Abstract
Dental caries is caused by acids released from bacterial biofilms. However, the in vivo formation of initial biofilms in relation to caries remains largely unexplored. The aim of this study was to compare the oral microbiome during the initial phase of bacterial colonization for individuals with (CC) and without (NC) cavitated dentin caries lesions. Bovine enamel slabs on acrylic splints were worn by the volunteers (CC: 14, NC: 13) for in situ biofilm formation (2 h, 4 h, 8 h, 1 ml saliva as reference). Sequencing of the V1/V2 regions of the 16S rRNA gene was performed (MiSeq). The relative abundances of individual operational taxonomic units (OTUs) were compared between samples from the CC group and the NC group. Random forests models were furthermore trained to separate the groups. While the overall heterogeneity did not differ substantially between CC and NC individuals, several individual OTUs were found to have significantly different relative abundances. For the 8 h samples, most of the significant OTUs showed higher relative abundances in the CC group, while the majority of significant OTUs in the saliva samples were more abundant in the NC group. Furthermore, using OTU signatures enabled a separation between both groups, with area-under-the-curve (AUC) values of ~0.8. In summary, the results suggest that initial oral biofilms provide the potential to differentiate between CC and NC individuals.
Introduction
Dental caries is a major health problem. Recent data demonstrate its high relevance for both the global prevalence of chronic asymptomatic caries as well as for the incidence of acute-caries associated tooth pain1,2. Dental caries is a process, which is mainly determined by lifestyle and can be delayed or even prevented by measures of oral hygiene and the use of fluorides3. However, even in developed countries young adults still display a remarkably high prevalence of decayed and filled tooth-surfaces4,5.
The caries process leads to the destruction of enamel and dentin by acids of microbial origin formed in dental biofilms6. Biofilm development is characterized by several typical stages irrespective of caries activity. These stages include the formation of an abacterial acquired pellicle, adhesion of saliva-borne bacterial early colonizers, and the formation of an extracellular matrix7,8. Finally, detached biofilm components are taken up by saliva and form the basis for the settlement on further surfaces. The cariogenicity of the biofilms can be influenced by diet and systemic diseases6. A diet rich in carbohydrates potentially leads to an increase of aciduric and acidogenic bacteria in biofilm and in saliva9. The availability of mono- and disaccharides in the food leads to a short-term decrease of the pH in plaque. This acidic attack dissolves calcium hydroxyapatite from the tooth’s hard substance. A high frequency of such acidic attacks over a prolonged period leads to a transition of biofilms from a healthy to a pathogenic state8, accelerating the dynamics of the caries process and followed by the formation of cavities10,11. Since caries is a process strongly influenced by microorganisms, the analysis of the microbiomes of biofilms in all stages and in saliva is crucial to further advance the understanding of this process and to provide tools for caries prediction and prevention6,12.
Biofilm in situ models offer a promising and standardizable option for obtaining biofilm samples over strictly defined periods of time7,13. These in situ models can be used to investigate initial bioadhesion processes, basic microbiological and molecular genetic questions, as well as the influences of food and oral hygiene products13–15. Depending on the requirements, glass, polymers, dental materials or dental hard substances can be used as the basic substrate13,16–18. Bovine enamel is particularly suitable for simulating the natural formation of biofilm on tooth enamel. The cattle are BSE-free and their meat is approved for human consumption. This procedure avoids the use of human enamel and appears more tolerable for many test persons. Furthermore, it is much easier to produce samples of homogeneous quality.
Conventional microbiology approaches have been used successfully and repeatedly for the analysis of the oral microbiome19–21. In addition, recent advances in culture-independent approaches, e.g., using high-throughput random shotgun sequencing (Next Generation Sequencing, NGS), provide improved turn-around times, reduced costs, and the potential to resolve hitherto unculturable microorganisms22. Meanwhile, about 700 prokaryote species have been detected in the oral cavity23. A comprehensive overview of oral bacterial taxa is available in the human oral microbiome database (eHOMD)23,24. NGS is also used for specific questions on the composition of the oral microbiome in healthy and diseased individuals25–27. With NGS methods, differences in the oral microbiome composition of individuals with severe, as well as those with moderate caries, were identified28–33. The general process of initial colonization of bacteria on tooth surfaces has been investigated in vivo and in situ with conventional approaches34–39. However, only few recent studies have characterized the microbiome during initial bacterial colonization in healthy adults in vivo or in situ using high-throughput sequencing approaches40–42. Therefore, the aim of the present study was to evaluate and to compare the microbiome of the initial phase of bacterial colonization on an in situ enamel biofilm model systematically after 2 hours, 4 hours and 8 hours for young adults with and without cavitated caries for the first time.
Results
Subjects
Groups of individuals without clinical signs of current caries activity (NC; 13 individuals) or with cavitated caries (CC; 14 individuals) were comparable with respect to their age (NC: median age 25 y, min-max: 19 to 33 y; CC: median 25 y, min-max 20 to 30 y), oral hygiene (approximal plaque index: NC: 61.6 ± 29.4%, CC: 70.8 ± 27.7%) and oral inflammation (modified sulcus bleeding index: NC: 35.2 ± 24.2%, CC: 44.6 ± 23.8%). All subjects showed physiological salivary flow rates of >0.25 ml saliva per minute. NC individuals presented no open decayed surfaces whereas CC individuals had a mean of 4.4 ± 1.8 cavitated carious surfaces reaching into the dentin. No open caries lesions were observed in the NC group (Supplemental Table 1).
