Abstract
Over the past decades, consistent studies have shown that race/ethnicity have a great impact on cancer incidence, survival, drug response, molecular pathways and epigenetics. Despite the influence of race/ethnicity in cancer outcomes and its impact in health care quality, a comprehensive understanding of racial/ethnic inclusion in oncological research has never been addressed. We therefore explored the racial/ethnic composition of samples/individuals included in fundamental (patient-derived oncological models, biobanks and genomics) and applied cancer research studies (clinical trials). Regarding patient-derived oncological models (n = 794), 48.3% have no records on their donor’s race/ethnicity, the rest were isolated from White (37.5%), Asian (10%), African American (3.8%) and Hispanic (0.4%) donors. Biobanks (n = 8,293) hold specimens from unknown (24.56%), White (59.03%), African American (11.05%), Asian (4.12%) and other individuals (1.24%). Genomic projects (n = 6,765,447) include samples from unknown (0.6%), White (91.1%), Asian (5.6%), African American (1.7%), Hispanic (0.5%) and other populations (0.5%). Concerning clinical trials (n = 89,212), no racial/ethnic registries were found in 66.95% of participants, and records were mainly obtained from Whites (25.94%), Asians (4.97%), African Americans (1.08%), Hispanics (0.16%) and other minorities (0.9%). Thus, two tendencies were observed across oncological studies: lack of racial/ethnic information and overrepresentation of Caucasian/White samples/individuals. These results clearly indicate a need to diversify oncological studies to other populations along with novel strategies to enhanced race/ethnicity data recording and reporting.
Introduction
Oncological research encompasses many aspects which can be categorized into fundamental and applied research. Fundamental research intends to understand the principles of cancer biology. This has been studied using tumor samples, which have been used to develop patient-derived oncological models (cell lines and xenografts)1, biobanks2 and genomic-based molecular classifications3. The knowledge acquired in basic research has been exploited for anti-cancer drug development and further clinical trials.
Cancer cell lines have historically been used as an essential tool for the investigation of cancer-related biological processes. They also have been used for anticancer drug testing over the past 50 years4,5. For instance, the United States (U.S.) National Cancer Institute 60 human tumor cell lines (NCI-60) panel, developed in the late 1980s, has been exploited to study ~400,000 compounds5. Since then, hundreds of cancer cell lines have been developed to study the molecular basis of cancer1. Nowadays, the NCI has decided to refocus its drug screening strategy on patient-derived models, which maintain a more realistic tumor-stroma enviroment. The NCI Patient-Derived Models Repository (PDMR) includes patient-derived xenografts (PDXs) and in vitro patient-derived cell cultures (PDCs)6.
Another important aspect of basic research is the identification of tumor biomarkers, which can be used for early cancer detection, diagnosis and prognosis. Cancer biomarkes are usually studied using body liquid biopsies or tissue samples deposited in biobanks2,7,8. One of these biobanks has been developed by the NCI Tissue Array Research Program (TARP). TARP develops tissue microarrays (TMA) from paraffin embedded tumoral tissues collected by the Cooperative Human Tissue Network (CHTN). Several studies have made use of these TMAs to analyze the expression of various tumor-associated markers9–15.
Racial/ethnic differences have been observed in cancer biomarker levels16,17. For instance, Preat et al.16 showed that Ki-67 labeling index, a biomarker of invasiveness in breast cancer, was higher in Arab/Moroccan patients compared with European individuals16. Similarly, Yamoah et al.17 found differential expression of six prostate cancer-associated biomarkers (AMACR, ERG, SPINK1, NKX3-1, GOLM1 and AR) between African American and European American patients. These markers predicted the risk of clinic-pathologic outcomes in an ethnicity-dependent manner17.
The advances in “omics” technologies have led to an improved understanding of cancer. Cancer multi-omics has facilitated the molecular characterization of a wide range of human cancers. For instance, The Cancer Genome Atlas (TCGA) and Therapeutically Applicable Research To Generate Effective Treatments (TARGET) efforts aim to identify molecular alterations, at the protein, RNA, DNA and epigenetic levels, to establish tumor classifications with improved accuracy3. These genomic signatures allow clinicians to administer personalized cancer treatments13,18–21. Similarly, The OncoArray Consortium aims to lay the genetic groundwork of breast, ovarian, prostate, colorectal, and lung cancers22. Additionally, cancer-related genome-wide association studies (GWAS)23 are also improving our understanding of cancer biology24. However, Popejoy & Fullerton25 showed by analyzing 2,511 GWAS (35 million samples) that the majority of these samples (81%) were isolated from European descents25. These data were obtained using the publicly available GWAS catalog; however, analysis focused on cancer GWAS studies has never been performed.
Basic oncological research aims to identify the underlying biological processes involved in cancer. From such understanding, several anti-cancer drugs have been developed and tested in clinical trials. Racial/ethnic differences in drug response have also been reported in many studies26,27. Among the most prominent examples of race/ethnicity-based drug response is found in Asian populations with lung adenocarcinoma (AD). Chinese, Korean, and Japanese female non-smokers with AD presented a higher prevalence of mutations in the epidermal growth factor receptor (EGFR) gene compared to Whites.
