Abstract
Background/Aims
Syndecan-2 (SDC2) methylation was previously reported as a sensitive serologic biomarker for the early detection of colorectal cancer (CRC). The purpose of this study was to investigate whether SDC2 methylation is detectable in precancerous lesions and to determine the feasibility of using SDC2 methylation for the detection of CRC and precancerous lesions in bowel lavage fluid (BLF).
Methods
A total of 190 BLF samples were collected from the rectum at the beginning of colonoscopy from patients with colorectal neoplasm and healthy normal individuals. Fourteen polypectomy specimens were obtained during colonoscopy. A bisulfite pyrosequencing assay and quantitative methylation-specific polymerase chain reaction were conducted to measure SDC2 methylation in tissues and BLF DNA.
Results
SDC2 methylation was positive in 100% of villous adenoma (VA) and high-grade dysplasia, and hyperplastic polyp samples; 88.9% of tubular adenoma samples; and 0% of normal mucosa samples. In the BLF DNA test forSDC2 methylation, the sensitivity for detecting CRC and VA was 80.0% and 64.7%, respectively, at a specificity of 88.9%. The BLF of patients with multiple tubular adenomas, single tubular adenoma and hyperplastic polyps showed 62.8%, 26.7% and 28.6% rates of methylation-positive SDC2, respectively.
Conclusions
Our results demonstrated that SDC2 methylation was a frequent event in precancerous lesions and showed high potential in BLF for detecting patients with colorectal neoplasm.
Keywords: Syndecan-2, Methylation, Feces, Colorectal neoplasms, Adenoma
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer globally and its incidence is rapidly increasing in South Korea, and it is one of main causes of cancer-related death.1 In CRC, molecular level alterations occur before morphological alterations begin, and it progresses slowly through adenoma to dysplasia. Thus, it is possible to detect precancerous lesions by screening.2 Early screening and detection of colorectal tumors can reduce CRC mortality. Despite the importance of a CRC screening test, the fact that only 40% of population over 50 years of age receive a colonoscopy examination indicates the need for improved practicability and accessibility of a screening test.3
The fecal occult blood test (FOBT) is the most widely-used noninvasive CRC screening test. It has been demonstrated to decrease CRC mortality in a randomized prospective study.4 Yet, the test has limited sensitivity in detecting early stage CRC. The second-generation FOBT is an immunochemical test. It has improved sensitivity. Still, for advanced adenoma and early stage CRC, which are less likely to bleed, the diagnostic sensitivity is only around 20%. This has meant the continued use of the invasive colonoscopy examination to detect early stage colorectal tumor that can be completely cured by colonoscopic treatment. Colonoscopy is the gold standard for CRC diagnosis due to high sensitivity and specificity, whereas it is less preferred due to its invasiveness and difficulty of preparation. If it could be accurately predicted whether asymptomatic people harbor a tumor or not by using a tumor specific biomarker aside from the FOBT, fear of CRC as well as unnecessary medical procedure can be reduced. Redundant medical procedures and their cost could be reduced if a noninvasive molecular diagnostic technology using characteristics of tumors, such as DNA methylation, could sensitively detect colorectal tumors instead of colonoscopy.
Harada et al.5 reported that DNA methylation was detectable in bowel lavage fluid (BLF) collected from the patients with colorectal tumors during colonoscopy. BLF samples from invasive CRC revealed significantly higher levels of methylation than noninvasive tumors. BLF from CRC patients contained many exfoliated tumor cells and use of BLF could be an effective approach to detect CRC.5,6
Recently, syndecan-2 (SDC2) has been identified as a novel potential epigenetic biomarker for the detection of CRC using a genome-wide Media CpG microarray approach. While SDC2 methylation was detected at all CRC stages, even early stage showed very high frequency of SDC2 methylation. Thus, it suggested that SDC2 methylation can be potential biomarker for the detection of early stage of CRC.7
To date, SDC2 methylation has not been studied in adenomas that are prodromal lesions of CRC. It is also necessary to confirm if SDC2 methylation exfoliated from colon tumor can be detectable in BLF from patients with colon tumors before confirming detection in the stool.5,7 Therefore, in this study, we performed bisulfite-pyrosequencing on an independent group of patients with precancerous lesions to confirm the high frequency of aberrant SDC2 methylation in precancerous biopsies with various stages compared to normal tissues. For feasibility test of SDC2 methylation in BLF samples for early CRC detection, we assessed SDC2 methylation in patients with CRC and various precancerous lesions, and healthy individuals by quantitative methylation-specific polymerase chain reaction (PCR) assay. The results suggest that SDC2 methylation test in BLF could be a useful biomarker for early CRC detection.