General sequence data
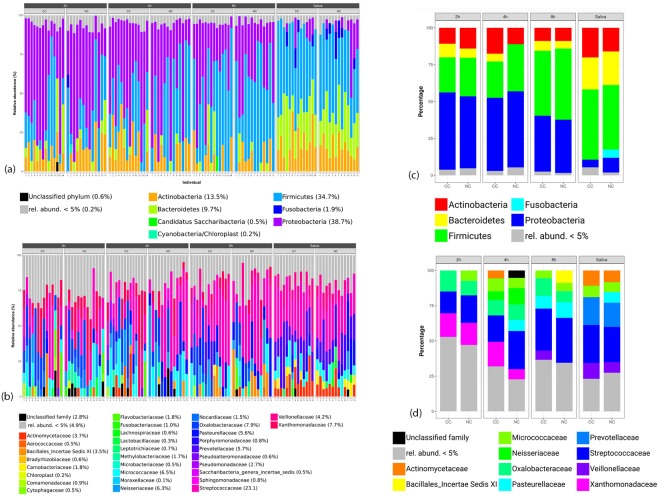
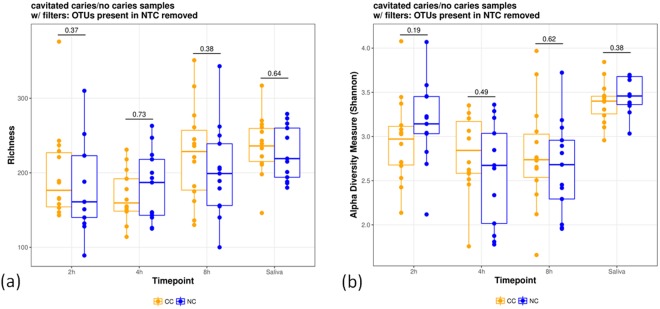
A total of 2,192,964 sequences were obtained from all 81 biofilm samples (2 h, 4 h, and 8 h) and from all 27 saliva samples after processing using LotuS (Supplemental Fig. S1). Additionally, 160,542 sequences were found in the DNA preparations of non-orally exposed enamel specimens (enamel control) or water (PCR control), and 9,350 sequences in the non-template control (NTC). OTUs were defined at the 97% identity level and 1,875 OTUs remained after discarding OTUs present in any of the controls (Supplemental Table 2). The identified OTUs were derived from members of the Proteobacteria (28%), Bacteroidetes (15%), Firmicutes (15%), Actinobacteria (10%), Fusobacteria (3%), Candidatus Saccharibacteria (2%), and Parcubacteria (2%) with other phyla covering less than 2% of the remaining OTUs according to taxonomic assignments created by LotuS. Nineteen percent of OTUs could not be assigned to a phylum. An overview of the taxonomic composition of OTUs in all samples as well as of the excluded OTUs present in controls is provided in the supplements (Supplemental Fig. S2a,b). Furthermore, per-sample compositions on phylum and family levels over all OTUs are presented in Fig. 1a,b. A high heterogeneity of the individual microbiomes is apparent (Fig. 1a,b). While a pronounced difference can be observed for the saliva samples compared to the biofilm samples, the biofilm samples are mutually overlapping (Figs 1c,d and 2, Supplemental Fig. S3a,d). Performing the ordination only on biofilm samples, i.e., excluding the saliva samples, did not lead to a markedly improved separation of the biofilm time points (Supplemental Fig. S4). Moreover, no statistically significant differences of the alpha-diversity nor of the richness were found when comparing the CC and NC groups per time point or in saliva (Wilcoxon-Mann-Whitney test, alpha = 0.05, Fig. 3a,b).
Figure 1.
Relative abundances of individual taxa per sample at phylum (a) and family (b) level, and summarized per indication at phylum (c) and family (d) level. The abundances were computed over all OTUs before filtering out OTUs present in controls. Samples were grouped by indication (CC: cavitated caries, NC: no cavitated caries) and time point (2 h-, 4 h- and 8 h-biofilms, and saliva) in (a) and (b); in (c) and (d), the indication is shown per time point. Taxa with a relative abundance below 5% per sample (a,b) and indication (c,d) were grouped into one category (“rel. abund. <5%”). In panels (a) and (b), the relative taxon abundance across all non-control samples and OTUs is shown in brackets next to the taxon name in the legend. It was computed as the sum of OTU counts corresponding to the respective taxon divided by the total OTU count across all non-control samples.
Figure 2.
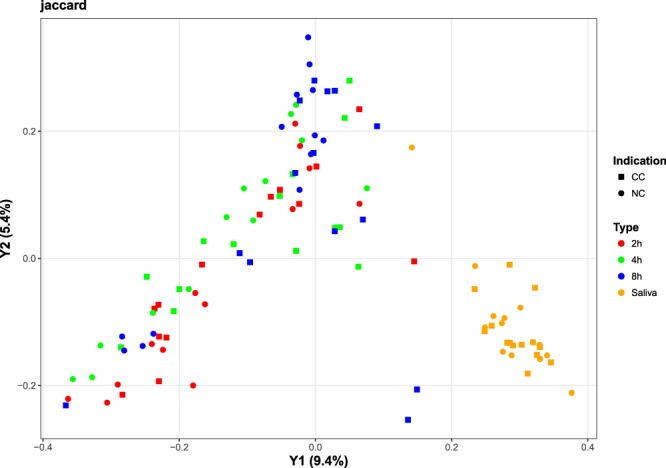
PCoA-based ordination of the Jaccard distance matrix. Colors reflect the sample type (2 h, 4 h, 8 h, and saliva) and shapes reflect the CC and NC groups. The percentage of variation explained is included in the axis labels.
