Abstract
MicroRNA (miRNA) dysregulation is associated with the tumorigenesis and development of numerous human cancers. The defect in miRNA biogenesis is the main cause of miRNA dysregulation. We previously demonstrated that ERK-induced phosphorylation of XPO5 followed by peptidyl-prolyl cis/trans isomerase Pin1-mediated isomerization downregulates miRNA expression and contributes to hepatocellular carcinoma (HCC) development. However, how Pin1 precisely regulates miRNA biogenesis in HCC remains elusive. Here we reveal that Pin1 has a pivotal role in the miRNA maturation process by modulating phosphorylated Serine-Proline (pS-P) motif of XPO5 in a phosphorylation-dependent manner. By recognizing and binding to XPO5 via its WW domain, Pin1 catalyzes the conformation change of XPO5 and diminishes XPO5 ability to export pre-miRNAs from the nucleus to the cytoplasm, resulting in the reduced mature miRNA levels and promoted HCC development. Furthermore, downregulation of Pin1 by shRNA restores XPO5-dependent pre-miRNA export and effective biogenesis of mature miRNAs, leading to both in vitro and in vivo HCC inhibition. Therefore, our research discloses a new posttranscriptional regulatory mechanism of miRNA biosynthesis and provides the experimental basis for a novel HCC therapy by targeting Pin1.
Introduction
MicroRNA (miRNA) is a class of non-coding single-stranded RNAs with key roles in cellular post-transcriptional regulation [1]. By inducing target mRNA degradation and/or translational inhibition, miRNAs participate extensively in cell proliferation, differentiation, apoptosis, and other biological processes. It is well recognized that miRNAs are generally downregulated in many malignancies [2], and their altered expression is causally involved in different steps of tumor development [3–5]. So far, compelling evidence has revealed that the defective miRNA biogenesis, rather than enhanced miRNA degradation, drives aberrant miRNA expression and cancer development [6]. Thus, investigating the mechanism of miRNA biogenesis is crucial to understand miRNA dysregulation in human cancers.
miRNA biogenesis can be summarized as the following steps: (1) transcription of miRNA gene to primary miRNA (pri-miRNA) by RNA polymerase II; (2) cleavage of pri-miRNA by Drosha and its cofactor (such as DGCR8) into precursor miRNA (pre-miRNA) with short hairpin structure; (3) export of pre-miRNA from the nucleus to the cytoplasm by the Ran/GTP-dependent transporter exportin-5 (XPO5); and (4) processing of pre-miRNA by Dicer to produce mature miRNA and subsequent assembly of the miRNA-associated RNA-induced silencing complex by AGO2 [7]. This process is precisely regulated at multiple levels, particularly the posttranslational modifications of involved proteins. For example, the phosphorylated TRBP protein stabilizes the Dicer–TRBP complex and promotes miRNA biosynthesis [8], whereas the phosphorylated AGO2 protein cannot effectively bind to Dicer, thereby inhibiting miRNA generation [9].
One of the key proteins participating in miRNA maturation is XPO5, which belongs to the karyopherin-β family and uses the Ran/GTP complex to control cargo association [10, 11]. The pre-miRNA export by XPO5 is a rate-limiting step in miRNA biogenesis [10]. Notably, little is known about the post-translational regulation of XPO5. In this regard, we recently discovered new phosphorylation modifications of XPO5. To be specific, ERK-activated serine/threonine phosphorylation coupled with peptidyl-prolyl isomerase Pin1-mediated regulation of XPO5 inhibits miRNA expression and contributes to hepatocellular carcinoma (HCC) development [12]. However, the precise mechanism how Pin1 interacts with XPO5 and impairs miRNA biogenesis has not been fully elucidated yet.
The peptidyl-prolyl cis/trans isomerase Pin1 is a parvulin-type enzyme containing an N-terminal WW domain and a C-terminal PPIase domain. The WW domain specifically recognizes and binds to the phosphorylated serine/threonine-proline (pS/T-P) motif and the PPIase domain catalyzes the cis/trans isomerization [13, 14]. Pin1-mediated switch of protein conformation affects stability, activity, and phosphorylation status of the substrates [15]. Prevalent overexpression of Pin1 occurs in several tumors including HCC and correlates with poor clinical prognosis [16]. Mechanistically, overexpressed Pin1 has been found to activate multiple oncogenes and inactivate several tumor suppressors. For example, Pin1 regulates β-catenin protein turnover and subcellular localization by interfering with its interaction with adenomatous polyposis coli protein (APC) [17]. Pin1 also interacts with Notch1 and potentiates Notch1 cleavage by γ-secretase, resulting in an increased release of the active intracellular domain and ultimately enhanced Notch1 transcriptional and tumorigenic activity in breast cancer [18]. In addition, Pin1 was found to downregulate tumor suppressor p27kip1 expression through inhibiting the transcriptional activity of FOXO4 [19]. These findings verify that Pin1 plays a central role in tumorigenesis and tumor development by activating and/or amplifying numerous cancer-driving pathways [16].
In this report, we demonstrate a novel posttranscriptional regulatory mechanism of miRNA biogenesis mediated by Pin1 in HCC. Briefly, Pin1 interacts with and catalyzes cis/trans isomerization of XPO5 in a phosphorylation-dependent manner, which impairs XPO5 activity for exporting pre-miRNA from the nucleus to the cytoplasm, leading to decreased miRNA production. Moreover, the involvement of Pin1 in HCC development is achieved, at least in part, by downregulating miRNA expression. Knockdown of Pin1 restores XPO5 function and miRNA biogenesis, thereby inhibiting HCC proliferation and migration. Our findings highlight the importance of posttranscriptional regulation of miRNA biogenesis in tumors and provide Pin1 as a potential target for HCC treatment.