MATERIALS AND METHODS
1. Patients
Two hundred and eight patients agreed to participate in this study, those screened for CRC using a colonoscopy at Eulji General Hospital of Eulji University July 1, 2015 to May 30, 2016. Among them 18 patients were excluded due to incidentally combined disease such as infectious colitis or inflammatory bowel disease. A total of 190 patients were enrolled in this study. Before colonoscopy, patients were pretreated with 2 L of polyethylene glycol lavage solution and 10 mL of BLF specimens were collected from the rectum at the beginning of the colonoscopy. Based on colonoscopic and histologic findings, the BLF samples were divided into six groups: patients with CRC, villous adenoma (VA), multiple tubular adenomas (MTA), single tubular adenoma (STA), hyperplastic polyp (HP), and patients without colorectal lesions (N).
MTA were defined as being 1.0 cm or more in diameter, and more than three adenomas. STA was defined small adenomas less than three, and one tubular adenoma (TA) being 1.0 cm or more in diameter. The details of characteristics for enrolled patients are provided in Table 1.
Table 1.
Clinicopathologic Features of the Subjects in This Study
| Number | Male sex | Age, yr | |
|---|---|---|---|
| Patients without colorectal lesion | 54 | 15 (27.8) | 56.6±13.6 |
| Patients with hyperplastic polyp | 21 | 10 (47.6) | 55.7±12.1 |
| Patients with single tubular adenoma | 45 | 27 (60.0) | 60.4±11.5 |
| Patients with multiple tubular adenomas | 43 | 32 (74.4) | 64.4±10.4 |
| Patients with villous adenoma and high-grade dysplasia | 17 | 11 (64.7) | 61.7±9.8 |
| Patients with colorectal cancer | 10 | 6 (60.0) | 61.1±15.4 |
| Total | 190 | 101 (53.2) | 59.9±12.3 |
Data are presented as number (%) or mean±SD.
2. Methods
1) Reagents
All chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. Oligonucleotides and fluorescent probes were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
2) Clinical specimens
Fourteen biopsy specimens were obtained from colorectal tumor during colonoscopy examination. Their identity as colorectal tumor was verified by an expert pathologist. Pathologic results were VA and/or high-grade dysplasia (n=2), TA (n=9), HP (n=3). Also five genomic DNA samples from normal mucosa without any history of malignancy were purchased from Bio-Chain Institute (Hayward, CA, USA).
A total of 190 BLF samples from patients with CRC (n=10), VA and/or high-grade dysplasia (n=17), MTA (n=43), STA (n=45), HP (n=21) and N (n=54) were obtained by collecting 10 mL of remnant fluids in the rectum during insertion of the colonoscope. BLF were immediately frozen and stored at −80°C until used.
This study was approved by the Institutional Review Board of Eulji University (EMCS 2015-06-011-001). Written informed consent was obtained from all study participants, adhering to local ethics guidelines.
3) DNA isolation
Genomic DNA was isolated from tissue specimens using DNA Mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. BLF DNA was isolated using QIAamp DNA Stool Mini kit (Qiagen) according to manufacturer’s instructions. The genomic DNA was finally eluted in 50 to 100 μL of TE buffer and frozen until use.
4) Bisulfite treatment
Genomic DNA was chemically modified using sodium bisulfite which converts all unmethylated cytosine to uracil, while leaving methylated cytosine unmodified, using the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. In brief, genomic DNA was treated with sodium bisulfite for 2.5 hours at 65°C and desulfonation was performed for 20 minutes at room temperature. Bisulfite-converted DNA was purified using a Zymo-Spin IC column (Zymo Research) and eluted with 10 μL of distilled water. The eluted DNA was either used immediately for methylation analysis or was stored at −20°C until further use.