Figure 3.
Richness (a) and alpha diversity (b) measures of different time points (2 h-, 4 h-, and 8 h-biofilms) and for saliva and indications (CC: cavitated caries, NC: no cavitated caries). OTUs present in the NTC were removed and richness and Shannon’s alpha diversity measures are shown. The individual points represent individual diversity estimates.
Statistical pairwise comparisons of individual OTUs
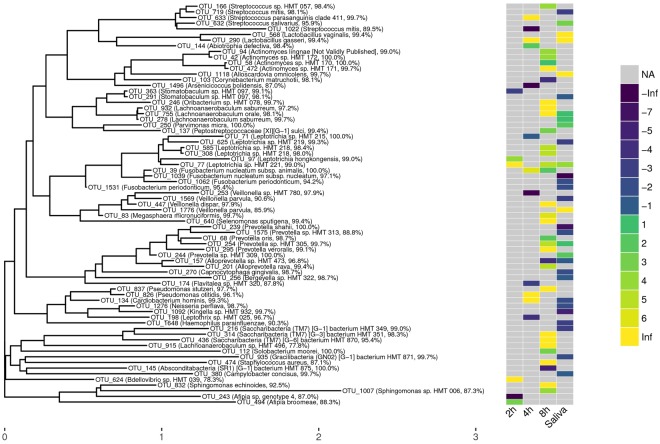
While the overall compositions at the phylum level as well as at other taxonomic levels were largely consistent between samples from individuals with cavitated caries and individuals without cavitated caries, individual OTUs demonstrate statistically significant differential relative abundances when comparing NC to CC samples (Fig. 4). Altogether, independent of the time point, statistically significant differences were found for 71 distinct OTUs (Fig. 4, Supplemental Table 3). At the 2 h time point, six significantly different OTUs were identified (genus according to eHOMD: 2x Leptotrichia, Stomatobaculum, 2x Afipia, and Bdellovibrio). At the 4 h time point, 12 significant hits were found (2x Streptococcus, Lactobacillus, Abiotrophia, Arsenicococcus, Leptotrichia, Fusobacterium, Veillonella, Flavitalea, Pseudomonas, Cardiobacterium, and Leptothrix). Moreover, 31 significant hits were detected at the 8 h time point (Streptococcus, 4x Actinomyces, Corynebacterium, Oribacterium, 3x Lachnoanaerobaculum, Peptostreptococcaceae, 3x Leptotrichia, Fusobacterium, Veillonella, Megasphera, Selenomonas, 3x Prevotella, 2x Alloprevotella, Pseudomonas, 2x Saccharibacteria, Solobacterium, Staphylococcus, Absconditabacteria, and 2x Sphingomonas). Finally, for the saliva samples, 30 significant hits were found additionally (2x Streptococcus, 2x Lactobacillus, Alloscardovia, Stomatobaculum, 2x Lachnoanaerobaculum, Parvimonas, 2x Leptotrichia, 3x Fusobacterium, 2x Veillonella, 4x Prevotella, Alloprevotella, Capnocytophaga, Bergeyella, Cardiobacterium, Neisseria, Kingella, Haemophilus, Saccharibacteria, Gracilibacteria, and Campylobacter). While individual OTU hits occurred on more than one time point and with consistent direction-of-change (i.e., more or less abundant), e.g., OTU 77 (Leptotrichia sp. according to eHOMD) and OTU 254 (Prevotella sp. according to eHOMD) were more abundant in the CC groups of the 8 h time points and saliva, no single OTU was found to have significantly different relative abundance in all groups or time points (Supplemental Fig. S5). Furthermore, it was observed that the majority of OTU hits for the 8 h samples showed a positive log fold-change. Thus, they were more abundant in the CC group, whereas the majority of the OTU hits for the saliva samples showed a negative log fold-change.
Figure 4.
Phylogenetic OTU tree created by LotuS with taxonomic assignment from the eHOMD database. Only OTUs with significantly different relative abundance are depicted with a heat map showing the log2(fold change) within the respective groups used for comparison. The values were highlighted as missing (“NA”), or colored w.r.t. their rounded value. A positive log fold-change indicates a higher relative abundance in the cavitated-caries group, a negative log fold-change stands for higher abundances in the no cavitated-caries group. Match identities according to eHOMD are given following the taxonomic name.
The mean relative OTU abundances were additionally compared over time between individuals with and without cavitated caries. The absolute difference in the correlation values for the groups was computed, possibly reaching a maximal value of 2, showing strongly contrasting behavior over time (exemplary OTU 100 in Fig. 5). A total of 109 OTUs were found to have an absolute difference in their correlation values greater or equal to 1.5 (Supplemental Table 4).
Figure 5.