Results
Pin1 induces cell proliferation, migration, and invasion in HCC cells
Pin1 is overexpressed in several human malignancies including HCC and has a crucial role in tumorigenesis [16]. Comparing Pin1 levels in human HCC tissues with their adjacent normal tissues, we found upregulated Pin1 expression in human HCC tissues (Fig. 1a,b), consistent with previous findings [20–22]. Both SK-Hep1 and Huh-7 cell lines were chosen to study Pin1 function in HCC, because they displayed the highest and lowest phospho-ERK level, leading to different phosphorylation status of XPO5 (Supplementary Figure 1a, 1b, and 1c). To investigate cellular function of Pin1, we established the SK-Hep1-stable transfectants expressing shPin1 (SK-Hep1 shPin1) and scramble vector (SK-Hep1 shCtrl) (Fig. 1c). Cell proliferation of SK-Hep1 shPin1 cells was suppressed compared to shCtrl cells (Fig. 1d). Moreover, Pin1 knockdown significantly reduced cell migration and invasion (Fig. 1e and Supplementary Figure 1d). To further examine the phenotype of Pin1, Huh-7 MEKDD stable transfectants expressing constitutively active MEK to activate ERK were established and verified by increased pXPO5 and pERK compared with the control cells (Supplementary Figure 1e). After the knockdown of Pin1 expression in Huh-7 MEKDD cells (Fig. 1f), cell proliferation, migration, and invasion were significantly inhibited (Fig. 1g,h and Supplementary Figure 1f). Besides the above loss-of-function experiments, we also took advantage of gain-of-function strategy to confirm the cellular function of Pin1. As LM3 cells displayed relatively low expression of Pin1 (Fig. 1i), we overexpressed Pin1 in LM3 cells (Fig. 1j). As expected, upregulation of Pin1 expression promoted cell proliferation, migration, and invasion (Fig. 1k,l). In addition, the decreased cell proliferation, migration, and invasion in SK-Hep1-shPin1 and Huh7-MEKDD-shPin1 cells were rescued by the overexpression of short hairpin RNA-resistant Pin1 (R-WT) (Supplementary Figure 1g-1l). Therefore, these results strongly indicate that Pin1 promotes cell proliferation, migration and invasion in HCC cells.
Fig. 1.
Pin1 promotes cell growth, migration and invasion in HCC cells. a Western blotting and b quantification of Pin1 expression in human HCC tumors and their adjacent normal tissues. CB: Coomassie blue staining. c, f, i, j Pin1 expression measured by western blotting in SK-Hep1-stable transfectants c or Huh-7 MEKDD stable transfectants f expressing shPin1 and shCtrl, in LM3, SK-Hep 1, and Huh-7 cells i, or in LM3 cells with or without Pin1 overexpression j. d, g, k MTT assays for SK-Hep1 shPin1 and shCtrl cells d, Huh7 MEKDD shPin1 and shCtrl cells g, or LM3 Pin1 and LM3 Ctrl cells k. e, h, l Transwell migration and invasion assays for SK-Hep1 shPin1 and shCtrl cells e, Huh7 MEKDD shPin1 and shCtrl cells h, or LM3 Pin1 and LM3 Ctrl cells l. Error bars indicate ± SD. Statistical significance was determined by the Student’s t-test. *P < 0.01. Experiments were repeated three times.
Pin1 interacts with XPO5 in a phosphorylation-dependent manner
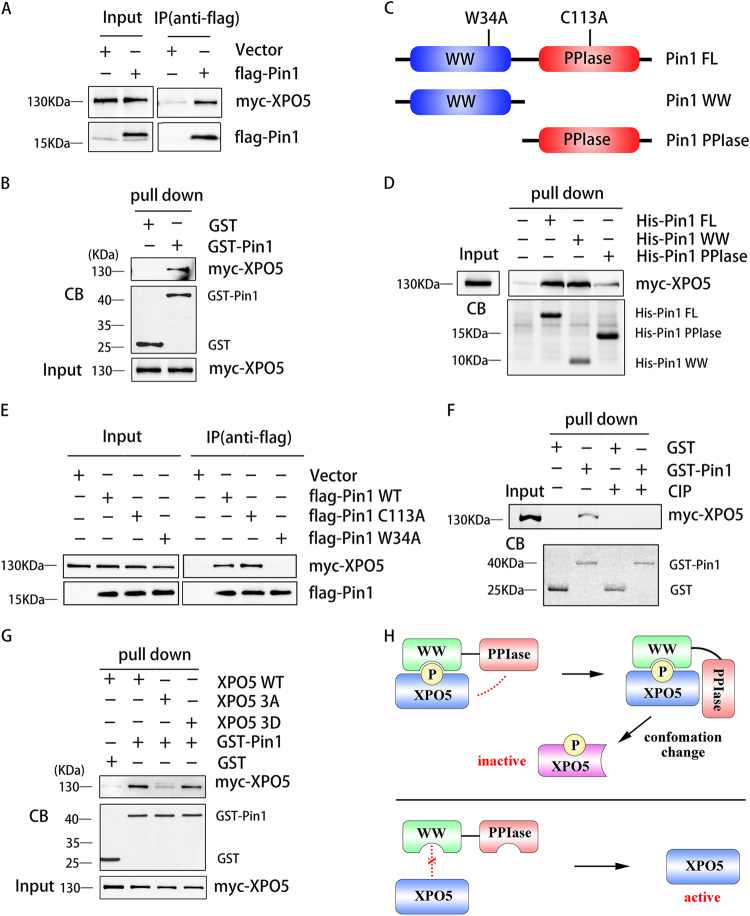
Recently, we demonstrated that Pin1 participates in HCC development by regulating XPO5 function and miRNA expression [12]. To test whether Pin1 interacts with XPO5, co-immunoprecipitation was performed. SK-Hep1 cells were transfected with plasmids expressing myc-tagged XPO5 and flag-tagged Pin1, and protein complexes recovered by immunoprecipitation with monoclonal antibody to the flag tag were analyzed by western blotting. Clearly, XPO5 was specifically recovered with Pin1 (Fig. 2a). Moreover, we expressed and purified glutathione S-transferase (GST)-tagged Pin1 and GST in Escherichia coli, and incubated with XPO5 for GST pull-down assay. As shown in Fig. 2b, XPO5 could be pulled down by GST-Pin1, indicating the direct interaction between XPO5 and Pin1. Moreover, co-localization of endogenous Pin1 and XPO5 in the nucleus was observed by immunofluorescence (Supplementary Figure 2a). The interaction between XPO5 and Pin1 was further confirmed by co-immunoprecipitation using mice tumor lystaes (Supplementary Figure 2b).
Fig. 2.
