Abstract
The authors have developed a Web-based system that provides summary information about clinical trials being conducted throughout the United States. The first version of the system, publicly available in February 2000, contains more than 4,000 records representing primarily trials sponsored by the National Institutes of Health. The impetus for this system has come from the Food and Drug Administration (FDA) Modernization Act of 1997, which mandated a registry of both federally and privately funded clinical trials “of experimental treatments for serious or life-threatening diseases or conditions.” The system design and implementation have been guided by several principles. First, all stages of system development were guided by the needs of the primary intended audience, patients and other members of the public. Second, broad agreement on a common set of data elements was obtained. Third, the system was designed in a modular and extensible way, and search methods that take extensive advantage of the National Library of Medicine's Unified Medical Language System (UMLS) were developed. Finally, since this will be a long-term effort involving many individuals and organizations, the project is being implemented in several phases.
We have developed a database of clinical trials information that provides summary information about clinical trials being conducted throughout the United States. The first version of the system, publicly available in February 2000, contains more than 4,000 records representing primarily trials sponsored by the National Institutes of Health. Each record contains the title of the trial, a brief statement about the purpose of the trial (e.g., what intervention is being tested for what disease), the criteria that are relevant for patient participation in a trial (e.g., age range of the participants and certain characteristics of the disease for which the intervention is being developed) and, importantly, the place where the trial is being conducted together with telephone and other contact information. Some additional information may also be included, such as significant results if the trial has already been concluded or references to the research that led to a study that is currently under way.
In the following sections we first describe the motivation for developing the clinical trials database and the ways in which it differs from some other attempts to establish clinical trials registries. Next, we discuss the principles that guided our design of the system, including the importance of developing a system that would be readily accessible to patients and other members of the public. We discuss in some detail the standard set of data elements that underlie the system, and then turn to a fuller description of the system implementation. We conclude with some remarks on the lessons we have learned and some of the challenges we expect to face in the future.
Background
The impetus for the clinical trials database came from legislation passed in late 1997 that mandated a registry of clinical trials for both federally and privately funded trials “of experimental treatments for serious or life-threatening diseases or conditions.”1 Patient groups have demanded ready access to information about clinical research studies so that they might be more fully informed about a range of potential treatment options, particularly for very serious diseases. The law emphasizes that the information in such a registry, or databank, must be easily accessible, stating that it shall be “available to individuals with serious or life-threatening diseases and conditions, to other members of the public, to health care providers, and to researchers” and that it “shall be in a form that can be readily understood by members of the public.” The database should provide a sort of “one-stop shopping” for information on clinical trials, thereby minimizing the frustration that patients and others often face when they attempt to find and then search a wide range of disparate information sources, in some cases finding no information at all.
Many attempts have been made over the last several decades to establish clinical trials information systems, although most have focused not on patient access but on clinician and researcher access and use. An important concern has been that if the results of a trial are not available, either because the researcher has neglected to publish them or because the study has been stopped for some reason, then it is possible that, at best, a study is unnecessarily carried out multiple times or, at worst, a potiential treatment is administered when it has already been shown to be harmful. The Cochrane Collaboration was formed some years ago with the goal of performing and sharing systematic reviews of randomized controlled trials as these have been reported in the literature.2,3 If relevant data about trials are not published or are poorly reported, then this could lead to severe publication bias and, ultimately, poor care. Attempts to practice evidence-based medicine will necessarily fail. Prospective trial registries would address part of the problem, since trials would be listed as they were initiated, allowing tracking of all trials, even if the results were never published. Several researchers argue that it is critical that such trial registries, whether prospective or retrospective, capture data in a standard format, to facilitate meta-analysis and systematic reviews of the large number of trials.4,5,6,7 Some have pointed out the value of clinical trials registries to enhance patient recruitment, and others have been concerned with developing automated methods specifically for determining patient eligibility for particular trials.8,9,10,11 Other investigators have developed planning tools for clinical trial protocols, with the hope that this will lead to greater accuracy and efficiency in the conduct of the clinical trials themselves.12,13 Some work has been done in the development of systems that are intended to manage the entire life cycle of a clinical trial.14,15,16 These systems assist clinicians and researchers as they are carrying out the trial, and they may also yield valuable data that are useful for subsequent analyses. All these cases, although the goals may differ, have a common concern with making clinical trials information more readily available in order to improve the human condition.17
Design Objectives
We were guided by several principles as we began the design of our system, in September 1998. First, in all stages of the design and implementation we needed to be guided by the needs of our primary intended audience, patients and other members of the public. Second, the only way to successfully effect a project of this scope was to get broad agreement on a common set of data elements with a standard syntax and semantics. Third, since we needed to build a system as quickly as possible, it seemed clear to us that our requirements would evolve over time as we gained additional experience. Therefore, it would be important for us to design the system in a modular and extensible way. Finally, since this would be a long-term effort involving many individuals and organizations with varying backgrounds, technical expertise, and data sets, it would be necessary to implement the project in phases.