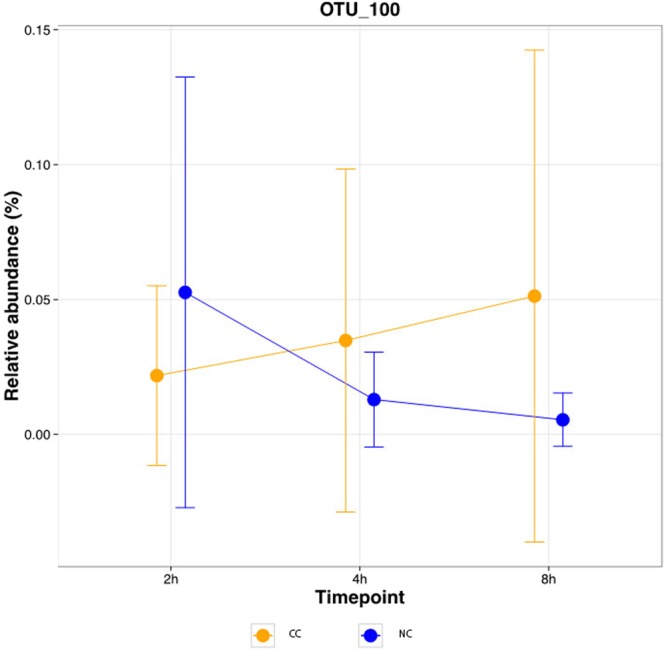
Relative OTU abundance over time per indication for an exemplary OTU. The mean +/− standard deviation of OTU 100 (Moryella sp. according to LotuS, Stomatobaculum longum according to eHOMD); absolute difference of the correlation of the mean values = 2 are shown.
OTU signatures for specimen classification
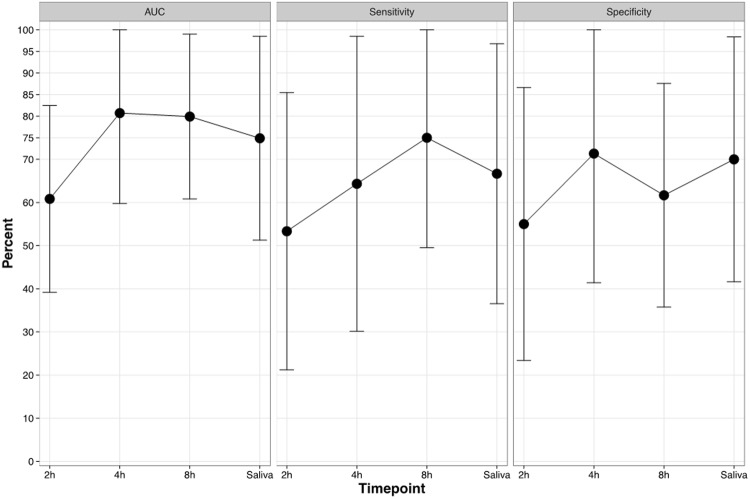
In addition to the study of individual OTUs, OTU signatures, i.e., combinations of individual OTUs, were used to train in silico classifiers for each time point and the saliva samples. The classification of samples obtained at 2 h exhibited the lowest mean values of AUC, sensitivity, and specificity (Fig. 6). At 4 h and 8 h, the mean AUCs were ~0.8. While the specificity was higher for the earlier time point, the later time point showed higher sensitivity in identifying samples from the CC group. The mean sensitivity for the initial biofilm samples increased over time, reaching its maximum at 8 h. In contrast, the 4 h samples showed the highest mean specificity values compared to all other samples (Fig. 6). The classification performance of saliva samples was below that of the 4 h and 8 h time points in terms of AUC, and between 4 h and 8 h in terms of sensitivity and specificity.
Figure 6.
Classification performances. The cross-validated (5-fold, 10 repetitions) means +/− standard deviations (capped at 100%) of AUC, sensitivity, and specificity are shown per time points for the classification of samples as “cavitated caries”.
Discussion
For the first time, the present in situ study evaluated and compared initial bacterial colonization on a biofilm model in adult individuals of different caries status using next generation sequencing. The results offer new information for basic research in oral microbiology concerning the process of bacterial colonization on dental enamel. In this context oral microbiomes were investigated, which consisted of biofilms formed over the periods of 2 h, 4 h and 8 h. The present in situ biofilm model enables the reproducible placement of identical biofilm-free enamel specimens in different individuals. This biofilm model thus overcomes the limitations of obtaining biofilms from natural tooth surfaces, as a well-defined and reproducible cleaning of tooth surfaces is only possible with great effort. Bovine enamel provides comparable properties as human enamel with respect to bioadhesion18. Tooth enamel from extracted teeth is of an unknown age. Furthermore, it is difficult to prepare larger samples from human teeth. The limitations of the in situ biofilm model are possible DNA contaminations from human salivary bacteria or from cattle oral microbiota. Saliva contamination was minimized by rinsing the specimens after removal from the oral cavity. Bacterial contamination from the bovine enamel itself was controlled by removing of all dentin and extensive cleaning of the enamel samples and by excluding all OTUs contained in control samples. It should also be noted that the NTC and the other no-biofilm controls showed contaminations. The corresponding OTUs were also systematically removed43.
The biofilms investigated in the present study represent typical oral microbiomes14,25,27–33. The 2 h and 4 h biofilms displayed high similarity; for the 8 h biofilms in particular the proportion of Firmicutes increased from 15% to 19%. Larger differences can be observed in comparison to the saliva samples. Here, the Firmicutes attained a share of 35%. The proportion of Bacteroidetes was 35% compared to 15% in biofilms. The relative abundance of Bacteroidetes was 35%. The proportion of Fusobacteria, Candidatus Saccharibacteria and Spirochaetes also increased, while the proportion of proteobacteria in biofilm samples dropped from more than 30% to 9% in saliva. A comparison of the microbiomes of the NC groups and the CC groups revealed remarkable differences. While in the NC group non-cariogenic taxa were significantly more frequent relatively, in the CC group members of the cariogenic microbiome were significantly more frequent with Actinomyces, Lachnoaerobaculum from the biofilms and Lactobacillus and Lachnoaerobaculum from the saliva, which is in line with previous reports33. Currently, caries risk prediction is mainly based on the inspection of existing caries lesions. This study presents a standardized in situ biofilm model which allowed, albeit not perfectly, to differentiate between young adults with and without cavitated caries using microbial signatures of early dental biofilms. Thus, this model represents a compelling new means for caries risk assessment, providing predictive readouts after 4 h or 8 h.