Interaction between Pin1 and XPO5 occurs in a phosphorylation-dependent manner. a SK-Hep1 cells were co-transfected with myc-XPO5 and flag-Pin1 or empty vector expression plasmid. After cell lysis, flag-Pin1 was immunoprecipitated with anti-flag antibody. Immunoprecipitates were immunoblotted with anti-myc and anti-flag antibodies. b Lysates from SK-Hep1 cells transfected with myc-XPO5 expression plasmid were incubated with GST or GST-Pin1. GST pull-down complexes were subjected to SDS-PAGE and immunoblotted with anti-myc antibody or stained by Coomassie blue (CB). c Schematic diagram of full length Pin1 and its mutants (W34A and C113A), Pin1 WW domain, and Pin1 PPIase domain. d Lysates from SK-Hep1 cells transfected with myc-XPO5 expression plasmid were incubated with His-tagged full-length Pin1 (His-Pin1 FL), WW domain (His-Pin1 WW), or PPIase domain (His-Pin1 PPIase), respectively. The pull-down complexes by Ni-NTA agarose beads were subjected to SDS-PAGE and immunoblotted with anti-myc antibody or stained by Coomassie blue (CB). e SK-Hep1 cells were co-transfected with myc-XPO5 and flag-Pin1 (WT, C113A, or W34A) or empty vector expression plasmid. After cell lysis, flag-Pin1 was immunoprecipitated with anti-flag antibody. Immunoprecipitates were immunoblotted with anti-myc and anti-flag antibodies. f Lysates from SK-Hep1 cells transfected with myc-XPO5 expression plasmid were incubated with or without CIP before the incubation with GST or GST-Pin1. GST pull-down complexes were subjected to SDS-PAGE and immunoblotted with anti-myc antibody or stained by Coomassie blue (CB). g Lysates from SK-Hep1 cells transfected with myc-XPO5 (WT, 3A, or 3D) or empty vector expression plasmid were incubated with GST or GST-Pin1. GST pull-down complexes were subjected to SDS-PAGE and immunoblotted with anti-myc antibody or stained by Coomassie blue (CB). 3A: T345A, S416A and S497A mutations; 3D: T345D, S416D and S497D mutations; h Graphic abstract of the mechanism how Pin1 interacts with XPO5
Pin1 protein consists of an N-terminal WW domain and a C-terminal PPIase domain (Fig. 2c). To identify the domain of Pin1 responsible for its interaction with XPO5, we expressed and purified His-tagged WW domain and PPIase domain for His-tag pull-down assay. Western blotting indicated that WW domain, not PPIase domain, specifically interacts with XPO5 (Fig. 2d and Supplementary Figure 2c). Importantly, W34A mutant within the Pin1 WW domain, which inactivates the binding of Pin1 to its substrate, failed to bind XPO5 in both SK-Hep1 (Fig. 2e) and Huh-7 MEKDD cells (Supplementary Figure 2d), further confirming that Pin1 WW domain is responsible for the binding of XPO5.
Pin1 is the only peptidyl prolyl isomerase that recognizes and catalyzes pS/T-P motif [16]. We previously showed that XPO5 has phosphorylatable S416-P417 and S497-P498 motifs that are critical for its function [12]. To investigate whether phosphorylation of XPO5 is indispensable for XPO5-Pin1 association, we performed an in vitro GST pull-down assay with or without the treatment of calf intestine phosphatase (CIP). Dephosphorylation of XPO5 by CIP abolished the interaction between XPO5 and Pin1 (Fig. 2f and Supplementary Figure 2e). Moreover, non-phosphorylatable 3A mutant of XPO5 (3A), but not phosphomimetic 3D mutant (3D), failed to be pulled down by GST-Pin1 (Fig. 2g and Supplementary Figure 2f), strongly demonstrating that this interaction was highly dependent on XPO5 phosphorylation. Taken together, these results exhibit that Pin1 directly interacts with XPO5 in a phosphorylation-dependent manner (Fig. 2h).
Pin1 mediates conformation change of XPO5
Given that Pin1 recognizes a pS/T-P substrate and catalyzes its cis–trans isomerization, we further tested whether XPO5 could be isomerized by Pin1. Partial proteolytic cleavage assays were performed on myc-XPO5 purified from transiently transfected SK-Hep1 or Huh-7 MEKDD cell lysates. As shown in Fig. 3a,b, subtilisin, a protease sensitive to substrate conformation [14], is able to degrade XPO5 (lane 4), but wild-type Pin1 protected XPO5 from subtilisin-mediated proteolysis (lane 5), indicating that Pin1 mediated protein conformation change of XPO5. Furthermore, inactive Pin1 with C113A mutation in the PPIase domain has no effect on subtilisin-mediated proteolysis of XPO5 (lane 6), suggesting that XPO5 conformation change was catalyzed by Pin1 PPIase, rather than the steric hindrance caused by Pin1 binding. To further explore whether the pS-P motif of XPO5 is a bona fide Pin1 client, we customized two XPO5 polypeptides (494–499 residues) containing phosphorylated (SVFpSPS-pNa) or non-phosphorylated (SVFSPS-pNa) S497 and performed in vitro PPIase activity assay. As shown in Fig. 3c, wild-type Pin1 isomerized the phosphorylated S494–S499 peptide rather than the nonphosphorylated peptide, indicating Pin1 is specific to pS-P motif. Moreover, treatment by juglone, a Pin1 inhibitor, significantly impeded isomerization of phosphorylated peptide, but had little impact on the nonphosphorylated counterpart (Fig. 3d). These results strongly demonstrate that Pin1 specifically recognizes and isomerizes phosphorylated XPO5 via the pS-P motif.
Fig. 3.