5) Quantitative bisulfite-pyrosequencing analysis
To quantify the methylation levels of SDC2 in colorectal tissues, quantitative bisulfite-pyrosequencing8 was performed. Specific bisulfite PCR and pyrosequencing primers were designed to analyze 149 bp of 5′ untranslated region (UTR) including 4 CpG dinucleotides sites (+456, +460, +466, +473 bp) of SDC2 gene using PyroMark Assay Design Software version 2.0 (Qiagen). Primer sequences were: forward, 5′-GGGAGTAGGAGTAGGAGGAGGAA-3′; reverse, 5′-Biotin-ACCAAAACAAAACCAAACCTCCTACCCA-3′; sequencing, 5′-AGTAGGAGTAGGAGGAGGAA-3′. Bisulfite-modified DNA (20 ng) was amplified in a 25 μL reaction volume with gene-specific primers using PyroMark PCR kit (Qiagen). Samples were heated to 94°C for 10 minutes and then amplified for 45 cycles at 94°C for 30 seconds, 61°C for 45 seconds and 72°C for 40 seconds, followed by a final extension at 72°C for 10 minutes. Pyrosequencing was performed using the PyroMark Gold Q96 reagent and PyroMark ID96 instrument (Qiagen) as instructed by the manufacturer. Briefly, 25 μL of each biotinylated PCR product was immobilized on streptavidin-coated Sepharose HP beads (Amersham Biosciences, Piscataway, NJ, USA) and sequenced using automatically generated nucleotide dispensation order for “sequence to analyze” corresponding to each reaction. Each CpG site was assigned a percentage (%) of methylation by evaluating the C to T ratio as methylation indexes (MtI). The average % of methylation across four CpG sites was obtained. Methylated non-CpG cytosines were used for internal controls, and to check the fidelity of bisulfite conversion. If MtI of each sample is greater than 5% of the detection limitation of pyrosequencing,9 it considered as methylation-positive.
6) Two-step quantitative methylation-specific PCR
For measurement of SDC2 methylation in BLF DNA, we used two-step fluorescence-based quantitative methylation-specific PCR (qMSP) method. For this assay, two methylation-specific primers and probe were designed to bind to bisulfite-converted methylated DNA for the 5′ untranslated region (124 bp) of the SDC2 gene by slight modification of primers described previously (Table 2).7 To confirm PCR adequacy and quality of bisulfite-converted stool DNA, COL2A1 (86 bp) was used as control gene (Table 2). In the first step of amplification, a total of 20 μL of reaction mixture contained 20 ng of bisulfite-converted stool DNA, reverse methylation-specific primer (0.05 μM) for SDC2, reverse specific primer (0.05 μM) for control COL2A1 and 4 μL of 5× AptaTaq PCR master mix (Roche Diagnostics, Basel, Swiss). Linear amplification was performed as follows: one cycle at 95°C for 5 minutes and 35 cycles at 95°C for 15 seconds and at 60°C for 60 seconds.
Table 2.
Sequences of Primers and Probes Used in Two-Step qMSP
| Gene | Primers or probes | Sequences (5′→3′) |
|---|---|---|
| SDC2 | Forward* | GTAGAAATTAATAAGTGAGAGGGC |
| Reverse* | ACGACTCAAACTCGAAAACTCG | |
| Probe* | FAM-TTCGGGGCGTAGTTGCGGGCGG-Iowa Black | |
| COL2A1 | Forward† | GTAATGTTAGGAGTATTTTGTGGITA |
| Reverse† | CTAICCCAAAAAAACCCAATCCTA | |
| Probe | FAM-AGAAGAAGGGAGGGGTGTTAGGAGAGG-Iowa Black |
qMSP, quantitative methylation-specific polymerase chain reaction.
Underlining indicates CpG nucleotides;
I represents inosine nucleotide.
Adapted from Oh T, et al. J Mol Diagn 2013;15:498–507.7
After the first step amplification, reaction mixture was divided into two portions and each 16 μL and 4 μL was used for SDC2 and COL2A1 amplification, respectively. For SDC2 amplification, a total of 40 μL reaction mixture contained each 0.25 μM of methylation-specific forward and reverse primers, 0.125 μM of fluorescent probes (FAM) and 8 μL of 5× PCR master mix. A total of 40 μL reaction mixture for COL2A1 amplification was contained each 0.25 μM of gene-specific forward and reverse primers, 0.1 μM of fluorescent probes (FAM) and 8 μL of 5× AptaTaq PCR master mix (Roche). qMSP was performed on a Rotor-Gene Q real time PCR system (Qiagen, Hilden Germany). The thermal cycling conditions were as follows: one cycle at 95°C for 5 minutes and 40 cycles at 95°C for 15 seconds and at 60°C for 60 seconds. All reactions were performed in duplicate. For each run, methylated DNA (Qiagen, Hilden, Germany) and unmethylated DNA (Qiagen, Hilden, Germany) were used as methylation controls. Nontemplate control was also included. The CT (cycle threshold) value was calculated by using Rotor Gene Q software.
