typing of the intragenic rs3142493 single nucleotide polymorphism (SNP) confirmed the Ln82 congenic interval contained the B6 origin Nfkbid allele (Fig. 1A). We also subsequently found by qPCR analyses that Nfkbid is expressed at ~2-fold higher levels in flow cytometrically sorted equalized numbers of DP thymocytes from NOD-AI4 than NOD.Ln82-AI4 mice (Fig. 2F).
Nfkbid (aka IκBNS) encodes a non-conventional modulator of the transcription factor NF-κB (17) and ironically, was first discovered as a gene expressed in thymocytes undergoing negative, but not positive selection (31). Depending on cell type and physiological context, Nfkbid can stimulate or inhibit NF-κB pathway activity (17). Low to moderate levels of NF-κB activity (primarily the p50/p65 heterodimeric form) reportedly inhibits thymic negative selection of CD8+ lineage-destined T-cells (32). On the other hand, high NF-κB activity levels can elicit thymic negative selection of CD8+ T-cells (32). Thus, Nfkbid-mediated modulation of NF-κB activity could potentially contribute to the extent of diabetogenic CD8+ T-cell thymic deletion. Hence, we annotated which genes identified by our microarray analyses to be differentially expressed in thymocytes from gp33 treated NOD.LCMV and NOD.Ln82-LCMV mice were subject to NF-κB mediated regulation. As noted in Supplementary Table 1, we found 51 direct NF-κB-target genes, described in Dr. Thomas Gilmore’s curated database (http://www.bu.edu/nf-kb/gene-resources/target-genes/, accessed 3/26/2018), were upregulated versus none that were repressed in NOD.LCMV compared to congenic NOD.Ln82-LCMV thymocytes following gp33 antigen stimulation. This indicated higher Nfkbid levels may be critical for upregulating NF-κB-mediated gene expression in antigen stimulated thymocytes. For this reason, we hypothesized that the higher Nfkbid expression levels characterizing antigen stimulated NOD thymocytes may elicit up-regulation of NF-κB target genes rendering these cells less prone to negative selection than those from B6 mice.
Generation of NOD-Nfkbid deficient mice
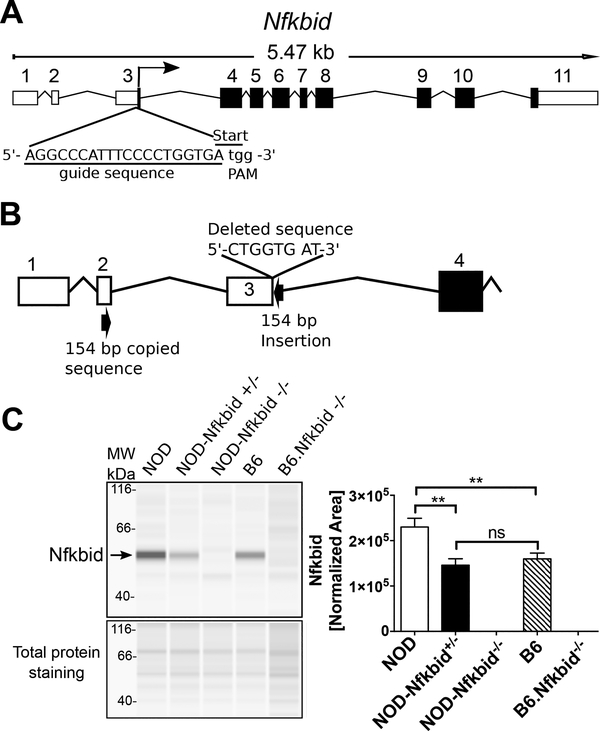
To test whether the higher Nfkbid expression in NOD mice contributes to thymic retention of diabetogenic CD8+ T-cells, we utilized a CRISPR/Cas9 approach to ablate this gene. The mouse Nfkbid gene encodes four mRNA transcripts, three of which generate the same 327 amino acid protein, with a fourth representing a non-coding transcript (33). Based on the reference sequence (NM_172142.3), Nfkbid consists of 11 exons, with the start codon located at the end of exon 3 (Fig. 3A). We utilized a single guide RNA (sgRNA) (Fig. 3A) designed to disrupt the start codon in exon 3 and thereby eliminate translation of any potential transcripts. A resultant mutation consisting of an 8-nucleotide deletion coupled with an inverted insertion of a 154-nucleotide sequence copied from exon 2 was identified (Fig. 3B, Supplemental Fig. 1) and subsequently fixed to homozygosity in a new stock officially designated NOD/ShiLtDvs-Nfkbid<em3Dvs>/Dvs and for simplicity abbreviated hereafter as NOD-Nfkbid-/−. Western blot analyses using total thymus lysates indicated Nfkbid was clearly present in NOD controls while being completely absent in both NOD-Nfkbid−/− and previously generated (34, 35) B6.Nfkbid−/− mice (Fig. 3C). Quantitative western blot analyses correlating with the earlier described microarray studies indicated thymic Nfkbid levels are significantly higher in NOD than B6 mice (Fig. 3C, right panel). Of potential physiological significance thymic Nfkbid protein levels in heterozygous NOD-Nfkbid+/− mice were similar to that of the B6 strain. We did find transcription from the targeted allele is possible but does not produce Nfkbid protein, as shown in homozygous mutants (Supplementary Figure 1C, Figure 3C).
Figure 3: Targeting Nfkbid by CRISPR/Cas9 in NOD mice.
(A) Representation of mouse Nfkbid gene showing the translation start site at the end of exon 3 and the sgRNA (underlined) designed to target this region by CRISPR/Cas9. The PAM sequence is indicated in lower case. (B) Schematic of the resulting targeted mutation consisting of an 8-nucleotide deletion and an insertion of 154 nucleotides originated by copy and inversion of a segment from Nfkbid exon 2 (For more detail, see Supplementary Figure 1). (C) Nfkbid protein abundance was assessed in total thymus lysates from the indicated strains by Simple Wes automated western blot analysis. B6.Nfkbid−/− thymus samples were provided by Dr. Ingo Schmitz. Nfkbid was detected as a 55 kDa band. The difference in molecular weight compared to the theoretical 35 kDa, could be due to posttranslational modifications. Lower panel indicates the corresponding total protein staining for each sample. Right panel shows the Nfkbid-normalized area for NOD (n=6), NOD-Nfkbid+/− (n=9), NOD-Nfkbid−/− (n=8), B6 (n=6), and B6.Nfkbid−/− (n=4) thymus samples. Nfkbid-peak area was normalized as follows: . Bars represent the mean ± SEM. ns p>0.05, ** p<0.01, Mann-Whitney analysis.