To reach the broadest audience with the fewest barriers to access, we designed a Web-based system that would be easy even for a novice user to use and yet would have extensive functionality. The goal was to make it simple for users to formulate their queries and then obtain results that would guide them to further relevant, “just-in-time” information. We involved patients and patient advocates in the early testing of the system, and we identified and then tested our site for accessibility using several readily available tools, also making sure that the system performs reasonably on a wide range of Web browsers.
We decided to begin the project by working with our colleagues at the National Institutes of Health (NIH), making NIH-sponsored trials available first. This first phase has involved working with 21 NIH institutes, each of which have had varying approaches to data management and collection and varying levels of technical expertise. Some institutes have a large number of ongoing clinical trials, while others have only a few. When we began the project, some institutes had well-established databases for managing their clinical trials, others had Web pages that described their trials but no back-end database support, and yet others were still managing their data in paper form. As a first step, we convened representatives from all 21 institutes and discussed and then agreed on a common set of data elements for the clinical trials data. Several groups at NIH as well as other groups in the clinical trials community had already given a good deal of thought to this, and their insights, together with the requirements of the law, allowed us to arrive at a common set of elements in the first few months of the project. We decided on just over a dozen required data elements and another dozen or so optional elements. The elements fall into several high-level categories—descriptive information such as titles and summaries; recruitment information, which lets patients know whether it is still possible to enroll in a trial; location and contact information, which lets patients and their doctors discuss further details with the persons who are actually conducting the trials; administrative data, such as trial sponsors and identification (ID) numbers; and optional supplementary information, such as literature references and key words.▶ lists the required and optional data elements for the system.
Table 1.
Required and Optional Data Elements in Current System
| Required Data Elements | Optional Data Elements |
|---|---|
| Study identification number | NIH grant or contract number |
| Study sponsor | Investigator |
| Brief title | Official title |
| Brief summary | Detailed description |
| Location of trial | Study start date |
| Recruitment status | Study completion date |
| Contact information | References for background citations |
| Eligibility criteria | References for completed studies |
| Study type | Results |
| Study design | Keywords |
| Study phase | Supplementary information |
| Condition | URL for trial information |
| Intervention | |
| Data provider | |
| Date last modified |
|
| Note: NIH indicates National Institutes of Health; URL, universal resource locator. | |
The study ID number is a unique number assigned by the data provider, which is critical for tracking the trial in the system. In some cases our data providers already had developed methods for assigning IDs to their trials. Those who did not have IDs have since developed and assigned them. In addition to the primary ID, there may be secondary IDs, such as NIH grant or contract numbers, and these are also accommodated. Once a trial record comes into our system, we assign it a number that functions much like a MEDLINE unique identifier. Its form is “NCT” followed by eight digits.
The study sponsor is the primary institute, agency, or organization responsible for conducting and funding the clinical study. There may be additional sponsors, and these may also be listed in the database. Investigator names are included at the discretion of the data provider. Study titles and summaries are important because they give a patient or other user of the system a quick indication of the purpose of the trial. We have asked our data providers to provide us with brief, readily understood titles and summaries. The summaries should provide background information, including why the study is being performed, what drugs or other interventions are being studied, which populations are being targeted, how participants are assigned to a treatment design, and what primary and secondary outcomes are being examined for change (e.g., tumor size, weight gain, quality of life). More detailed descriptions may be provided, and these are often somewhat more technical descriptions of the clinical study intended for health professionals.
Location information includes geographic locations, contact information, and status of a clinical trial at a specific location. Many trials are being conducted at multiple locations, sometimes dozens of sites. It is important that the contact information and recruitment status for all sites be accurate and current. We have established six categories into which the recruitment status might fall—not yet recruiting (the investigators have designed the study but are not yet ready to recruit patients); recruiting (the study is ready to begin and is actively recruiting and enrolling subjects); no longer recruiting (the study is under way and has completed its recruiting and enrollment phase); completed (the study has ended, and the results have been determined); suspended (the study has stopped recruiting or enrolling subjects, but may resume recruiting); and terminated (the study has stopped enrolling subjects and there is no potential to resume recruiting). Sometimes information about the exact start and completion dates of the study is available and, if so, it is added to the status information. Contact information needs to be provided for each trial and includes the name of a contact person and a telephone number for further inquiries. For large multicenter trials, a single coordinating center may handle and then refer the calls.