Pin1 catalyzes cis/trans conformation change of XPO5. a, b Partial proteolytic cleavage assay of XPO5 immunoprecipitated from SK-Hep-1 lysates a or Huh-7 MEKDD lysates b using GST, GST-Pin1, or GST-Pin1 C113A. c In vitro isomerization assay of wild-type Pin1 using SVFSPS-pNa and SVFpSPS-pNa as substrates. d In vitro isomerization assay of wild-type Pin1 using SVFSPS-pNa and SVFpSPS-pNa as substrates. Before the assay, wild-type Pin1 was incubated with or without juglone. Assays were repeated three times. Error bars represent ± SD. Statistical significance was determined by the Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001
Pin1 diminishes the nucleus-to-cytoplasm export of XPO5 and pre-miRNAs
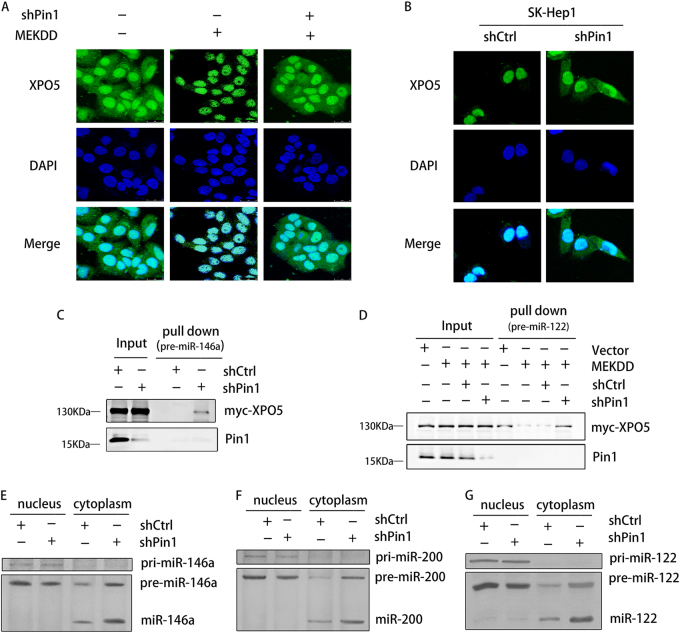
XPO5 is a transport protein that shuttles between the nucleus and the cytoplasm in normal cells and we previously found that phosphorylated XPO5 by ERK kinase was retained in the nucleus of HCC cells. Thus, we investigated whether Pin1-induced isomerization could change the subcellular distribution of XPO5. In Huh-7 cells with low XPO5 phosphorylation, XPO5 was located in both the nucleus and the cytoplasm (Fig. 4a). Increased XPO5 phosphorylation caused by MEKDD-induced ERK activation led to the retention of XPO5 in the nucleus, whereas Pin1 knockdown restored the nucleus–cytoplasm distribution of XPO5, implying that Pin1 regulated the nucleus-to-cytoplasm export of XPO5 in a phosphorylation-dependent manner (Fig. 4a). The similar phenomenon was observed in SK-Hep1 cells (Fig. 4b).
Fig. 4.
Pin1 impairs pre-miRNA binding capacity of XPO5. a, b Confocal imaging of XPO5 (Green) subcellular localization in Huh-7 non-activated, MEKDD-activated, and MEKDD-activated shPin1 cells a, or in SK-Hep1 shCtrl and shPin1 cells b. c Lysates from SK-Hep1 shCtrl or shPin1 cells were incubated with 3′-biotin-labeled pre-miR-146a. Biotin pull-down proteins were subjected to Western blotting. d Lysates from Huh-7-stable transfectants expressing empty vector, MEKDD, MEKDD plus shCtrl, or MEKDD plus shPin1 were incubated with 3′-biotin-labeled pre-miR-122. Biotin pull-down proteins were subjected to western blotting. e, f, g RNA solution hybridization assays to examine pri-miRNA, pre-miRNA, and mature miRNA levels of miR-146a e, miR-200b f, and miR-122 g
Considering that XPO5 is an indispensable transport vehicle for pre-miRNAs, we subsequently investigated if Pin1 could influence the pre-miRNA binding capacity of XPO5. Based on the abundance and importance of miR-122 and miR-146a during HCC development and our previous results that SK-Hep1 and Huh-7 cells highly express miR-146a and miR-122, respectively, we chose them as study objects and synthesized 3′-biotin-labeled pre-miR-146a and pre-miR-122 for RNA pull-down assay. As expected, Pin1 downregulation in SK-Hep1 cells contributed to the interaction between pre-miR-146a and XPO5 (Fig. 4c). Huh-7 cells accord with normal hepatic cells in highly expressing miR-122 [23]; thus, a strong combination of pre-miR-122 and XPO5 was observed in Huh-7 cells (Fig. 4d). Consistent with XPO5 distribution in Fig. 4a, phosphorylation of XPO5 impeded its capacity to bind pre-miR-122, whereas the lost function was rescued by Pin1 knockdown (Fig. 4d), suggesting that binding of XPO5 to pre-miRNA depends not only on the phosphorylation of XPO5, but also on the following conformation change catalyzed by Pin1.
To explore whether Pin1-induced dissociation of XPO5 and pre-miRNAs could affect the location and biogenesis of pri-/pre-/mature miRNAs, RNA solution hybridization was performed in HCC cells. As shown in Fig. 4e,f,g, high Pin1 expression resulted in nuclear retention of pre-miRNAs. Pin1 knockdown promoted the nucleus-to-cytoplasm export of pre-miRNAs and the mature miRNA biogenesis in shPin1 cells was consequently increased compared to shCtrl cells. Notably, the pri-miRNAs levels were almost unchanged after Pin1 knockdown, suggesting that Pin1 has no effect on pri-miRNA transcription. Therefore, conformational change of XPO5 mediated by Pin1 impaired its binding capacity to pre-miRNAs, resulting in the pre-miRNAs retention in the nucleus and the decreased mature miRNAs biogenesis.
