Abstract
After decades of directed research, there remains no effective regenerative therapy for the injured human heart. The epicardium, a layer of mesothelial tissue that envelops all vertebrate hearts, has emerged as a recent player in cardiac repair and regeneration. The epicardium is essential for muscle regeneration in the zebrafish model of innate heart regeneration, and it also participates in fibrotic responses of mammalian hearts. This structure serves as a source of key cells like vascular smooth muscle, pericytes, and fibroblasts during heart development and repair. It also secretes factors that are essential for proliferation and survival of cardiomyocytes. Here, we describe recent advances in our understanding of the biology of the epicardium and the impact of these findings on its candidacy as a therapeutic target for heart repair.
1. Introduction
One of the most significant challenges of regenerative medicine is devising how to replace millions of cardiomyocytes (CMs) that are lost after myocardial infarction (MI). In the past two decades, researchers have identified several potential strategies for cardiac repair, such as inducing the proliferation of existing CMs, administering cell therapy by injecting stem cells or stem cell-derived CMs, reprogramming non-muscle cells into CMs, applying a hypoxic environment, and applying patches seeded with pro-regenerative factors (Cahill et al., 2017; Galdos et al., 2017; Tzahor and Poss, 2017; Uygur and Lee, 2016; Zhang et al., 2015). However, no clinically meaningful regenerative therapy is currently available for human heart disease. Diverse approaches that combine efforts from multiple fields and use different model systems are required.
The epicardium is a thin mesothelial tissue comprising the outermost layer of vertebrate hearts. It represents a multipotent cardiac progenitor tissue and a signaling center for heart development. Recent studies have indicated the epicardium may be a key target for cardiac repair strategies. The findings obtained in investigations exploring the epicardium of adult mouse hearts, which are poorly regenerative, have mirrored those obtained by studying the highly regenerative zebrafish (Huang et al., 2012; Kikuchi et al., 2011a; Lepilina et al., 2006; Smart et al., 2011; Smart et al., 2007; Wang et al., 2013; Zhou et al., 2011). In this review, we discuss the models currently available to explore innate heart regeneration, the cellular and molecular contributions of epicardial cells to cardiac regeneration and scarring, the regenerative capacity of the epicardium itself, and the development of strategies to harness the properties of the epicardium to improve mammalian heart repair.
2. Models of heart regeneration
Functional regeneration of an injured heart is expected to involve clearance of dead tissue, restoration of lost muscle, revascularization, electrical coupling of new CMs, and resolution of inflammation and collagen/fibrin (Gonzalez-Rosa et al., 2011; Kikuchi et al., 2010; Lai et al., 2017; Marin-Juez et al., 2016; Poss et al., 2002). Of these events, the key endpoint is manifestation of new, healthy CMs. Although low-level CM proliferation has been reported in adult mammalian hearts, there are ostensibly too few such natural events to have a meaningful impact on heart repair (Mollova et al., 2013; Senyo et al., 2013; Zhang et al., 2010). In 2011, Porrello et al. performed experiments in which they removed 10% of the ventricular apex from neonatal mice of various ages. Their results were the first to demonstrate that neonatal mice could, in the very first days of post-natal life, mount a significant regenerative response after resection injury to yield full size ventricles with limited scarring. Genetic fate-mapping indicated that this repair is mediated through the proliferation of existing CMs (Porrello et al., 2011). Neonatal mouse heart regeneration has also been demonstrated after ligation of the left anterior descending artery (LAD), applied to one‐day‐old mice (Haubner et al., 2012; Porrello et al., 2013). In a cryoinjury model, in which a cooled probe was applied to the ventricular surface in one‐day‐old mice to induce localized cell death, hearts regenerate after non-transmural but not transmural injury (Darehzereshki et al., 2015). It is not surprising that the regenerative capacity relies on the severity of injury. Mice lose this regenerative capacity by 7 days of age, after which injury results in scarring. It is unlikely to be purely coincidence that the ability to regenerate is linked closely with a period of massive cardiac growth by CM proliferation (Soonpaa and Field, 1998). Additionally, at early post-natal stages most murine CMs are diploid; recent studies have associated CM polyploidy with the capacity to injury-induced regeneration (Gonzalez-Rosa et al., 2018; Patterson et al., 2017).
By contrast with mammals, lower vertebrate model systems like zebrafish and certain urodele amphibians possess an elevated ability to regenerate injured heart muscle as adults (Cano-Martinez et al., 2010; Chablais et al., 2011; Mercer et al., 2013; Poss et al., 2002; Wang et al., 2011). Among these, the zebrafish has been most extensively studied because of the consistent and extensive replacement of heart muscle, and the advantages of the system for molecular genetics.
The first model of zebrafish heart regeneration was introduced over 15 years ago (Poss et al., 2002). Within 2 months of surgical removal of ~20% of the ventricular apex, the resected tissue is replaced by a wall of new muscle. This regenerative process involves rapid clotting, CM proliferation, and neovascularization. Collagen is evident during regeneration but there is little or no permanent scarring. Resection injuries deplete muscle by direct removal as well as death at the injury site; therefore a cryoinjury model was developed in 2011 to induce localized cell death in ~25% of the ventricle (Chablais et al., 2011; Gonzalez-Rosa et al., 2011). This model better mimics MI and involves cell death, a strong inflammatory response, CM proliferation and transient collagen deposition, with much of the collagen eventually clearing in 3–4 months. Coincidentally, Wang and colleagues established a genetic model to induce immediate destruction of up to ~60% of ventricular (and atrial) CMs by expression of diphtheria toxin A (Wang et al., 2011). This injury is limited to CMs and has several experimental advantages: 1) a non-surgical, tamoxifen-activated injury with high consistency; 2) rapid regeneration within one month of ablation, likely facilitated by preservation of non-myocardial scaffolding like endocardium and ECM; 3) a diffuse injury throughout the heart that activates a regenerative response from most or all of the spared myocardium, facilitating large-scale tissue collection and profiling experiments (Goldman et al., 2017; Wang et al., 2013); and 4) affected animals develop heart failure that resolves during innate muscle regeneration (Wang et al., 2011). The cellular and molecular injury responses can vary somewhat between methods of injury (reviewed and compared in (Gonzalez-Rosa et al., 2017; Uygur and Lee, 2016; Vivien et al., 2016)).
Studies in zebrafish have provided concepts and mechanisms germane to heart regeneration. For instance, new muscle is generated from spared CMs, which transiently dedifferentiate and proliferate – not stem cells, a mechanism that also occurs during neonatal mouse heart regeneration. Also, CM proliferation is promoted by the activities of non-muscle cells like endocardium, nerves, and immune cells (Choi et al., 2013; Gonzalez-Rosa et al., 2012; Hui et al., 2017; Jopling et al., 2010; Kikuchi et al., 2011a; Kikuchi et al., 2011b; Kikuchi et al., 2010; Kim et al., 2010; Lepilina et al., 2006; Mahmoud et al., 2015; Poss et al., 2002; Wang et al., 2011). Most relevant to the topic of this review, the epicardium was first implicated as a participant in heart regeneration through studies with adult zebrafish (Lepilina et al., 2006).
3. The epicardium as a progenitor tissue
3.1. Formation of the epicardium during heart development
The epicardium was first described in a published study of dissected chick embryos by Kurkiewicz (Kurkiewicz, 1909). This observation was confirmed by electron microscopy ~60 years later (Manasek, 1968, 1969). Further studies found that the epicardial cells are derived from a transient embryonic cell cluster called the proepicardial organ (PEO). The PEO was first described in the chick as pericardial villi that form at the venous pole of the embryonic heart tube (Manner, 1992, 1993). The PEO has been investigated in many additional species, including zebrafish, Xenopus, axolotl, mouse, rat and human (Fransen and Lemanski, 1990; Hirakow, 1992; Jahr et al., 2008; Komiyama et al., 1987; Nesbitt et al., 2006; Risebro et al., 2015; Serluca, 2008). The formation and cellular components of the PEO have been reviewed elsewhere (Maya-Ramos et al., 2013). Proepicardial (PE) cells were reported to translocate to the ventricular surface by direct PEO-chamber contact, and/or by release of PE cells into the pericardial cavity and adherence to the ventricle, depending on the species and approaches that were used (Fransen and Lemanski, 1990; Jahr et al., 2008; Komiyama et al., 1987; Nahirney et al., 2003; Peralta et al., 2013; Plavicki et al., 2014; Rodgers et al., 2008). By live surveillance of PEO formation and PE cell translocation in zebrafish embryos, Peralta and colleagues defined three clusters of epicardial precursor cells, located at the level of the atrioventricular canal, adjacent to the venous pole, and one that directly contacts the myocardium (located at a region of the pericardial mesothelium close to the arterial pole) (Peralta et al., 2013). Cells from the first two PE clusters adhere to the ventricle between 60 to 72 hours post fertilization (hpf). After translocation to the myocardial surface, PE cells expand over the surfaces of ventricle, atrium and outflow tract (or bulbous arteriosus, BA) to form a contiguous epicardial sheet. During development, a subset of epicardial cells delaminates from the epicardium and undergoes epithelial-to-mesenchymal transition (EMT). During EMT, these cells migrate to the subepicardial space, where they give rise to a variety of cell types often referred to collectively as epicardium-derived cells (EPDCs) (Lie-Venema et al., 2007).
3.2. Cellular contributions of the epicardium in higher vertebrates
Early studies in chick embryos used dyes to label epicardial cells, employed cell transplantation in chimeras containing quail cells, and tagged epicardial cells with retroviral vectors. These approaches all indicated that the epicardium contributes to vascular smooth muscle cells and cardiac fibroblasts during avian heart development (Table 1). More controversial findings indicated contributions to endothelial and endocardial cells in chick embryos (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Guadix et al., 2006; Manner, 1999; Mikawa and Fischman, 1992; Mikawa and Gourdie, 1996; Perez-Pomares et al., 2002a; Perez-Pomares et al., 2002b). These approaches used in avians might have limitations in efficiency, specificity, and interpretation. A contemporary approach to assess the cell fates in several model systems is genetic fate-mapping, which requires regulatory sequences to induce a permanent label within a candidate progenitor cell population and its progeny in transgenic animals. In these experiments, a regulatory sequence-driven cassette that harbors a tamoxifen-inducible Cre recombinase (CreER) is often used in conjunction with a fluorescent reporter gene that is normally kept inactive by an upstream loxP-flanked stop sequence separating the reporter gene from a constitutive promoter. Importantly, there are no gene regulatory sequences known to direct expression that is absolutely restricted to the epicardium. Yet, several markers of the epicardium (and other tissues) have been employed to permanently label and trace epicardial cells in mice. Transcription factor 21 (Tcf21), Wilms’ tumor 1 (Wt1), T-Box 18 (Tbx18), Scleraxis (Scx), and Semaphorin 3D (Sema3D) are each expressed in the epicardium, but their regulatory sequences also mark additional populations (Braitsch et al., 2013; Braitsch and Yutzey, 2013; Cai et al., 2008; Christoffels et al., 2009; Katz et al., 2012; Rudat and Kispert, 2012; Zhou et al., 2008a; Zhou et al., 2008b). The reliance on a set of regulatory sequences to infer cell lineage relationships is a pitfall of using Cre/CreER lines for tracing epicardial cell fates. Other caveats include the persistence of the inducer tamoxifen within animals; the toxicity of Cre lines in some cell types; the fact that Cre knockins typically disrupt one allele by coopting its endogenous regulatory sequences; and that the constitutive promoter might not be expressed and reveal the trace in all progeny of a labeled progenitor cell (Bersell et al., 2013; Lafontant et al., 2013; McLellan et al., 2017; Patel et al., 2017; Smith, 2011; Song and Palmiter, 2018).
Table 1.