Diminution of Nfkbid expression decreases thymic mRNA transcript levels of both some NF-κB subunit and target genes associated with enhanced negative selection of AI4 and NY8.3 diabetogenic CD8+ T-cells in NOD mice.
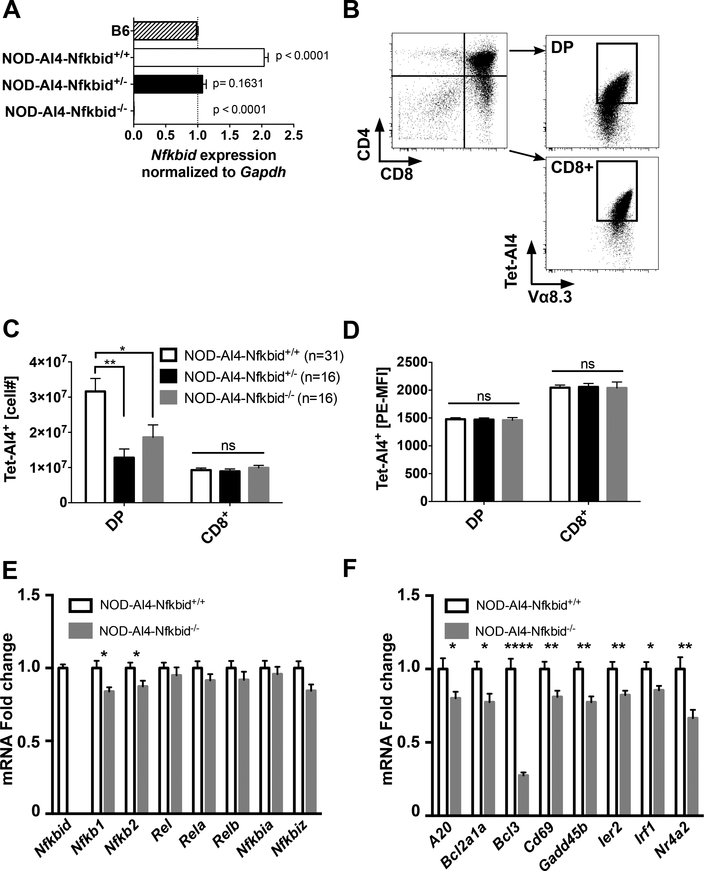
We next evaluated whether decreasing Nfkbid expression diminished thymic levels of diabetogenic CD8+ T-cells in NOD mice. To initially do this, the inactivated Nfkbid gene was transferred in both a heterozygous and homozygous state to the NOD-AI4 strain. Quantitation analyses of Nfkbid mRNA transcripts utilized primers that only detected expression of the wild type allele. As expected, analysis of Nfkbid gene expression by qPCR showed respective high and absent Nfkbid wild type mRNA transcript levels in thymocytes from NOD-AI4 and NOD-AI4-Nfkbid−/− mice (Fig. 4A). Also, as expected, wild type Nfkbid mRNA transcript levels were significantly higher in thymocytes from NOD-AI4 than B6 control mice (Fig. 4A). In agreement with analyses of protein levels, a finding of potential physiological relevance was that thymic wild type Nfkbid mRNA transcript expression in NOD-AI4-Nfkbid+/− heterozygous mice was reduced to that characterizing B6 controls (Fig. 4A). Numbers of DP, but not CD8+ SP thymocytes positively staining with the H2-Db/MimA2 tetramer capable of binding the AI4 TCR (Tet-AI4) (gating strategy shown in Fig. 4B) were significantly less in both heterozygous and homozygous Nfkbid knockout NOD mice, compared to gene-intact controls (Fig. 4C). The same result was obtained by staining with Vα8.3 and Vβ2 specific antibodies binding the AI4 TCRα- and β-chains (results not shown). As described earlier, AI4 T-cells developing in the thymus are antigenically exposed. Thus, the differing numbers of AI4 DP thymocytes in NOD-AI4 and NOD-AI4-Nfkbid−/− mice likely represents a varying capacity to undergo antigen induced negative selection. Previously described data shown in Fig. 1D indicated TCR expression was significantly higher on DP thymocytes from NOD.Chr7B6Ln82-AI4 congenic mice than in controls in which the proximal Chr. 7 region was of NOD origin. In contrast, partial or complete ablation of Nfkbid did not alter AI4 TCR expression levels on either DP or CD8+ SP thymocytes (Fig. 4D). This finding indicated some proximal Chr. 7 region gene(s) other than Nfkbid regulates thymic TCR expression levels. However, these results do not completely disprove an earlier hypothesis (8) that a mechanism through which a proximal Chr. 7 region gene(s) controls +the thymic deletion efficiency of diabetogenic CD8+ T-cell is by modulating TCR expression levels. This is because the extent that AI4 DP thymocytes were diminished by genetic ablation of Nfkbid was less than that previously observed by introduction of the Chr7B6FL congenic interval (8). Hence, in addition to Nfkbid-mediated effects, another gene(s) within the original Chr7B6FL congenic region may also contribute to the negative selection efficiency of diabetogenic CD8+ T-cells by modulating TCR expression levels.
Figure 4: Genetic attenuation of Nfkbid expression controls the numbers of AI4 DP thymocytes.