Patients with these alterations, which are clustered between exons 18 and 21 of EGFR, respond very well to EGFR tyrosine kinase inhibitors, such as erlotinib or gefitinib28,29. In addition, clinical trials of bevacizumab (to treat stomach cancer) and cetuximab (to treat non-small cell lung cancer) have shown effectiveness only in White patients27. Also, genetic-based pharmacoethnic differences in drug pharmacokinetics and pharmacodynamics have been reported for many compounds30–34.
In addition, a series of reports have shown that race/ethnicity has a great impact on cancer incidence26,35–37, survival38–41, drug response42,43 cancer molecular pathways44–46 and even on cancer-related epigenetic phenomena47–49. For instance, a lower incidence of glioma has constantly been reported in African Americans compared to Caucasians: 3.6 vs. 6.7 per 100,000 adults50,51. Additionally, it is largely known that non-Hispanic Black women and Hispanic women with breast cancer have a higher risk of cancer mortality compared to non-Hispanic White women52.
Concerning ethnic differences in cancer molecular mechanisms, a well-reported example is found in the TP53-mediated apoptosis pathway. TP53 is a well-known tumour suppressor that controls growth arrest and apoptosis. Chen et al.53 showed that non-Caucasian adult patients with glioma were five times more likely to present TP53 mutations in exons 5–8 compared with other populations53. Concomitantly, Hill & Sommer54 reported that TP53 mutational pattern differs among 15 geographically and ethnically populations54.
Regarding cancer epigenetics, ethnic differences in DNA methylation patterns have been described in several cancer types (e.g. lung, prostate, breast and colorectal)47–49. For instance, in lung squamous cell carcinomas (SCCs), Piyathilake et al.55 suggested that DNA hypomethylation is involved in the progression of SCCs in Caucasians but not in African Americans55.
Although the reasons underlying these observations have not been fully understood, it seems that differences in tumor genomics, health care access, disease detection, quality of treatment and lack of participation in health research may contribute to these outcomes17,26,50,56–64. For example, it has been estimated that approximately 1 per each 20 adult cancer patients participate in clinical trials; this rate persist over the time65 and is significant lower among racial/ethnic minority groups57,65. Despite several U.S initiatives lead by the Federal Drug Administration (FDA), the National Institutes of Health (NIH) and the Centers for Medicare and Medicaid Services, minorities are still underrepresented in clinical trials56.
Along with many other factors, racial/ethnic disparities produce inequalities in health care quality66–68. Eliminating these disparities is of a great interest because population demographics are constantly evolving and medical expenditures could be reduced66–68. For example, it is projected that by 2050 the majority group will no longer be non-Hispanic whites in the United States69. Additionally, it is estimated that the U.S. cost of this disparity to be in excess of $245 billion dollars annually70,71. Despite the clear influence of race/ethnicity in cancer outcomes and its impact in health care quality, an overview of racial/ethnic inclusion in oncological research has not been addressed yet. To have a complete understanding of racial/ethnic inclusion in cancer research, we have studied these demographic characteristics in several aspects of oncological research, from cell lines and patient-derived xenografts to biobanking, genomics and clinical trials.
Results
Fundamental cancer research
Patient-derived oncological models
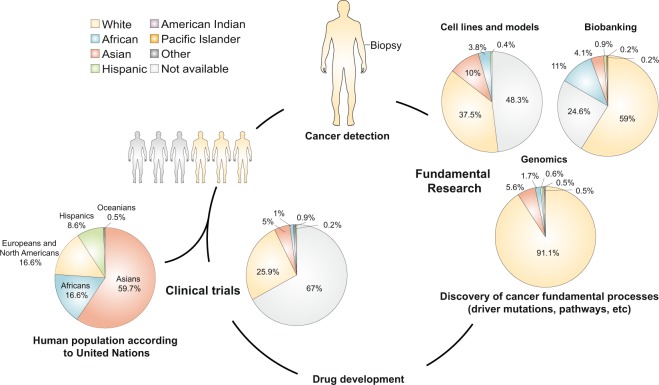
Racial/ethnic status was collected from the most common cancer cell lines and tumor samples available at the NCI PDMR. The majority of cancer cell lines (n = 689) have no records on race/ethnicity (46.1%). The rest were isolated manly from Whites (37.7%), followed by Asians (11.6%), African Americans (4.2%) and Hispanics (0.4%). The same tendency is observed in tumor samples available at the PDMR (n = 105). We found no racial/ethnic data for the majority of samples (62.86%). Of those with race/ethnicity reported, 36.19% were obtained from Whites and 0.95% were obtained from African Americans. Overall (n = 794), we found that 48.3% of samples have no records on their donor’s race/ethnicity. The remaining specimens were isolated mainly from White patients (37.5%), followed by Asian (10%), African American (3.8%) and Hispanic (0.4%) donors (Fig. 1). Supplementary tables 1 and 2 have detailed information of both datasets.
Figure 1.
Racial/Ethnic disparities in cancer research. Racial/ethnic inclusion was studied in several aspects of oncological research, from cell lines and patient-derived xenografts to biobanking, genomics and clinical trials.