Eligibility criteria are the conditions that an individual must meet to participate in a clinical study. Both inclusion and exclusion criteria are often relevant. For example, patients who enroll in the study must have a specific disease, may need to be in a certain age range (e.g., under 3 months or over 65 years old), and may need to have already undergone a specific therapy regimen, such as chemotherapy. Exclusion criteria are those conditions that may prevent an individual from participating in a clinical study. For example, in a study involving women, perhaps a participant cannot be pregnant or nursing. In other types of studies, a participant cannot, perhaps, have a history of heart disease.
While many clinical trials are designed to investigate new therapies, there are several other study types as well. We have categorized these into nine types—diagnostic, genetic, monitoring, natural history, prevention, screening, supportive care, training, and treatment. Study design types include the familiar randomized control trial as well as others whose usage and frequency we are in the process of reviewing. The current list includes terms for clinical trial and observational study designs as well as methods (e.g., double-blind method) and other descriptors (e.g., multi-center site).
We have required certain items as separate data elements specifically to ensure optimal search capabilities. These include the study phase, the condition under study, and the intervention being tested. The phrase of the study is important information for patients who are considering enrolling in a particular trial. Phase I trials are the most preliminary and include the initial introduction of an investigational new drug into human use. Phase II trials include studies conducted to evaluate the effectiveness of drugs for particular indications and to determine common short-term side effects and risks. Phase III trials generally involve large numbers of patients and are performed after preliminary evidence suggesting effectiveness of a treatment has been obtained. Phase IV studies are generally post-market studies that seek to gain additional information about a drug's risks, benefits, and use. We have requested that data providers name the condition and intervention being studied using the Medical Subject Headings (MeSH) of the Unified Medical Language System (UMLS), if at all possible. Sometimes, of course, the investigational drug is too new to appear in MeSH, but in other cases the drug, procedure, or vaccine is already well established and the trial may be investigating new combinations of drugs or new uses of established procedures.
Some optional information that may be available for a particular study includes references for publications that either led to the design of a study or that report on the study results. In these cases, we have asked our data providers to provide us with a MEDLINE unique identifier (UI) so that we can link directly to a MEDLINE citation record. (In some cases, we have mapped the citations to UIs for our data providers.) A summary of the results can also be prepared specifically for inclusion in the database, and the use of MeSH keywords is also encouraged. Supplementary information may include URLs of Web sites related to the clinical trial. For example, a trial record on mild cognitive impairment, in addition to linking to NIH's National Institute on Aging, might also link to an Alzheimer's organization.
With the important step of agreeing on a common set of data elements completed by the end of 1998, we were able to devote the next six months to working with each institute individually on methods for receiving their data for inclusion in our centralized database at the National Library of Medicine (NLM). To move the project forward rapidly, we assisted those NIH data providers who had little technical infrastructure in a variety of ways. We developed a Web-based data entry system and offered it to anyone who preferred using it to developing a system of their own. If this system is used, the control of the data still resides with the institute, but we manage the process for them. In other cases, we assisted groups who already had databases by helping them redesign aspects of their databases for the purposes of this project or by writing scripts that would extract data from their databases and prepare them in the standard format. Some institutes were able to provide the data with minimal assistance from us, although some iteration was generally necessary before the data could be fully validated.
In all cases, data are sent to us in extensible markup language (XML) format according to a document type definition (DTD) that we have created. XML has been developed to address some of the deficiencies of HTML and at the same time provide a more streamlined version of SGML for use in Web applications.18,19 Its use has several advantages in our application. It is a standard, structured language that can be readily understood by both computers and people, and it provides a simple, verifiable method of exchanging data regardless of the underlying system that may have produced those data. Therefore, data providers are free to use whatever technology they prefer and can change their database and Web site designs at their discretion. The only requirement is that they capture the required data elements and that they can produce a report in the specified XML format. A portion of our DTD is shown in ▶.
Figure 1.
Portion of document type definition (DTD) in current system.
A study collection consists of one or more clinical study records, which themselves consist of a number of required and optional data elements, as described earlier. In the segment shown in ▶, the study ID is required, but additional administrative numbers, such as an NIH grant number, are optional. Titles and summaries consist of free text (textblocks), while dates need to adhere to a standard date format. Notice that, for intervention names (and for disease names, although this is not shown in this portion of the DTD), we allow not just a single name, but also any available synonyms.