No universal biomarkers for caries prevalence could be detected herein, i.e., no single OTU was statistically significantly more or less abundant in the CC group compared to the NC group at all three time points (2 h, 4 h, 8 h) and saliva. This result is in accordance with the current state of knowledge and indirectly confirms the extended ecological plaque hypothesis, according to which aciduric and acidogenic bacteria gain a quantitative overweight in the biofilm under certain conditions, triggering a progression of the caries process44. Interestingly, hits in the 8 h samples were dominated by OTUs that were more abundant in CC samples, while the inverse was observed for the saliva samples, i.e., most OTU hits were more abundant in NC samples. As a clinical consequence resulting from this study, it might be useful to combine information obtained from microbiome samples of 8 h biofilms and of saliva to improve caries risk determination in the future.
The present results on OTU signature-based classifiers (instead of individual OTUs) demonstrated that, while the mean AUC values were between 75% and 80% (4 h, 8 h, saliva), and the sensitivity and specificity values increased over time, pronounced standard deviations could be observed. In future studies, a long-term follow-up of these moderate carious lesions presenting individuals should be carried out to link the oral microbiome compositions to the long-term development of caries lesions.
The use of a culture independent, 16S rRNA gene-based NGS approach limited our analyses to taxonomic assignments at the genus-level. However, individual species or even strains are expected to play crucial roles in the caries process. Thus, additional taxonomic analysis was performed using the eHOMD database providing species level resolution when possible (Supplemental Table 2, Fig. 4). However, considering the OTUs with significantly different relative abundances between the groups with and without cavitated caries, it is important to note that some of these OTUs have an assignment with identity percentage below 98.7% considered as gold-standard threshold for species classification43. Moreover, we want to emphasize that the taxonomic information is not used in our analyses which were all performed on OTU level regardless of any taxonomic assignment and its confidence.
Future studies, using metagenomic sequencing rather than amplicon sequencing, are expected to offer increased taxonomic resolution as well as to provide functional information which could be used for metabolic reconstructions. Another limitation of our study is the low biomass which is expected in the 2 h samples and, albeit to a much lesser extent, in the 4 h specimens. In this context, the sensitivity of the employed molecular approach bears challenges to be considered, e.g., contaminant DNA in the preparation kits or from environmental sources especially in the case of low biomass samples45. Though we attempted to remove potential contamination by strictly discarding all OTUs present in negative controls some contaminating sequences may still be present. Concretely, the phylum Cyanobacteria/Chloroplast is found in a single sample for the 2 h time point with a relative abundance >= 5% before filtering out OTUs present in controls (Fig. 1a). This indeed represents very likely a contamination. In any case, no chloroplast-related OTU was found to be statistically significantly differentially abundant. Whereas the 8 h biofilms seem to be preferable in order to limit the impact of possibly contaminating sequences, the 4 h biofilms could be considered as suitable only to a certain extent.
The present study confirms the results of a previous paper46 where pellicle enzymes had been investigated in the same subjects. Activities of human enzymes in the pellicle layer were of high similarity in both (CC and NC) groups. However, there was a tendency towards higher levels of bacterial glycosyltransferase D GTFD in subjects with open caries lesions. This finding is in accordance with the present findings with respect to differentially abundant taxa46.
In summary, by using a standardized in situ biofilm model and controlling for potentially confounding factors during 8 h of biofilm formation, we were able to detect differences in the oral microbiome compositions between individuals with and without cavitated caries. Moreover, when combined with standardized sample processing and sequencing procedures, the herein presented approach could be used to predict caries risk based on the composition of the oral microbiome. While further research is needed to make this approach more broadly applicable, e.g., to improve the classification performance, microbial signatures combined from early biofilms and saliva might provide practitioners with helpful information for advising habit changes to reduce caries risk.
Methods
Subjects
Twenty-seven healthy adults were enrolled in this study. Thirteen of them presented no cavitated caries lesions (NC group) and no signs of current clinical caries activity according to the Nyvad criteria for caries lesion activity47. Fourteen volunteers had by minimum three caries lesions reaching the dentine (CC group) and requiring restorative therapy. All participants were non-smokers and had no general diseases or diseases of the salivary glands. During a clinical oral examination, the following parameters were recorded: approximal plaque index (API) for oral hygiene, modified sulcus bleeding index (mSBI) for oral inflammation and, number of open caries lesions reaching the dentine for caries activity, caries indices DMFS (decayed, missing, filled surfaces) and ICDAS (International Caries Detection and Assessment System)48. In addition, information about age was registered and salivary flow rate was calculated in ml/min. The study design was reviewed and approved by the Ethics Committees of the Saarland (Sn52/05/2009) and Dresden (EK275092012) Universities. All experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants.