The SDC2 methylation results for individual samples were scored as CT value, as detected by the instrument software. A sample in SDC2 methylation test was considered as positive if the value of SDC2 CT was less than 40 cycles when COL2A1 CT was less than 36 cycles. It was considered as negative if the CT of SDC2 was not detected when COL2A1 CT was less than 36 cycles. Stool samples from patients were classified as positive if one out of two PCR replicates were called methylation-positive and as negative if none of two PCR replicates called methylation-negative (1 out of 2 algorithm).
7) Statistical analysis
All statistical analysis was performed using MedCalc version 9.3.2.0 (MedCalc Software, Ostend, Belgium). A p-value <0.05 was considered statistically significant. Receiver operating characteristic (ROC), area under ROC (AUC) and 95% confidence intervals (CIs) were calculated.
RESULTS
1. SDC2 methylation in tissue samples from patients with colorectal neoplasia
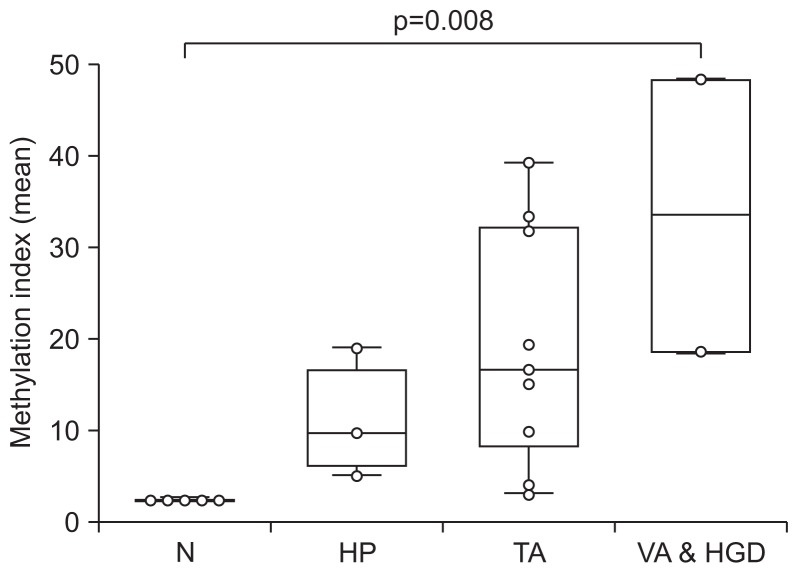
Previous study has reported that aberrant methylation of SDC2 is not observed in normal mucosa tissues, while frequently detected in colorectal tumor tissues.6 Presently, SDC2 methylation in precancerous lesions was examined using bisulfite-pyrosequencing in tissue specimens from VA or high-grade dysplasia (AA, n=2), TA (n=9), HP (n=3) and normal mucosa (N, n=5). SDC2 methylation was positive in 100% of AA, 88.9% of TA and 100% of HP tissue, and was negative in all of normal mucosa. The level of SDC2 methylation was significantly increased according to the malignant potential of precancerous lesions (p=0.008) (Fig. 1).
Fig. 1.
Methylation assessment of SDC2 gene in colorectal tissues by bisulfite pyrosequencing. The methylation level of the SDC2 gene was evaluated in normal mucosa (N), hyperplastic polyp (HP), tubular adenoma (TA) and villous adenoma and high-grade dysplasia tissues (VA & HGD). The methylation indexes (MtIs) of each sample are represented with box-and-whisker plots. The difference in the MtI of SDC2 was statistically significant at p<0.01 in VA & HGD vs TA vs HP vs N by the Kruskal-Wallis test.