System Implementation
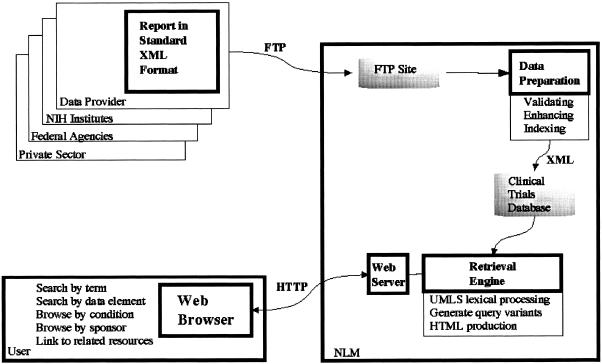
The approach we have taken for implementing the clinical trials system is to collect trials records from the NIH institutes and store them in a central data-base at NLM. The generic term “data provider” currently represents only the NIH data providers, but in the near future it will include other providers as well. ▶ shows the high-level flow of data from data providers through NLM to users.
Figure 2.
System design.
The data provider creates the report in the required XML format and peridocially sends that report to NLM via file transfer protocol (FTP). Our goal is to receive nightly updates from each data provider, but for some this is not yet possible. Whenever an update is sent to us, the data are subjected to various validations, including adherence to the specified format and inclusion of the required common data elements. After the data have been validated, they are enriched by mapping condition names to appropriate MeSH terms, adding cross-references to the literature, and adding links to related material in MEDLINEplus, NLM's consumer health site. Finally, the data are made available to users via the Web. Users' queries are processed by the retrieval engine. The engine checks the query for spelling errors, performs query expansion, and generates HTML for Web browsers.
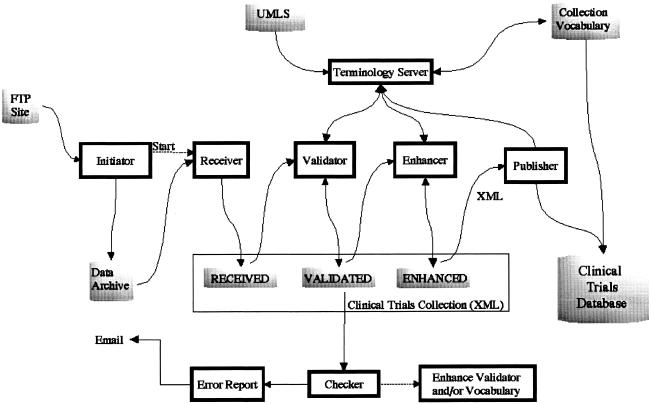
Data Preparation
▶ shows the flow of data in the data preparation subsystem. This subsystem consists of a number of components that have the overall task of receiving, validating, enhancing, and publishing the data.
Figure 3.
Data preparation subsystem.
Each clinical trial record is stored in a single XML document. The clinical trials collection holds all the XML documents. The collection comprises three logical sections, and each section contains the results of one stage of processing. The “received” area of the collection contains each record as an XML document as sent by the data provider. These XML documents are created by the receiver process which breaks a collection of studies into individual studies and saves them one per file. To ease the burden on the data providers, some minor complexities of the XML format were relaxed. These complexities include the handling of alternative character sets and handling of special characters such as “<” and “&.” The receiver process performs the minor translation required to create fully compliant XML.
The “validated” area of the collection contains each record that has been validated. The validator process performs a number of checks on each record. Each XML document is parsed and checked for adherence to the DTD. Adherence to the DTD identifies structural errors in the document. Assuming the XML document is structurally correct, a Java object is created to facilitate content validation. In general, content validation can be performed on any data elements that do not contain free text. For example, the address data elements must contain correctly spelled country and state names, and the study design type must contain one of the specified enumerated values. The implementation is flexible in allowing some variations in controlled fields. For example, USA, US, and United States are all recognized as equivalent. Likewise, the implementation recognizes “N/A” and “Not Applicable” as equivalent. The validation process is expected to evolve over time, as the system processes additional data and as constraints on data quality are tightened. A document may fail the automated validation process on the basis of the current algorithm, but on further review the conclusion may be that the validation process was in error. This might apply, for example, to unknown synonyms of terms in controlled fields. As we discover these, we will add them to the system in an iterative manner. A similar feedback loop is expected as requirements for data quality are refined and tightened. Initially, the necessity of collecting data dictated some permissiveness in quality. As the system matures and data quality problems are identified, the validation process will be strengthened.