Biobanks
Race/ethnicity information was collected from TARP repository, the Penn-CHOP Tumor Tissue Bank, the Children’s Brain Tumor Tissue Consortium (CBTTC) and the Komen Tissue Bank. Overall (n = 8,293), no data on race/ethnicity was found in 24.56% of samples. The rest were isolated from White people (59.03%), followed by African Americans (11.05%), Asian/Pacific Islanders (4.29%), Hispanics (0.87%) and American Indians (0.2%). Supplementary Table 3 contains detailed racial/ethnic information of all biobanks analyzed.
Cancer genomics
We collected racial/ethnic records from four major cancer genomic projects: TCGA, TARGET, cancer-related GWAS and OncoArray Consortium. NCI TCGA and TARGET projects (n = 12,980) did not report racial/ethnic data on 11.6% of their individuals. Specimens were donated mainly by White individuals (73.3%), followed by African American (8.9%), Asian (5.6%) and other donors (0.6%). Supplementary Table 4 contains detailed racial/ethnic information of all cancer types studied by TCGA and TARGET projects.
Racial/ethnic registers of 416 cancer-related GWAS (n = 6,375,784) showed that 0.64% of individuals have no data on race/ethnicity. The majority of individuals are Whites/Europeans (91.56%), followed by Asians (5.45%) and Hispanics (0.55%). Other race/ethnicities were represented by 0.33%. Supplementary Table 5 comprises detailed racial/ethnic information of this dataset. Additionally, race/ethnic information of the OncoArray Consortium (n = 314,268) was reported as follows: Whites (83.43%), Asians (8.97%), African Americans (4.59%) and other races/ethnicities (3.01%) (Supplementary Table 6).
In toto (n = 6,765,447), no racial/ethnic data was reported in 0.6% of individuals. Tumor samples were collected predominantly from Whites (91.1%), followed by Asians (5.6%), African Americans (1.7%), Hispanics (0.5%) and other populations (0,5%) (Fig. 1).
Applied cancer research
Clinical trials
We studied the racial/ethnic status of all patients involved in clinical trials of melanoma (2015 to 2017, n = 15,356), breast (2016 to 2017, n = 60,746) and lung cancer (2016 to 2017, n = 13,110). Overall (n = 89,212), no racial/ethnic registries were found in 66.95% of patients. Records were mainly obtained from Whites (25.94%), followed by Asians (4.97%), African Americans (1.08%), Hispanics (0.16%) and other minorities (0.9%). Supplementary Tables 7–9 contain detailed information of clinical trials analyzed in this study.
Discussion and Future Perspectives
On the basis of these select studies, we observe that some aspects of basic cancer research (patient-derived models, biobanks and genomics) and clinical trials have failed to record and/or report racial/ethnic information, as well as to include ethnically diverse populations. Thus, our analysis of racial/ethnic representation in select basic and applied cancer research studies revealed two tendencies: lack of racial/ethnic information and an overrepresentation of Caucasian/White samples/individuals.
Basic cancer understanding and initial drug screening have been accomplished using cell lines isolated mainly from White and unknown patients. For instance, the majority of the NCI-60 panel (33 out 60) have no records on their donor’s ethnicity, a tendency also observed throughout the entire dataset. Importantly, these 689 cell lines are the most studied in cancer research72 and constitute a representative sample (64.39%) of known cancer cell lines catalogued to date (n = 1070)1. Despite the proven importance of racial/ethnic inclusion in cancer research, these observations persist in modern patient-derived oncological models available at the PDMR6. In addition, a recent report showing the development of a comprehensive melanoma PDX collection does not provide any racial/ethnic information73, and this data is also missing from the Cancer Cell Line Encyclopedia (CCLE)1 and the Genomics of Drug Sensitivity in Cancer (GDSC) cell line collection74.
Similarly, a significant proportion of specimens available at biobanks lack racial/ethnic information or were isolated from White individuals. Since only biobanks that provide public data access were selected for this study, biorepositories were reduced to those present only in United Sates.
Racial/ethnic registers of these U.S. biospecimens may reflect therefore the U.S. population (White 73.60%, African American 12.60%, Asian 5.10% and Other 8.7%)75 and not an overrepresentation of White individuals; nonetheless, a significant proportion of these samples (24.56%) lack racial/ethnic information. Racial/ethnic registers of these U.S. biospecimens should reflect the U.S. population broadly (White 73.60%, African American 12.60%, Asian 5.10% and Other 8.7%)75 and not over represent Whites.
These tendencies persist in two major genomic efforts to understand the molecular basis of cancer: TCGA and TARGET projects, which are vastly used by medical and non-medical scientific communities. Also, cancer-related GWAS and OncoArray Consortium database are overrepresented by White/European-descendant populations. Interestingly, these results differ from the world population76: Asians represent 59.7%, Africans 16.6%, Europeans and North Americans 14.6%, Hispanics (Latin America and the Caribbean) 8.6% and Oceanians 0.5% (Fig. 1.). Since these international projects include cancer samples from all over the world, no limitations were found to globally address racial/ethnic status in cancer genomics. For example, the 416 cancer GWAS23 include genomic projects from China, India, Japan, Canada, among others. Similarly, the OncoArray Consortium is formed by a network including several European countries, the United States, Australia, China, Korea and Canada22. With more than six million individuals studied, we consider that these databases3,22,23 vastly represent cancer genomics globally.