(A) Wild type Nfkbid gene expression was analyzed by qPCR of total thymus RNA from 6-week-old B6 (n=2), NOD-AI4 (n=4), NOD-AI4-Nfkbid+/− (n=4) and NOD-AI4-Nfkbid−/−(n=3) mice. Gapdh was used as endogenous control and B6 as the reference group. Graph shows the mean ± SEM of relative fold change of Nfkbid expression normalized to Gapdh. Statistical significance was determined by one-way ANOVA and Bonferroni’s multiple comparison test. The p-value of samples compared to the reference group is indicated. Note: We did not use B6.H2g7-AI4 mice as a reference in this study because the numbers of thymocytes is extremely low and would not provide a fair comparison for gene expression analysis. (B) Illustration of gating to enumerate DP and CD8+ SP thymocytes staining with the AI4 TCR specific tetramer. Depicted profiles are for thymocytes from an NOD-AI4 control. (C) Numbers of AI4 TCR expressing DP and CD8+ SP thymocytes from 6-week-old NOD-AI4, NOD-AI4-Nfkbid+/−, and NOD-AI4-Nfkbid−/− female mice. Data represent the mean ± SEM of DP or CD8+ SP thymocytes staining positive with Tet-AI4. (D) Geometric MFI of Tet-AI4 staining of the same DP and CD8+ SP thymocytes evaluated in panel B. (E) Quantification of mRNA fold change expression of NF-κB family members in sorted DP NOD-AI4 vs NOD-AI4-Nfkbid−/− thymocytes. Individual gene expression was first normalized to Gapdh, and then fold change in comparison to expression in NOD-AI4 was calculated. Data is combined from 13 female mice, 5–6 weeks of age combined from two cohorts. (F) NF-κB target gene expression analysis was performed with the same samples as in (E). Bars represent the mean ± SEM. Statistical significance for A-D was analyzed by 2-way ANOVA and Bonferroni’s multiple comparison test. Statistical significance for E and F were analyzed by Mann-Whitney. ns p>0.05, * p<0.05, ** p<0.01, **** p<0.0001.
Subsequent studies evaluated whether differential Nfkbid expression might regulate thymic negative selection efficiency by modulating the mRNA transcript levels of NF-κB component members or downstream target genes we found they may control in the microarray study (NF-κB regulated genes annotated in Supplementary Table 1). We initially measured the expression of NF-κB family member genes in sorted DP thymocytes from NOD-AI4 and NOD-AI4-Nfkbid−/− mice. As shown in Figure 4E, Nfkb1 and Nfkb2 gene expression was modestly decreased in Nfkbid−/− mice, but other pathway members - Rel, Rela and Relb – were unchanged, as were two other nuclear inhibitors of NF-κB, Nfkbia and Nfkbiz. Additionally, we measured the expression levels of a subset of the 51 NF-κB target genes found to be upregulated to a greater extent in thymocytes from NOD.LCMV than NOD.Ln82-LCMV mice injected with gp33 peptide (Supplementary Table 1) to determine if they were also altered by ablation of Nfkbid. Expression levels of a panel of NF-κB target genes were significantly lower in sorted DP thymocytes from NOD-AI4-Nfkbid−/− than NOD-AI4 mice (Fig. 4F). These results indicated Nfkbid likely acts as an enhancer of NF-κB pathway activity in this cellular context.
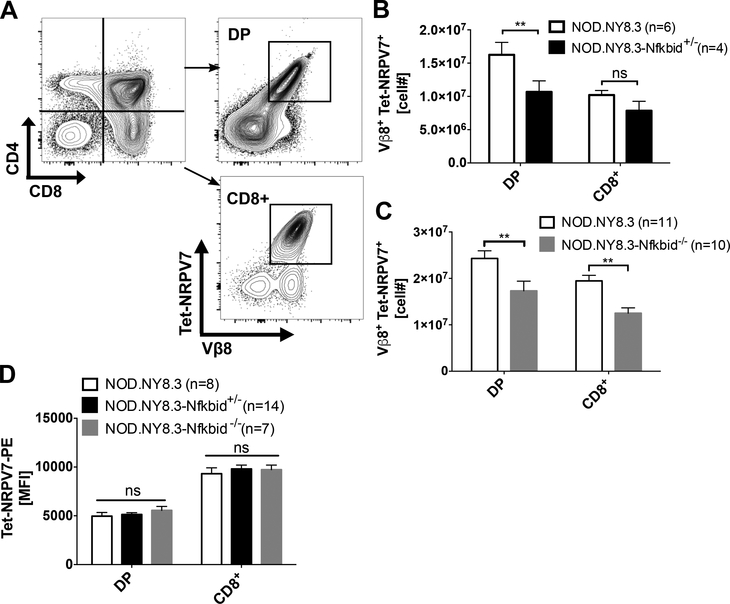
We next tested whether the apparent ability of varying Nfkbid expression levels to modulate thymic deletion efficiency of diabetogenic CD8+ T-cells extended to clonotypes beyond AI4. Thus, we introduced the Nfkbid knockout allele into a NOD stock with CD8+ T-cells transgenically expressing the NY8.3 TCR (NOD.NY8.3) that recognizes a peptide derived from the ß-cell protein islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) (21, 36). As observed for the AI4 clonotype, numbers of DP thymocytes staining with a tetramer (Tet-NRPV7) recognized by the NY8.3 TCR (Fig. 5A) were significantly less in 6-week-old NOD.NY8.3-Nfkbid+/− heterozygous mice than in controls (Fig. 5B). When examined at 4 weeks of age, numbers of both NY8.3 DP and CD8+ SP thymocytes were significantly less in NOD.NY8.3-Nfkbid−/− homozygous mice than in controls (Fig. 5C). The basis for the varying effect of eliminating Nfkbid expression on numbers of AI4 and NY8.3 CD8 SP thymocytes in NOD mice is currently unknown. However, this may possibly result from differing TCR avidity engagement with selecting thymic antigenic ligands by the two T-cell populations. Also, similar to the case for the AI4 clonotype, expression levels of the NY8.3 TCR on DP or CD8+ SP thymocytes was not altered by the presence or absence of Nfkbid (Fig. 5D). Thus, based on assessments of two separate clonotypes, we conclude that the Nfkbid expression variant characterizing NOD mice represents the proximal Chr. 7 region gene contributing to impaired thymic deletion of diabetogenic CD8+ T-cells in this strain.
Figure 5: Nfkbid expression levels modulate thymic negative selection efficiency of diabetogenic NY8.3 clonotypic CD8+ T-cells.