The “enhanced” area of the collection contains each record in a normalized XML format. The enhancer process enriches the data by adding URL links to related material, by standardizing state, country, and sponsor names, and by adding MeSH identifiers to condition fields. This last task is done to allow us to build a useful “browse-by-condition” capability. This means that users, in addition to being able to search for diseases, can choose a condition name from a list of disease categories. Because in many cases the conditions given to us by our data providers are not MeSH terms, we attempt to map them to MeSH through the UMLS. The UMLS Metathesarus currently contains some 730,000 concepts from approximately 50 vocabularies. The concepts are interrelated both hierarchically and nonhierarchically through relationships that are defined in the UMLS Semantic Network.20
The algorithm for discovering the best MeSH term proceeds in three steps. First, the condition name is mapped to the UMLS. If this is not successful, lexical normalization techniques are invoked.21 If this still fails to yield a match, then certain modifiers (e.g., acquired, acute, chronic, mild) are removed. For example, the term chronic neutropenia does not map directly to the UMLS, but neutropenia does. In those few cases where a term fails to map to the UMLS at all, it is set aside for human review and resolution. The second step involves an algorithm for mapping the UMLS concept to a MeSH term.22 In some cases there will be a direct mapping through synonymy; in other cases it will be through interconcept relationships. For example, the condition term cancrum oris is directly mapped to the MeSH noma through synonymy, and the condition term neurogenic hypertension is mapped to the MeSH term hypertension.
The third step maps MeSH terms to high-level MeSH categories. In MeSH, each term has both a unique identifier and one or more tree numbers, which reflect the hierarchic structure of the vocabulary. Once the MeSH term has been identified, its mapping to a relevant category is relatively straightforward. For example, the MeSH term adrenal gland neoplasms appears in both the C4 and C19 trees and is, thus, categorized as both a neoplasm (C4) and an endocrine disease (C19). The success of the mapping process is highly dependent on the quality of the data we receive. We continue to refine the process and are also working with our data providers to make them more sensitive to the value of vocabulary control.
The terminology server is software that provides vocabulary-based functionality. It builds a collection-specific vocabulary from the clinical trials documents. The collection vocabulary includes all words from the clinical trials collection as well as those terms from the UMLS vocabulary that have synonyms that occur in the collection. This vocabulary is then used by the retrieval engine to assist the user by correcting spelling errors and expanding queries with appropriate synonyms and lexical variants.
The final step in data preparation is the “publisher” process, which creates the clinical trials database from the clinical trials collection and the collection vocabulary. The database is then made available to the retrieval engine.
Retrieval Engine
The retrieval engine is responsible for managing the user's Web browser session, responding to queries, and, finally, presenting the data. The retrieval engine is implemented as a Java servlet using the Apache JServ module from the Java Apache Project.23 The servlet has three major components. The terminology server performs lexical processing and query expansion before passing the queries on to the search engine, which performs traditional query processing. The browse processor provides hierarchic browsing of the data, and the document renderer retrieves clinical trial records and converts the XML to HTML for presentation in a Web browser.
User queries are checked for spelling errors by extracting each phrase and word, generating the lexical variants, and checking these against the collection vocabulary. If a word is not found in the collection vocabulary, it might be because the user has made a spelling error or because the word is legitimate but not present in the data. In either case, a search for this word is unproductive, so the user is given appropriate feedback and provided with alternatives. The alternative words are generated by applying a spelling correction algorithm that takes into account common misspellings and algorithmically searches for words with similar spellings. For example, if a user performs a search on the misspelling osteoparosis, the system gives the message “osteoparosis was not found. Select an alternative below or change your query.” The alternatives offered at this point are the terms osteoporosis and osteopetrosis, both of which occur in the document collection. After being checked for spelling errors, the user's query is expanded to include lexical variants and synonyms. The LVG21 system is used to generate the lexical variants, and synonyms are found in the UMLS. If, for example, a user performs a search on heart attacks, ace inhibitors, the system will search not only those phrases but also their inflectional variants and synonyms, myocardial infarction and angiotensin-converting enzyme inhibitor.
Web search engines do not have consistent syntax, and the users of our system will have varying levels of Web expertise. Requiring them to formulate their queries using Boolean logic operators such as AND and OR can be particularly confusing. Heavy reliance on a specific syntax such as a comma separator may result in frustrated users who do not understand their search results. To address these issues, our retrieval engine processes queries in as many as four sequential steps. Essentially, we create four variants of each query, with each subsequent variant less restrictive than the previous one, thereby allowing iterative relaxing of the original query.