2. SDC2 methylation in bowel lavage fluid from patients with colorectal neoplasia
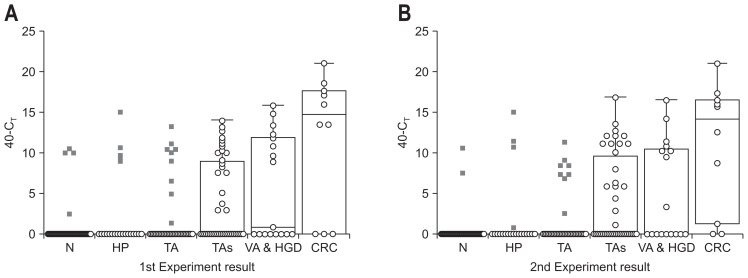
To determine whether SDC2 methylation can detect CRC and precancerous lesions, two-step qMSP was performed twice in BLF from patients with CRC (n=10), VA & HGD (n=17), TAs (n=43), TA (n=45), HP (n=21) and N (n=54) (Fig. 2). The degree of SDC2 methylation in each sample was determined as a CT by qMSP and expressed as 40-CT. A higher 40-CT value represents a higher methylation level of SDC2 and represented as 0, if CT of SDC2 was undetectable.
Fig. 2.
Results of SDC2 methylation analysis in bowel lavage fluid by two-step quantitative methylation-specific polymerase chain reaction (qMSP) test. (A, B) Two-step qMSP was performed twice in DNA from bowel lavage fluid. The distribution of SDC2 methylation was expressed in CT (threshold cycle) values as 40-CT for each sample. The methylation status of the SDC2 gene is shown as box-and-whisker plots. N, normal colonoscopy; HP, hyperplastic polyps; TA, single tubular adenoma; TAs, multiple tubular adenomas; VA & HGD, villous adenoma and/or high-grade dysplasia; CRC, colorectal cancer.
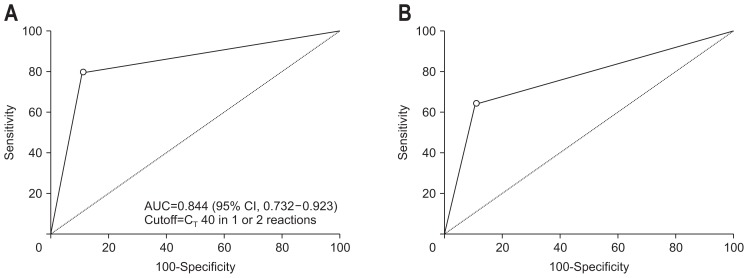
Overall methylation positivity was gradually increased depending on severity of lesion. ROC analysis was conducted to measure sensitivity and specificity of SDC2 methylation for diagnosis of CRC and VA & HGD using 1 out of 2 algorithm (Fig. 3). Sensitivities for detecting CRC and VA & HGD were 80.0% (8/10; 95% CI, 44.4% to 96.9%) and 64.7% (11/17; 95% CI, 38.4% to 85.7%), respectively with a specificity of 88.9% (6/54; 95% CI, 77.4% to 95.8%). Methylation positivity for MTA was 62.8% (27/43), significantly higher than that of STA 26.7% (12/45). Interestingly, HP also showed the methylation positivity of 28.6% (6/21) (Table 3). These results suggest that SDC2 methylation can be detected in patients with precancerous lesion and had a great role in CRC prevention as a time indicator for colonoscopic polypectomy of precancerous lesions.
Fig. 3.
Receiver operating characteristic (ROC) for SDC2 in detecting colorectal cancer (CRC). ROC curves were plotted as CRC vs healthy normal controls (A) and VA & HGD vs healthy normal controls (B). The cutoff values for methylation-positive, p-values, area under ROC (95% confidence interval [CI]), sensitivity (95% CI) and specificity (95% CI) are indicated at the bottom.
AUC, area under curve; VA & HGD, villous adenoma and/or high-grade dysplasia.
Table 3.
Methylation Positivity of SDC2 in Bowel Lavage Fluids from Patients with Colorectal Neoplasia
| Algorithm | Methylation positivity | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| N | HP | TA | TAs | VA & HGD | CRC | |
| 1 out of 2 | 11.1 (6/54) | 28.6 (6/21) | 26.7 (12/45) | 62.8 (27/43) | 64.7 (11/17) | 80.0 (8/10) |
Data are presented as percent (number/number).
N, normal colonoscopy; HP, hyperplastic polyp; TA, single tubular adenoma; TAs, multiple tubular adenomas; VA & HGD, villous adenoma and/or high-grade dysplasia; CRC, colorectal cancer.
SDC2 methylation positivity was also significantly increased depending on the number of detected adenomas (p<0.01, linear-by-linear association test). Positivity was 26.7% (12/45) for a single adenoma, 57.9% (22/38) for 2–5 adenomas and 100% (5/5) for 6 or more adenomas (Table 4).