(A) Illustration of gating to enumerate DP and CD8+ thymocytes staining with the NY8.3 TCR specific Tet-NRPV7. Depicted profiles are for thymocytes from an NOD.NY8.3 control. (B) Numbers of NY8.3 TCR expressing DP and CD8+ thymocytes from 6-week-old female NOD.NY8.3 and NOD.NY8.3-Nfkbid+/− heterozygous mice. Data represent the mean±SEM of DP or CD8+ SP thymocytes staining positive with the NY8.3 specific Tet-NRPV7 reagent. (C) Numbers of NY8.3 TCR expressing DP and CD8+ SP thymocytes from 4-week-old female NOD.NY8.3 and NOD.NY8.3-Nfkbid−/− homozygous mice. Bars represent the mean±SEM of 3 independent experiments. (D) Geometric MFI of Tet-NRPV7 staining of DP and CD8+ SP thymocytes from NOD.NY8.3, NOD.NY8.3-Nfkbid+/−, and NOD.NY8.3-Nfkbid−/− mice. Bars represent mean±SEM. Statistical significance was analyzed by 2-way ANOVA and Bonferroni’s multiple comparison test. ns p>0.05, ** p<0.01.
Nfkbid deficiency accelerates T1D in NOD mice associated with impaired development of regulatory lymphocyte populations.
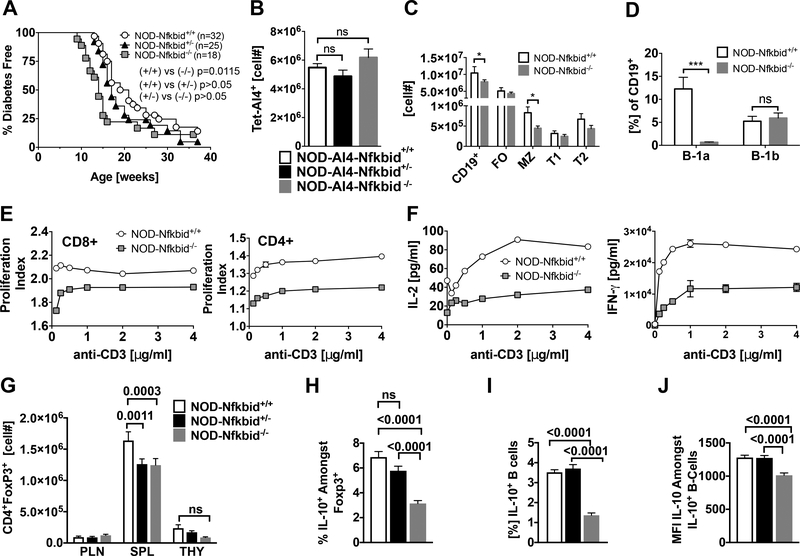
We previously reported that while numerically reduced in the thymus, the AI4 T-cells that did reach the periphery of B6.H2g7 and NOD.Chr7B6FL background mice were still able to expand and mediate T1D development (7, 8). Similarly, despite being characterized by enhanced thymic deletion of pathogenic T cells T1D development was actually accelerated in NOD-Nfkbid−/− mice (Fig. 6A). Such T1D acceleration did not occur NOD-Nfkbid+/− heterozygous mice (Fig. 6A). These results indicated T1D acceleration is dependent on complete ablation of Nfkbid expression. As previously observed upon introduction of the Chr7B6FL congenic interval, ablation of Nfkbid did not alter numbers of peripheral splenic AI4 T-cells in NOD mice (Fig. 6B). A similar effect was seen for diabetogenic NY8.3 CD8+ T-cells (data not shown). Hence, the thymic Nfkbid mediated genetic effect characterizing the NOD strain does not limit the ability of diabetogenic effectors that do reach the periphery to expand and/or survive and mediate disease development. We also assessed if ablation of the Nfkbid gene in NOD mice induced any numerical or functional differences in peripheral leukocyte populations that could possibly affect diabetogenic T-cell activity. As previously observed for the autoimmune resistant B6 background stocks (34, 35, 37), numbers of splenic CD4+ and CD8+ T-cells in NOD-Nfkbid−/− mice did not differ from wild type controls (data not shown). Also as previously observed in B6 (38), marginal zone (MZ) B-lymphocytes in the spleen and B1a cells in the peritoneum were both significantly reduced in Nfkbid deficient NOD mice (Fig. 6C, D). The proliferative capacity of CD4+ and CD8+ T-cells and their ability to secrete the cytokines IL-2 and IFNγ was reported to be diminished in Nfkbid deficient B6 mice (34). Similar phenotypes characterized CD4+ and CD8+ T-cells from NOD-Nfkbid−/− mice (Fig. 6E, F). Ablation of Nfkbid has also been reported to diminish numbers of FoxP3 expressing Tregs in B6 background mice (35). A similar numerical reduction in Tregs was elicited by both homozygous and heterozygous ablation of Nfkbid in the NOD strain (Fig. 6G). Hence, a diminution of Nfkbid expression to B6-like levels is sufficient to decrease Treg numbers in otherwise NOD genetic background mice. Previous reports indicated Nfkbid is a direct regulator of IL-10 (27). Homozygous, but not heterozygous ablation of Nfkbid also significantly reduced the proportions of FoxP3+ Tregs in NOD mice producing immunosuppressive IL-10 upon ex vivo stimulation (Fig. 6H). We recently found that IL-10 producing Bregs can inhibit autoimmune T1D development in NOD mice (39, 40). Homozygous, but not heterozygous ablation of Nfkbid also significantly reduced the proportions of IL-10 producing Bregs in NOD mice, as well as reducing in the remaining positive cells expression levels of this immunosuppressive cytokine (Fig. 6I, J). Among these alterations in potential immunoregulatory lymphocyte populations induced by complete ablation of Nfkbid, the numerical decrease in splenic Tregs was recapitulated in NOD.Ln82 congenic mice, albeit for unknown reasons a higher proportion of those that remained produced IL-10 than in NOD controls (Supplemental Fig. 2). Similar to heterozygous ablation of Nfkbid, presence of the Ln82 congenic interval did not alter proportions of IL-10 expressing B-lymphocytes or their production level of this cytokine (Supplemental Fig. 2). Hence, a complete ablation of Nfkbid expression is required to diminish IL-10 production by Tregs and Bregs. These collective data indicate that while reducing Nfkbid expression enhances thymic negative selection of diabetogenic CD8+ T-cells in NOD mice, the coincidental decrease in numbers of immunoregulatory lymphocyte populations that occurs when this gene is completely ablated may account for why any pathogenic effectors which do reach the periphery still aggressively expand and induce disease development.