In vivo epicardial cell fates during heart development and regeneration.
| Context | Species | Cell fates | Approaches and/or strains | References |
|---|---|---|---|---|
| Development | Chick | SMC, Fibro, EC | Dye labelling, retroviral labelling, and cell transplantation | (Mikawa and Gourdie, 1996) |
| SMC, Fibro, Endo | Dye labelling, retroviral labelling, and cell transplantation (quail-chick chimeras) | (Dettman et al., 1998) | ||
| SMC, Fibro, EC | Dye labelling, retroviral labelling, and cell transplantation | (Perez-Pomares et al., 2002a) | ||
| SMC, Fibro, EC, Endo | Cell transplantation (quail-chick chimeras) | (Gittenberger-de Groot et al., 1998) | ||
| SMC, Fibro, EC, Endo | (Manner, 1999) | |||
| SMC, Fibro, EC | (Perez-Pomares et al., 2002b) | |||
| SMC, Fibro, EC | (Guadix et al., 2006) | |||
| Mouse | SMC, EC, CM |
Wt1Cre;Rosa26fsLz or Z/Red Wt1CreERT2;Rosa26fsLz or Z/Red (Cre or CreERT2 knock-in) |
(Zhou et al., 2008a) | |
| SMC, Fibro, CM, Pericytes |
Tbx18Cre;R26RlacZ (Cre knock-in) |
(Cai et al., 2008) | ||
| SMC, Fibro |
Tbx18Cre;Rosa26mT/mG (Cre knock-in) |
(Grieskamp et al., 2011) | ||
| Fibro* |
Wt1GFPCre;R26RmT/mG (Wt1 BAC transgene) |
(Wessels et al., 2012) | ||
| Fibro* |
Tcf21iCre;R26RYFP or R26RtdT (Cre knock-in) |
(Acharya et al., 2012) | ||
| SMC, Fibro, CM, EC, Endo SMC, Fibro, CM, EC |
ScxGFPCre;R26RlacZ (Scx BAC transgene) Sema3DGFPCre;R26RlacZ (GFPCre knock-in) |
(Katz et al., 2012) | ||
| Fibro* |
Tbx18Cre;R26RmT/mG (Cre knock-in) |
(Ali et al., 2014) | ||
| Adipocyte* |
Tbx18Cre;R26RYFP (Cre knock-in) |
(Yamaguchi et al., 2015) | ||
| Zebrafish | Perivascular cells |
tcf21:CreER;gata5:RnG (BAC transgenes) |
(Kikuchi et al., 2011a) | |
| After injury | Mouse | SMC, Fibro, Pericytes |
Wt1CreERT2;R26RmT/mG (CreERT2 knock-in) |
(Zhou et al., 2011) |
| SMC, Fibro, EC, CM |
Wt1GFPCre;R26R (Wt1 BAC transgene) |
(van Wijk et al., 2012) | ||
| Adipocyte* |
Wt1CreERT2;R26RmT/mG or R26RRFP (CreERT2 knock-in) |
(Liu et al., 2014) | ||
| Adipocyte* |
Wt1CreERT2;R26RtdT (CreERT2 knock-in) |
(Zangi et al., 2017) | ||
| Pericytes* |
Wt1CreERT2;R26RYFP (CreERT2 knock-in) |
(Dube et al., 2017) | ||
| CM* | Tβ4 treatment Wt1CreERT2;R26RYFP (CreERT2 knock-in) |
(Smart et al., 2011) | ||
| SMC, Fibro | Tβ4 treatment Wt1CreERT2;R26RmT/mG (CreERT2 knock-in) |
(Zhou et al., 2012) | ||
| SMC, EC, CM | VEGFA modRNA treatment Wt1CreERT2;R26RmT/mG (CreERT2 knock-in) |
(Zangi et al., 2013) | ||
| Zebrafish | Perivascular cells |
tcf21:CreER;gata5:RnG (BAC transgenes) |
(Kikuchi et al., 2011a) | |
| Perivascular cells, Fibro | wt1b:EGFP, transplantation | (Gonzalez-Rosa et al., 2012) |
SMC, vascular smooth muscle cells; Fibro, fibroblasts; EC, endothelial cells; Endo, endocardial cells. Inconsistent cell fates are shaded.
Study was focused on a particular cell fate and other cell types were not reported.
In mice, fate-mapping studies have indicated that epicardial cells are a major source of cardiac fibroblasts during heart development (Table 1) (Acharya et al., 2012; Ali et al., 2014; Cai et al., 2008; Grieskamp et al., 2011; Katz et al., 2012). Murine epicardial cells also differentiate into vascular smooth muscle cells and pericytes that help construct the coronary vasculature (Table 1) (Cai et al., 2008; Grieskamp et al., 2011; Katz et al., 2012; Volz et al., 2015; Zhou et al., 2008a). Epicardial contributions to cardiac endothelium and endocardium were reported when assessing the lineages of Scx and Sema3D expressing cells in mouse (Katz et al., 2012), although such contributions were suggested to be rare, if at all present, in other studies (Cai et al., 2008; Grieskamp et al., 2011; Zhou et al., 2008a). In addition, a lineage tracing study using a Tbx18Cre mouse strain found that the epicardium is the origin of the cardiac adipose tissue (Yamaguchi et al., 2015). An intriguing question over the past decade has been whether the epicardium contributes CMs during development or regeneration. Whereas initial fate-mapping studies supported this idea (Cai et al., 2008; Zhou et al., 2008a), the tools involved for fate-mapping from Wt1 or Tbx18 regulatory sequences were subsequently reported to label other cardiac cells including CMs themselves (Christoffels et al., 2009; Rudat and Kispert, 2012). Katz et al. also reported rare myocardial contributions by the lineages of the Scx and Sema3D expressing epicardial cells (Katz et al., 2012). The current consensus view is that the contribution of epicardium to myocardium is minimal, if it occurs at all, in mice.
Following cardiac injury, lineage tracing studies in mice have demonstrated that adult epicardial cells contribute to vascular smooth muscle cells and pericytes within the infarct; fibroblasts, which proliferate extensively and generate scar tissue, are the principal progeny (Table 1, Figure 1) (Dube et al., 2017; van Wijk et al., 2012; Zangi et al., 2013; Zhou et al., 2011; Zhou et al., 2012). Besides epicardium, diverse cellular sources of cardiac fibroblasts, such as the endothelium, hematopoietic cells, and perivascular cells, have been proposed in different injury contexts. Although the accurate origin of cardiac fibroblast after injury is debatable and under investigation, it is widely accepted that they are heterogeneous with multiple origins in injury contexts, with the epicardium as a confirmed source (reviewed in (Fang et al., 2016; Moore-Morris et al., 2015; Travers et al., 2016)). The properties and predominance of fibroblasts in MIs have suggested innovative approaches designed to reprogram them into CM-like cells (Ieda et al., 2010; Qian et al., 2012; Song et al., 2012; Zhao et al., 2015). Current reprogramming approaches infect fibroblasts in the infarct with retroviruses containing cardiac transcription factors like Gata4, Tbx5, Hand2, and Mef2C, with or without small molecules that may increase efficiency (Figure 1) (Cao et al., 2016b). The chief hurdle of in vivo reprogramming is its very low efficiency, although rapid maturation and incorporation of de novo CMs is an additional challenge. Future translational advances may also require the identification of specific fibroblast subsets with high reprogramming potential, and/or supplementation with CM mitogens. This topic is reviewed by colleagues elsewhere in this issue (Ref.).
Figure 1. Cellular contributions of epicardial cells during zebrafish heart regeneration and mammalian heart repair.
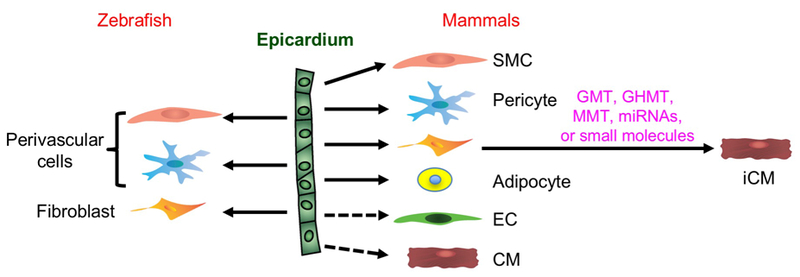
CM, cardiomyocyte; iCM, induced cardiomyocyte through reprogramming; SMC, smooth muscle cell; EC, endothelial cell; GMT, Gata4 + Mef2c + Tbx5; GHMT, GMT + Hand2; MMT, Mef2c + Myocd + Tbx5.
Most fate mapping studies using Cre or CreER lines driven by Wt1 regulatory sequences have indicated that epicardial cells are progenitors of smooth muscle cells (Table 1) (van Wijk et al., 2012; Zangi et al., 2013; Zhou et al., 2011; Zhou et al., 2012) and pericytes (Dube et al., 2017; Zhou et al., 2011) after MI. Epicardial contribution to coronary endothelial cells was reported in one study using a Wt1 BAC (bacterial artificial chromosome) reporter (Wt1GFPCre) (van Wijk et al., 2012), but not detected in others using transgenic knock-in strains (Wt1CreERT2)(Zhou et al., 2011; Zhou et al., 2012). Later studies using in situ hybridization, antibody staining, and lineage tracing (Wt1CreGFP or Wt1CreERT2 knock-in strains) identified Wt1 expression in coronary endothelial cells during development, and in adult hearts before and after MI (Duim et al., 2015; Rudat and Kispert, 2012), undermining evidence that epicardial cells are precursors to endothelial cells. However, the endothelial lineage could be promoted in one study by intramyocardial injection of VEGFA into adult mouse hearts after MI (detailed below) (Zangi et al., 2013). In addition, Wt1+ cells were reported to give rise to cardiac adipose tissue after MI in mice, an intriguing potential function for adult epicardial cells that might recapitulate earlier roles in development (Liu et al., 2014; Zangi et al., 2017).
Epicardial cells are not an innate source of CMs during cardiac repair, but it is conceivable that myogenic properties could be induced. Interestingly, Thymosin β4 (Tβ4) was reported as a regeneration factor that can reactivate mouse epicardial cells, causing them to differentiate into myocytes in rare circumstances (Smart et al., 2011; Smart et al., 2007). In a 2011 study, Tβ4-activated Wt1+ cells proliferated and expressed Isl1 (a marker of postnatal cardioblasts) and Nkx2–5 (an early marker of CM progenitors), with a subset further differentiated into CMs (Smart et al., 2011). Importantly with respect to potential translation, Tβ4 induced CM potency only when applied before MI, whereas another study that applied Tβ4 after MI did not identify epicardial-to-myocardial transitions (Zhou et al., 2012). Additional epicardial fate mapping experiments with a new generation of fate mapping tools will aid the interpretation of such rare transdifferentiation events (He et al., 2017).
3.3. Cellular contributions of the epicardium in zebrafish
In zebrafish, the regulatory sequences of tcf21, wt1 or tbx18 have been widely used to visualize epicardial cells in transgenic reporter strains (Gonzalez-Rosa et al., 2011; Kikuchi et al., 2011a; Schnabel et al., 2011). Among these, tcf21 is most widely and preferentially expressed in epicardial cells during development, at the adult stage, and during regeneration. By contrast, tbx18- or wt1-driven reporters do not label epicardial cells at all stages and show some expression in CMs during development and regeneration (Kikuchi et al., 2011a). Additionally, caveolin 1 regulatory sequences label epicardial cells and EPDCs in addition to endocardial and endothelial cells (Cao et al., 2016a), and fibronectin 1a regulatory sequences label a subset of epicardial cells after cardiac injury (Wang et al., 2013). Finally, the gene-trap line Et(krt4:EGFP)sqet27 displays reporter expression in epicardium and BA cells (Poon et al., 2010; Wu et al., 2016); however, the full tissue spectrum of expression has not been reported. Genetic lineage tracing experiments performed using the tcf21:CreER transgenic zebrafish indicated that epicardial cells contribute to perivascular support cells in the developing and regenerating zebrafish heart (Table 1, Figure 1) (Kikuchi et al., 2011a). In a later study using cell transplantation, wt1b:GFP labeled epicardial cells were found to contribute to myofibroblasts and perivascular fibroblasts in cryoinjured zebrafish hearts (Gonzalez-Rosa et al., 2012). Zebrafish show a rapid, extensive vascularization of regenerating muscle after injury (Lepilina et al., 2006; Marin-Juez et al., 2016), and there is evidence that epicardial generation of support cells facilitates or stabilizes this response (Kikuchi et al., 2011a; Kikuchi et al., 2011b). Notably, lineage tracing data indicated no evidence that zebrafish epicardial cells can generate CMs during heart development or regeneration, even in rare events (Gonzalez-Rosa et al., 2012; Kikuchi et al., 2011a). Thus, innate heart regeneration does not appear to involve transdifferentiation from epicardium to myocardium.
The fibroblasts in the zebrafish heart have not been extensively studied but are less prominent than in thick-walled mammalian hearts (Lafontant et al., 2013). In the cryoinjury model of zebrafish, a cell population(s) staining for intermediate filament vimentin (Vim), extracellular de-adhesive protein Tenascin-C (TNC), alpha smooth muscle actin (α-SMA), and myosin light chain kinase (MLCK) was described to appear transiently during regeneration (Chablais et al., 2011; Gonzalez-Rosa et al., 2011). The results of these studies suggest that fibroblasts deposit collagen after heart injury, although these markers can label other cardiac cell types (Lane et al., 1983; Moore-Morris et al., 2015; Tournoij et al., 2010; Zeisberg et al., 2007). Cell transplantation of wt1b+ EPDCs suggested epicardial contributions to fibroblasts expressing the markers col1a2 and periostin in addition to perivascular cells (Gonzalez-Rosa et al., 2012).
It is becoming apparent that epicardial cells have high cellular plasticity. Some debate over the precise contribution of epicardial lineages in higher and lower vertebrates will continue as more specific makers are identified, although for the most part a consensus has been reached on to what cell types epicardial cells do (perivascular cells) and do not (CMs) give rise. Enhancing the perivascular lineage while restraining the fibroblast lineage represents one direction to benefit cardiac repair, a goal that would be aided by further knowledge of key factors in epicardial lineage commitment.