The first variant is the most restrictive form. Words separated by spaces are treated as a single phrase. A comma or other separator is treated as a Boolean AND operator. The second variant is slightly less restrictive. Phrases are broken into the conjunction of their words. And, like the first variant, a comma is treated as a Boolean AND operator. The third variant relaxes the second variant by treating a comma as a Boolean OR instead of as a Boolean AND. The least restrictive and final variant treats all spaces and separators as Boolean OR operators. The most precise search is achieved by the first version of the query. If no results are found, the next, less restrictive version of the query is used, and so on. Users are given feedback about which form of the query found results, so that they will know how to modify their search if necessary. For example, suppose the user issues a query of lupus california, having in mind trials that are being conducted for lupus in the state of California. The system responds with: “There is no match for lupus california. Searching lupus AND california found 4 studies.” A search of heart attack, on the other hand, results in: “Searching heart attack found 38 studies.” In the first case, lupus california is not found in the collection as a phrase. When applying the second query variant, the system correctly splits the phrase and searches for the conjunction of the words. In the second case, the system finds a match for the phrase and does not need to use the less restrictive query form of heart AND attack.
As noted above, our system also enables users to browse the collection on the basis of a hierarchy of conditions. For example, suppose a clinical trial record contains the condition knee osteoarthritis. In the data preparation cycle, this condition is recognized as a MeSH term, which is classified under both osteoarthritis and arthritis. Users of our system will, therefore, find this record when browsing alphabetically under “A” for arthritis, under “O” for osteoarthritis, and under “K” for knee osteoarthritis.
Once users have identified clinical trial records of interest on the basis of their titles, they will want to view the records themselves. Since few browsers currently provide support for XML documents, the document renderer converts the XML document to standard HTML. This rendering process is done using an XSL Transformations processor and specifying an XSL stylesheet.24,25
The user interface to the system has been designed to be as simple and intuitive as possible, allowing for both simple and focused searching as well as browsing. Nielsen26 points out that his usability studies have shown that “more than half of all users are search-dominant, about a fifth of the users are link-dominant, and the rest exhibit mixed behavior.” Those who are search-dominant usually “go straight for the search button when they enter a website: they are not interested in looking around the site; they are task-focused and want to find specific information as fast as possible.” These types of users will most likely use the simple search window that appears on our home page (▶).
Figure 4.
Portion of the clinical trials home page.
In addition to performing the simple search, it is possible for the user to browse both by condition and by sponsor. Several links to resource information are also available, including a user's guide and access to other NLM and NIH resources.
A simple search for stage iv prostate cancer would yield 167 studies, as shown in ▶. The user is able to see a number of things at a glance. The titles themselves are informative, often including phase information as well as the intervention being studied. To the left of each title is an indication of whether the trial is recruiting patients, and under the title are the conditions being studied. Thus, before users even click on a single study record, they will already have a good idea of the nature of the available studies.
Figure 5.
Results of a simple search.
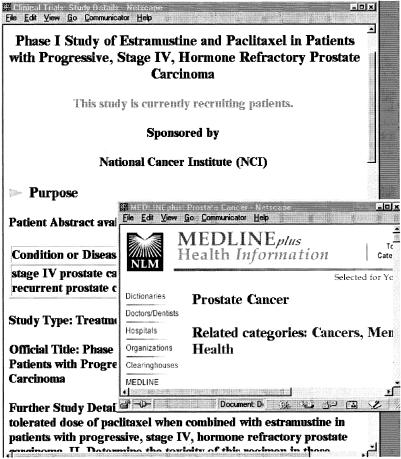
▶ shows a portion of the record for the first study listed in ▶. Each study record consists of four major sections—the purpose of the trial, the eligibility criteria for participating, the location and contact information, and further information, often providing links to more in-depth information about the particular trial or links to related sites. When the user clicks on the disease name, stage IV prostate cancer, the MEDLINEplus page for prostate cancer appears. From there users can find extensive information about the disease, including its diagnosis and therapy. If related references are available, users are also able to click on those and get directly to the specific MEDLINE citations.
Figure 6.
Portion of a clinical trials record.
Once there, they can take advantage of the related citations link as well.
Some users may decide that they would like to focus their search, to get more precise results. The focused search page is shown in ▶. If a user is interested in, for example, any phase III trials for breast cancer that are sponsored by NIH, that are being conducted in New York City, and that are currently recruiting patients, she would be able to formulate her search quite precisely using the focused search capability.