Figure 6: Nfkbid deficiency accelerates T1D onset in NOD mice associated with impaired development of regulatory lymphocyte populations.
(A) T1D development was analyzed in a cohort of NOD, NOD-Nfkbid+/− and NOD-Nfkbid−/− female mice. Survival curve comparisons were analyzed through Log-rank (Mantel-Cox) test. (B) Numbers of splenic AI4 T-cells in 6-week-old NOD (n=31), NOD-Nfkbid+/− (n=12) and NOD-Nfkbid−/− (n=16) female mice. (C) Splenocytes from 7-week-old female NOD (n=3) and NOD-Nfkbid−/−(n=5) mice were labeled with antibody for CD19, CD21 and CD23 and analyzed by flow cytometry. Singlet, DAPI- CD19+ cells were identified as total B-cells. Based on expression of CD23 and CD21, B-cells were classified as follicular (FO: CD23+CD21lo), marginal zone (MZ: CD23-CD21hi) transitional 1 (T1: CD23-CD21-) and transitional 2 (T2: CD23int CD21hi) (D) Peritoneal cavity B-cells were isolated by lavage with cold HBSS from the same group of mice described in (C), and stained for CD19, B220, CD5 and analyzed by flow cytometry. Total B-cells (Singlet, DAPI- CD19+) were classified based in the expression of B220 and CD5 as B1-a (CD5+ B220int/lo) and B1-b (CD5- B220lo) (E). The proliferation capacity of purified total T-cells from NOD and NOD-Nfkbid−/− mice was assessed as described in methods section. Briefly, MACS purified total T-cells were labeled with cell tracer eFluor670 and stimulated in vitro with increasing amounts of anti-CD3 monoclonal antibody. After 48 hours of culture the cells were labeled with antibody for CD4 and CD8. Proliferation in CD4+ and CD8+ T-cell subsets was analyzed by flow cytometric analyses of cell tracer eFluor670 dilution. Proliferation index was determined using the Flow Jo cell proliferation tool. Data represent the mean±SEM of 3 technical replicates. (F) Cell culture supernatants of purified T-cells used in the proliferation assay were assessed for secretion of the cytokines IL-2 and IFN-γ by ELISA. Data represent mean±SEM of 3 technical replicates. (G) Enumeration of CD4+ FoxP3+ Tregs in 6–11-week-old combined male and female NOD, NOD-Nfkbid+/−, and NOD-Nfkbid−/− mice (n=7–10 mice per group per organ) from pancreatic lymph node (PLN), spleen (SPL) and thymus (THY). (H) Identification by intracellular staining and subsequent flow cytometric analyses of proportions of Foxp3+ splenic CD4+ T-cells from 6–11-week-old female NOD, NOD-Nfkbid+/−, and NOD-Nfkbid−/− mice producing IL-10 (n=8–10 mice per group). (I) Identification by intracellular staining and subsequent flow cytometric analyses of proportions of IL-10 producing splenic B-lymphocytes in 6–11-week-old combined male and female NOD, NOD-Nfkbid+/−, and NOD-Nfkbid−/− mice (n=8–10 mice per group). (J) Median fluorescence intensity (MFI) of IL-10 staining amongst IL-10+ B-lymphocytes in 6–11-week-old combined male and female NOD, NOD-Nfkbid+/−, and NOD-Nfkbid−/− mice mice (n=8–10 mice per group). For G-J, extracellular staining was followed by fixation/permeabilization and then intracellular staining for IL-10 and/or Foxp3. For H-J, splenocytes were stimulated for 5hrs with LPS 10μg/mL, PMA 50ng/mL, ionomycin 500ng/mL and monensin 2μM prior to extracellular staining. All bar graphs represent mean±SEM. Statistical significance was determined by 2-way ANOVA and Bonferroni’s multiple comparison test, or 1-way ANOVA and Bonferroni’s multiple comparison test for H-J. ns p>0.05, * p<0.05, ** p<0.01, *** p<0.001.
Discussion
Previous work from our group demonstrated the common class I variants expressed by NOD mice (H2-Kd, H2-Db) have an aberrant lessened ability to mediate the thymic negative selection of diabetogenic CD8+ T-cells, primarily due to interactions with some non-MHC gene(s) on proximal Chr. 7 (7). In this study, we demonstrate an elevated Nfkbid expression allele is likely a proximal Chr. 7 region gene contributing to diminished thymic deletion of diabetogenic CD8+ T-cells in NOD mice. It remains unclear if Nfkbid is a component of the Chr. 7 Idd7 locus contributing to T1D susceptibility or resistance. Forty-three SNPs distinguish the NOD and B6 Nfkbid alleles. The majority of them found in introns, the 5’ and 3’ UTR with only three synonymous variants present within coding regions of Nfkbid. Some subset of the non-coding region SNPs likely regulates varying expression levels of the NOD and B6 Nfkbid alleles.
Nfkbid is a mostly nuclear protein containing 7 ankyrin repeat domains originally reported as an inhibitor of NF-κB signaling (31). However, due to the lack of DNA binding and transactivation domains, Nfkbid function is ultimately mediated through protein-protein interactions with NF-κB family members. Accordingly, Nfkbid can function as an inhibitor or enhancer of NF-κB activities depending on the protein partner and cell type (17). This includes previously reported interactions of Nfkbid with the NF-κB subunit p50 (Nfkb1) in thymocytes undergoing negative selection (31). Our current results indicate Nfkbid is not required for thymocytes destined for the CD8+ lineage to undergo negative selection, and instead its particular expression level in NOD mice appears to diminish the efficiency of this process. Both our microarray analysis of NOD.LCMV thymocytes, as well as qPCR analysis of sorted NOD-AI4 DP thymocytes, indicate that in the context of negative selection of autoreactive CD8+ T-cells, Nfkbid likely acts as an enhancer of NF-κB activity. Nfkbid can be incorporated into NF-κB heterodimers with the p50, p52, or c-Rel binding partners (17). As we observed in this study with ablation of Nfkbid, an acceleration of T1D development concomitant with a decrease in Treg numbers characterizes NOD mice made deficient in c-Rel expression (41). Thus, it is possible that in both cases T1D acceleration resulted from a decrease in c-Rel/Nfkbid heterodimer levels. The future dissection of such possible pathways in our NOD-AI4 model system is warranted to better understand how Nfkbid/NF-κB signaling contributes to CD8+ T-cell selection.