3.4. Cellular heterogeneity of the epicardium
Because of the different progeny cell types revealed by fate mapping of epicardial cells in mice, chicks, and zebrafish, most agree that the epicardium is likely to be a heterogeneous grouping of multiple subpopulations. Bollini et al. recently revealed that in mice, Tβ4-reactivated epicardial-derived cells labeled with a Wt1 lineage trace are heterogeneous (Bollini et al., 2014). By characterizing Tβ4-primed EPDCs sorted from MI injuries using quantitative PCR and immunostaining, the authors found that adult reactivated GFP+ EPDCs are a heterogeneous population with a variety of cardiovascular potentials. These cells restored expression of some embryonic genes but had an expression profile distinct from that of embryonic epicardial cells analyzed at E12.5. The stem cell antigen-1 positive (Sca1+) subset had the most strongly activated developmental program (expressing Wt1, Tbx18, Retinaldehyde dehydrogenase 2 (Raldh2) and Pdgfrβ) and cardiac progenitor marker expression (such as Isl1, Flk1, Gata4, Sma, Fapα, Sm22α and Pecam). The results of this study suggested that identifying particular subpopulations of EPDCs might be clinically important for manipulating the epicardium.
Contemporary technology to assess RNA levels genome-wide in individual cells has enabled the delineation of cell subpopulations in many tissues and species (Athanasiadis et al., 2017; Haber et al., 2017; Jaitin et al., 2014; Macosko et al., 2015; Tang et al., 2009). A recent single cell transcriptome study in zebrafish revealed some heterogeneity of tcf21-expressing cells purified from uninjured adult hearts (Cao et al., 2016a). This analysis suggested the presence of multiple cell subsets, each defined by a specific expression signature, as well as genes that may represent subset-specific markers. Further validation of markers from studies like this may provide new reporters and tools that will benefit lineage-tracing experiments. Moreover, whereas this study examined the transcriptomes of only 39 cells, it is now possible to economically assess tens of thousands of cells, albeit at low read depth, for a much higher resolution assessment of heterogeneity. Additionally, profiling epicardial cells collected from injured hearts can answer the question of whether there are notable changes in cell heterogeneity during cardiac regeneration.
4. Epicardial signals during heart regeneration
4.1. Activation of the epicardium by injury
A growing number of studies over the past decade have employed animal model systems to identify mechanisms by which epicardial tissue influences heart repair. Following cardiac injury, epicardial cells induce the expression of genes that mark the embryonic epicardium, proliferate, and cover the injury site, a process referred to as “activation”. In zebrafish, virtually the entire zebrafish epicardium is activated within a day or two of injury (Figure 2A), expressing markers like raldh2, tbx18, wt1 and fibronectin 1 (fn1) (Gonzalez-Rosa et al., 2011; Kikuchi et al., 2011b; Lepilina et al., 2006; Schnabel et al., 2011; Wang et al., 2013; Wang et al., 2011; Wills et al., 2008). In a resection injury model, developmentally activated epicardial becomes confined to the injured area by 7 days post amputation (dpa, Figure 2A) (Kikuchi et al., 2011b; Lepilina et al., 2006). In neonatal mice, apical resection was similarly associated with rapid epicardial activation, indicated by the induction of Wt1 and Aldh1a2 (raldh2) in an organ-wide manner (Porrello et al., 2011; Porrello and Olson, 2014). In adult mice, cardiac injury also provokes re-expression of embryonic epicardial markers (Huang et al., 2012; van Wijk et al., 2012; Zhou et al., 2011; Zhou et al., 2012).
Figure 2. Epicardial signals in heart repair and regeneration.
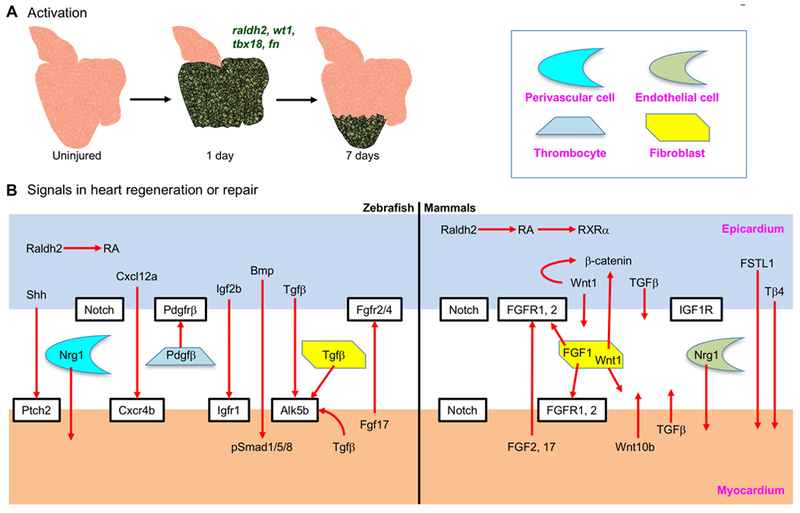
(A) Organ-wide activation of zebrafish epicardial cells one day after resection of the ventricular apex. Activation (induction of embryonic epicardial markers such as raldh2, wt1, tbx18 and fn) is restricted to the injury site by 7 days after injury. Green color represents the re-expression of embryonic genes. (B) Proposed epicardium-myocardium signals in zebrafish heart regeneration and mammalian heart repair.
The molecular mechanisms by which heart injury activates the epicardium (especially in an organ-wide manner) are likely to be fascinating but still are poorly understood. Huang et al. approached this question by assessing conservation of potential DNA regulatory elements near genes that mark the embryonic epicardium. They identified the C/EBP family of transcription factors as regulators of Raldh2 and Wt1 expression in embryonic hearts and in adult hearts after MI, through binding to enhancer elements near these genes (Huang et al., 2012). A recent study by Vieira and colleagues found Brahma-related gene 1 (BRG1), a catalytic subunit of the SWI/SNF chromatin–remodeling complex that has epicardial expression during heart development and after heart injury, as a regulator of Wt1 expression and epicardial activation (Vieira et al., 2017). They identified evolutionary conserved regions in the Wt1 locus that are responsible for Wt1 expression. BRG1 is recruited by C/EBPβ to these conserved regulatory elements in the Wt1 locus, and it is required for Wt1 expression. Interestingly, Tβ4 interacts with BRG1 and augments Wt1 expression post-MI (Vieira et al., 2017). Additional evidence has suggested that epicardial activation amplifies the inflammatory response. After MI/reperfusion injury, neutrophils were observed on the epicardial surface, in the subepicardial space, and in the infarct area. Viral expression of a dominant negative C/EBP in the adult epicardium decreased the number of neutrophils, suggesting a function in regulating neutrophil infiltration (Huang et al., 2012). Mice deficient in epicardial Yap/Taz exhibited profound pericardial inflammation, caused by a decrease in the number of T-regulatory (Treg) cells in the injured myocardium (Ramjee et al., 2017). In addition, the close connection of epicardium with macrophages suggests a link to epicardial cell activation (Pinto et al., 2014; Stevens et al., 2016). Identification of intrinsic programs in epicardial cells and extrinsic factors (e.g. from macrophages) that activate the epicardium will stimulate new discovery biology and potential therapeutic strategies.
4.2. Signaling interactions with myocardium
Once activated, the epicardium secretes signals, including potential mitogenic factors for CMs, with the potential to influence heart regeneration (Foglia and Poss, 2016; Karra and Poss, 2017; Zhou et al., 2011). The epicardium has been proposed to stimulate CM proliferation during both embryonic heart development and adult heart regeneration. It also contributes extracellular components and growth factors to maintain cardiac tissue structure and electrophysiological properties (Furtado et al., 2016; Moore-Morris et al., 2015). Multiple signaling pathways mediate interactions between epicardium and myocardium, reviewed elsewhere for the context of heart development (Olivey and Svensson, 2010; Smart and Riley, 2012). Here, we provide snapsnots for signals that have been reported to be involved in cardiac repair (Table 2, Figure 2B).
Table 2:
Epicardial signals relevant to heart regeneration or repair
| Signaling | Models | Gene induction domains upon injury * | Functional evidence in heart regeneration | References |
|---|---|---|---|---|
| Retinoic Acid (RA) | Zebrafish | raldh2: epicardium, endocardium | Expression of a dominant-negative RARα impairs CM proliferation and heart regeneration. | (Kikuchi et al., 2011b; Lepilina et al., 2006) |
| Mouse |
Raldh2: epicardium (RXRα: epicardium) |
(Limana et al., 2010; Merki et al., 2005; Porrello et al., 2011; van Wijk et al., 2012) | ||
| Fibroblast Growth Factors (FGFs) | Zebrafish |
fgf17b: CMs fgfr2 and fgfr4: EPDCs |
Expression of a dominant-negative fgfr1 blocked epicardial cell EMT, coronary neovascularization, and muscle regeneration. | (Lepilina et al., 2006) |
| Rat |
FGF1: inflammatory cells and fibroblast-like cells FGF2: endothelial cells, CM FGFR: CM |
(Zhao et al., 2011) | ||
| Transforming Growth Factor-beta (TGFβ) | Zebrafish |
tgfb1, 2, and 3: epicardial cells, fibroblasts and CMs Alk5b: CMs and fibrotic cells pSmad3: CMs and non-CMs in the infarct area |
Inhibition of the TGFβ pathway impaired ECM deposition, CM proliferation, and heart regeneration. | (Cao et al., 2016a; Chablais and Jazwinska, 2012; Choi et al., 2013) |
| Rat, Pig | Tgfb1, 2, and 3: infarcted myocardium | (Deten et al., 2001; Hao et al., 1999; Vilahur et al., 2011) | ||
| Notch | Zebrafish |
notch1a, notch1b, and notch2: endocardium notch1a and notch2: epicardium notch1b, notch2, notch3, and Dll4: endocardium |
Suppression of Notch signaling decreased CM proliferation, induced scar formation and impaired heart regeneration. Overexpression of NICD suppressed CM proliferation and heart regeneration in the resection model while augmenting CM proliferation in the cryoinjury model. | (Zhao et al., 2014) (Munch et al., 2017) |
| Mouse | Notch activity reporter: CMs, epicardial cells/EPDCs NICD1: CM |
Overexpression of NICD1 in adult CM increased the number of Ki67+ CMs. | (Kratsios et al., 2010; Russell et al., 2011) | |
| NF-κB | Zebrafish |
nfkb1, nfkbiaa and nfkbiab: CMs |
NF-κB blockade in CMs impaired epicardial cell infiltration into the wound, CM proliferation and heart regeneration. | (Karra et al., 2015) |
| Platelet-Derived Growth Factors (PDGFs) | Zebrafish |
pdgfrβ: epicardium and fibrin clots pdgfb: thrombocytes at the wound site |
Pharmacological inhibition of PDGF decreased epicardial cell proliferation and coronary blood vessel formation. | (Kim et al., 2010) |
| Mouse |
PDGFB, PDGFRα and PDGFRβ: the infarct phosphorylated PDGFRβ: perivascular cells in the infarct |
PDGFRβ blockade impaired collagen deposition and vasculature maturation in the infarct. PDGFRα blockade impaired collagen deposition. | (Zymek et al., 2006) | |
| Wnt | Mouse |
Wnt1: epicardium, cardiac fibroblasts Wnt10b: CMs in the peri-infarct area. |
Deleting β-catenin in the epicardium disrupted epicardial cell expansion and EMT and decreased heart function after MI; deleting β-catenin in cardiac fibroblasts led to acute cardiac dilatation and cardiac dysfunction. Wnt10b overexpression increased neovascularization of scar tissue and attenuated fibrosis. |
(Duan et al., 2012) (Paik et al., 2015) |
| Hedgehog (Hh) | Zebrafish |
shha: epicardial tissue adjacent to and within the injury site ptch2: CMs in the injury site |
Pharmacological inhibition of Hh or epicardial specific deletion of shha decreased CM proliferation, while an Hh agonist increased proliferation. | (Choi et al., 2013; Sugimoto et al., 2017) |
| Insulin-like Growth Factor (IGF) | Zebrafish |
Igf2b: epicardium and endocardium igfr1: CMs at the injury site |
Expression of a dominant-negative igf1r impaired CM proliferation and heart regeneration. Inhibiting Igf decreased CM proliferation, an Igf agonist increased CM proliferation. |
(Huang et al., 2013) (Choi et al., 2013) |
| Mouse | IGF1R: epicardium | Inhibition of IGF1R in Wt1+ lineages after MI reduced adipogenic differentiation. | (Zangi et al., 2017) | |
| BMP | Zebrafish |
bmp2b, bmp7: epicardium, endocardium bmpr1aa: wound border zone id2b: endocardium pSmad1/5/8: CM |
Overexpression of noggin3 decreased CM dedifferentiation and proliferation, and increased scar tissue, while overexpression of bmp2b did the opposite. bmpr1aa mutants had fewer CM proliferation. | (Wu et al., 2016) |
| Mouse |
Bmp4: whole heart (RT-PCR) |
Overexpression of noggin, pharmacological inhibition of BMP, or using the Bmp4+/− mice demonstrated reduced infarct size and CM apoptosis in a MI and reperfusion mouse model. | (Pachori et al., 2010) | |
| Hippo/Yap | Mouse | (Yap, Taz, Tead1–3, Lats1 and Lats2 are expressed in the proepicardium and epicardium during development.) | Deficiency in epicardial Yap/Taz caused pericardial inflammation, myocardial fibrosis, cardiomyopathy, and death after MI. Deletion of Lats1/2 in embryonic (E11.5) epicardium reduced the epicardial differentiation to fibroblast. |
(Lin and Pu, 2014; Ramjee et al., 2017) (Xiao et al., 2018) |
| Chemokines (Cxcl12a-Cxcr4b) | Zebrafish |
cxcl12a: epicardium cxcr4: CMs |
An antagonist that blocks Cxcr4 function disrupted heart regeneration by impairing CM migration to the injury site. | (Itou et al., 2012) |
| Neuregulin 1 (Nrg1) | Zebrafish | nrg1: epicardial-derived perivascular cells, regulatory T cells | Overexpression of Nrg1 promotes CM proliferation. | (Gemberling et al., 2015; Hui et al., 2017; Kang et al., 2016) |
| Mammals | Nrg1: endothelial cells | Overexpression of Nrg1 promotes proliferation of mononucleated CMs. | (Bersell et al., 2009; Parodi and Kuhn, 2014) | |
| Follistatin-like-1 (FSTL1) | Mouse, swine | FSTL1: CMs (FSTL1 is expressed in the epicardium in uninjured hearts, and is induced in CMs and decreased in the epicardium.) FSTL1: fibroblast in the infarct |
Application of epicardially derived FSTL1 stimulated CM proliferation, diminished infarct size and improved heart function after MI. Intravenous delivery of FSTL1 protein or adenovirus-driven FSTL1 expression before MI reduced the infract size. FSTL1 promotes cardiac fibroblast activation and protects the heart from rupture |
(Wei et al., 2015) (Ogura et al., 2012; Oshima et al., 2008) (Maruyama et al., 2016) |
| Thymosin β4 (Tβ4) | Mouse | Tβ4: epicardium, endocardium and capillaries | Treatment with Tβ4 prior to MI activated the adult epicardium, stimulated vascular growth, and induced epicardial to CM differentiation in very rare cases. Treatment with Tβ4 after MI increased the thickness of the epicardium and coronary capillary density. Deletion of Tβ4 in heart diminished neovascularization in the infarct border zone. | (Bock-Marquette et al., 2009; Dube et al., 2017; Smart et al., 2011; Smart et al., 2007; Zhou et al., 2012) |
| Caveolin 1 (cav1) | Zebrafish | (cav1 is expressed in epicardial cells, EPDCs, and coronary vascular endothelial cells.) | cav1 deletion decreased CM proliferation and impaired heart regeneration. | (Cao et al., 2016a) |
Non-injury related expression and downregulation are listed in brackets.