Table 4.
Methylation Positivity of SDC2 in Bowel Lavage Fluid According to the Number of Detected Adenomas
| No. of adenoma | 1 | 2–5 | >5 | p-value* |
|---|---|---|---|---|
| Methylation positivity | 26.7 (12/45) | 57.9 (22/38) | 100 (5/5) | <0.01 |
Data are presented as percent (number/number).
Calculated by linear-by-linear association test.
DISCUSSION
The results of this study demonstrated that SDC2 methylation was frequently occurred in precancerous lesions in tissues and BLF samples. The incidence of CRC is rapidly increasing worldwide,1 and the development of screening has increased the survival rate of CRC over the past 20 years. A FOBT, computed tomography (CT) colonography, double-contrast barium enema, flexible sigmoidoscopy, and colonoscopy are commonly used to screen CRC.10 However, the sensitivity and specificity of the FOBT and barium enema for CRC are low,11–15 and a CT colonoscopy has a limitation with CT-related radiation exposure. Flexible sigmoidoscopy cannot explore the entire colon, and a screening colonoscopy is invasive and can be vary by the patients’ status and doctor’s ability. Recent research suggests that CRC may be detectable using DNA markers.4,6,8,9 Colorectal epithelial cells are continuously exfoliated, usually more often in colorectal tumors than in normal tissue. Validation of this method as a screening test for colorectal tumors has been explored.16 Other studies have sought genetic and epigenetic biomarkers for CRC detection.17,18 DNA mutations including K-ras, p53, adenomatous polyposis coli (APC), microsatellite instability (MSI) markers and unfragmented L-DNA have been used as genetic biomarkers for CRC diagnosis.19–23 However, a large-scale prospective study using 21 combinations of genetic marker (K-ras, APC, p53, MSI, BAT and L-DNA) in stool reported a diagnostic sensitivity of CRC and colorectal adenoma of 52% and 41%, respectively, which was superior to the 13% and 14% respective sensitivity of FOBT.24 These studies imply that the DNA panel has better sensitivity and adequate specificity in colorectal tumor diagnosis compared to the FOBT. However, the performance is not satisfactory considering its cost.
The limitation of genetic biomarkers has prompted studies using epigenetic biomarkers. Approximately 60% of human genes have CpG islands, where cytosine and guanine are concentrated at their promoters. Epigenetic biomarkers refer to hypermethylation in the promoter region, which is observed early in the development of human cancer. This gene methylation marker has been used in attempts to improve the diagnostic sensitivity of CRC and advanced adenoma.25,26 Typical epigenetic markers are SFRP2, CpG island methylator phenotype, intestinal stem cell marker DCLK1 and SEPT9. Sensitivity of CRC increased by over 90%, but adenoma sensitivity and specificity were both was 70%, which is a limitation to use as a general CRC test.27–32
The clinical trial of multitarget stool DNA testing as a CRC screening test compared to the FOBT has been explored.31 The DNA test included the KRAS mutations along with NDRG4 and BMP3 methylations, which are epigenetic markers. The study represented sensitivity of 92.3% for the DNA test and 73.8% for fecal immunochemical test (FIT) in detecting CRC. For those with advanced precancerous lesions, sensitivities of the DNA test and FOBT were only 42.4% and 23.8%.33 Among patients in a subclinical stage who were at average risk for CRC, multi-target stool DNA testing had higher sensitivity compared to FIT, while the false positive rate was still significantly high. These prior studies suggested that the epigenetic biomarkers were still unsatisfactory and need for more sensitive and specific epigenetic biomarkers.
Syndecan-2 is a cell membrane protein that is involved in cell proliferation, cell migration and in the interaction between cells and intercellular substances. SDC2 is not expressed in normal intestinal endothelial cells, but is expressed in mesenchymal cells.34 It was previously reported that SDC2 was aberrantly methylated in tumor tissues of most CRC patients and demonstrated a high potential of quantification of SDC2 methylation in blood for early detection of CRC.7 SDC2 methylation has been described in more than 95% of CRC tissue and no difference with respect to the CRC stage has been found.7 This implies that SDC2 methylation may begin early in tumor development and continue thereafter.