Concerning clinical trials of melanoma, lung and breast cancer, racial/ethnic information is frequently unreported despite the fact that genetic-based pharmacoethnic differences in drug response have been well documented30–34,77. This raises serious concerns for future cancer clinical and drug development guidelines. Lung and breast cancer were selected for this study because they are diagnosed with the greatest frequency worldwide78. Similarly, melanoma is the most commonly diagnosed cancer in western countries and its treatment changed importantly when BRAF/MEK inhibitors and immunotherapy became the new standard therapy79. However, more research is needed to globally address racial/ethnic status in all cancer types.
Recent comprehensive analyses have provided a solid groundwork of human genetic variation that may possibly contribute to the race/ethnicity-related differences observed in cancer outcomes80–82. The 1000 Genomes Project Consortium has analyzed 2,504 genomes of different ancestry (26.4% African, 20.1% East Asian, 20.1% European, 19.5% South Asian and 13,9% Latin American) across five continental regions. This consortium identified a massive number of 88 million variants among 26 human populations81. Similarly, the Exome Aggregation Consortium (ExAC), analyzing 60,706 exomes of diverse ancestries (60.4% European, 13.6% South Asian, 9.5% Latin American, 8.6% African, 7.1% East Asian and 0.7% Other) has identified 7.4 million variants82. These results underscore the relevance of considering racial/ethnic-based human genetic variation as a critical factor in oncological research83.
Some initiatives have taken place over the last years to increase underrepresented minorities in cancer research84. For instance, the Hoy y Mañana project aims to increase biospecimen donation of ethnically diverse populations8. Similarly, the Geographic Management Program (GMaP) and the Minority Biospecimen/Biobanking - Geographic Management Program (BMaP) aim to reduce cancer related racial disparities by implementing a multi-institutional network infrastructure in the United States85. In this regard, BMaP for region 3 (Southeastern United States and Puerto Rico) developed and validated TMAs of invasive ductal carcinoma from ethnically diverse populations86. In addition, the U.S-based National Institute on Minority Health and Health Disparities (NIMHD) leads scientific research to reduce health disparities and improve minority health focusing on cardiovascular diseases, diabetes and cancer. Also, several studies have analyzed cancer-related genes of underrepresented human populations, such as Native Americans and Mestizo populations87–91.
Samples collected by the aforementioned strategies should be predominantly included in basic aspects of oncological research, such as patient-derived oncological models, initial drug screening and cancer genomics. This will alleviate racial/ethnic disparities in fundamental cancer research and further drug development. This should be enhanced by legal regulations in health policies. For example, the NIH Revitalization Act of 1993 should establish inclusion of minorities not only in clinical trials but also in fundamental cancer research, such as development of patient-derived cancer models (PDXs and PDCs), biobanks and genomics. Also, other legal initiatives should endorse race/ethnicity recording and reporting in all aspects of fundamental and applied oncological research.
To improve minority representation in cancer research, research agencies worldwide should promote fundamental projects to develop patient-derived models, biobanks and cancer genomics projects based on their populations. Also, clinical trials of new anti-cancer drugs should be extended to other countries and supported by international collaborations. Racial/ethnical disparities could also be reduced by increasing the participation of minorities in research projects.
In this concern, several studies have been performed and many strategies have been suggested to increase participation of underrepresented populations56,57,63,64.
Additionally, race/ethnicity should be determined by more accurate approaches such as genetic-based ancestry identification methods; for instance, race/ethnicity of genomic samples could be determined in silico using ancestry markers92.
Methods
Racial/ethnic categories
Since many studies analyzed in this work were performed by U.S. initiatives, we decided to standardized our data using the U.S. federal register 62 FR 5878293 to classify race and ethnicity.
Cancer cell lines and modern oncological models
Cell lines dataset were constructed as follow: the NCI-60 panel was merged with the 675 most frequently used cancer cell lines72, giving a total number of 689 cell lines. Racial/ethnic information was obtained from Klijn et al.72 and updated using: ExPASy-Cellosaurus (https://web.expasy.org/cellosaurus/), COSMIC Cell Lines Project v83 (http://cancer.sanger.ac.uk/cell_lines), HyperCLDB94 (http://bioinformatics.hsanmartino.it/hypercldb/indexes.html), the catalogue of Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) human and animal cell lines (https://www.dsmz.de/catalogues/catalogue-human-and-animal-cell-lines.html), European Collection of Authenticated Cell Cultures (ECACC) (https://www.phe-culturecollections.org.uk/collections/ecacc.aspx) and the Japanese Collection of Research Bioresources Cell Bank (JCRB) (http://cellbank.nibiohn.go.jp/). To increase our analysis on patient-derived oncological models, we include the NCI PDMR, which represents an improved version of the NCI-60 panel95. Racial/ethnic registries from PDMR (n = 105) was obtained from https://pdmr.cancer.gov/.