Retinoic Acid (RA).
RA signaling is critical for zebrafish heart regeneration. Raldh2 is the rate-limiting enzyme for RA synthesis, and its expression is induced by injury in both the epicardium and endocardium (Lepilina et al., 2006). Inhibition of RA signaling by overexpressing a dominant-negative RA receptor α (dnRARα) or the RA-degrading enzyme cyp26a1 blocks CM proliferation and heart regeneration (Kikuchi et al., 2011b). This study inhibited RA signaling throughout the animal; thus, how directly RA acts on CMs remains unclear. In studies of neonatal mouse heart regeneration, Raldh2 was also induced by resection injury, presumably in the epicardium although the expression domain was not assessed by the authors (Porrello et al., 2011). In adult mouse hearts, Raldh2 is induced by MI in the epicardium but not in the endocardium (Limana et al., 2010; van Wijk et al., 2012). Retinoid X receptor alpha (RXRα) signaling in mouse epicardium is required for myocardial growth and coronary artery formation (Merki et al., 2005), suggesting a potential role of RA signaling in mammalian heart regeneration that has not been directly explored.
Fibroblast Growth Factors (FGFs).
Fgf signaling is also required for zebrafish heart regeneration. Following amputation injury, the ligand gene fgf17b is induced in CMs, while fgfr2 and fgfr4 are induced in EPDCs. The induced expression of a dominant-negative fgfr1 construct blocked epicardial cell EMT, coronary neovascularization, and muscle regeneration (Lepilina et al., 2006). After MI is induced in rats, FGF-1 is upregulated in inflammatory cells and fibroblast-like cells in the border zone of infarcted myocardium, whereas FGF-2 is induced in both the border zone (primarily in endothelial cells) and the infarct (in CMs) (Zhao et al., 2011). Fgf receptor signaling in muscle is required for the normal response to MI in rat (Zhao et al., 2011), and FGF has been widely examined for potential therapeutic applications to cardiovascular disease (reviewed in (Itoh and Ohta, 2013; Itoh et al., 2016) ), but the roles of this pathway in the mammalian epicardial response to heart injury requires further investigation.
Transforming Growth Factor-beta (TGFβ).
Following injury in zebrafish, tgfb1, 2, and 3 are broadly induced in epicardial cells, fibroblast-like cells, and CMs, while the receptor Alk5b is induced in CMs and fibroblast-like cells (Chablais and Jazwinska, 2012). Phosphorylated-Smad3, an indicator of active signaling, is evident in CMs and non-CMs in the infarct area (Cao et al., 2016a; Chablais and Jazwinska, 2012). Pharmacologically inhibiting the TGFβ pathway impaired ECM deposition and CM proliferation, consequently disrupting regeneration (Chablais and Jazwinska, 2012). Upon MI in adult rats or pigs, Tgfb1, 2, and 3 are sharply induced in infarcted myocardium (Deten et al., 2001; Hao et al., 1999; Vilahur et al., 2011). Considering the multifarious roles of TGFβ signaling in cardiovascular development and disease (Frangogiannis, 2017), this pathway must be explored methodically for cell-type and isoform-specific functions in the response to cardiac injury. Recent work by Dogra et al. identified two Activin type 2 receptor ligands of the TGFβ family, Myostatin b (Mstnb) and Inhibin beta Aa (Inhbaa), which exerted opposing effects on CM proliferation and heart regeneration. Inhbaa increased indices of CM proliferation when overexpressed. By contrast, overexpression of mstnb or deletion of inhbaa suppressed CM proliferation and impaired heart regeneration (Dogra et al., 2017).
Notch Signaling.
In zebrafish, notch1a, notch1b, notch2, and notch3 are expressed in the endocardium (Zhao et al., 2014). Upon resection injury, notch1a, notch1b, and notch2 are predominantly upregulated in the endocardium, and notch1a and notch2 are also sharply induced in the epicardium (Zhao et al., 2014). Expression of notch3 was not changed. After cryoinjury, notch1b, notch2, notch3, and the Notch ligand Delta-like 4 (Dll4) are induced in endocardial cells adjacent to the wound (Munch et al., 2017). Induced transgenic inhibition of Notch signaling impaired heart regeneration, coincident with decreased CM proliferation. Interestingly, transgenic hyperactivation of Notch signaling (overexpression of the Notch intracellular domain NICD) suppressed CM proliferation and heart regeneration in the resection model, while augmenting CM proliferation in the cryoinjury model (Munch et al., 2017; Zhao et al., 2014). It is unclear whether this difference is attributable to different injury models, the expression levels of NICD, or other factors. Similarly, Notch signaling was activated in CMs and epicardial cells/EPDCs following MI in mice (Kratsios et al., 2010; Russell et al., 2011). The inducible overexpression of NICD1 in adult CMs after MI increased the percentage of Ki67+, but not phospho-Histone H3+ CMs, and the authors proposed a role in promoting CM survival (Kratsios et al., 2010). Perhaps unsurprisingly, these studies from both zebrafish and mice suggest that Notch signaling activity must be fine-tuned for positive effects on heart repair.
NF-κB signaling.
In zebrafish, nfkb1, nfkbiaa and nfkbiab are induced in CMs after resection injury (Karra et al., 2015). Suppressing NF-κB signaling by induced expression in CMs of a super-repressor IκB (IκBSR) impaired epicardial cell colonization of wounds, CM proliferation, and heart regeneration. The authors suggested that NF-κB contributes to crosstalk between CMs and epicardial cells, which has yet to be explored at the molecular level (Karra et al., 2015).
Platelet-Derived Growth Factors (PDGFs).
Upon resection injury in zebrafish, pdgfrβ is induced in the epicardium and in cells within fibrin clots at the wound site, while the ligand gene pdgfb is induced in thrombocytes (Kim et al., 2010). PDGF signaling induced epicardial cell proliferation in vitro, and pharmacologically inhibiting PDGF signaling in vivo suppressed coronary blood vessel formation during heart regeneration (Kim et al., 2010). Following MI in mice, PDGFB, PDGFRα and PDGFRβ are induced in infarcts, and PDGFRβ is phosphorylated, an indicator of active signaling, in perivascular cells within the MI (Zymek et al., 2006). Antibody blockade of PDGFRα and PDGFRβ decreased collagen deposition and impaired vascular maturation in the infarct (Zymek et al., 2006).
Wnt/β-catenin Signaling.
Following MI injury in mice, Wnt1 ligand gene expression is induced in the epicardium and cardiac fibroblasts in the region of injury (Duan et al., 2012). Deleting β-catenin in epicardial cells disrupted epicardial cell expansion and EMT and decreased heart function after MI. Moreover, deleting β-catenin in cardiac fibroblasts led to acute cardiac dilatation and cardiac dysfunction (Duan et al., 2012). In another report, Paik et al. found that Wnt10b is transiently induced in CMs in the peri-infarct area after MI, a response that the authors suggest coordinates arterial formation and attenuates fibrosis (Paik et al., 2015). It will be intriguing to dissect the functions of this family of genes in zebrafish and neonatal mouse regeneration models.
Hedgehog (Hh) Signaling.
This pathway was implicated for heart regeneration in a screen for CM mitogens performed in zebrafish embryos using a FUCCI-based transgenic reporter system. In zebrafish, shha regulatory sequences are activated in epicardial tissue adjacent to and within the injury site at 7 days after partial ventricular resection, while the expression of a reporter that reflects expression of the Hh target gene ptch2 is present in CMs within the area of regeneration (Choi et al., 2013). The results of both pharmacological manipulations of Hh and of epicardial-specific deletion of shha (by combining a Cre-dependent invertible gene-trap cassette with the tcf21:CreER line mentioned above) suggested that Hh signaling promotes CM proliferation during regeneration (Choi et al., 2013; Sugimoto et al., 2017). As indicated below, Hh signaling has also been reported to regulate epicardial cell regeneration (Wang et al., 2015). Hh signaling has been implicated in mammalian epicardial biology during cardiac development and homeostasis, where it was suggested to be a potential therapeutic target for ischemic heart disease (Dunaeva and Waltenberger, 2017; Lavine and Ornitz, 2007, 2008).
Insulin-like Growth Factor (IGF).
In the same screen for CM mitogens in zebrafish described above, treatment with the Igf signaling agonist NBI-31772 increased CM proliferation in the developing heart (Choi et al., 2013). The expression of the ligand gene igf2b was further found to be induced in adult endocardial cells and epicardial cells in the injury site by 7 days post-injury, with igfr1 expression in CMs. Pharmacologically inhibiting Igf signaling decreased CM proliferation and impaired heart regeneration, whereas an Igf signaling agonist increased proliferation (Choi et al., 2013). Similar results were reported by Huang et al., who also used a dominant-negative form of Igf1 receptor (dn-Igf1r) to block Igf signaling and heart regeneration in zebrafish (Huang et al., 2013). In addition, IGF1R is expressed in murine Wt1+ epicardial cells, and inhibition of IGF1R in Wt1+ lineages after MI reduced adipogenic differentiation (Zangi et al., 2017).
Bone Morphogenetic Protein (BMP).
The function of BMP signaling in cardiac repair might differ between mice and zebrafish. By using a spatially resolved RNA sequencing technique called tomo-seq, Wu et al. generated a spatiotemporal landscape of gene expression in the regenerating zebrafish heart following cryoinjury, revealing multiple genes in the BMP pathway with differential expression at the injury (Wu et al., 2016). bmp2b and bmp7 were induced in the epicardium and endocardium, id2b in the endocardium and bmpr1aa in the wound border zone. Phosphorylated Smad 1/5/8, an indicator of signal activation, was detected in CMs in the wound area (Wu et al., 2016). Overexpression of the BMP antagonist noggin3 decreased CM dedifferentiation and proliferation, and increased scarring, whereas overexpression of bmp2b did the opposite. In addition, bmpr1aa mutants showed reduced CM proliferation upon injury (Wu et al., 2016). By contrast, inhibition of BMP signaling (overexpressing of Noggin or pharmacological treatment) or using Bmp4+/− mice demonstrated reduced infarct size and CM apoptosis in a mouse model of MI /eperfusion (Pachori et al., 2010). These studies suggest that species-specific responses to the same activated signaling pathways dictate the primary outcome of cardiac injury, an inference that needs further investigation.
Hippo/Yap Signaling.