In-situ-formation of oral biofilms
Enamel slabs (diameter 5 mm × 1 mm) were prepared from the labial surfaces of bovine incisors of BSE-negative 2-year old cattle. The surfaces of all samples were polished by wet grinding with abrasive paper (320–4000 grit). The smear layer on the slabs was removed by ultrasonication in 3% NaOCl for 3 min. Then, samples were washed twice in distilled water for 5 min and disinfected in 70% ethanol for 10 min under ultrasonication, respectively. Finally, the slabs were stored in fresh distilled water for 24 h before oral exposure18,49. Alginate impressions (Blueprint cremix®, Dentsply DeTrey, Konstanz, Germany) were taken from the upper jaws of all volunteers. Transparent custom made individual splints were fabricated as carriers of the bovine enamel specimens. Enamel slabs were fixed onto the splints in the left and right buccal position in the molar and premolar regions with polyvinyl siloxane impression material (Aquasil, Denstply De-Trey, Konstanz, Germany). The splints were exposed intraorally for 2, 4 and 8 h, respectively (Supplemental Fig. S6). Two slabs per time point were used per subject, one on the left side and one on the right side. Two hours before wearing the splints, the subjects brushed their teeth without toothpaste, flossing was optional. No meals or tooth brushing were allowed during exposure times. After intraoral exposure, specimens were rinsed for 10 s with sterile water. One ml of unstimulated saliva was obtained from each participant after 4 h by continuously collecting saliva in a sterile tube. No individuals with hypo- and hypersalivation were included in the study. All samples were stored at −20 °C until further processing.
DNA preparation and next generation sequencing
DNA was extracted from biofilm samples and saliva samples using a commonly used DNA preparation kit (Qiagen DNA Blood and tissue kit, Qiagen, Hilden, G). Briefly, enamel slabs were placed in a 2 ml microcentrifuge tube and vortexed for 30 s at 2,500 rpm in 180 µl lysozyme-Triton TE-solution (20 mM Tris-Cl, pH 8.0, 2 mM sodium EDTA, 1.2% Triton® X-100, lysozyme 20 mg/ml) and stored at 37 °C for 30 min. Then, 20 µl of Proteinase K were added and the samples were thoroughly vortexed again for 30 s. After adding 200 µl Buffer AL and incubation at 56 °C for 60 min samples were placed on ice for 2 min. The next steps followed the bacterial and tissue protocol exactly. DNA was suspended in 20 µl of water and the DNA prepared from the two corresponding enamel plates was pooled. After preparation, DNA concentration was measured photometrically (Nanodrop 2000c, Thermo Fisher Scientific, Wilmington, DE, USA). PCRs of the 16S rRNA gene V1 and V2 variable regions were performed (5–25 ng template, 80 mM Tris-HCL, 20 mM (NH4)2SO4, 0.2% Tween-20, 2.5 mM MgCl2, 0.2 mM of each dNTP, 2.5 U HotFirePol (Solis BioDyne, Tartu, Est) using 200 pmol of each primer (forw16S_27F: 5′-agagtttgatcmtggctcag-3′, rev16S_338R: 5′-tgctgcctcccgtaggagt-3′) with Illumina universal adaptor sequences attached at the 5′-end. PCRs were performed in a thermocycler starting with 15 min at 95 °C followed by 33 cycles 95 °C for 1 min, 54 °C for 1 min, 72 °C for 1 min and a 5 min final extension at 72 °C. Amplicons were purified with Agencourt Ampure XP beads (BeckmanCoulter, Krefeld, Germany), diluted, pooled and sequenced (v3 chemistry: 2 × 300 bp paired-end) on the Illumina MiSeq (Illumina, San Diego, Ca, USA) following the manufacturer´s instructions aiming at 30,000 reads per sample.
Sequencing control samples
For control purposes, enamel slabs without biofilms were fixed on acrylic splints and incubated at 37 °C for 2, 4 and 8 h in a sterile glass beaker under dry conditions. DNA was prepared from these specimens as described above. Additionally, DNA preparation was carried out with sterile water (PCR control) and a non-template control (NTC) were generated and sequenced.
Sequence analysis
In a parallel study, children-derived CC and NC samples were generated and sequenced. Sequencing datasets from the present, adult samples and children samples of the parallel study were processed using LotuS, as described below. This joint processing was performed in order to generate common OTUs. OTUs uniquely present in the parallel samples were removed before further analysis in the present study. The sequencing reads in FASTQ-format were processed with LotuS version 1.47 including primer removal50. The default parameters of the sdm-MiSeq protocol were used, in particular, minSeqLength 170, minAvgQuality 27, TruncateSequenceLength 170, maxAccumulatedError 0.75. Operational taxonomic units (OTUs) were defined based on 97% sequence identity. In addition to the taxonomic assignments created by LotuS, the expanded Human Oral Microbiome Database (eHOMD23,24) was used to classify the OTUs. BLAST search (v. 2.2.23) was performed using the provided webserver (http://www.homd.org/?name = RNAblast&link = upload) in the database eHOMD 16S rRNA RefSeq version 15.1 with start at position 28 (parameters were set to default values). Top hits were extracted and saved including hit taxon, identity values and the number of mismatches. Relative OTU abundances were computed by dividing OTU read counts per sample by the respective total read count. OTUs present in any of the control samples (NTC, enamel, PCR) were discarded: OTUs detected in the NTC were discarded before computing the OTUs’ relative abundances, OTUs present in the other controls (PCR, enamel) were discarded after computing the relative abundances. PCR and enamel controls are considered as experiment-specific background. To account for this, we included these controls also during the bioinformatic steps and corrected for them following the computational analyses. Krona plots51 were created to provide an overview of the taxonomic composition of the dataset for all OTUs, for OTUs of 2 h, 4 h, and 8 h biofilm samples as well as from saliva samples. In addition, a Krona plot was prepared for the OTUs present in NTC, enamel and PCR controls. For the ordination analysis, the vegdist function and the Jaccard distance from the vegan R package, and the cmdscale function in the stats R package were used.