Biobanks
We initially consider 15 international cancer biobanks96 in addition to 3 U.S. biorepositories and 1 published data from the Komen Tissue Bank97 (also from U.S.). Since only biobanks that provide public data access96 were selected for this study, 15 biobanks were excluded from our analysis: Australasian Biospecimen Network (Australia), Australian Prostate Cancer BioResource, BancoADN (Spain), Canadian Tumor Repository Network, Centro National de Investigationes Oncologicas Tumor Bank Network (Spain), Chernobyl Thyroid Tissue Bank (Russian Federation), Confederation of Cancer Biobanks (UK), Cooperative Human Tissue Network (USA), Kathleen Cuningham Consortium for Research into Familial Breast Cancer (kConfab; Australia), onCore UK (UK), Singapore Tissue Network, European Human Tumor Frozen Tissue Bank, UK Biobank, Victorian Cancer Research Tissue Bank (Australia) and Wales Cancer Bank. Thus, we studied race/ethnicity of all samples available at the TARP repository (n = 1,203), the Penn-CHOP Tumor Tissue Bank (n = 1,815) and the Children’s Brain Tumor Tissue Consortium (CBTTC) (n = 2,302). We also included recent data from the Komen Tissue Bank (n = 2,973), which harbors normal breast tissue for cancer research97. Racial/ethnic data from TARP biorepository was obtained at https://ccrod.cancer.gov/confluence/display/CCRTARP/Home. Racial/ethnic information of The Penn-CHOP Tumor Tissue Bank and the CBTTC were obtained through the Biorepository Portal Toolkit2.
Genomics
We initially selected 5 major cancer genomics projects: 1) TCGA3, 2) TARGET (https://ocg.cancer.gov/programs/target), 3) GWAS related to cancer23, 4) OncoArray Consortium22 and 5) The Chinese Cancer Genome Consortium (CCGC)98. To our knowledge, these projects vastly represent cancer genomics globally3,22,23. CCGC (n = 260) was excluded from our analysis because the raw data is still not publicly available. Thus, demographic characteristics (race and ethnicity data) were obtained from 33 TCGA projects and 6 TARGET projects (Supplementary Table 4) through the NCI’s Genomic Data Commons (GDC, https://gdc.cancer.gov/). 416 cancer-related GWAS23 (Supplementary Table 5) were selected from the GWAS catalog comprising 6,375,784 samples (https://www.ebi.ac.uk/gwas/). Racial/ethnic information was obtained from the OncoArray Consortium database22 (n = 314,268) through https://epi.grants.cancer.gov/oncoarray/ (Supplementary Table 6).
Clinical trials
All clinical trials (randomized or not) were systematically selected from PubMed, associated with lung (from December 2016 to December 2017, n = 13,110 participants), breast cancer (from December 2016 to December 2017, n = 60,746 participants) and melanoma (from January 2015 to March 2017, n = 15,356 participants), which are related with active treatments (oncospecific drugs, radiotherapy and surgery). Racial/ethnic information was obtained from 55 studies in melanoma, 71 in breast cancer and 82 in lung cancer (Supplementary Tables 7–9).
Electronic supplementary material
Acknowledgements
We are grateful to Dr. Fátima Gebauer, Group leader at Centre for Genomic Regulation (Barcelona, Spain) for critical reading and constructive suggestions on the manuscript.
Author Contributions
S.G. conceived the subject and wrote the manuscript. CPyM supervised the project and provided conceptual advice and valuable scientific input. Data analysis was contributed as follows S.G., A.L.C. and A.I., Figure 1; S.G., A.L.C., A.I., J.M.G., A.K.Z., D.A.G., A.C.A., P.G.R. and P.E.L., Supplementary data.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Santiago Guerrero, Andrés López-Cortés and Alberto Indacochea contributed equally.
Contributor Information
Santiago Guerrero, Email: santiago.guerrero@ute.edu.ec.
César Paz-y-Miño, Email: cesar.pazymino@ute.edu.ec.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32264-x.
References
- 1.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felmeister AS, Masino AJ, Rivera TJ, Resnick AC, Pennington JW. The biorepository portal toolkit: an honest brokered, modular service oriented software tool set for biospecimen-driven translational research. BMC Genomics. 2016;17:434. doi: 10.1186/s12864-016-2797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson (Chairperson) TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery — past, present and future. Nat. Rev. Drug Discov. 2014;13:588–602. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 5.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 6.NCI Patient-Derived Models Repository (PDMR). Available at: https://pdmr.cancer.gov. (Accessed: 20th October 2017)
- 7.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez EM, et al. Engaging diverse populations in biospecimen donation: results from the Hoy y Mañana study. J. Community Genet. 2016;7:271–277. doi: 10.1007/s12687-016-0275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reis-Filho JS, et al. Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP-4 multi-tumor tissue microarray. Virchows Arch. 2003;443:122–132. doi: 10.1007/s00428-003-0859-2. [DOI] [PubMed] [Google Scholar]
- 10.Traicoff JL, et al. Expression of EIF3-p48/INT6, TID1 and Patched in cancer, a profiling of multiple tumor types and correlation of expression. J. Biomed. Sci. 2007;14:395–405. doi: 10.1007/s11373-007-9149-3. [DOI] [PubMed] [Google Scholar]