Control of signaling by Hippo pathway components in CMs has been proposed to regulate their proliferation during mammalian life. Indeed, multiple reports now indicate that derepression of YAP and its ability to bind target genes can have dramatic effects on CM proliferation, even at the adult stage (Leach et al., 2017; Morikawa et al., 2017; von Gise et al., 2012; Xin et al., 2013; Xin et al., 2011). During murine heart development, Hippo pathway genes (Yap, Taz, Tead1–3, Lats1 and Lats2) are also expressed in the proepicardium and epicardium, and Yap and Taz are required in the epicardium for coronary vasculature development (Singh et al., 2016). Most recently, Xiao et al. deleted Lats1 and 2 in mouse embryonic (E11.5) epicardium using the Wt1CreERT2 knock-in allele, an effect that reduced epicardial-to-fibroblast differentiation events (Xiao et al., 2018). Single-cell RNA-seq suggested that Lats1/2 deletion preserved epicardial gene expression profiles and elevated the expression of Yap targets like the negative regulator of retinoic acid synthesis Dhrs3 and the extracellular matrix regulator Dpp4. Whether epicardial Hippo/Yap signaling components are regulated by cardiac injury has not been investigated. As described above, deletion of Yap/Taz in the mouse epicardium induced profound pericardial inflammation, myocardial fibrosis, cardiomyopathy, and death after MI (Ramjee et al., 2017). This further supports the concept that the epicardium mediates inflammatory responses after MI.
Chemokines.
Following resection injury in zebrafish, Itou et al. reported that the expression of the marker C-X-C motif chemokine 12 (cxcl12a), which encodes a chemokine ligand, is induced in epicardial tissue, while the receptor C-X-C chemokine receptor type 4b (cxcr4b) is expressed in CMs (Itou et al., 2012). An antagonist that blocks Cxcr4 function disrupted heart regeneration, and it was suggested that this effect was the result of impaired CM migration to the injury site (Itou et al., 2012). This study was the first to propose that CM migration is an essential mechanism in heart regeneration. It is worth noting that Harrison et al. reported that myocardial Cxcl12b and endothelial Cxcr4a signaling were important in guiding coronary vessel development, and that a cxcr4a mutant showed defects in heart regeneration (Harrison et al., 2015). The regeneration phenotype was proposed to be a secondary outcome of defects in coronary vessel patterning. These studies suggest that the zebrafish heart may harness different Cxcl12-Cxcr4 systems for development and regeneration.
Neuregulin 1 (Nrg1).
Nrg1 is an extracellular factor implicated as an endothelial cell-derived mitogen for mammalian CMs (Bersell et al., 2009; Parodi and Kuhn, 2014). tcf21+ epicardial-derived perivascular cells induce nrg1 after injury in the adult zebrafish heart, though levels remain overall low (Cao et al., 2016a; Gemberling et al., 2015). Inducing pharmacological blockade of its ErbB receptors decreased injury-induced CM proliferation, whereas overexpressing Nrg1 in CMs increased CM proliferation and caused cardiomegaly in the absence of injury (Gemberling et al., 2015). Injury-induced, transgenic overexpression of Nrg1 in endocardium and epicardium, by way of a leptinb-linked enhancer element, also boosted CM proliferation in zebrafish (Kang et al., 2016). In addition, regulatory T cells (Treg-like cells) that home to injury sites were identified as a source of Nrg1 during zebrafish heart regeneration (Hui et al., 2017). Thus, Nrg1 is a potent, inducible mitogen in zebrafish CMs, and the epicardium and Treg-like cells appear to be its primary sources. It is important to note that Nrg1 might only effectively induce cell division in mononucleated CMs (Bersell et al., 2009), potentially limiting its therapeutic potential for the injured human heart. In addition, current evidence does not exclude the possibility ErbB pathway ligands other than Nrg1 may act as ancillary or primary mitogens during heart regeneration.
Other factors.
As discussed above and below, Tβ4 and follistatin-like 1 (FSTL1) are reported to be involved in epicardium-myocardium interactions that benefit heart repair (Bock-Marquette et al., 2009; Smart et al., 2011; Smart et al., 2007; Wei et al., 2015; Zhou et al., 2012). In addition, caveolin 1 (cav1), a component of caveolae that is predominantly expressed in epicardial cells and EPDCs, but also in coronary vascular endothelial cells, was shown to be required for CM proliferation and heart regeneration in adult zebrafish (Cao et al., 2016a). The results of that study suggested that signal transduction via caveolae are likely to be critical for epicardium-CM interactions.
4.2. Production of ECM components
The extracellular matrix (ECM) is a non-cellular component secreted by cells to provide chemical and mechanical cues to surrounding cells (Frantz et al., 2010). ECM supports cell proliferation and maturation in addition to tissue organization during heart development (George et al., 1997; Ieda et al., 2009; Magnusson and Mosher, 1998; Trinh and Stainier, 2004). Accumulating evidence suggests that epicardial and fibroblast ECM remodeling is involved in heart development and regeneration, and that manipulating the deposition of epicardial ECM might influence regeneration. For example, during mouse heart development, inhibiting α4-integrin signaling stimulated epicardial EMT and altered epicardial lineages (Dettman et al., 2003). The developing and adult murine epicardium is surrounded by ECM components (such as Fn, collagen IV and hyaluronic acid (HA)), which are broken down and reform following MI (Balmer et al., 2014).
In zebrafish, following resection injury, genes encoding the ECM components fn1 and fibronectin-1b (fn1b) are induced in epicardial cells (Wang et al., 2013). fn1, as well as Fibronectin protein deposition, are more dynamically expressed - first chamber-wide, but then becoming localized to the injury site. The Fibronectin receptor (integrin-β3, itgb3) is induced in CMs near the injury site, suggesting a possible epicardial-myocardial interaction. Mutation of fn1 or induced expression of a dominant-negative fibronectin cassette disrupted heart regeneration and led to scar formation. Further analyses suggested that CM proliferation was not affected in either of these experiments, and the authors speculated on possible roles in CM patterning or displacement (Wang et al., 2013). Similarly, in a newt heart regeneration model, ECM deposition (tenascin-C, hyaluronic acid (HA), and fibronectin) was induced organ-wide in epicardial cells after resection injury before becoming restricted to the injury site (Mercer et al., 2013). In this study, the authors hypothesized that unspecified proliferating cells within the heart migrate to the ECM-rich epicardial sheath and then to the regenerating ventricular apex (Mercer et al., 2013). Although the mechanisms underlying these processes need further clarification, the results of these two studies suggest that the epicardium helps establish an ECM environment that supports heart regeneration.
Additional studies have identified epicardial-derived ECM components that may be involved in heart regeneration. Missinato et al. reported that the extracellular component HA and its receptor, Hyaluronan-mediated motility receptor (Hmmr), are required in zebrafish for epicardial cell EMT and heart regeneration (Missinato et al., 2015). The expression patterns of both HA and HMMR were similar in the post-MI infarct area in rats (Missinato et al., 2015). col12a1b, which encodes non-fibrillar type XII collagen, is expressed in epicardial cells in intact zebrafish hearts and boosted in cryoinjury wounds (Marro et al., 2016). During larval zebrafish spinal cord regeneration, both col12a1a and col12a1b expression and the deposition of Collagen XII are necessary for axon regeneration. Overexpressing col12a1a accelerated spinal cord regeneration (Wehner et al., 2017). Permanent collagen-enriched scar tissue is a hallmark of mammalian cardiac injury and a roadblock for regeneration, whereas collagen deposition is transient, and potentially beneficial, in injured zebrafish. The reports described here support the possibility that manipulating epicardial ECM components like collagens could improve heart regeneration.
5. High regenerative capacity of the epicardium
After a heart injury, adult epicardial cells proliferate and migrate toward the injury site. In a recent study, Wang and colleagues applied a genetic ablation system (tcf21:NTR) to ablate up to 90% of the epicardial cell population in adult zebrafish while sparing the myocardium (Wang et al., 2015), and the epicardium quickly regenerated. Combining their ablation tool with an ex vivo culture system allowed them to monitor epicardial cell behavior during regeneration in real time (Figure 3) (Cao and Poss, 2016). During regeneration, spared epicardial cells proliferate and migrate as a sheet to repopulate the exposed ventricular surface from the ventricular base toward its apex (Figure 3A) (Wang et al., 2015). These regeneration events are likely relevant to mechanisms by which epicardial cells respond to injury cues. Unexpectedly, the authors found that the outflow tract (or BA) is essential for regeneration of the ventricular epicardium ex vivo, and that it can guide the direction of regeneration when grafted ectopically. They further identified Hh ligand derived from the BA as a likely molecular regulator of these processes. The detailed molecular mechanisms underlying these events are unclear, as is the role of BA in in vivo epicardial regeneration. It is possible that a signaling gradient forms along the BA/ventricular base-to-apex that guides the regeneration process. Although the regenerative capacity of mammalian epicardium has not been assessed, an analogous strain in mice, Wt1CreERT2; Rosa26DTA, was used to ablate epicardial cells in fetal hearts, an injury that decreased fetal cardiac macrophage numbers (Stevens et al., 2016). It will be intriguing to investigate the regenerative capacity and the requirement of the epicardium in adult vertebrates using genetic ablation tools.
Figure 3. Ex vivo epicardial regeneration.
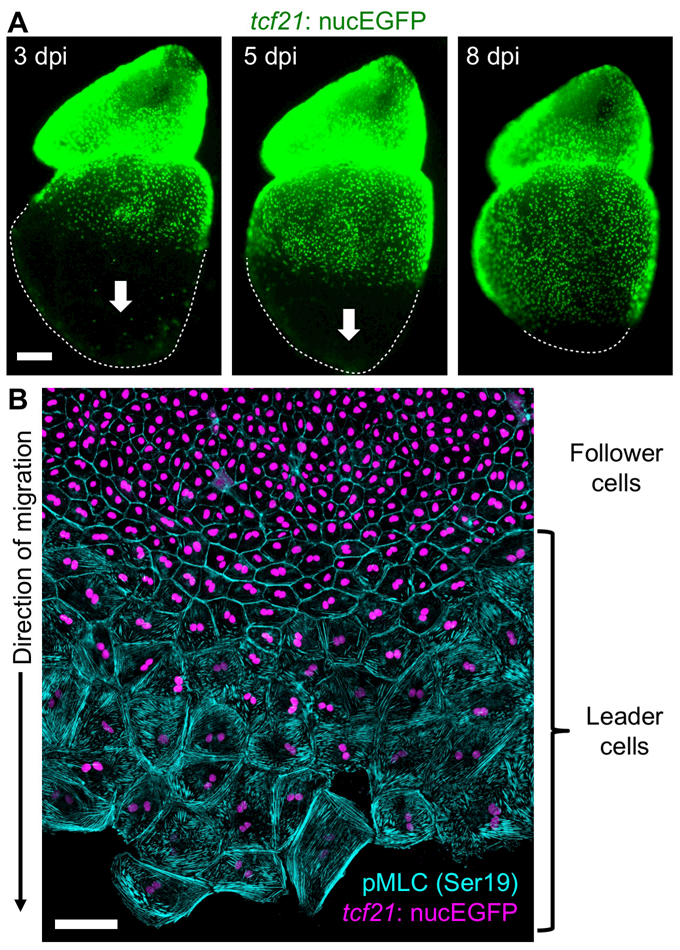
(A) Whole-mount images of explanted zebrafish heart showing epicardial regeneration along the ventricular surface in a base-to-apex direction (arrows). tcf21:nucEGFP visualizes epicardial cell nuclei (green). dpi, days post Mtz incubation. (B) The epicardium generates a leading edge of large, multinucleate (leader, bottom) cells during migration ex vivo. Trailing (follower, top) cells are small and mononucleate. Epicardial nuclei are indicated in violet (tcf21:nucEGFP) and phosphorylated myosin light chain 2 (Ser19), an indicator of mechanical tension, is cyan.
To further address cellular mechanisms of epicardial regeneration in zebrafish, Cao et al. live-imaged epicardial regeneration at higher resolution, finding that a subpopulation of polyploid epicardial cells appears at the front of regenerating tissue (Figure 3B). These leader cells are established and maintained by endoreplication and ostensibly eliminated via apoptosis after regeneration (Cao et al., 2017). Mechanical tension was much higher at the leading edge (Figure 3B), estimated by extrapolating the speed of recoil after laser damage ex vivo to cell membranes, and stretching of epicardial tissue sheets ex vivo could increase the frequency of endoreplication. However, it is unclear how relevant the ex vivo conclusions on tension are to the in vivo context, or what factors (e.g. ECM components) might regulate cellular tension. Small molecules, like the PKC/Akt inhibitor GSK1007102 and the Hedgehog pathway inhibitor Cyclopamine, were identified in screens that either augmented or blocked regenerative responses in explanted hearts and cultured epicardial tissue sheets (Cao et al., 2017; Wang et al., 2015). Compounds from these screens that modulate epicardial regeneration are potentially useful in dissecting the epicardial injury response and in modulating in vivo cardiac repair.