Pin1 promotes cell proliferation, migration through regulating miRNA biogenesis
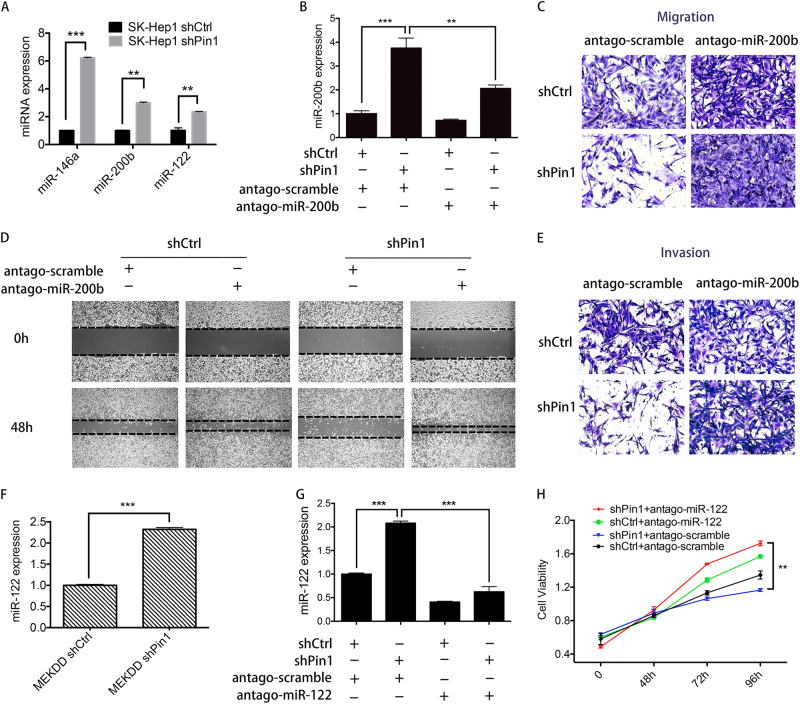
As Pin1 regulates miRNA expression through impairing the nucleus-to-cytoplasm transport of pre-miRNAs, we explored whether Pin1 promotes cell proliferation and migration through downregulating miRNAs biogenesis. As expected, the miRNAs acting as tumor suppressors in human, such as miR-122, miR-200b, and miR-146a, were significantly increased upon the downregulation of Pin1 in SK-Hep1 cells (Fig. 5a). To test whether such miRNAs were involved in Pin1-mediated tumorigenesis, we transfected antago-miR-200b into SK-Hep1 cells to reduce miR-200b expression (Fig. 5b) and performed wound-healing experiment, Transwell migration, and invasion assays. As shown in Fig. 5c,d,e, the differences of cell migratory and invasive ability between antago-miR-200b- and antago-scramble-transfected SK-Hep1 shPin1 cells were larger than those in the SK-Hep1 shCtrl cells, indicating that Pin1-induced cell migration and invasion are, at least in part, mediated by downregulation of miR-200b expression. We also assessed the miRNA expression in Huh-7 MEKDD-activated cells. MiR-122, which dominates the miRNA content in normal liver and is frequently reduced in HCC [24], was significantly increased in shPin1 cells compared to the shCtrl cells (Fig. 5f). Moreover, overexpression of short hairpin-resistant Pin1 (R-WT) in Huh-7 MEKDD shPin1 cells reduced miR-122 expression (Supplementary Figure 3a). Upon an inhibition of miR-122 expression by antago-miR-122, cell proliferation was increased, indicating that Pin1-induced cell proliferation is attributed to miR-122 downregulation (Fig. 5g,h). It is well known that some miRNAs act as oncogenic miRNAs in tumors. We further tested the effect of Pin1 on expression of such miRNAs, and found that Pin1 knockdown had negligible effect on some well-recognized oncomiRNAs, such as miR-21, miR-92a, and miR-221 (Supplementary Figure 3b). We cannot rule out the possibility that Pin1 affects the expression of some oncogenic miRNAs. In views of the fact that miR-122 accounts for ~ 70% of the liver’s total miRNAs [24], change of miR-122 expression level has an overwhelming effect and may counteract the opposite impact caused by oncogenic miRNAs with low abundance. Taken together, Pin1-increased cell proliferation, migration, and invasion in HCC cells are in part mediated by its downregulation of some tumor suppressive miRNA expression.
Fig. 5.
Involvement of Pin1 in HCC progression is partially mediated by its regulated miRNA. a MiRNA expression in SK-Hep1 shCtrl and shPin1 cells was examined by qPCR. b SK-Hep1 shCtrl and shPin1 cells were transfected with antago-scramble or antago-miR-200b. After the transfection, miR-200b expression was examined by qPCR. c, d, e Transwell migration assays c, wound-healing assays d, and Matrigel invasion assays e for SK-Hep1 shCtrl and shPin1 cells transfected with antago-scramble or antago-miR-200b. f miR-122 expression in Huh-7 MEKDD shCtrl and MEKDD shPin1 cells was examined by qPCR. g miR-122 expression measured by qPCR in Huh-7 MEKDD shCtrl and MEKDD shPin1 cells transfected with antago-scramble or antago-miR-122. h MTT assays for Huh-7 MEKDD shCtrl and MEKDD shPin1 cells transfected with antago-scramble or antago-miR-122. In a, b, f, g, and h, assays were repeated three times. Error bars represent ± SD. Statistical significance was determined by the Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001
Pin1 induces HCC development in vivo by modulating miRNA expression
We further tested our findings in vivo upon establishing xenograft models in nude mice. As shown in Fig. 6a,b, the tumor growth of SK-Hep1 shPin1 tumor was significantly slower than that of the shCtrl group, exhibiting the tumor-promoting role of Pin1 in vivo. Consistent with the disclosed in vitro mechanism, tumor suppressive miRNAs were also increased in SK-Hep1 shPin1 tumor (Fig. 6c,d). We also evaluated the Pin1 function in Huh-7 xenografts models. In agreement with the SK-Hep1 model, Huh-7 shPin1 tumor displayed lower growth rate and upregulated miR-122 expression when compared with the Huh-7 shCtrl group (Supplementary Figure 4). These data demonstrated that Pin1 promotes HCC tumor growth in vivo, at least in part, by modulating some miRNA expression.
Fig. 6.
Pin1 promotes HCC development by modulating miRNA expression in vivo. a Photograph of SK-Hep1 shCtrl and shPin1 xenografts in nude mice. b Tumor volumes of SK-Hep1 shCtrl and shPin1 xenografts in nude mice. Graphic data shown as the means ± SD. c Pin1 expression in SK-Hep1 shCtrl and shPin1 xenografts was examined by western blotting. d miRNA expression in SK-Hep1 shCtrl and shPin1 xenografts was measured by qPCR. Assays were repeated three times. Error bars represent ± SD. In b and d, statistical significance was determined by the Student’s t-test. **P < 0.01, ***P < 0.001
Discussion
Aberrant expression of miRNAs widely occurs in human malignancies and the impaired miRNA biogenesis is the main cause of miRNA dysregulation. During miRNA biogenesis, XPO5-mediated nucleus-to-cytoplasm export of pre-miRNAs is an indispensable step for miRNA maturation [25, 26]. With the assistance of Ran-GTP, XPO5 binds to the 3′-overhang of pre-miRNAs by hydrogen bonds and ionic interaction, and encapsulates pre-miRNAs in a baseball mitt-like structure to protect it from degradation in a sequence-independent manner [27]. Increasing evidences show that genetic disorders of XPO5 gene are implicated in human cancers. For example, XPO5-inactivating mutation in a subset of colorectal tumors traps pre-miRNA in the nucleus and reduces miRNA expression [28], and the single nucleotide polymorphisms in XPO5 gene are associated with prognosis and survival rate of HCC [29]. However, little has been reported regarding XPO5 regulation at protein level in human cancers. Although our recent work suggests the involvement of Pin1 in miRNA biogenesis, the detailed mechanism how Pin1 regulates XPO5 still keeps obscure. In this study, we present that Pin1 directly binds to the phosphorylated XPO5, mediates the change of XPO5 protein conformation and impairs its nucleus-to-cytoplasm transport ability, thus leading to decreased miRNA expression and increased cell proliferation, migration, and invasion in HCC cells.