Statistical analysis
All statistical analyses were performed using R52. Plots were mainly generated using ggplot253. The biom-format output from LotuS, including the OTU relative abundance values, sample information, and taxonomy information, was used as input for phyloseq54. The Shannon Diversity index was used to estimate the alpha-diversity and the number of unique OTUs to estimate the richness. The two-tailed Wilcoxon-Mann-Whitney test (alpha = 0.05) was used to compare the diversity, the richness, and the relative abundance distributions of the individual OTUs between the cavitated-caries and no-cavitated-caries groups for each time point (2 h, 4 h, 8 h, saliva) after removal/correction of likely-contaminant OTUs. Raw p-values were adjusted using the Benjamini & Hochberg method for multiple comparisons55. Moreover, for each OTU, the area under the curve (AUC) was computed using the rocdemo.sca function from the made4 package56. The randomForest function from the randomForest package was used to build classification models in the case of OTU signatures, i.e., OTU sets, for the individual time points (2 h, 4 h, 8 h, saliva)57. Furthermore, the Spearman correlation coefficient was computed using the core function in R.
Ethics approval and consent to participate
The study design was reviewed and approved by the Ethics Committees of the Saarland (Sn52/05/2009) and Dresden (EK275092012) Universities. All experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants.
Electronic supplementary material
Acknowledgements
The study was supported by grants of the Deutsche Forschungsgemeinschaft (DFG) HA 5192/7-1, HA 2718/11-1 and RU 866/2-1.
Author Contributions
M.H., C.H. and S.R. designed the study. N.U., J.K. and S.R. performed the sample collection. A.E., S.T., C.L.P., J.W. performed sample processing. C.C.L., C.B., S.R. and V.G. analyzed the data. S.R., C.C.L. and A.K. wrote the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request basis.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Stefan Rupf, Cedric C. Laczny, Matthias Hannig and Christian Hannig contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32361-x.
References
- 1.Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 386, 743–800 (2015). [DOI] [PMC free article] [PubMed]
- 2.Kassebaum NJ, et al. Global burden of untreated caries: a systematic review and metaregression. J. Dent. Res. 2015;94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- 3.Marthaler TM. Changes in dental caries 1953–2003. Caries Res. 2004;38:173–181. doi: 10.1159/000077752. [DOI] [PubMed] [Google Scholar]
- 4.Mejàre I, Stenlund H, Zelezny-Holmlund C. Caries incidence and lesion progression from adolescence to young adulthood: a prospective 15-year cohort study in Sweden. Caries Res. 2004;38:130–141. doi: 10.1159/000075937. [DOI] [PubMed] [Google Scholar]
- 5.Hugoson A, Koch G, Helkimo AN, Lundin SA. Caries prevalence and distribution in individuals aged 3–20 years in Jönköping, Sweden, over a 30-year period (1973–2003) Int. J. Paediatr. Dent. 2008;18:18–26. doi: 10.1111/j.1365-263X.2007.00874.x. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi N. Oral microbiome metabolism: From “Who are they?” to “What are they doing?”. J. Dent. Res. 2015;94:1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- 7.Hannig C, Hannig M. The oral cavity – a key system to understand substratum-dependent bioadhesion on solid surfaces in man. Clin. Oral Invest. 2009;13:123–139. doi: 10.1007/s00784-008-0243-3. [DOI] [PubMed] [Google Scholar]
- 8.Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian J, et al. Microbiome interaction with sugar plays an important role in relapse of childhood caries. Biochem. Biophys. Res. Commun. 2015;468:294–299. doi: 10.1016/j.bbrc.2015.10.110. [DOI] [PubMed] [Google Scholar]
- 10.Sheiham A, James WP. A new understanding of the relationship between sugars, dental caries and fluoride use: implications for limits on sugars consumption. Public. Health. Nutr. 2014;17:2176–2184. doi: 10.1017/S136898001400113X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llena C, Leyda A, Forner L, Garcet S. Association between the number of early carious lesions and diet in children with a high prevalence of caries. Eur. J. Paediatr. Dent. 2015;16:7–12. [PubMed] [Google Scholar]
- 12.Marsh PD. Dental plaque as a biofilm and a microbial community – implications for health and disease. BMC Oral Health. 2006;6:S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prada-López I, Quintas V, Vilaboa C, Suárez-Quintanilla D, Tomás I. Devices for In situ Development of Non-disturbed Oral Biofilm. A Systematic Review. Front. Microbiol. 2016;7:1055. doi: 10.3389/fmicb.2016.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wake N, et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. npj Biofilms Microbiomes. 2016;2:16018. doi: 10.1038/npjbiofilms.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kensche A, et al. Impact of customary fluoride rinsing solutions on the pellicle’s protective properties and bioadhesion in situ. Sci. Rep. 2017;7:16584. doi: 10.1038/s41598-017-16677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Ahmad, et al. Bacterial adhesion and biofilm formation on yttria-stabilized, tetragonal zirconia and titanium oral implant materials with low surface roughness - an in situ study. J. Med. Microbiol. 2016;65:596–604. doi: 10.1099/jmm.0.000267. [DOI] [PubMed] [Google Scholar]
