Abstract
Skeletal muscle differentiation is controlled by multiple cell signaling pathways, however, the JNK/MAPK signaling pathway dominating this process has not been fully elucidated. Here, we report that the JNK/MAPK pathway was significantly downregulated in the late stages of myogenesis, and in contrast to P38/MAPK pathway, it negatively regulated skeletal muscle differentiation. Based on the PAR-CLIP-seq analysis, we identified six elevated miRNAs (miR-1a-3p, miR-133a-3p, miR-133b-3p, miR-206-3p, miR-128-3p, miR-351-5p), namely myogenesis-associated miRNAs (mamiRs), negatively controlled the JNK/MAPK pathway by repressing multiple factors for the phosphorylation of the JNK/MAPK pathway, including MEKK1, MEKK2, MKK7, and c-Jun but not JNK protein itself, and as a result, expression of transcriptional factor MyoD and mamiRs were further promoted. Our study revealed a novel double-negative feedback regulatory pattern of cell-specific miRNAs by targeting phosphorylation kinase signaling cascade responsible for skeletal muscle development.
Introduction
Proliferation and differentiation are tightly controlled during skeletal muscle formation (myogenesis) and organ development, dysregulation of these processes results in muscle dysplasia and deficiency [1–3]. A complex interplay of cytokines and signaling cascades, including their downstream transcriptional effectors, orchestrates the balance between proliferation and differentiation [4, 5].
The mitogen-activated protein kinase (MAPK) signaling pathway is a phosphorylation kinase signaling cascade that regulates many cell processes, such as cell division, differentiation, and release of inflammatory mediators [6, 7]. Many genes participate in the signal transduction of the MAPK cascade [8, 9]. For example, there are multiple upstream signals in the c-Jun N-terminal kinases (JNK/MAPK) cascade, containing MAP3K1 (MEKK1), MAP3K2 (MEKK2), MAP2K7 (MKK7), and MAP2K4 (MKK4), activate JNK and its downstream effector c-Jun by transferring a phosphate group [10]. Phosphorylated c-Jun, together with the Fos family proteins or a subset of ATF proteins, forms a dimeric compound that is called activating protein-1 (AP-1) [11], but the role of AP-1 in muscle cells is equivocal. Some reports have indicated that the function of AP-1 during myogenesis is dependent on its subunit composition, for the reason that Fra-2 containing AP-1 complexes could transactivate the MyoD enhancer/promoter [12], revealing that AP-1 is a positive role for myogenesis. Others have implicated AP-1 is inhibitory for this process [13, 14], and reported that AP-1 binds to a putative cAMP response element of the MyoD promoter and negatively modulates MyoD expression [15]. In particular, big data sorting study reported c-Jun and MyoD coordinate muscle enhancers, indicating a more complex role for AP-1 in muscle than previously anticipated [16]. Recent publications have shown that JNK/MAPK signaling mediates the activation of skeletal muscle glycogen synthase by insulin [17]. However, emerging researches indicate that JNK/MAPK signaling plays multiple roles in the regulation of insulin resistance [18, 19]. Additionally, the fact that myostatin activates the JNK/MAPK signaling reveals JNK/MAPK signaling can act as a potential negative regulator of muscle development [20]. Overall, these contradictory studies mirror the JNK/MAPK signaling in skeletal muscle development needs to be elaborated further.
Recently, microRNAs (miRNAs) have emerged as important regulators of development, and can fine-tune the activity of canonical signaling cascades [21, 22]. Tissue-specific miRNAs have been shown to play essential roles in cell fate determination [23–25]. Several miRNAs exclusively expressed in muscle have been labeled as myomiRs, including miR-1, miR-133a/b, miR-206, miR-208a/b, miR-486, and miR-499 [26, 27], which play significant roles during muscle proliferation, differentiation, and regeneration [28]. For example, miR-1 regulated skeletal satellite cell proliferation and differentiation by repressing Pax7 and YY1, and YY1 indeed repressed muscle miRNAs expression in myoblasts [29, 30]. Remarkably, previous studies have reported that miR-133 enhanced myoblasts proliferation by repressing serum response factor [25]. On the contrary, we elucidated a negative feedback circuit of miR-133 that represses IGF-1R expression to modulate the IGF-1R signaling pathway, which consequently promotes myogenesis [28]. Thus, the regulatory network of miRNAs has not been fully mapped and it is unclear how multiple miRNAs can interact with one signaling pathway to induce an integrated phenotype.