- 11.Mobasheri A, Airley R, Hewitt SM, Marples D. Heterogeneous expression of the aquaporin 1 (AQP1) water channel in tumors of the prostate, breast, ovary, colon and lung: a study using high density multiple human tumor tissue microarrays. Int. J. Oncol. 2005;26:1149–1158. [PubMed] [Google Scholar]
- 12.Abdul M, Hoosein N. Expression and activity of potassium ion channels in human prostate cancer. Cancer Lett. 2002;186:99–105. doi: 10.1016/S0304-3835(02)00348-8. [DOI] [PubMed] [Google Scholar]
- 13.Bass AJ, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SX, Hewitt SM, Steinberg SM, Liewehr DJ, Swain SM. Expression levels of eIF4E, VEGF, and cyclin D1, and correlation of eIF4E with VEGF and cyclin D1 in multi-tumor tissue microarray. Oncol. Rep. 2007;17:281–7. [PubMed] [Google Scholar]
- 15.Darash-Yahana M, et al. The Chemokine CXCL16 and Its Receptor, CXCR6, as Markers and Promoters of Inflammation-Associated Cancers. PLoS One. 2009;4:e6695. doi: 10.1371/journal.pone.0006695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preat F, Simon P, Noel J-C. Differences in breast carcinoma immunohistochemical subtypes between immigrant Arab and European women. Diagn. Pathol. 2014;9:26. doi: 10.1186/1746-1596-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamoah K, et al. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. J. Clin. Oncol. 2015;33:2789–2796. doi: 10.1200/JCO.2014.59.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbani R, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrar JE, et al. Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res. 2016;76:2197–2205. doi: 10.1158/0008-5472.CAN-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amos CI, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol. Biomarkers Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat. Rev. Cancer. 2017;17:692–704. doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 25.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma BB, Hui EP, Mok TS. Population-based differences in treatment outcome following anticancer drug therapies. Lancet Oncol. 2010;11:75–84. doi: 10.1016/S1470-2045(09)70160-3. [DOI] [PubMed] [Google Scholar]
- 27.Saijo N. The Role of Pharmacoethnicity in the Development of Cytotoxic and Molecular Targeted Drugs in Oncology. Yonsei Med. J. 2013;54:1. doi: 10.3349/ymj.2013.54.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paez JG, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science (80-.). 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 29.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 30.Schwab M, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J. Clin. Oncol. 2008;26:2131–8. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 31.Petros WP, et al. Associations Between Drug Metabolism Genotype, Chemotherapy Pharmacokinetics, and Overall Survival in Patients With Breast Cancer. J. Clin. Oncol. 2005;23:6117–6125. doi: 10.1200/JCO.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 32.Lal S, et al. Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics. 2007;8:567–75. doi: 10.2217/14622416.8.6.567. [DOI] [PubMed] [Google Scholar]
- 33.Innocenti F, et al. Comprehensive Pharmacogenetic Analysis of Irinotecan Neutropenia and Pharmacokinetics. J. Clin. Oncol. 2009;27:2604–2614. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renbarger JL, McCammack KC, Rouse CE, Hall SD. Effect of race on vincristine-associated neurotoxicity in pediatric acute lymphoblastic leukemia patients. Pediatr. Blood Cancer. 2008;50:769–71. doi: 10.1002/pbc.21435. [DOI] [PubMed] [Google Scholar]
- 35.Weir HK, et al. Annual report to the nation on the status of cancer, 1975-2000, featuring the uses of surveillance data for cancer prevention and control. J. Natl. Cancer Inst. 2003;95:1276–99. doi: 10.1093/jnci/djg040. [DOI] [PubMed] [Google Scholar]
- 36.Coupland VH, et al. Ethnicity in relation to incidence of oesophageal and gastric cancer in England. Br. J. Cancer. 2012;107:1908–14. doi: 10.1038/bjc.2012.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner WA, et al. Associations among ancestry, geography and breast cancer incidence, mortality, and survival in Trinidad and Tobago. Cancer Med. 2015;4:1742–53. doi: 10.1002/cam4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer-Chammard A, Taylor TH, Anton-Culver H. Survival differences in breast cancer among racial/ethnic groups: a population-based study. Cancer Detect. Prev. 1999;23:463–73. doi: 10.1046/j.1525-1500.1999.99049.x. [DOI] [PubMed] [Google Scholar]
- 39.Barnholtz-Sloan JS, Sloan AE, Schwartz AG. Racial differences in survival after diagnosis with primary malignant brain tumor. Cancer. 2003;98:603–609. doi: 10.1002/cncr.11534. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Qiu M, Xu R, Dobs AS. Comparison of survival and clinicopathologic features in colorectal cancer among African American, Caucasian, and Chinese patients treated in the United States: Results from the surveillance epidemiology and end results (SEER) database. Oncotarget. 2015;6:33935–43. doi: 10.18632/oncotarget.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadan-Lottick NS, Ness KK, Bhatia S, Gurney JG. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA. 2003;290:2008–14. doi: 10.1001/jama.290.15.2008. [DOI] [PubMed] [Google Scholar]
- 42.Patel JN. Cancer pharmacogenomics: implications on ethnic diversity and drug response. Pharmacogenet. Genomics. 2015;25:223–230. doi: 10.1097/FPC.0000000000000134. [DOI] [PubMed] [Google Scholar]