Pin1 is the only enzyme that catalyzes the cis/trans isomerization of pS/T-P motif and its prevalent overexpression in tumors is closely related to tumor development [16, 30]. In HCC, Pin1 expression is elevated, contributing to hepatic carcinogenesis [20, 21]. Moreover, higher Pin1 expression is significantly associated with shorter overall survival and higher recurrence rate in HCC patients [22]. MiRNA expression is generally downregulated in human tumors and has a significant negative correlation with Pin1 expression. For instance, miR-296 functions as tumor suppressive miRNA by targeting Pin1 in prostate cancer [31] and decreased expression of miR-200 family contributes to Pin1 overexpression and promotes cancer stem-like cell traits in breast cancer [32, 33]. However, how Pin1 affects miRNA expression has not been elucidated. In this study, we found that Pin1 can in turn inhibit the expression of tumor suppressor miRNAs, including miR-200b (Figs. 4f, 5a, and 6d), which may serve as a positive feedback loop of Pin1 overexpression in cancer.
By performing partial proteolytic cleavage assay and in vitro isomerization assay, we proved that Pin1 recognizes and catalyzes cis/trans isomerization of pS-P motif of XPO5 (Fig. 3). In fact, both the cis- and trans-conformation exist naturally owing to spontaneous equilibration, although trans-conformation is a more beneficial one [34]. We also exhibited that, via GST pull-down and co-IP assays (Fig. 2d,e), Pin1 directly binds at its WW domain to XPO5. It has been proposed that the WW domain of Pin1 interacts with the trans-conformation of pS/T-P motif [35], thus, we speculated that Pin1 isomerizes pS-P motif on XPO5 from the trans-conformation to the cis-conformation. Moreover, we have found that PP2A, a phosphatase specific for the trans-pS/T-P isomer [36], catalyzes XPO5 dephosphorylation, which can be prevented by Pin1-induced isomerization (data not shown), indicating a trans-to-cis conformation change of XPO5 mediated by Pin1. However, further structural and functional researches are still needed to fully elucidate the dynamic isomerization process.
The isomerization of XPO5 impedes the nucleus-to-cytoplasm export of several tumor suppressor miRNAs (miR-146a, miR-200b, and miR-122) in HCC (Fig. 4e,f,g). Among the tested miRNAs, miR-146a inhibits invasion and metastasis by down-regulating VEGF in HCC [37], miR-200b suppresses breast cancer metastasis in a moesin-dependent manner [38], and the anti-HCC function of miR-122 is unequivocally confirmed in miR-122-deletion mouse model [39]. Our work demonstrated that Pin1-mediated conformation change of XPO5 reduces the biogenesis of these tumor suppressive miRNAs. Whereas it is undeniable that not all miRNA biogenesis is regulated through Pin1/XPO5 axis, as XPO5 is the major but not the only transporter of pre-miRNAs. A subset of miRNAs are generated in a XPO5-independent manner [40]. Thus, our findings provides a novel insight into the understanding of post-transcriptional regulation of miRNA biogenesis, but the whole process of miRNA biogenesis in human cancer needs continued profiling in future.
In summary, Pin1 modulates the biogenesis of tumor suppressive miRNAs and promotes HCC development by catalyzing the conformational change of pS-P motif of XPO5. Our study gains an insight into the novel post-transcriptional mechanism of miRNA biogenesis modulated by Pin1/XPO5 axis and suggests that Pin1 is a potential promising target for HCC treatment.
Materials and methods
Antibodies and reagents
Anti-Pin1 and anti-XPO5 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA) and anti-myc tag antibody was obtained from Santa Cruz (CA, USA). Anti-flag tag antibody was provided by Huaan Biotechnology (Hangzhou, China). Anti-XPO5 antibody for immunofluorescence was obtained from Novus Biologicals (USA). Anti-Flag M2 agarose beads and anti-c-Myc agarose beads were purchased form Sigma-Aldrich (St. Louis, MO, USA). Glutathione-Sepharose and Ni-NTA agarose were obtained from GE Healthcare (USA).
Plasmids
The full-length Pin1 (#19027, Addgene) cDNA was subcloned into the pCMV5-flag vector. Pin1 W34A and C113A mutants were generated from pCMV5-Flag-Pin1 by QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. His-Pin1 FL (full-length), His-Pin1 WW, and His-Pin1 PPIase were kindly provided by Dr. Yih-Cherng Liou at Department of Biological Science, National University of Singapore, Singapore. The plasmids myc-XPO5 (#12552, Addgene), GST-Pin1 (#19027, Addgene), and siPin1 (TRCN0000010577, sigma) were bought from companies. The plasmids myc-XPO5 3A, myc-XPO5 3D, and MEKDD were kindly provided by Hui-Lung Sun at Department of Biochemistry and Molecular Biology, Institute for Biophysical Dynamics, Howard Hughes Medical Institute, University of Chicago, Chicago, Illinois.
Cell culture
SK-Hep-1, Huh-7, LM3, HepG-2, and SNU-423 cells were cultured in Dulbecco’s modified Eagle's medium (DMEM) medium supplemented with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator.
Cell transfection
Cells were seeded overnight and transfected with Lipofectamine 2000 in accordance with the manufacturer’s instructions. The ratio of DNA to Lipofectamine 2000 is 1 μg: 2.5 μl. Briefly, DNA and Lipofectamine 2000 were first diluted into Opti-MEM medium (Invitrogen) separately and stand at room temperature for 5 min. Then gently combined the diluted DNA and Lipofectamine 2000 together, and incubated at room temperature for 20 min. Lastly, the mixture was added onto cells and incubated for certain time for experiments. For lentivirus preparation, 293T cells were transfected with pLKO.1-derived plasmid with siPin1, packaging plasmid pCMV-dR8.2 dvpr, and envelope plasmid pCMV-VSVG (System Bioscience). The resultant lentivirus infected SK-Hep1 or Huh7 cells, and the stable transfectants were obtained after selection by puromycin.