Up to date, SDC2 methylation has not been addressed in pre-cancerous lesions, thus this study was conducted to confirm the SDC2 methylation in precancerous biopsies at various stages. It is new that SDC2 methylation appears 100% in the stage of advanced adenoma and 88.9% of non-advanced adenomas, while did not occur in normal tissues. Thus it provides good evidence for SDC2 methylation as a potential biomarker for the early detection of CRC and prevention to CRC development.
The usefulness of SDC2 as a CRC biomarker for the first time was studied from blood samples and showed high sensitivity and specificity in the serum of CRC patients, and almost no methylation in serum DNA of normal healthy population, that resulted in suggesting the possibility as a circulating biomarker.7 If these epigenetic markers can detect tumor specific DNA methylation separated from the tumor at stool before being detected in the serum, the epigenetic marker can be noninvasive and more sensitive biomarkers in CRC screening. Thus, we tried to confirm SDC2 methylation in BLF before confirming the detection in the stool.
In this study, SCD2 methylation test by qMSP in BLF showed sensitivities of 80.0%, 64.7% and 62.8% for CRC, VA and MTA, respectively at the specificity of 88.9%. Meanwhile, Harada et al.5 analyzed the methylation of 15 genes in BLF of patients with CRC and advanced adenoma and identified three highly sensitive genes. Sensitivities of miR-124-3, LOC386758, and SFRP1 were 71.8%, 79.5% and 4.4%, respectively, for CRC detection. Combination of three genes revealed 82% of sensitivity and 79% of specificity with the AUC of 0.834, which are comparable to our data.
The SDC2 methylation in the BLF displayed high sensitivity and specificity as a marker for the detection of CRC patients. Also, it is encouraging that sensitivity and specificity increased to the expected levels of clinically important precancerous lesion such as VA and/or high-grade dysplasia. Sensitivity of SDC2 methylation significantly increased with more than three adenomas than 1 or 2 adenomas. The false positive rate in the normal group was 11.1% and positive rate in STA or HP was less than 30%. This is an acceptable limitation for the use of single biomarker in BLF. Thus, a large scale of clinical validation will be warranted in voided stool samples to evaluate the clinical utility of stool-based SDC2 methylation test to early detect CRC.
In conclusion, the SDC2 DNA methylation was detected in the tissue of almost colorectal neoplasm. The SDC2 DNA methylation in BLF showed a high sensitivity and specificity in patients with CRC and precancerous lesions. Also it has potential value in detection of the patients with precancerous lesion.
ACKNOWLEDGEMENTS
This study was supported by Eulji University Research Fund in 2015.
Footnotes
See editorial on page 479.
CONFLICTS OF INTEREST
S.A. and T.J.O. are employees and shareholders of Genomic-tree, Inc. The other authors declare that they have no conflicting interests.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2006;15:389–394. doi: 10.1158/1055-9965.EPI-05-0678. [DOI] [PubMed] [Google Scholar]
- 3.Ahluwalia IB, Mack KA, Murphy W, Mokdad AH, Bales VS. State-specific prevalence of selected chronic disease-related characteristics: Behavioral Risk Factor Surveillance System, 2001. MMWR Surveill Summ. 2003;52:1–80. [PubMed] [Google Scholar]
- 4.Duffy MJ, van Dalen A, Haglund C, et al. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39:718–727. doi: 10.1016/S0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 5.Harada T, Yamamoto E, Yamano HO, et al. Analysis of DNA methylation in bowel lavage fluid for detection of colorectal cancer. Cancer Prev Res (Phila) 2014;7:1002–1010. doi: 10.1158/1940-6207.CAPR-14-0162. [DOI] [PubMed] [Google Scholar]
- 6.Kamimae S, Yamamoto E, Yamano HO, et al. Epigenetic alteration of DNA in mucosal wash fluid predicts invasiveness of colorectal tumors. Cancer Prev Res (Phila) 2011;4:674–683. doi: 10.1158/1940-6207.CAPR-10-0214. [DOI] [PubMed] [Google Scholar]
- 7.Oh T, Kim N, Moon Y, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Dejeux E, El abdalaoui H, Gut IG, Tost J. Identification and quantification of differentially methylated loci by the pyrosequencing technology. Methods Mol Biol. 2009;507:189–205. doi: 10.1007/978-1-59745-522-0_15. [DOI] [PubMed] [Google Scholar]