- 43.O’Donnell PH, Dolan ME. Cancer Pharmacoethnicity: Ethnic Differences in Susceptibility to the Effects of Chemotherapy. Clin. Cancer Res. 2009;15:4806–4814. doi: 10.1158/1078-0432.CCR-09-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat. Rev. Cancer. 2004;4:79–84. doi: 10.1038/nrc1257. [DOI] [PubMed] [Google Scholar]
- 45.Guo Y, Sigman DB, Borkowski A, Kyprianou N. Racial differences in prostate cancer growth: apoptosis and cell proliferation in Caucasian and African-American patients. Prostate. 2000;42:130–6. doi: 10.1002/(SICI)1097-0045(20000201)42:2<130::AID-PROS7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 46.Khani F, et al. Evidence for Molecular Differences in Prostate Cancer between African American and Caucasian Men. Clin. Cancer Res. 2014;20:4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia Y, et al. Racial/ethnic disparities in human DNA methylation. Biochim. Biophys. Acta. 2014;1846:258–62. doi: 10.1016/j.bbcan.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Kader F, Ghai M. DNA methylation-based variation between human populations. Mol. Genet. Genomics. 2017;292:5–35. doi: 10.1007/s00438-016-1264-2. [DOI] [PubMed] [Google Scholar]
- 49.Song M-A, et al. Racial differences in genome-wide methylation profiling and gene expression in breast tissues from healthy women. Epigenetics. 2015;10:1177–1187. doi: 10.1080/15592294.2015.1121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curry WT, Barker FG. Racial, ethnic and socioeconomic disparities in the treatment of brain tumors. J. Neurooncol. 2009;93:25–39. doi: 10.1007/s11060-009-9840-5. [DOI] [PubMed] [Google Scholar]
- 51.Deorah, S., Lynch, C. F., Sibenaller, Z. A. & Ryken, T. C. Trends in brain cancer incidence and survival in the United States: Surveillance, Epidemiology, and End Results Program, 1973 to 2001. Neurosurg. Focus 20, E1 (2006). [DOI] [PubMed]
- 52.Noone, A. M. et al. SEER Cancer Statistics Review, 1975-2015, National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018.
- 53.Chen P, et al. Ethnicity Delineates Different Genetic Pathways in Malignant Glioma. Cancer Res. 2001;61:3949–3954. [PubMed] [Google Scholar]
- 54.Hill KA, Sommer SS. p53 as a mutagen test in breast cancer. Environ. Mol. Mutagen. 2002;39:216–27. doi: 10.1002/em.10065. [DOI] [PubMed] [Google Scholar]
- 55.Piyathilake CJ, et al. Race- and age-dependent alterations in global methylation of DNA in squamous cell carcinoma of the lung (United States) Cancer Causes Control. 2003;14:37–42. doi: 10.1023/A:1022573630082. [DOI] [PubMed] [Google Scholar]
- 56.George S, Duran N, Norris K. A Systematic Review of Barriers and Facilitators to Minority Research Participation Among African Americans, Latinos, Asian Americans, and Pacific Islanders. Am. J. Public Health. 2014;104:e16–e31. doi: 10.2105/AJPH.2013.301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giuliano AR, et al. Participation of minorities in cancer research: the influence of structural, cultural, and linguistic factors. Ann. Epidemiol. 2000;10:S22–34. doi: 10.1016/S1047-2797(00)00195-2. [DOI] [PubMed] [Google Scholar]
- 58.Ademuyiwa FO, et al. Breast cancer racial disparities: unanswered questions. Cancer Res. 2011;71:640–4. doi: 10.1158/0008-5472.CAN-10-3021. [DOI] [PubMed] [Google Scholar]
- 59.Ward, E. et al. Cancer disparities by race/ethnicity and socioeconomic status. CA. Cancer J. Clin. 54, 78–93 [DOI] [PubMed]
- 60.Haiman CA, et al. Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans. PLoS Genet. 2011;7:e1001387. doi: 10.1371/journal.pgen.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang JJ, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat. Genet. 2011;43:237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhatia S, et al. Racial and ethnic differences in survival of children with acute lymphoblastic leukemia. Blood. 2002;100:1957–1964. doi: 10.1182/blood-2002-02-0395. [DOI] [PubMed] [Google Scholar]
- 63.Webb, F. J., Khubchandani, J., Striley, C. W. & Cottler, L. B. Black–White Differences in Willingness to Participate and Perceptions About Health Research: Results from the Population-Based HealthStreet Study. J. Immigr. Minor. Heal. 1–7, 10.1007/s10903-018-0729-2 (2018). [DOI] [PMC free article] [PubMed]
- 64.Heredia NI, et al. Community Perceptions of Biobanking Participation: A Qualitative Study among Mexican-Americans in Three Texas Cities. Public Health Genomics. 2017;20:46–57. doi: 10.1159/000452093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unger JM, Cook E, Tai E, Bleyer A. The Role of Clinical Trial Participation in Cancer Research: Barriers, Evidence, and Strategies. Am. Soc. Clin. Oncol. Educ. B. 2016;36:185–198. doi: 10.14694/EDBK_156686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fiscella K, Sanders MR. Racial and Ethnic Disparities in the Quality of Health Care. Annu. Rev. Public Health. 2016;37:375–394. doi: 10.1146/annurev-publhealth-032315-021439. [DOI] [PubMed] [Google Scholar]
- 67.Wheeler SM, Bryant AS. Racial and Ethnic Disparities in Health and Health Care. Obstet. Gynecol. Clin. North Am. 2017;44:1–11. doi: 10.1016/j.ogc.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Ayanian, J. Z. & Williams, R. A. Principles for Eliminating Racial and Ethnic Disparities in Health Care Under Healthcare Reform. In Healthcare Disparities at the Crossroads with Healthcare Reform. 421–432, 10.1007/978-1-4419-7136-4_23 (Springer US, 2011).