Three different Nfkbid deficient mouse models have been previously produced, all on the B6 background (34, 37, 42). These previously produced models have been useful to identify several functional aspects of Nfkbid. In the B-cell compartment, Nfkbid expression is necessary for development of the MZ and B1-a subsets as well as the generation of antibody responses to T-dependent and independent antigens (34, 38, 42–44). Nfkbid deficiency was also associated with an impairment in IL-10 secreting B-lymphocytes (45). Nfkbid deficient T-cells have impaired in vitro proliferation and cytokine secretion (IL-2, IFN-γ) (34). These phenotypes also characterize our Nfkbid deficient NOD mouse stock.
Interaction of Nfkbid with c-Rel reportedly contributes to Foxp3 expression in Tregs (35). Thus, we found it of interest that in addition to enhancing thymic deletion of diabetogenic CD8+ T-cells, diminution of Nfkbid expression also resulted in decreased numbers of peripheral Tregs in NOD mice. We recently reported that IL-10 producing Bregs can exert T1D protective immunoregulatory activity in NOD mice (39, 40). Ablation of Nfkbid expression also diminished levels of IL-10 producing Bregs in the periphery. Thus, we conjecture it is the loss of both Tregs and Bregs that at least in part accounts for the finding that T1D development is actually accelerated in Nfkbid deficient NOD mice despite their improved, albeit not completely restored, ability to mediate the thymic deletion of pathogenic CD8+ T-cells. Hence, Nfkbid expression levels appear to act as a two-edged sword, levels lower than in the thymus of NOD mice can result in better negative selection of autoreactive CD8+ T-cells, but may also contribute to autoimmunity in the periphery by numerically reducing regulatory lymphocyte populations. Determining whether Nfkbid expression- or activity-levels can be fine-tuned to improve autoreactive CD8+ T-cell negative selection without diminishing Treg and Breg populations is warranted.
In conclusion, Nfkbid expression levels appear to be an important contributory factor to how efficiently autoreactive diabetogenic CD8+ T-cells undergo thymic negative selection. However, this study also revealed a caveat that would have to be considered if it might ultimately become possible to diminish Nfkbid expression through pharmacological means. That is while diminishing Nfkbid expression can allow for increased thymic deletion of diabetogenic CD8+ T-cells, it does so at the price of numerically reducing levels of regulatory lymphocytes that may allow any pathogenic effectors that do reach the periphery to more rapidly induce disease onset. This may explain our previous findings that the proximal Chr. 7 region exerts opposing effects where the presence of B6-derived genome in the region reduced thymic levels of diabetogenic CD8+ T-cells, but in the periphery actually promotes disease development (8). Thus, when contemplating possible future interventions designed to diminish numbers of diabetogenic T-cells, a consideration that should be taken into account is whether the treatment might also impair regulatory cell numbers or activity potentially allowing the pathogenic effectors which do remain present to become more aggressive.
Supplementary Material
Acknowledgements
We would like to thank the Jackson Laboratory Research Animal Facility, Genetic Engineering Technology group, Flow Cytometry service, and Transgenic Genotyping core for technical support on this project. We thank the National Institutes of Health tetramer core facility for the tetramers used in this study. The authors would also like to thank Carl Stiewe and his wife, Maike Rohde, for their generous donation toward T1D research at The Jackson Laboratory, which contributed to this work.
Footnotes
MP was supported by JDRF Fellowship 3-PDF-2014–219-A-N. For parts of this work, JJR1 was supported by either NIH Fellowship 1F32DK111078 or JDRF Fellowship 3-PDF-2017–372-A-N. He was also supported by a grant from the Diabetes Research Connection DRC 006887 JR. DVS is supported by NIH grants DK-46266, DK-95735, and OD-020351–5022, as well as by Juvenile Diabetes Research Foundation grant 2018–568. This work was also partly supported by Cancer Center Support Grant CA34196. IS was supported by grants of the Deutsche Forschunggemeinschaft (SCHM1586/6–1 and project A23 of SFB854).
MP designed and conducted experimentation, interpreted data, and wrote the manuscript. JJR1, designed and conducted experimentation, contributed to data interpretation, and writing of the manuscript. JRD designed and conducted experiments, interpreted data and contributed to writing the manuscript. JJR2, DJL, JA and HDC conducted experimentation. VKS and TS contributed to statistical analyses. YGC, AG and IS contributed to experimental design. DVS contributed to study conception, supervised experimental effort, and writing of the manuscript.
C57BL/6J, (B6); CD4+CD8+ double positive, (DP); CD4+Foxp3+ regulatory T-cell, (Treg); chromosome, (Chr.); dendritic cell, (DC); Lymphocytic Choriomeningitis Virus, (LCMV); H2-Db/MimA2 tetramer capable of binding the AI4 TCR, (Tet-AI4); H2-Kd/NRPV7 tetramer capable of binding the NY8.3 TCR, (Tet-NRPV7); islet-specific glucose-6-phosphatase catalytic subunit-related protein, (IGRP); NOD/ShiLtDvs, (NOD); NOD/ShiLtDvs-Nfkbid<em3Dvs>/Dvs, (NOD-Nfkbid−/−); NOD.B6-(Gpi1-D7Mit346)/LtJ, (NOD.Chr7B6FL); NOD.Cg-Prkdcscid Emv30b/Dvs, (NOD.scid); pancreatic lymph node, (PLN); propidium iodide, (PI); regulatory B-cell, (Breg); single guide RNA, (sgRNA); type 1 diabetes, (T1D); spleen, (SPL); thymus, (Thy); type 1 diabetes susceptibility, (Idd).