- 9.Tsiatis AC, Norris-Kirby A, Rich RG, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson G, Cunningham CW, Parry S. The prevalence of colorectal adenomas in Maori and New Zealand Europeans parallels colorectal cancer rates. N Z Med J. 2010;123:45–49. [PubMed] [Google Scholar]
- 11.U.S. Preventive Services Task Force. Screening for colorectal cancer: recommendation and rationale. Ann Intern Med. 2002;137:129–131. doi: 10.7326/0003-4819-137-2-200207160-00014. [DOI] [PubMed] [Google Scholar]
- 12.Park DI, Ryu S, Kim YH, et al. Comparison of guaiac-based and quantitative immunochemical fecal occult blood testing in a population at average risk undergoing colorectal cancer screening. Am J Gastroenterol. 2010;105:2017–2025. doi: 10.1038/ajg.2010.179. [DOI] [PubMed] [Google Scholar]
- 13.Rozen P, Comaneshter D, Levi Z, et al. Cumulative evaluation of a quantitative immunochemical fecal occult blood test to determine its optimal clinical use. Cancer. 2010;116:2115–2125. doi: 10.1002/cncr.25012. [DOI] [PubMed] [Google Scholar]
- 14.Smith RA, Cokkinides V, Eyre HJ American Cancer Society. American Cancer Society guidelines for the early detection of cancer, 2003. CA Cancer J Clin. 2003;53:27–43. doi: 10.3322/canjclin.53.1.27. [DOI] [PubMed] [Google Scholar]
- 15.Winawer SJ, Stewart ET, Zauber AG, et al. A comparison of colonoscopy and double-contrast barium enema for surveillance after polypectomy. National Polyp Study Work Group. N Engl J Med. 2000;342:1766–1772. doi: 10.1056/NEJM200006153422401. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang DL, Chen JJ, Getzenberg RH, Schoen RE. Noninvasive testing for colorectal cancer: a review. Am J Gastroenterol. 2005;100:1393–1403. doi: 10.1111/j.1572-0241.2005.41427.x. [DOI] [PubMed] [Google Scholar]
- 17.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 18.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Boynton KA, Summerhayes IC, Ahlquist DA, Shuber AP. DNA integrity as a potential marker for stool-based detection of colorectal cancer. Clin Chem. 2003;49:1058–1065. doi: 10.1373/49.7.1058. [DOI] [PubMed] [Google Scholar]
- 20.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 21.Losi L, Roncucci L, di Gregorio C, de Leon MP, Benhattar J. K-ras and p53 mutations in human colorectal aberrant crypt foci. J Pathol. 1996;178:259–263. doi: 10.1002/(SICI)1096-9896(199603)178:3<259::AID-PATH473>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Mills AA. p53: link to the past, bridge to the future. Genes Dev. 2005;19:2091–2099. doi: 10.1101/gad.1362905. [DOI] [PubMed] [Google Scholar]
- 23.Traverso G, Shuber A, Levin B, et al. Detection of APC mutations in fecal DNA from patients with colorectal tumors. N Engl J Med. 2002;346:311–320. doi: 10.1056/NEJMoa012294. [DOI] [PubMed] [Google Scholar]
- 24.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 25.Simmer F, Brinkman AB, Assenov Y, et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7:1355–1367. doi: 10.4161/epi.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci. 2015;2:13. doi: 10.3389/fmolb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang KJ, Min BH, Ryu KJ, et al. The role of the CpG island methylator phenotype on survival outcome in colon cancer. Gut Liver. 2015;9:202–207. doi: 10.5009/gnl13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su XL, Wang YF, Li SJ, Zhang F, Cui HW. High methylation of the SEPT9 gene in Chinese colorectal cancer patients. Genet Mol Res. 2014;13:2513–2520. doi: 10.4238/2014.January.17.5. [DOI] [PubMed] [Google Scholar]
- 30.Vedeld HM, Andresen K, Eilertsen IA, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vedeld HM, Skotheim RI, Lothe RA, Lind GE. The recently suggested intestinal cancer stem cell marker DCLK1 is an epigenetic biomarker for colorectal cancer. Epigenetics. 2014;9:346–350. doi: 10.4161/epi.27582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Hu B, Choi AJ, et al. Unique DNA methylome profiles in CpG island methylator phenotype colon cancers. Genome Res. 2012;22:283–291. doi: 10.1101/gr.122788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 34.Park H, Kim Y, Lim Y, Han I, Oh ES. Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J Biol Chem. 2002;277:29730–29736. doi: 10.1074/jbc.M202435200. [DOI] [PubMed] [Google Scholar]