- 69.U.S. Census Bureau, P. D. Annual Estimates of the Resident Population by Sex, Race Alone or in Combination, and Hispanic Origin for the United States, States, and Counties: April 1, 2010 to July 1, 2016. (2017).
- 70.Ayanian, J. Z. The Costs of Racial Disparities in Health Care. Harv. Bus. Rev. (2015).
- 71.LaVeist TA, Gaskin D, Richard P. Estimating the Economic Burden of Racial Health Inequalities in the United States. Int. J. Heal. Serv. 2011;41:231–238. doi: 10.2190/HS.41.2.c. [DOI] [PubMed] [Google Scholar]
- 72.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 73.Krepler C, et al. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep. 2017;21:1953–1967. doi: 10.1016/j.celrep.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.2016 American Community Survey 1-Year Estimates. U.S. Census Fact Finder. American Community Survey. Retrieved June 12, 2018.
- 76.United Nations. Deparment of Economic and Social Affairs. Population Division. World Populations 2017. (2017).
- 77.Quinones LA, et al. Perception of the usefulness of drug/gene pairs and barriers for pharmacogenomics in Latin America. Curr. Drug Metab. 2014;15:202–8. doi: 10.2174/1389200215666140202220753. [DOI] [PubMed] [Google Scholar]
- 78.Ferlay J, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 79.Schadendorf D, et al. Melanoma. Nat. Rev. Dis. Prim. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 80.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erickson AK. Ethnicity puts clinical trials to the test. Nat. Med. 2003;9:983–983. doi: 10.1038/nm0803-983a. [DOI] [PubMed] [Google Scholar]
- 84.Hindorff LA, et al. Prioritizing diversity in human genomics research. Nat. Rev. Genet. nrg. 2017;2017:89. doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wells KJ, et al. Assessing needs and assets for building a regional network infrastructure to reduce cancer related health disparities. Eval. Program Plann. 2014;44:14–25. doi: 10.1016/j.evalprogplan.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seijo E, et al. Construction and Validation of a Multi-Institutional Tissue Microarray of Invasive Ductal Carcinoma From Racially and Ethnically Diverse Populations. Cancer Control. 2016;23:383–389. doi: 10.1177/107327481602300409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paz-Y-Miño C, Guillen Sacoto MJ, Leone PE. Genetics and genomic medicine in Ecuador. Mol. Genet. genomic Med. 2016;4:9–17. doi: 10.1002/mgg3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.López-Cortés, A., Guerrero, S., Redal, M. A., Alvarado, A. T. & Quiñones, L. A. State of art of cancer pharmacogenomics in Latin American populations. International Journal of Molecular Sciences18 (2017). [DOI] [PMC free article] [PubMed]
- 89.López-Cortés, A. et al. Genotyping the high altitude mestizo ecuadorian population affected with prostate cancer. BioMed Research International 2017 (2017). [DOI] [PMC free article] [PubMed]
- 90.López-Cortés A, et al. Breast cancer risk associated with gene expression and genotype polymorphisms of the folate-metabolizing MTHFR gene: a case-control study in a high altitude Ecuadorian mestizo population. Tumor Biol. 2015;36:6451–6461. doi: 10.1007/s13277-015-3335-0. [DOI] [PubMed] [Google Scholar]
- 91.López-Cortés, A. et al. Breast Cancer Risk Associated with Genotype Polymorphisms of the Aurora Kinase a Gene (AURKA): a Case-Control Study in a High Altitude Ecuadorian Mestizo Population. Pathol. Oncol. Res. 10.1007/s12253-017-0267-6 (2017). [DOI] [PubMed]
- 92.Hu Y, Willer C, Zhan X, Kang HM, Abecasis GR. Accurate local-ancestry inference in exome-sequenced admixed individuals via off-target sequence reads. Am. J. Hum. Genet. 2013;93:891–9. doi: 10.1016/j.ajhg.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Registrar F. Revisions to the standards for the classification of federal data on race and ethnicity. Fed. Regist. 1997;62:58781–58790. [Google Scholar]
- 94.Romano P, et al. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:D925–D932. doi: 10.1093/nar/gkn730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ledford H. US cancer institute to overhaul tumour cell lines. Nature. 2016;530:391–391. doi: 10.1038/nature.2016.19364. [DOI] [PubMed] [Google Scholar]
- 96.Vaught J, Kelly A, Hewitt R. A Review of International Biobanks and Networks: Success Factors and Key Benchmarks. Biopreserv. Biobank. 2009;7:143–150. doi: 10.1089/bio.2010.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCarty JR, et al. Abstract 5281: Komen Tissue Bank donors: Genetically determined ethnicity and race. Cancer Res. 2017;77:5281–5281. doi: 10.1158/1538-7445.AM2017-5281. [DOI] [Google Scholar]
- 98.Lv Y. Biobanks and cancer genome projects in China. J. Transl. Med. 2012;10:A51. doi: 10.1186/1479-5876-10-S2-A51. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.