Patient samples
Primary HCC tumor tissues and their adjacent normal tissues were collected from West China Hospital (China), which was approved by the Ethics Committee of West China Hospital of Sichuan University. Written informed consent for research purposes was provided for the patients. To examine Pin1 expression in patients’ tissues, total proteins were prepared from HCC tumor tissues and their adjacent normal tissues. Briefly, the patients’ tissues were dissected into small pieces, and grounded into powder in the liquid nitrogen. Subsequently, cell lysates were obtained by adding RIPA buffer into the powder and sonicated to efficiently lyse the cells, followed by centrifugation at 12,000 r.p.m. for 15 min at 4 °C. The resultant supernatants were subjected to western blotting analysis.
Western blotting
Total proteins were extracted in RIPA buffer (25 mM Tris·HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 × phosphotase inhibitor and protease inhibitor cocktail, and separated on SDS-polyacrylamide gel electrophoresis (PAGE) gel and electroblotted onto polyvinylidene difluoride membrane (Millipore). The membrane was blocked for 1 h at room temperature in 5% non-fat dry milk or bovine serum albumin (BSA) in TBST buffer (20 mM Tris-HCl, pH 7.5,150 mM NaCl, and 0.1% NP-40), then incubated with the primary antibody at 4 °C overnight, washed, and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. Blots were developed with SuperSignal chemiluminescent substrate (Pierce).
MTT assay
Cell viability was determined using the standard MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Briefly, 3,000 cells were seeded into 96-well plate and cultured for certain time. Then MTT (Sigma) was added and incubated for 3 h, the resultant purple formazan crystals were dissolved in 100 μl dimethyl sulfoxide, and the absorbance was measured by UV spectrophotometer (Thermal Fisher) at a wavelength of 570 nm.
Transwell migration assay, Matrigel invasion assay, and wound-healing assays
Cell migration assay was performed using Transwell chamber (Millipore) and cell invasion assay was done with chambers uniformly covered with Matrigel (BD Biosciences). Cells were suspended in serum-free DMEM medium and seeded in the top chambers, whereas DMEM supplemented with 10% FBS was added into the bottom chambers. After 24 h, the top chambers were fixed by 4% paraformaldehyde for 20 min and stained with 1% crystal violet for 15 min. For wound-healing assay, cells were cultured in the six-well plate to full confluency and scratched using a sterilized pipet tip. After washing three times with phosphate-buffered saline (PBS), the cells were cultured in DMEM medium containing 0.5% FBS. Images were recorded using microscope after certain time.
Production and purification of recombinant proteins
E. coli BL21(DE3) harboring GST-tag, or His-tag fusion construct, or empty vector was grown to A600nm = ~ 0.8 in Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin at 37 °C, then induced overnight by isopropyl β-D-thiogalactoside (IPTG) to produce target protein at 16 °C. The bacteria were collected and resuspended in PBS for sonication to lyse the cells. After centrifugation (11,000 × r.p.m., 4 °C, 15 min). the resultant supernatants were subjected to Glutathione Sepharose 4B (GE) for GST-tag protein purification, or Ni-NTA column for His-tag protein purification, in accordance with the manufacturer’s instructions.
Immunoprecipitation and pull-down assay
For co-IP assay, cells were co-transfected with myc-XPO5 and Flag-Pin1 or Pin1 mutant expression plasmids. Forty-eight hours later, cells were collected and suspended in the lysis buffer (100 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 0.1% NP-40, and protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). After incubation on ice for 15 min, cells were homogenized with 30 strokes of a homogenizer. Then the cell lysates were clarified by centrifugation at 2,000 × g for 15 min and immunoprecipitated with anti-Flag M2 agarose beads (Sigma). The bound proteins were subjected to SDS-PAGE and western blotting. For pull-down assay, cell lysates containing myc-XPO5 were incubated with purified GST-Pin1 or His-Pin1 protein at 4 °C for 4 h, and then with Glutathione Sepharose or Ni-NTA agarose beads for another 1 h at 4 °C. The pulled-down proteins were subjected to SDS-PAGE and immunoblotted with anti-myc antibody.
Partial proteolytic cleavage assay
Lysates from SK-Hep1 cells transfected with myc-XPO5 expression plasmid were incubated with anti-c-Myc agarose beads. Purified myc-XPO5 was incubated with GST, GST-Pin1, or GST-Pin1 (C113A) protein in the buffer containing 50 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM MgCl2, and phosphatase inhibitors at 20 °C for 30 min. Subtilisin A (Sigma-Aldrich) was added for proteolytic cleavage at 4 °C. One minute later, SDS sample buffer was added to stop the reaction and the supernatants were subjected to immunoblotting.
In vitro isomerization assay
GST-Pin1 protein (80 nmol/l) was pre-incubated with or without juglone (5 μM) for 10 min at room temperature and then added to the reaction buffer containing 50 mM HEPES (pH 7.5), ɑ-chymotrypsin (6 mg/ml, Sigma), and synthesized phosphorylated or non-phosphorylated polypeptide of S497P motif on XPO5 (50 μmol/l) at room temperature. The absorbance of the released 4-nitroaniline was immediately monitored at 390 nm for 180 s.
Immunofluorescence analysis
Cells after treatments were fixed in 4% paraformaldhehyde, permeabilized with 0.5% Triton X-100, blocked with 5% BSA, and incubated with primary anti-XPO5 or anti-Pin1 antibody at 4 °C overnight, followed by incubation with Alexa Fluor dye-conjugated secondary antibodies for 1 h. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature before mounting. Confocal fluorescence images were captured using Leica TCS SP5 II confocal spectral microscope.
RNA pull-down assay
Synthetic pre-miRNAs (Sigma-Aldrich) were denatured at 65 °C for 5 min and then cooled down to room temperature in the buffer (10 mM HEPES, 10 mM MgCl2, 0.1 M NaCl). Cells transfected with myc-XPO5 expression plasmid were lysed by sonication in the buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 0.5% NP-40, protease, and phosphatase inhibitor cocktail (Thermo Fisher Scientific) and RNase Inhibitor (Invitrogen). After centrifugation, the resultant supernatants were incubated with the above prepared pre-miRNAs at 4 °C for 4 h, followed by incubation with Dynabeads MyOne Streptavidin T1 (Invitrogen) for another 1 h at 4 °C. Then the pull-down proteins were subjected to western blotting.