Figure 7.
Focused search page.
In designing the search algorithms and the user interface, we have been concerned with ease of use and accessibility issues. Many of our users will be seriously ill or will have family members who are seriously ill. We have attempted to create a system that is both easy to use and at the same time is quite powerful. In the fall of 1999, we worked with the National Health Council, an organization that has as members some 40 voluntary health agencies, and asked for their assistance in testing the system. Sixty testers, including patients and patient advocates, from 19 member organizations of the council participated in a two-week test to evaluate our prototype Web site and provided valuable feedback on various components of the system. Overall, the comments were positive and supportive, with many people suggesting additional features to enhance the system. Some reported problems related to the search and browse functions, and others related to the data themselves (e.g., the titles and summaries were expressed in language that was too technical). Additional reported problems ranged from screen layout issues to better user support through online documentation and help. We have addressed the technical problems that were found and continue to work with our data providers to improve the readability of their records.
We have also made an effort to comply with technical and accessibility standards. These have been checked over the past two months through the W3C validation service and the Bobby system, which checks Web pages for accessibility on the basis of the W3C accessibility guidelines.27,28 We avoid special features that may cause problems for low-end browsers and continue to test our system on a variety of browsers and platforms.
Lessons Learned
The work on this project has been both technically and organizationally challenging. Perhaps the most important lesson we have learned is that the success of a project of this scope depends crucially on the willingness of people to contribute to a joint enterprise for a common good. In those cases where our collaborators faced certain problems or constraints, we found that when we were able to understand those problems, we could work together to resolve them. As mentioned earlier, in some cases we developed tools and programs specifically for particular groups; in other cases we helped groups modify their existing infrastructure; and in still other cases we manipulated their data for them and returned the data to them for their own use as well as ours.
Our system is only as good as the data contained in it. It is clear that the relevance of the records retrieved by a search is completely dependent on the quality of the underlying data records. Weakness in the data potentially results in users not finding the appropriate records, finding inappropriate records, or not understanding the records they do find. Certain data elements, such as the condition field, are crucial. Lexical processing, synonymy, and sophisticated search algorithms can help overcome some data inconsistencies, but they cannot correct errors and omissions in the underlying clinical trial records. Our challenge is to continually monitor and improve the quality of the records that come to us, even though we are the recipients of the data rather than their creators. A close and continuing relationship with our data providers is critical for accomplishing this task.
The XML format of the data records is sufficiently rigid that it requires data providers to have every detail correct. Our process is for data providers to use FTP to transmit the records to our facility. We subsequently process them, examine them for formatting errors and translate the sometimes cryptic error messages into understandable text, and then forward the messages back to the data providers. This data format iteration loop is a labor-intensive process. In retrospect, it might have been useful for us to create a software tool that data providers could use themselves to validate their data before sending it to us. We will probably make such a tool available in the future. Our data-entry tool effectively provides this capability already, and we may model a generic validation tool on this system.
Conclusions
In the last year and a half, we have designed, implemented, and deployed a system that we hope will be used by many patients, family members, and others who are interested in having integrated access to clinical trials information. The first phase of our project has involved building the core technologies for the system, working with NIH institutes to incorporate their data, conducting focused testing with selected members of the public, and releasing the first version of the system. The next phase will involve expanding the system to include trials sponsored by other federal agencies as well as by private organizations. It is important to note that no phase is ever completely finished, since we are developing a system containing data that continually change as studies are completed, new ones are started, and existing ones are modified in a variety of ways.
References
- 1.FDA Modernization Act of 1997, Public Law 105-115, 105th Congress. Section 113, Information Program on Clinical Trials for Serious or Life-threatening Diseases. Food and Drug Administration Web site. Available at: http://www.fda.gov/cder/guidance/105-115.htm . Accessed Jan 11, 2000.
- 2.Chalmers I. The Cochrane Collaboration: Preparing, maintaining, and disseminating systematic reviews of the effects of health care. Ann NY Acad Sci. 1993;703: 156-63. [DOI] [PubMed] [Google Scholar]
- 3.Dickersin K. Report from the panel on the case for registers of clinical trials at the eighth annual meeting of the society for clinical trials. Control Clin Trials. 1988;9: 76-81. [DOI] [PubMed] [Google Scholar]
- 4.Meinert CL. Toward prospective registration of clinical trials. Control Clin Trials. 1988;9: 1-5. [DOI] [PubMed] [Google Scholar]
- 5.Sim I. A trial bank model for the publication of clinical trials. Proc AMIA Annu Fall Symp. 1995: 863-7. [PMC free article] [PubMed]
- 6.Sim I. Trial banks: an informatics foundation for evidence-based medicine. University of California Trial Bank Project Web site. 1999. Available at: http://rctbank.ucsf.edu:8000/home/intro.html . Accessed Jan 11, 2000.