6. Therapeutic applications
6.1. Directing the adult epicardium for heart repair
Activating the adult epicardium for heart repair requires directing this tissue to promote neovascularization rather than scarring, and to function as pro-regenerative rather than pro-inflammatory (Figure 4). Tβ4 treatment was the first attempt to prime the epicardium (Smart et al., 2011; Smart et al., 2007), while other studies since have similarly implicated Prokineticin-1 (Prok1) and VEGFA (Urayama et al., 2008; Zangi et al., 2013). As mentioned above, Tβ4 was suggested to regulate epicardial activation through chromatin remodeling (Vieira et al., 2017). In other cases, these factors are not particularly efficient, and their mechanisms have not been fully explored. Explant culture and screening systems as described above provide an opportunity to identify additional pharmacological factors (Cao and Poss, 2016; Cao et al., 2017; Wang et al., 2015). It is likely that a combinatorial approach using multiple factors, ideally aided by a generation of viral vectors that might readily access the epicardium, will yield a consensus strategy for boosting pro-regenerative epicardial properties. This might take advantage of regulatory sequences that preferentially direct gene expression in injured tissue (Goldman et al., 2017; Kang et al., 2016), to amplify expression of secreted factors or beneficial ECM components.
Figure 4. An epicardium-derived strategy for heart repair.
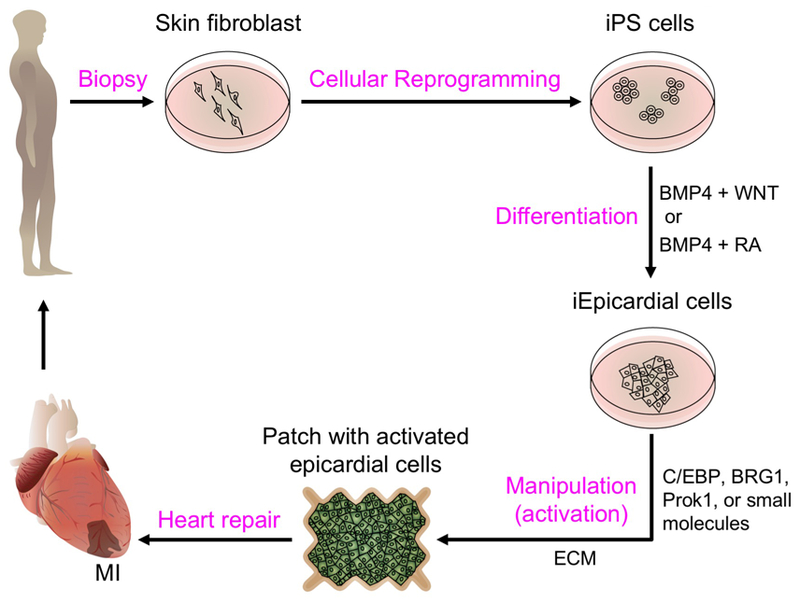
Skin fibroblasts from the MI patient could be reprogrammed into induced pluripotent stem cells (iPS cells) and further differentiated into induced epicardial cells (iEpicardial cells) by defined factors (see section 6.3). iEpicardial cells could be activated through treatment with small molecules or pro-regenerative factors (sections 4 and 6.1), and engineered with ECM components (section 4.2) into a patch (section 6.2) to be applied to the MI.
6.2. Epicardial patches for heart repair
An intriguing study performed by Wei et al. demonstrated that a bioengineered epicardial patch stimulated CM proliferation, diminished infarct size, and improved heart function after MI (Wei et al., 2015). In this study, Wei et al. co-cultured an epicardial mesothelial cell line (EMC) with immature CMs derived from mouse embryonic stem cells and found that myocardial cell proliferation was increased. Conditioned medium obtained from the EMC mimicked this effect. The authors then sutured a collagen nanofibrillar patch seeded with conditioned medium to the heart surface immediately after MI, finding improvement in cardiac function. The active factor in the conditioned medium was later identified as follistatin-like 1 (FSTL1), a secreted glycoprotein implicated in previous studies to suppress inflammation, reduce CM apoptosis, promote cardiac fibroblast activation, protect the heart from rupture after MI, and/or attenuate hypertrophy and cardiac failure after pressure overload (Maruyama et al., 2016; Ogura et al., 2012; Oshima et al., 2008; Shimano et al., 2011). Interestingly, collagen patches without conditioned medium or FSTL1 also improve the outcome after MI, suggesting a significant role of interventional physical support in heart repair. Wei et al. found that FSTL1, which is normally expressed in the epicardium in uninjured hearts, was strongly induced in CMs and decreased in the epicardium after MI. By contrast, Maruyama et al. reported that FSTL1 was predominantly expressed in fibroblasts of the MI (Maruyama et al., 2016). Interestingly, Wei et al. found that only epicardium-derived FSTL1 and not myocardial FSTL1 could evoke CM cell division. The authors speculated that the functional difference between the two sources might be attributable to differential glycosylation. The fact that pro-repair effects were also observed when the patch was applied at 1 week after injury has implications for therapies involving an expanded patient pool. The authors reported similar effects in a small-scale swine study (Wei et al., 2015). Of note, two previous studies in mice, showing reduced infarct size by intravenous delivery of human FSTL1 protein or adenovirus-driven FSTL1 expression before MI, did not assess protein modifications (Ogura et al., 2012; Oshima et al., 2008). In addition, CM-specific knock out of Fstl1 in mice was reported to promote hypertrophy in a pressure overload model induced by transverse aortic constriction, suggesting complex cardiac functions of Fstl1 (Shimano et al., 2011). Although the mechanisms require some clarification, a paradigm has emerged for how epicardium may be a source of mitogenic factors for delivery via engineered cardiac patches (Figure 4).
6.3. Primary or stem cell-derived epicardial cells in epicardial biology
Studies exploring epicardial cells of human origin are ultimately essential for translational goals. To this end, van Tuyn and coauthors cultured primary epicardial cells obtained from the right atrial appendages of adult humans and characterized their features (van Tuyn et al., 2007). They found that these cells could differentiate into smooth muscle cells when infected with adenovirus vectors expressing the transcription factor myocardin or treated with Tgfβ1 or BMP2. However, these cells also spontaneously underwent EMT during culture and attained a fibroblastic morphology. In a later study, Moerkamp et al. developed a protocol for culturing isolated human fetal and adult EPDCs from cardiac specimens (Moerkamp et al., 2016). These cells successfully maintained epithelial features and only underwent EMT upon stimulation with Tgfβ. The authors found that fetal EPDCs were less migratory but progressed faster through EMT than those obtained from adults. The authors further stated that fetal EPDCs responded to environmental changes more rapidly. Although the mechanism underlying these effects and their translational implications are unclear, this primary cell culture system allows activation and plasticity to be directly assessed in human epicardial cells.
Experiments involving primary human epicardial cells have technical challenges. As an alternative approach, Witty et al. developed a method to generate functional epicardial-like cells from human pluripotent stem cells, thus providing an unlimited source of these cells for functional, in vitro studies of the human epicardium (Witty et al., 2014). This method also presented the opportunity to develop precision therapies (Figure 4). Here, doses of BMP and WNT signaling were manipulated to control cardiovascular lineages of the differentiated human pluripotent stem cells (hPSCs). The authors found that increases in the dose of BMP4 were correlated with reductions in the CM lineage (which was eventually abolished), and an increase in the formation of cells expressing the epicardial markers Wt1 and Tbx18. After the cells were cultured and passaged for 4 days, the resulting epicardial-like cells formed a confluent monolayer with cobblestone morphology and expressing the tight junction protein ZO1 at cell borders. These cells underwent EMT and differentiated into cells that displayed characteristics of smooth muscle cells in response to treatment with TGFB1 and bFGF, and of fibroblasts when treated with only bFGF. Thus, the results of this study established a method for inducing epicardial cells in hPSC cultures.
Following up on that study, Iyer and coauthors developed chemical means to differentiate epicardium and epicardium-derived smooth muscle cells/fibroblasts from hPSCs (Iyer et al., 2015). hPSCs were first differentiated to form early mesoderm via a combined treatment including FGF2, BMP4 and Ly294002 (an inhibitor of phosphoinositide 3- kinases). They were then treated with FGF2 and BMP4 to induce their differentiation into lateral plate mesoderm and subsequently with BMP4, WNT3A and RA to induce differentiation into the epicardial lineage. The induced epicardial cells displayed epithelial cell morphology and expressed epicardial markers (i.e., TBX18, WT1 and TCF21). These cells underwent EMT and differentiated into smooth muscle cells and fibroblasts upon induction. Interestingly, the cells survived, were incorporated into the epicardium and even differentiated in vivo when injected into the chick extra-embryonic circulation. Similarly, Guadix et al. used RA and BMP4 to promote the differentiation of hESCs and hPSCs into proepicardial-like cells without using WNTs (Guadix et al., 2017). When grafted onto chick embryo hosts, the resulting proepicardial-like cells displayed functional properties including adhesion and spreading over the myocardium. In the Iyer et al. study, injected cells were predominantly found in the subepicardial space between the epicardium and myocardium; however, Guadix et al. found that the cells generated by their methods spread over the myocardial surface and incorporated in the epicardium itself, suggesting they possess greater epithelial properties. These studies reveal possibilities for transplanting epicardial cells and epicardial-derived lineages for treatment of cardiovascular disease. To avoid risks of pathogenicity and immunogenic contamination using albumin-rich medium, two recent studies used small molecular compounds to manipulate WNT and RA signaling in albumin-free media (Bao et al., 2016; Zhao et al., 2017). This approach may make these cells more suitable for clinical applications.
7. Conclusions
Therapeutic regeneration of the injured human heart is a huge challenge, and the epicardium is a relatively new player on this stage. One scenario is the generation of engineered patches with the appropriate ECM components and tension properties (Figure 4). Seeded with epicardial and EPDCs, or other cells genetically manipulated to possess pro-regenerative features of epicardium, these constructs could secrete mitogens and impact neovascularization in injured myocardium. To achieve this goal, more candidate and consensus CM and epicardial mitogens must be identified. Also, identification of pro-regenerative subsets of epicardial cells with specific markers will enable genetic methods to precisely manipulate the most appropriate cells after injury. An additional scenario could influence heart repair with evolved viral vectors that target epicardium and its progeny. Marrying more closely the models of high innate heart regeneration with stem cell-based strategies will help achieve the ultimate goals of epicardial cell research.
Key points.
The epicardium is surface mesothelium for all vertebrate hearts.
The epicardium contributes key cells and signals during heart development and regeneration.
The epicardium is a heterogeneous population and itself is highly regenerative.
The epicardium is required for normal myocardial regeneration in zebrafish.
Current research attempts to activate the adult epicardium to promote heart regeneration.
ACKNOWLEDGEMENTS
We thank A. Dickson for artwork, R. Karra and J. Kang for comments on the manuscript. We apologize to our colleagues whose work we could not discuss owing to space limitations. J.C. was supported by American Heart Association postdoctoral fellowships (14POST20230023 and 16POST30230005). K.D.P. acknowledges grant support from NHLBI (HL081674, HL131319, and HL136182), an American Heart Association Merit Award, and Fondation Leducq.
Footnotes
COMPETING INTERESTS
The authors declare they have no conflicts of interest.