RNA solution hybridization
Nuclear and cytoplasmic RNAs of SK-Hep1 or Huh-7 cells were extracted by PARIS kit (Ambion) according to the manufacturer’s protocol. Oligonucleotide probes (miR-146a: 5′-AACCCATGGAATTCAGTTCTCA-3′; miR-200b: 5′-TCATCATTACCAGGCAGTATTA-3′; miR-122: 5′-AAACACCATTGTCACACTCCA-3′) were labeled with [γ-32P]-ATP (PerkinElmer) by T4 polynucleotide kinase (NEB). RNA samples were hybridized with oligonucleotide probes for 3 h at 42 °C. Hybridized RNAs were subjected to non-denaturing-PAGE and transferred onto nitrocellulose membranes, and then examined by autoradiography.
miRNA quantification
Total RNAs were extracted by TRIzol reagent (Invitrogen) and the cDNAs were generated by M-MLV Reverse Transcriptase Kit (Invitrogen). The mature miRNA levels were examined by SYBR Green Master Mix Kit (Applied Biosystems) using U6 as control. Primers for miRNA quantifcation were purchased from GenePharma Company.
Xenografts in nude mice
The procedures involved in mouse experiments were approved by the Ethics Committee of West China Hospital of Sichuan University. SK-Hep1 cells were collected and suspended in saline solution containing 50% Matrigel (Corning) at 5 × 107/ml and Huh-7 cells were suspended at 3 × 107/ml. Then 100 μl of cells were subcutaneously injected into Balb/c nude mice. Tumor volume was examined and calculated using the formula: volume (mm3) = ab2/2 (a, long diameter; b, short diameter). After tumors were removed from mice, proteins and RNAs in tumors were isolated for Pin1 and miRNAs detection.
Electronic supplementary material
Acknowledgements
This work was supported by National Key R&D Program of China (2017YFA0504304 and 2016YFA0502204 to Y.P.), National Natural Science Foundation of China (81772960 and 81572739 to Y.P.), China Postdoctoral Science Foundation (2017M612976 to W.P.). We thank Dr Yih-Cherng Liou (Department of Biological Science, National University of Singapore) for providing the plasmids His-Pin1, His-Pin1 WW, and His-Pin1 PPIase.
Author contribution
J.L.,W.P., X.F., X.L., and Z.X. performed cell culture, biochemistry, cell biology, and xenograft experiments. Y.Z. prepared the recombinant proteins. H.L.S and J.K.Z. prepared the lentivirus and stable cell lines. H.L.S., J.K.Z., and J.H. constructed the plasmids. J.L., W.P., and Y.P. wrote the manuscript. L.L. provided patients’ samples. Y.P. and Y.Q.W. oversaw the experimental design and data analysis.
Compilance with ethical standards
Conflict of interests
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0065-z) contains supplementary material, which is available to authorized users.
References
- 1.de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol. 2017;35:872–8. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215–22. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–22. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, et al. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell. 2016;30:723–36. doi: 10.1016/j.ccell.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–7. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 14.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–16. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 15.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–14. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer. 2016;16:463–78. doi: 10.1038/nrc.2016.49. [DOI] [PubMed] [Google Scholar]
- 17.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 18.Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133–42. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- 19.Brenkman AB, de Keizer PL, van den Broek NJ, van der Groep P, van Diest PJ, van der Horst A, et al. The peptidyl-isomerase Pin1 regulates p27kip1 expression through inhibition of Forkhead box O tumor suppressors. Cancer Res. 2008;68:7597–605. doi: 10.1158/0008-5472.CAN-08-1059. [DOI] [PubMed] [Google Scholar]
- 20.Pang R, Yuen J, Yuen MF, Lai CL, Lee TK, Man K, et al. PIN1 overexpression and beta-catenin gene mutations are distinct oncogenic events in human hepatocellular carcinoma. Oncogene. 2004;23:4182–6. doi: 10.1038/sj.onc.1207493. [DOI] [PubMed] [Google Scholar]
- 21.Pang RW, Lee TK, Man K, Poon RT, Fan ST, Kwong YL, et al. PIN1 expression contributes to hepatic carcinogenesis. J Pathol. 2006;210:19–25. doi: 10.1002/path.2024. [DOI] [PubMed] [Google Scholar]
- 22.Shinoda K, Kuboki S, Shimizu H, Ohtsuka M, Kato A, Yoshitomi H, et al. Pin1 facilitates NF-κB activation and promotes tumour progression in human hepatocellular carcinoma. Br J Cancer. 2015;113:1323–31. doi: 10.1038/bjc.2015.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 24.Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012;9:137–42. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 26.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–9. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 28.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, An J, Lin J, Liu Y, Bao L, Zhang W, et al. Single nucleotide polymorphisms of microRNA processing machinery genes and outcome of hepatocellular carcinoma. PLoS ONE. 2014;9:e92791. doi: 10.1371/journal.pone.0092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev Mol Med. 2011;13:e21. doi: 10.1017/S1462399411001906. [DOI] [PubMed] [Google Scholar]
- 31.Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH, Tsai CH, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta. 2014;1843:2055–66. doi: 10.1016/j.bbamcr.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Zhang B, Gao J, Wang X, Liu Z. Regulation of the microRNA 200b (miRNA-200b) by transcriptional regulators PEA3 and ELK-1 protein affects expression of Pin1 protein to control anoikis. J Biol Chem. 2013;288:32742–52. doi: 10.1074/jbc.M113.478016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo ML, Gong C, Chen CH, Lee DY, Hu H, Huang P, et al. Prolyl isomerase Pin1 acts downstream of miR200c to promote cancer stem-like cell traits in breast cancer. Cancer Res. 2014;74:3603–16. doi: 10.1158/0008-5472.CAN-13-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer G, Aumüller T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol. 2003;148:105–50. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- 35.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–43. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 36.Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, et al. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–83. doi: 10.1016/S1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5. doi: 10.1186/1476-4598-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Roslan S, Johnstone CN, Wright JA, Bracken CP, Anderson M, et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene. 2014;33:4077–88. doi: 10.1038/onc.2013.370. [DOI] [PubMed] [Google Scholar]
- 39.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–97. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez I, Hayes KE, Barr JA, Harold AD, Xie M, Bukhari SIA, et al. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc Natl Acad Sci USA. 2017;114:E4961–E4970. doi: 10.1073/pnas.1618732114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.