Microarray data has been deposited at geo@ncbi.nlm.nih.gov. Accession # GSE115754.
References
- 1.Atkinson MA, Eisenbarth GS, and Michels AW. 2014. Type 1 diabetes. The Lancet 383: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ram R, Mehta M, Nguyen QT, Larma I, Boehm BO, Pociot F, Concannon P, and Morahan G. 2016. Systematic Evaluation of Genes and Genetic Variants Associated with Type 1 Diabetes Susceptibility. The Journal of Immunology 196: 3043–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driver J, Serreze D, and Chen Y-G. 2011. Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Seminars in Immunopathology 33: 67–87. [DOI] [PubMed] [Google Scholar]
- 4.Geenen V 2012. Thymus and type 1 diabetes: an update. Diabetes Res Clin Pract 98: 26–32. [DOI] [PubMed] [Google Scholar]
- 5.Jeker LT, Bour-Jordan H, and Bluestone JA. 2012. Breakdown in Peripheral Tolerance in Type 1 Diabetes in Mice and Humans. Cold Spring Harbor Perspectives in Medicine 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishimoto H, and Sprent J. 2001. A defect in central tolerance in NOD mice. Nat Immunol 2: 1025–1031. [DOI] [PubMed] [Google Scholar]
- 7.Choisy-Rossi C-M, Holl TM, Pierce MA, Chapman HD, and Serreze DV. 2004. Enhanced Pathogenicity of Diabetogenic T Cells Escaping a Non-MHC Gene-Controlled Near Death Experience. The Journal of Immunology 173: 3791–3800. [DOI] [PubMed] [Google Scholar]
- 8.Serreze DV, Choisy-Rossi CM, Grier AE, Holl TM, Chapman HD, Gahagan JR, Osborne MA, Zhang W, King BL, Brown A, Roopenian D, and Marron MP. 2008. Through Regulation of TCR Expression Levels, an Idd7 Region Gene(s) Interactively Contributes to the Impaired Thymic Deletion of Autoreactive Diabetogenic CD8+ T Cells in Nonobese Diabetic Mice. The Journal of Immunology 180: 3250–3259. [DOI] [PubMed] [Google Scholar]
- 9.von Boehmer H, and Melchers F. 2010. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol 11: 14–20. [DOI] [PubMed] [Google Scholar]
- 10.Varanasi V, Avanesyan L, Schumann DM, and Chervonsky AV. 2012. Cytotoxic Mechanisms Employed by Mouse T Cells to Destroy Pancreatic β-Cells. Diabetes 61: 2862–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight RR, Kronenberg D, Zhao M, Huang GC, Eichmann M, Bulek A, Wooldridge L, Cole DK, Sewell AK, Peakman M, and Skowera A. 2013. Human β-Cell Killing by Autoreactive Preproinsulin-Specific CD8 T Cells Is Predominantly Granule-Mediated With the Potency Dependent Upon T-Cell Receptor Avidity. Diabetes 62: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, and Todd JA. 2007. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, Lane JA, Lavant E, Rappner R, Louey A, Concannon P, Mychaleckyj JC, Erlich HA, and f. t. T. D. G. Consortium. 2010. HLA Class I and Genetic Susceptibility to Type 1 Diabetes: Results From the Type 1 Diabetes Genetics Consortium. Diabetes 59: 2972–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Reifsnyder P, Scheuplein F, Schott W, Mileikovsky M, Soodeen-Karamath S, Nagy A, Dosch M, Ellis J, Koch-Nolte F, and Leiter E. 2005. “Agouti NOD”: identification of a CBA-derived Idd locus on Chromosome 7 and its use for chimera production with NOD embryonic stem cells. Mammalian Genome 16: 775–783. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh S, Palmer SM, Rodrigues NR, Cordell HJ, Hearne CM, Cornall RJ, Prins JB, McShane P, Lathrop GM, Peterson LB, and et al. 1993. Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat Genet 4: 404–409. [DOI] [PubMed] [Google Scholar]
- 16.McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, Rodrigues NR, Prins J-B, Serreze DV, DeLarato NH, Wicker LS, Peterson LB, Schork NJ, Todd JA, and Leiter EH. 1995. Crosses of NOD Mice With the Related NON Strain: A Polygenic Model for IDDM. Diabetes 44: 1186–1195. [DOI] [PubMed] [Google Scholar]
- 17.Schuster M, Annemann M, Plaza-Sirvent C, and Schmitz I. 2013. Atypical IkappaB proteins - nuclear modulators of NF-kappaB signaling. Cell Communication and Signaling 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graser RT, DiLorenzo TP, Wang F, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, and Serreze DV. 2000. Identification of a CD8 T cell that can independently mediate autoimmune diabetes development in the complete absence of CD4 T cell helper functions. J Immunol 164: 3913–3918. [DOI] [PubMed] [Google Scholar]
- 19.Pircher H, Burki K, Lang R, Hengartner H, and Zinkernagel RM. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature 342: 559–561. [DOI] [PubMed] [Google Scholar]
- 20.Serreze DV, Johnson EA, Chapman HD, Graser RT, Marron MP, DiLorenzo TP, Silveira P, Yoshimura Y, Nathenson SG, and Joyce S. 2001. Autoreactive Diabetogenic T-Cells in NOD Mice Can Efficiently Expand From a Greatly Reduced Precursor Pool. Diabetes 50: 1992–2000. [DOI] [PubMed] [Google Scholar]
- 21.Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, and Santamaria P. 1997. Spontaneous Autoimmune Diabetes in Monoclonal T Cell Nonobese Diabetic Mice. The Journal of Experimental Medicine 186: 1663–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill JT, Demarest BL, Bisgrove BW, Su Y-C, Smith M, and Yost HJ. 2014. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Developmental Dynamics 243: 1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Kerr MK, Cui X, and Churchill GA. 2003. MAANOVA: A Software Package for the Analysis of Spotted cDNA Microarray Experiments In The Analysis of Gene Expression Data: Methods and Software Parmigiani G, Garrett ES, Irizarry RA, and Zeger SL, eds. Springer New York, New York, NY: 313–341. [Google Scholar]
- 24.Storey JD 2002. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 64: 479–498. [Google Scholar]