REFERENCES
- Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, et al. (2012). The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 139, 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, Kamran P, Muller AM, Volz KS, Tang Z, et al. (2014). Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res 115, 625–635. [DOI] [PubMed] [Google Scholar]
- Athanasiadis EI, Botthof JG, Andres H, Ferreira L, Lio P, and Cvejic A (2017). Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat Commun 8, 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer GM, Bollini S, Dube KN, Martinez-Barbera JP, Williams O, and Riley PR (2014). Dynamic haematopoietic cell contribution to the developing and adult epicardium. Nat Commun 5, 4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright A, et al. (2016). Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, and Kuhn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. [DOI] [PubMed] [Google Scholar]
- Bersell K, Choudhury S, Mollova M, Polizzotti BD, Ganapathy B, Walsh S, Wadugu B, Arab S, and Kuhn B (2013). Moderate and high amounts of tamoxifen in alphaMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech 6, 1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock-Marquette I, Shrivastava S, Pipes GC, Thatcher JE, Blystone A, Shelton JM, Galindo CL, Melegh B, Srivastava D, Olson EN, et al. (2009). Thymosin beta4 mediated PKC activation is essential to initiate the embryonic coronary developmental program and epicardial progenitor cell activation in adult mice in vivo. J Mol Cell Cardiol 46, 728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini S, Vieira JM, Howard S, Dube KN, Balmer GM, Smart N, and Riley PR (2014). Re-activated adult epicardial progenitor cells are a heterogeneous population molecularly distinct from their embryonic counterparts. Stem Cells Dev 23, 1719–1730. [DOI] [PubMed] [Google Scholar]
- Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, and Yutzey KE (2013). Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol 65, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch CM, and Yutzey KE (2013). Transcriptional Control of Cell Lineage Development in Epicardium-Derived Cells. J Dev Biol 1, 92–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ, Choudhury RP, and Riley PR (2017). Heart regeneration and repair after myocardial infarction: translational opportunities for novel therapeutics. Nat Rev Drug Discov 16, 699–717. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Martinez A, Vargas-Gonzalez A, Guarner-Lans V, Prado-Zayago E, Leon-Oleda M, and Nieto-Lima B (2010). Functional and structural regeneration in the axolotl heart (Ambystoma mexicanum) after partial ventricular amputation. Arch Cardiol Mex 80, 79–86. [PubMed] [Google Scholar]
- Cao J, Navis A, Cox BD, Dickson AL, Gemberling M, Karra R, Bagnat M, and Poss KD (2016a). Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development 143, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, and Poss KD (2016). Explant culture of adult zebrafish hearts for epicardial regeneration studies. Nat Protoc 11, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wang J, Jackman CP, Cox AH, Trembley MA, Balowski JJ, Cox BD, De Simone A, Dickson AL, Di Talia S, et al. (2017). Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev Cell 42, 600–615 e604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu JD, Nie B, Xie M, Zhang M, Wang H, et al. (2016b). Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220. [DOI] [PubMed] [Google Scholar]
- Chablais F, and Jazwinska A (2012). The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 139, 1921–1930. [DOI] [PubMed] [Google Scholar]
- Chablais F, Veit J, Rainer G, and Jazwinska A (2011). The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC developmental biology 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Gemberling M, Wang J, Holdway JE, Shen MC, Karlstrom RO, and Poss KD (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, and Kispert A (2009). Tbx18 and the fate of epicardial progenitors. Nature 458, E8–9; discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Darehzereshki A, Rubin N, Gamba L, Kim J, Fraser J, Huang Y, Billings J, Mohammadzadeh R, Wood J, Warburton D, et al. (2015). Differential regenerative capacity of neonatal mouse hearts after cryoinjury. Dev Biol 399, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deten A, Holzl A, Leicht M, Barth W, and Zimmer HG (2001). Changes in extracellular matrix and in transforming growth factor beta isoforms after coronary artery ligation in rats. J Mol Cell Cardiol 33, 1191–1207. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W Jr., Ordahl CP, and Bristow J (1998). Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol 193, 169–181. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Pae SH, Morabito C, and Bristow J (2003). Inhibition of alpha4-integrin stimulates epicardial-mesenchymal transformation and alters migration and cell fate of epicardially derived mesenchyme. Dev Biol 257, 315–328. [DOI] [PubMed] [Google Scholar]
- Dogra D, Ahuja S, Kim HT, Rasouli SJ, Stainier DYR, and Reischauer S (2017). Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat Commun 8, 1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gherghe C, Liu D, Hamlett E, Srikantha L, Rodgers L, Regan JN, Rojas M, Willis M, Leask A, et al. (2012). Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J 31, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube KN, Thomas TM, Munshaw S, Rohling M, Riley PR, and Smart N (2017). Recapitulation of developmental mechanisms to revascularize the ischemic heart. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duim SN, Kurakula K, Goumans MJ, and Kruithof BP (2015). Cardiac endothelial cells express Wilms’ tumor-1: Wt1 expression in the developing, adult and infarcted heart. J Mol Cell Cardiol 81, 127–135. [DOI] [PubMed] [Google Scholar]
- Dunaeva M, and Waltenberger J (2017). Hh signaling in regeneration of the ischemic heart. Cell Mol Life Sci 74, 3481–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Xiang FL, Braitsch CM, and Yutzey KE (2016). Epicardium-derived fibroblasts in heart development and disease. J Mol Cell Cardiol 91, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglia MJ, and Poss KD (2016). Building and re-building the heart by cardiomyocyte proliferation. Development 143, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG (2017). The role of transforming growth factor (TGF)-beta in the infarcted myocardium. J Thorac Dis 9, S52–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen ME, and Lemanski LF (1990). Epicardial development in the axolotl, Ambystoma mexicanum. Anat Rec 226, 228–236. [DOI] [PubMed] [Google Scholar]
- Frantz C, Stewart KM, and Weaver VM (2010). The extracellular matrix at a glance. J Cell Sci 123, 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado MB, Nim HT, Boyd SE, and Rosenthal NA (2016). View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 143, 387–397. [DOI] [PubMed] [Google Scholar]
- Galdos FX, Guo Y, Paige SL, VanDusen NJ, Wu SM, and Pu WT (2017). Cardiac Regeneration: Lessons From Development. Circ Res 120, 941–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M, Karra R, Dickson AL, and Poss KD (2015). Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EL, Baldwin HS, and Hynes RO (1997). Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood 90, 3073–3081. [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, and Poelmann RE (1998). Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 82, 1043–1052. [DOI] [PubMed] [Google Scholar]
- Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, Foglia MJ, Karra R, Dickson AL, Sun F, Tolstorukov MY, et al. (2017). Resolving Heart Regeneration by Replacement Histone Profiling. Dev Cell 40, 392–404 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Burns CE, and Burns CG (2017). Zebrafish heart regeneration: 15 years of discoveries. Regeneration (Oxf) 4, 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, and Mercader N (2011). Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138, 1663–1674. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Peralta M, and Mercader N (2012). Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev Biol 370, 173–186. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, and Burns CG (2018). Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell 44, 433–446 e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieskamp T, Rudat C, Ludtke TH, Norden J, and Kispert A (2011). Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ Res 108, 813–823. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Carmona R, Munoz-Chapuli R, and Perez-Pomares JM (2006). In vivo and in vitro analysis of the vasculogenic potential of avian proepicardial and epicardial cells. Dev Dyn 235, 1014–1026. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Orlova VV, Giacomelli E, Bellin M, Ribeiro MC, Mummery CL, Perez-Pomares JM, and Passier R (2017). Human Pluripotent Stem Cell Differentiation into Functional Epicardial Progenitor Cells. Stem Cell Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Ju H, Zhao S, Junaid A, Scammell-La Fleur T, and Dixon IM (1999). Elevation of expression of Smads 2, 3, and 4, decorin and TGF-beta in the chronic phase of myocardial infarct scar healing. J Mol Cell Cardiol 31, 667–678. [DOI] [PubMed] [Google Scholar]
- Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF, and Lien CL (2015). Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell 33, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubner BJ, Adamowicz-Brice M, Khadayate S, Tiefenthaler V, Metzler B, Aitman T, and Penninger JM (2012). Complete cardiac regeneration in a mouse model of myocardial infarction. Aging (Albany NY) 4, 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li Y, Li Y, Pu W, Huang X, Tian X, Wang Y, Zhang H, Liu Q, Zhang L, et al. (2017). Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med 23, 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakow R (1992). Epicardial formation in staged human embryos. Kaibogaku Zasshi 67, 616–622. [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, et al. (2012). C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 338, 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, and Lien CL (2013). Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS One 8, e67266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui SP, Sheng DZ, Sugimoto K, Gonzalez-Rajal A, Nakagawa S, Hesselson D, and Kikuchi K (2017). Zebrafish Regulatory T Cells Mediate Organ-Specific Regenerative Programs. Dev Cell 43, 659–672 e655. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, and Srivastava D (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, and Srivastava D (2009). Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell 16, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, and Ohta H (2013). Pathophysiological roles of FGF signaling in the heart. Front Physiol 4, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ohta H, Nakayama Y, and Konishi M (2016). Roles of FGF Signals in Heart Development, Health, and Disease. Front Cell Dev Biol 4, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, and Kawakami Y (2012). Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 139, 4133–4142. [DOI] [PubMed] [Google Scholar]
- Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Talasila A, and Sinha S (2015). Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development 142, 1528–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr M, Schlueter J, Brand T, and Manner J (2008). Development of the proepicardium in Xenopus laevis. Dev Dyn 237, 3088–3096. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, and Belmonte JC (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, and Poss KD (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra R, Knecht AK, Kikuchi K, and Poss KD (2015). Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci U S A 112, 13255–13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra R, and Poss KD (2017). Redirecting cardiac growth mechanisms for therapeutic regeneration. J Clin Invest 127, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, and Tabin CJ (2012). Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 22, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, and Poss KD (2011a). tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 138, 2895–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, and Poss KD (2011b). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, and Poss KD (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, et al. (2010). PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A 107, 17206–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M, Ito K, and Shimada Y (1987). Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 176, 183–189. [DOI] [PubMed] [Google Scholar]
- Kratsios P, Catela C, Salimova E, Huth M, Berno V, Rosenthal N, and Mourkioti F (2010). Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ Res 106, 559–572. [DOI] [PubMed] [Google Scholar]
- Kurkiewicz TO (1909). Histogenezie miesna sur cowego zwierzat kregowych — Zur Histogenese des Herzmuskels der Wilbertiere. Bull Int Acad Sci Cracov, 148–191. [Google Scholar]
- Lafontant PJ, Behzad AR, Brown E, Landry P, Hu N, and Burns AR (2013). Cardiac myocyte diversity and a fibroblast network in the junctional region of the zebrafish heart revealed by transmission and serial block-face scanning electron microscopy. PLoS One 8, e72388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Marin-Juez R, Moura PL, Kuenne C, Lai JKH, Tsedeke AT, Guenther S, Looso M, and Stainier DY (2017). Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane EB, Hogan BL, Kurkinen M, and Garrels JI (1983). Co-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryo. Nature 303, 701–704. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, and Ornitz DM (2007). Rebuilding the coronary vasculature: hedgehog as a new candidate for pharmacologic revascularization. Trends Cardiovasc Med 17, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, and Ornitz DM (2008). Fibroblast growth factors and Hedgehogs: at the heart of the epicardial signaling center. Trends Genet 24, 33–40. [DOI] [PubMed] [Google Scholar]
- Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, and Martin JF (2017). Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, and Poss KD (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619. [DOI] [PubMed] [Google Scholar]
- Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, and Gittenberger-de Groot AC (2007). Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. ScientificWorldJournal 7, 1777–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana F, Bertolami C, Mangoni A, Di Carlo A, Avitabile D, Mocini D, Iannelli P, De Mori R, Marchetti C, Pozzoli O, et al. (2010). Myocardial infarction induces embryonic reprogramming of epicardial c-kit(+) cells: role of the pericardial fluid. J Mol Cell Cardiol 48, 609–618. [DOI] [PubMed] [Google Scholar]
- Lin Z, and Pu WT (2014). Harnessing Hippo in the heart: Hippo/Yap signaling and applications to heart regeneration and rejuvenation. Stem Cell Res 13, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang X, Oh JH, Lin RZ, Duan S, Yu Y, Yang R, Qiu J, Melero-Martin JM, Pu WT, et al. (2014). Epicardium-to-fat transition in injured heart. Cell Res 24, 1367–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson MK, and Mosher DF (1998). Fibronectin: structure, assembly, and cardiovascular implications. Arterioscler Thromb Vasc Biol 18, 1363–1370. [DOI] [PubMed] [Google Scholar]
- Mahmoud AI, O’Meara CC, Gemberling M, Zhao L, Bryant DM, Zheng R, Gannon JB, Cai L, Choi WY, Egnaczyk GF, et al. (2015). Nerves Regulate Cardiomyocyte Proliferation and Heart Regeneration. Dev Cell 34, 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasek FJ (1968). Embryonic development of the heart. I. A light and electron microscopic study of myocardial development in the early chick embryo. J Morphol 125, 329–365. [DOI] [PubMed] [Google Scholar]
- Manasek FJ (1969). Embryonic development of the heart. II. Formation of the epicardium. J Embryol Exp Morphol 22, 333–348. [PubMed] [Google Scholar]
- Manner J (1992). The development of pericardial villi in the chick embryo. Anat Embryol (Berl) 186, 379–385. [DOI] [PubMed] [Google Scholar]
- Manner J (1993). Experimental study on the formation of the epicardium in chick embryos. Anat Embryol (Berl) 187, 281–289. [DOI] [PubMed] [Google Scholar]
- Manner J (1999). Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec 255, 212–226. [DOI] [PubMed] [Google Scholar]