- 7.Strang WN, Cucherat M, Yzebe D, Boissel J-P. Trial summary software. Comput Methods Prog Biomed. 2000;61(1): 49-60. [DOI] [PubMed] [Google Scholar]
- 8.Afrin LB, Kuppuswamy V, Slater B, Stuart RK. Electronic clinical trial protocol distribution via the World Wide Web: a prototype for reducing costs and errors, improving accrual, and saving trees. J Am Med Inform Assoc. 1997;4(1): 25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu SW, Kemper CA, Lane NM, Carlson RW, Musen MA. A methodology for determining patients' eligibility for clinical trials. Methods Inform Med. 1993;32: 317-25. [PubMed] [Google Scholar]
- 10.Rubin DL, Gennari JH, Srinivas S, et al. Tool support for authoring eligibility criteria for cancer trials. Proc AMIA Symp. 1999: 369-73. [PMC free article] [PubMed]
- 11.Breitfeld PP, Weisburd M, Overhage JM, Sledge G, Tierney WM. Pilot study of a point-of-use decision support tool for cancer clinical trials eligibility. J Am Med Inform Assoc. 1999;6(6): 466-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haag U. Knowledge representation for computer-aided planning of controlled clinical trials: the PATriCIa project. Methods Inf Med. 1997;36: 172-8. [PubMed] [Google Scholar]
- 13.Wyatt JC, Altman DG, Healthfield HA, Pantin CFA. Development of Design-A-Trial, a knowledge-based critiquing system for authors of clinical trial protocols. Comput Methods Prog Biomed. 1994;43: 283-91. [DOI] [PubMed] [Google Scholar]
- 14.Musen MA, Carlson CW, Fagan LM, Deresinski SC, Shortliffe EH. T-HELPER: automated support for community-based clinical research. Proc Annu Symp Comput Appl Med Care. 1992: 719-23. [PMC free article] [PubMed]
- 15.Nadkarni PM, Brandt C, Frawley S, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system. J Am Med Inform Assoc. 1998;5(2): 139-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva J, Wittes R. Role of clinical trials informatics in the NCI's cancer informatics infrastructure. Proc AMIA Symp. 1999: 950-4. [PMC free article] [PubMed]
- 17.McCray AT. A national resource for information on clinical trials. Nat Forum. 1999: 19-21.
- 18.The World Wide Web Consortium (W3C). Extensible Markup Language (XML). Available at: http://www.w3.org/XML/ . Accessed Jan 11, 2000.
- 19.Bosak J, Bray T. XML and the second-generation Web. Sci Am. May 1999. Available at: http://www.sciam.com/1999/0599issue/0599bosak.html . Accessed Jan 11, 2000.
- 20.McCray AT, Nelson SJ. The representation of meaning in the UMLS. Methods Inf Med. 1995;34: 193-201. [PubMed] [Google Scholar]
- 21.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc AMIA Symp. 1994: 235-9. [PMC free article] [PubMed]
- 22.Bodenreider O, Nelson SJ, Hole WT, Chang HF. Beyond synonymy: exploiting the UMLS semantics in mapping vocabularies. Proc AMIA Symp. 1998: 815-9. [PMC free article] [PubMed]
- 23.The Apache Group. Java Apache Project Web site. Available at: http://java.apache.org/ . Accessed Jan 11, 2000.
- 24.The World Wide Web Consortium (W3C). XSL Transformations (XSLT), version 1.0. Available at: http://www.w3.org/TR/1999/PR-xslt-19991008 . Accessed Jan 11, 2000.
- 25.The World Wide Web Consortium (W3C). Extensible Style Sheet Language (XSL). Available at: http://www.w3.org/Style/XSL/ . Accessed Jan 11, 2000.
- 26.Nielsen J. Search and you may find. Useit.com : Jakob Nielsen's Web site. Jul 1997. Available at: http://www.useit.com/alertbox/9707b.html . Accessed Jan 11, 2000.
- 27.The World Wide Web Consortium (W3C). HTML Validation Service. Available at: http://validator.w3.org/ . Accessed Jan 11, 2000.
- 28.Center for Applied Special Technology (CAST). Bobby 3.1.1 Available at: http://www.cast.org/bobby/ . Accessed Jan 11, 2000.