- 25.Julius MH, and Herzenberg LA. 1974. ISOLATION OF ANTIGEN-BINDING CELLS FROM UNPRIMED MICE. Demonstration of Antibody-Forming Cell Precursor Activity and Correlation between Precursor and Secreted Antibody Avidities 140: 904–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman SM, Takaki T, Han B, Santamaria P, Serreze DV, and DiLorenzo TP. 2004. Individual Nonobese Diabetic Mice Exhibit Unique Patterns of CD8+ T Cell Reactivity to Three Islet Antigens, Including the Newly Identified Widely Expressed Dystrophia Myotonica Kinase. The Journal of Immunology 173: 6727–6734. [DOI] [PubMed] [Google Scholar]
- 27.Annemann M, Wang Z, Plaza-Sirvent C, Glauben R, Schuster M, Ewald Sander F, Mamareli P, Kühl AA, Siegmund B, Lochner M, and Schmitz I. 2015. IκBNS Regulates Murine Th17 Differentiation during Gut Inflammation and Infection. The Journal of Immunology 194: 2888–2898. [DOI] [PubMed] [Google Scholar]
- 28.Taylor SC, and Posch A. 2014. The Design of a Quantitative Western Blot Experiment. BioMed Research International 2014: 361590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fosang AJ, and Colbran RJ. 2015. Transparency Is the Key to Quality. Journal of Biological Chemistry 290: 29692–29694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamont D, Mukherjee G, Kumar PR, Samanta D, McPhee CG, Kay TWH, Almo SC, DiLorenzo TP, and Serreze DV. 2014. Compensatory Mechanisms Allow Undersized Anchor-Deficient Class I MHC Ligands To Mediate Pathogenic Autoreactive T Cell Responses. The Journal of Immunology 193: 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiorini E, Schmitz I, Marissen WE, Osborn SL, Touma M, Sasada T, Reche PA, Tibaldi EV, Hussey RE, Kruisbeek AM, Reinherz EL, and Clayton LK. 2002. Peptide-Induced Negative Selection of Thymocytes Activates Transcription of an NF-kB Inhibitor. Molecular cell 9: 637–648. [DOI] [PubMed] [Google Scholar]
- 32.Jimi E, Strickland I, Voll RE, Long M, and Ghosh S. 2008. Differential Role of the Transcription Factor NF-κB in Selection and Survival of CD4+ and CD8+ Thymocytes. Immunity 29: 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T, Howe K, Kähäri A, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel J-H, White S, Zadissa A, Flicek P, and Searle SMJ. 2016. The Ensembl gene annotation system. Database 2016: baw093-baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Touma M, Antonini V, Kumar M, Osborn SL, Bobenchik AM, Keskin DB, Connolly JE, Grusby MJ, Reinherz EL, and Clayton LK. 2007. Functional Role for IκBNS in T Cell Cytokine Regulation As Revealed by Targeted Gene Disruption. The Journal of Immunology 179: 1681–1692. [DOI] [PubMed] [Google Scholar]
- 35.Schuster M, Glauben R, Plaza-Sirvent C, Schreiber L, Annemann M, Floess S, Kühl Anja A., Clayton Linda K., Sparwasser T, Schulze-Osthoff K, Pfeffer K, Huehn J, Siegmund B, and Schmitz I. 2012. IκBNS Protein Mediates Regulatory T Cell Development via Induction of the Foxp3 Transcription Factor. Immunity 37: 998–1008. [DOI] [PubMed] [Google Scholar]
- 36.Amrani A, Verdaguer J, Serra P, Tafuro S, Tan R, and Santamaria P. 2000. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature 406: 739–742. [DOI] [PubMed] [Google Scholar]
- 37.Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, Koga R, and Takeda K. 2006. IκBNS Inhibits Induction of a Subset of Toll-like Receptor-Dependent Genes and Limits Inflammation. Immunity 24: 41–51. [DOI] [PubMed] [Google Scholar]
- 38.Touma M, Keskin DB, Shiroki F, Saito I, Koyasu S, Reinherz EL, and Clayton LK. 2011. Impaired B Cell Development and Function in the Absence of IκBNS. The Journal of Immunology 187: 3942–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratiu JJ, Racine JJ, Hasham MG, Wang Q, Branca JA, Chapman HD, Zhu J, Donghia N, Philip V, Schott WH, Wasserfall C, Atkinson MA, Mills KD, Leeth CM, and Serreze DV. 2017. Genetic and Small Molecule Disruption of the AID/RAD51 Axis Similarly Protects Nonobese Diabetic Mice from Type 1 Diabetes through Expansion of Regulatory B Lymphocytes. The Journal of Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Racine JJ, Ratiu JJ, Wang S, Ettinger R, Wasserfall C, Atkinson MA, and Serreze DV. 2017. Transient BAFF Blockade Inhibits Type 1 Diabetes Development in Nonobese Diabetic Mice by Enriching Immunoregulatory B Lymphocytes Sensitive to Deletion by Anti-CD20 Cotherapy. The Journal of Immunology 199: 3757–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishnan P, Yui MA, Tomalka JA, Majumdar D, Parameswaran R, and Baltimore D. 2016. Deficiency of Nuclear Factor-κB c-Rel Accelerates the Development of Autoimmune Diabetes in NOD Mice. Diabetes 65: 2367–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB, and Beutler B. 2012. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proceedings of the National Academy of Sciences 109: 12286–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen GK, Àdori M, Khoenkhoen S, Dosenovic P, Beutler B, and Karlsson Hedestam GB. 2014. B-1a transitional cells are phenotypically distinct and are lacking in mice deficient in IκBNS. Proceedings of the National Academy of Sciences 111: E4119–E4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen GK, Ádori M, Stark j. M., Khoenkhoen S, Arnold C, Beutler B, and Karlsson Hedestam GB. 2016. Heterozygous mutation in IκBNS leads to reduced levels of natural IgM antibodies and impaired responses to T-independent type 2 antigens. Frontiers in Immunology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miura M, Hasegawa N, Noguchi M, Sugimoto K, and Touma M. 2016. The atypical IκB protein IκBNS is important for Toll-like receptor-induced interleukin-10 production in B cells. Immunology 147: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.