- 17.Feng J, et al. In situ antibiofilm effect of glass-ionomer cement containing dimethylaminododecyl methacrylate. Dent. Mater. 2015;31:992–1002. doi: 10.1016/j.dental.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Hannig C, et al. Fluorescence microscopic visualization and quantification of initial bacterial colonization on enamel in situ. Arch. Oral Biol. 2007;52:1048–1056. doi: 10.1016/j.archoralbio.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 20.Marsh PD. Dental plaque as a microbial biofilm. Caries Res. 2004;38:204–211. doi: 10.1159/000077756. [DOI] [PubMed] [Google Scholar]
- 21.Colombo AP, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaura E. Next-generation sequencing approaches to understanding the oral microbiome. Adv. Dent. Res. 2012;24:81–85. doi: 10.1177/0022034512449466. [DOI] [PubMed] [Google Scholar]
- 23.Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch. Microbiol. 2018;200:525–540. doi: 10.1007/s00203-018-1505-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen, T. et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. baq013 (2010) [DOI] [PMC free article] [PubMed]
- 25.Mason MR, Chambers S, Dabdoub SM, Thikkurissy S, Kumar PS. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome. 2018;6:67. doi: 10.1186/s40168-018-0443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Freitas A. R. et al. Oral bacterial colonization on dental implants restored with titanium or zirconia abutments: 6-month follow-up. Clin. Oral. Investig. 10.1007/s00784-018-2334-0 (2018). [DOI] [PubMed]
- 27.Keijser BJF, et al. The impact of maltitol-sweetened chewing gum on the dental plaque biofilm microbiota composition. Front. Microbiol. 2018;9:381. doi: 10.3389/fmicb.2018.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ximenes M, Armas RD, Triches TC, Cardoso M, Vieira RS. Structure of the bacterial community in different stages of early childhood caries. Oral Health Prev. Dent. 2018;1:1–5. doi: 10.3290/j.ohpd.a39691. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, et al. Research on oral microbiota of monozygotic twins with discordant caries experience - in vitro and in vivo study. Sci. Rep. 2018;8:7267. doi: 10.1038/s41598-018-25636-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lif Holgerson P, Öhman C, Rönnlund A, Johansson I. Maturation of oral microbiota in children with or without dental caries. PLoS One. 2015;10:e0128534. doi: 10.1371/journal.pone.0128534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agnello M, et al. Microbiome Associated with severe caries in canadian first nations children. J. Dent. Res. 2017;96:1378–1385. doi: 10.1177/0022034517718819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksson L, Lif Holgerson P, Esber A, Johansson I. Microbial complexes and caries in 17-year-olds with and without Streptococcus mutans. J. Dent. Res. 2018;97:275–282. doi: 10.1177/0022034517731758. [DOI] [PubMed] [Google Scholar]
- 33.Johansson I, Witkowska E, Kaveh B, Lif Holgerson P, Tanner AC. The Microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 2016;95:80–86. doi: 10.1177/0022034515609554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 35.Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 37.Diaz PI, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dige I, Nilsson H, Kilian M, Nyvad B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur. J. Oral. Sci. 2007;115:459–467. doi: 10.1111/j.1600-0722.2007.00494.x. [DOI] [PubMed] [Google Scholar]
- 39.Al-Ahmad A, et al. Bacterial colonization of enamel in situ investigated using fluorescence in situ hybridization. J. Med. Microbiol. 2009;58:1359–1366. doi: 10.1099/jmm.0.011213-0. [DOI] [PubMed] [Google Scholar]
- 40.Hertel S, et al. Initial microbial colonization of enamel in children with different levels of caries activity: An in situ study. Am. J. Dent. 2017;3:171–176. [PubMed] [Google Scholar]
- 41.Heller D, et al. Microbial diversity in the early In vivo-formed dental biofilm. Appl. Environ. Microbiol. 2016;82:1881–1888. doi: 10.1128/AEM.03984-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer RJ, et al. Interbacterial adhesion networks within early oral biofilms of single human hosts. Appl. Environ. Microbiol. 2017;83:pii: e00407–17. doi: 10.1128/AEM.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today. 2006;8:6–9. [Google Scholar]
- 44.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 45.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirsch J, et al. Enzymology and ultrastructure of the in situ pellicle in caries-active and caries-inactive patients. Caries Res. 2017;51:109–118. doi: 10.1159/000452226. [DOI] [PubMed] [Google Scholar]
- 47.Nyvad B, Baelum V. Nyvad criteria for caries lesion activity and severity assessment: A validated approach for clinical management and research. Caries Res. 2018;52:397–405272. doi: 10.1159/000480522. [DOI] [PubMed] [Google Scholar]
- 48.Pitts N. “ICDAS”—an international system for caries detection and assessment being developed to facilitate caries epidemiology, research and appropriate clinical management. Community Dent. Health. 2004;21:193–198. [PubMed] [Google Scholar]
- 49.Jung DJ, et al. Visualization of initial bacterial colonization on dentine and enamel in situ. J. Microbiol. Methods. 2010;81:166–174. doi: 10.1016/j.mimet.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Hildebrand F, Tadeo R, Voigt AY, Bork B, Raes J. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome. 2014;2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0 (2008).
- 53.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer New York. (2009).
- 54.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 56.Culhane AC, Thioulouse J, Perriere G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21:2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- 57.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request basis.