- Marin-Juez R, Marass M, Gauvrit S, Rossi A, Lai SL, Materna SC, Black BL, and Stainier DY (2016). Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc Natl Acad Sci U S A 113, 11237–11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro J, Pfefferli C, de Preux Charles AS, Bise T, and Jazwinska A (2016). Collagen XII Contributes to Epicardial and Connective Tissues in the Zebrafish Heart during Ontogenesis and Regeneration. PLoS One 11, e0165497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Nakamura K, Papanicolaou KN, Sano S, Shimizu I, Asaumi Y, van den Hoff MJ, Ouchi N, Recchia FA, and Walsh K (2016). Follistatin-like 1 promotes cardiac fibroblast activation and protects the heart from rupture. EMBO Mol Med 8, 949–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Ramos L, Cleland J, Bressan M, and Mikawa T (2013). Induction of the Proepicardium. J Dev Biol 1, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan MA, Rosenthal NA, and Pinto AR (2017). Cre-loxP-Mediated Recombination: General Principles and Experimental Considerations. Curr Protoc Mouse Biol 7, 1–12. [DOI] [PubMed] [Google Scholar]
- Mercer SE, Odelberg SJ, and Simon HG (2013). A dynamic spatiotemporal extracellular matrix facilitates epicardial-mediated vertebrate heart regeneration. Dev Biol 382, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Izpisua Belmonte JC, et al. (2005). Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A 102, 18455–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, and Fischman DA (1992). Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci U S A 89, 9504–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, and Gourdie RG (1996). Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol 174, 221–232. [DOI] [PubMed] [Google Scholar]
- Missinato MA, Tobita K, Romano N, Carroll JA, and Tsang M (2015). Extracellular component hyaluronic acid and its receptor Hmmr are required for epicardial EMT during heart regeneration. Cardiovasc Res 107, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerkamp AT, Lodder K, van Herwaarden T, Dronkers E, Dingenouts CK, Tengstrom FC, van Brakel TJ, Goumans MJ, and Smits AM (2016). Human fetal and adult epicardial-derived cells: a novel model to study their activation. Stem Cell Res Ther 7, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, et al. (2013). Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A 110, 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore-Morris T, Guimaraes-Camboa N, Yutzey KE, Puceat M, and Evans SM (2015). Cardiac fibroblasts: from development to heart failure. J Mol Med (Berl) 93, 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Heallen T, Leach J, Xiao Y, and Martin JF (2017). Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Grivas D, Gonzalez-Rajal A, Torregrosa-Carrion R, and de la Pompa JL (2017). Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development 144, 1425–1440. [DOI] [PubMed] [Google Scholar]
- Nahirney PC, Mikawa T, and Fischman DA (2003). Evidence for an extracellular matrix bridge guiding proepicardial cell migration to the myocardium of chick embryos. Dev Dyn 227, 511–523. [DOI] [PubMed] [Google Scholar]
- Nesbitt T, Lemley A, Davis J, Yost MJ, Goodwin RL, and Potts JD (2006). Epicardial development in the rat: a new perspective. Microsc Microanal 12, 390–398. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Ouchi N, Ohashi K, Shibata R, Kataoka Y, Kambara T, Kito T, Maruyama S, Yuasa D, Matsuo K, et al. (2012). Therapeutic impact of follistatin-like 1 on myocardial ischemic injury in preclinical models. Circulation 126, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivey HE, and Svensson EC (2010). Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res 106, 818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Ouchi N, Sato K, Izumiya Y, Pimentel DR, and Walsh K (2008). Follistatin-like 1 is an Akt-regulated cardioprotective factor that is secreted by the heart. Circulation 117, 3099–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachori AS, Custer L, Hansen D, Clapp S, Kemppa E, and Klingensmith J (2010). Bone morphogenetic protein 4 mediates myocardial ischemic injury through JNK-dependent signaling pathway. J Mol Cell Cardiol 48, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Paik DT, Rai M, Ryzhov S, Sanders LN, Aisagbonhi O, Funke MJ, Feoktistov I, and Hatzopoulos AK (2015). Wnt10b Gain-of-Function Improves Cardiac Repair by Arteriole Formation and Attenuation of Fibrosis. Circ Res 117, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi EM, and Kuhn B (2014). Signalling between microvascular endothelium and cardiomyocytes through neuregulin. Cardiovasc Res 102, 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SH, O’Hara L, Atanassova N, Smith SE, Curley MK, Rebourcet D, Darbey AL, Gannon AL, Sharpe RM, and Smith LB (2017). Low-dose tamoxifen treatment in juvenile males has long-term adverse effects on the reproductive system: implications for inducible transgenics. Sci Rep 7, 8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, et al. (2017). Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 49, 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta M, Steed E, Harlepp S, Gonzalez-Rosa JM, Monduc F, Ariza-Cosano A, Cortes A, Rayon T, Gomez-Skarmeta JL, Zapata A, et al. (2013). Heartbeat-driven pericardiac fluid forces contribute to epicardium morphogenesis. Curr Biol 23, 1726–1735. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, and Munoz-Chapuli R (2002a). Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol 46, 1005–1013. [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, and Wessels A (2002b). Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Dev Biol 247, 307–326. [DOI] [PubMed] [Google Scholar]
- Pinto AR, Godwin JW, and Rosenthal NA (2014). Macrophages in cardiac homeostasis, injury responses and progenitor cell mobilisation. Stem Cell Res 13, 705–714. [DOI] [PubMed] [Google Scholar]
- Plavicki JS, Hofsteen P, Yue MS, Lanham KA, Peterson RE, and Heideman W (2014). Multiple modes of proepicardial cell migration require heartbeat. BMC Dev Biol 14, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon KL, Liebling M, Kondrychyn I, Garcia-Lecea M, and Korzh V (2010). Zebrafish cardiac enhancer trap lines: new tools for in vivo studies of cardiovascular development and disease. Dev Dyn 239, 914–926. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, and Sadek HA (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, and Sadek HA (2013). Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A 110, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, and Olson EN (2014). A neonatal blueprint for cardiac regeneration. Stem Cell Res 13, 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, and Keating MT (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, and Srivastava D (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjee V, Li D, Manderfield LJ, Liu F, Engleka KA, Aghajanian H, Rodell CB, Lu W, Ho V, Wang T, et al. (2017). Epicardial YAP/TAZ orchestrate an immunosuppressive response following myocardial infarction. J Clin Invest 127, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risebro CA, Vieira JM, Klotz L, and Riley PR (2015). Characterisation of the human embryonic and foetal epicardium during heart development. Development 142, 3630–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers LS, Lalani S, Runyan RB, and Camenisch TD (2008). Differential growth and multicellular villi direct proepicardial translocation to the developing mouse heart. Dev Dyn 237, 145–152. [DOI] [PubMed] [Google Scholar]
- Rudat C, and Kispert A (2012). Wt1 and epicardial fate mapping. Circ Res 111, 165–169. [DOI] [PubMed] [Google Scholar]
- Russell JL, Goetsch SC, Gaiano NR, Hill JA, Olson EN, and Schneider JW (2011). A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ Res 108, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel K, Wu CC, Kurth T, and Weidinger G (2011). Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PloS one 6, e18503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, and Lee RT (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serluca FC (2008). Development of the proepicardial organ in the zebrafish. Dev Biol 315, 18–27. [DOI] [PubMed] [Google Scholar]
- Shimano M, Ouchi N, Nakamura K, van Wijk B, Ohashi K, Asaumi Y, Higuchi A, Pimentel DR, Sam F, Murohara T, et al. (2011). Cardiac myocyte follistatin-like 1 functions to attenuate hypertrophy following pressure overload. Proc Natl Acad Sci U S A 108, E899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ramesh S, Cibi DM, Yun LS, Li J, Li L, Manderfield LJ, Olson EN, Epstein JA, and Singh MK (2016). Hippo Signaling Mediators Yap and Taz Are Required in the Epicardium for Coronary Vasculature Development. Cell Rep 15, 1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, et al. (2011). De novo cardiomyocytes from within the activated adult heart after injury. Nature 474, 640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, and Riley PR (2012). The epicardium as a candidate for heart regeneration. Future Cardiol 8, 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, and Riley PR (2007). Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445, 177–182. [DOI] [PubMed] [Google Scholar]
- Smith L (2011). Good planning and serendipity: exploiting the Cre/Lox system in the testis. Reproduction 141, 151–161. [DOI] [PubMed] [Google Scholar]
- Song AJ, and Palmiter RD (2018). Detecting and Avoiding Problems When Using the Cre-lox System. Trends Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, and Field LJ (1998). Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res 83, 15–26. [DOI] [PubMed] [Google Scholar]
- Stevens SM, von Gise A, VanDusen N, Zhou B, and Pu WT (2016). Epicardium is required for cardiac seeding by yolk sac macrophages, precursors of resident macrophages of the adult heart. Dev Biol 413, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Hui SP, Sheng DZ, and Kikuchi K (2017). Dissection of zebrafish shha function using site-specific targeting with a Cre-dependent genetic switch. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6, 377–382. [DOI] [PubMed] [Google Scholar]
- Tournoij E, Weber GJ, Akkerman JW, de Groot PG, Zon LI, Moll FL, and Schulte-Merker S (2010). Mlck1a is expressed in zebrafish thrombocytes and is an essential component of thrombus formation. J Thromb Haemost 8, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers JG, Kamal FA, Robbins J, Yutzey KE, and Blaxall BC (2016). Cardiac Fibrosis: The Fibroblast Awakens. Circ Res 118, 1021–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, and Stainier DY (2004). Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell 6, 371–382. [DOI] [PubMed] [Google Scholar]
- Tzahor E, and Poss KD (2017). Cardiac regeneration strategies: Staying young at heart. Science 356, 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urayama K, Guilini C, Turkeri G, Takir S, Kurose H, Messaddeq N, Dierich A, and Nebigil CG (2008). Prokineticin receptor-1 induces neovascularization and epicardial-derived progenitor cell differentiation. Arterioscler Thromb Vasc Biol 28, 841–849. [DOI] [PubMed] [Google Scholar]
- Uygur A, and Lee RT (2016). Mechanisms of Cardiac Regeneration. Dev Cell 36, 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuyn J, Atsma DE, Winter EM, van der Velde-van Dijke I, Pijnappels DA, Bax NA, Knaan-Shanzer S, Gittenberger-de Groot AC, Poelmann RE, van der Laarse A, et al. (2007). Epicardial cells of human adults can undergo an epithelial-to-mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem Cells 25, 271–278. [DOI] [PubMed] [Google Scholar]
- van Wijk B, Gunst QD, Moorman AF, and van den Hoff MJ (2012). Cardiac regeneration from activated epicardium. PLoS One 7, e44692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira JM, Howard S, Villa Del Campo C, Bollini S, Dube KN, Masters M, Barnette DN, Rohling M, Sun X, Hankins LE, et al. (2017). BRG1-SWI/SNF-dependent regulation of the Wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat Commun 8, 16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilahur G, Juan-Babot O, Pena E, Onate B, Casani L, and Badimon L (2011). Molecular and cellular mechanisms involved in cardiac remodeling after acute myocardial infarction. J Mol Cell Cardiol 50, 522–533. [DOI] [PubMed] [Google Scholar]
- Vivien CJ, Hudson JE, and Porrello ER (2016). Evolution, comparative biology and ontogeny of vertebrate heart regeneration. Npj Regenerative Medicine 1, 16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, and Red-Horse K (2015). Pericytes are progenitors for coronary artery smooth muscle. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. (2012). YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A 109, 2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao J, Dickson AL, and Poss KD (2015). Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 522, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Karra R, Dickson AL, and Poss KD (2013). Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol 382, 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner D, Tsarouchas TM, Michael A, Haase C, Weidinger G, Reimer MM, Becker T, and Becker CG (2017). Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat Commun 8, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, Zhu W, Fajardo G, Noseda M, Nakamura K, et al. (2015). Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 525, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, Briggs LE, Norris RA, van Wijk B, Perez-Pomares JM, et al. (2012). Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol 366, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Holdway JE, Major RJ, and Poss KD (2008). Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development 135, 183–192. [DOI] [PubMed] [Google Scholar]
- Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li RK, Kattman SJ, and Keller G (2014). Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol 32, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, Grun D, Berezikov E, Engel FB, et al. (2016). Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev Cell 36, 36–49. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Hill MC, Zhang M, Martin TJ, Morikawa Y, Wang S, Moise AR, Wythe JD, and Martin JF (2018). Single cell transcriptomics reveal an essential 1 role for Hippo signaling in cell state transitions during cardiac fibroblast development. Developmental Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. (2013). Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A 110, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, and Olson EN (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal 4, ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Cavallero S, Patterson M, Shen H, Xu J, Kumar SR, and Sucov HM (2015). Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARgamma activation. Proc Natl Acad Sci U S A 112, 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, et al. (2013). Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 31, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L, Oliveira MS, Ye LY, Ma Q, Sultana N, Hadas Y, Chepurko E, Spater D, Zhou B, Chew WL, et al. (2017). Insulin-Like Growth Factor 1 Receptor-Dependent Pathway Drives Epicardial Adipose Tissue Formation After Myocardial Injury. Circulation 135, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al. (2007). Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 13, 952–961. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li TS, Lee ST, Wawrowsky KA, Cheng K, Galang G, Malliaras K, Abraham MR, Wang C, and Marban E (2010). Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One 5, e12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mignone J, and MacLellan WR (2015). Cardiac Regeneration and Stem Cells. Physiol Rev 95, 1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Cao H, Tian L, Huo W, Zhai K, Wang P, Ji G, and Ma Y (2017). Efficient Differentiation of TBX18+/WT1+ Epicardial-Like Cells from Human Pluripotent Stem Cells Using Small Molecular Compounds. Stem Cells Dev 26, 528–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, Burns CG, and Burns CE (2014). Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci U S A 111, 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhao W, Chen Y, Ahokas RA, and Sun Y (2011). Acidic and basic fibroblast growth factors involved in cardiac angiogenesis following infarction. Int J Cardiol 152, 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, et al. (2015). High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun 6, 8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, et al. (2011). Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 121, 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, Ma Q, Oh JH, Lin RZ, Melero-Martin JM, von Gise A, Zhou P, Hu T, He L, et al. (2012). Thymosin beta 4 treatment after myocardial infarction does not reprogram epicardial cells into cardiomyocytes. J Mol Cell Cardiol 52, 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. (2008a). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Rivera-Feliciano J, and Pu WT (2008b). Nkx2–5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun 375, 450–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zymek P, Bujak M, Chatila K, Cieslak A, Thakker G, Entman ML, and Frangogiannis NG (2006). The role of platelet-derived growth factor signaling in healing myocardial infarcts. J Am Coll Cardiol 48, 2315–2323. [DOI] [PubMed] [Google Scholar]