The definition of the biological targets of miRNAs is fundamental for understanding their functions. Over the past decade, by combination of computational prediction with high-throughput experiments, hundreds of genes have been identified as being regulated by miRNAs [31]. However, relative to the growing number of candidate targets, only a small percentage of miRNAs is well-characterized and contribute to skeletal muscle development [32]. Determining whether these predicted interactions exist and the importance of different targets for a given miRNA remains a challenge. Fortunately, newly developed high-throughput RNA capturing and sequencing technologies have made it possible. By sequencing of Argonaute (Ago)-bound RNAs isolated by crosslinking immunoprecipitation (CLIP-Seq, also known as HITS-CLIP), researchers have successfully identified tens of thousands of miRNA-target interactions in various systems [31, 33–36]. And after, modified forms of this method, such as photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) [37], individual-nucleotide resolution crosslinking and immunoprecipitation (iCLIP) [38], and some other variants [39, 40], have been successfully used in decoding RNA–protein interactions.
Here, we deciphered the functional linkage of myogenesis-associated miRNAs, which named as “mamiRs”, and demonstrated how multi-miRNA cooperates to create a common phenotype. Integrative analysis of global small RNA expression profiling, PAR-CLIP sequencing data, miRNA targeting interactions, and pathway architecture, we explored that JNK/MAPK signaling blocks muscle differentiation and demonstrated these mamiRs form an interaction network in which the combined activity of all mamiRs converged to counteract the JNK/MAPK signaling cooperatively by repressing multiple factors for it’s phosphorylation and to amplify P38/MAPK signaling. A shift in the balance between these central pathways favors skeletal muscle development. As a result, the expression of transcriptional factor MyoD and mamiRs were further promoted. Thus, we explored a mamiRs-MAPK axis responsible for skeletal muscle development.
Results
JNK/MAPK signaling pathways negatively regulates skeletal muscle differentiation
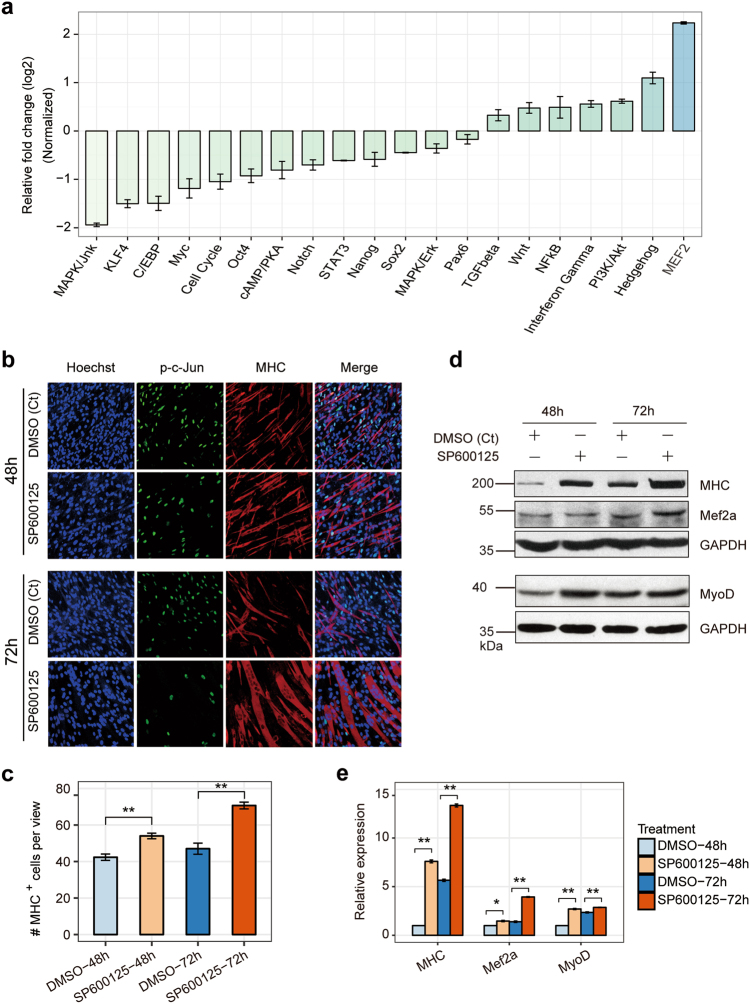
Mouse C2C12 myoblast cells are used to mimic skeletal muscle differentiation [41]. C2C12 myoblast cells proliferate in the presence of serum and differentiate upon serum withdrawal, which results in significant morphology changes (Supplementary Fig. S1a, b) and upregulating specific myogenenic markers such as MyoD, myogenin (MyoG), Mef2 family, and myosin heavy chain (MHC) (Supplementary Fig. S1c, d). To investigate the regulatory signaling pathway changes during myogenesis, we performed a pathway reporter array to identify the predominant signaling pathways that are regulated during C2C12 differentiation. In the 20 selected pathways we assayed, the activity of JNK/MAPK signaling was the most downregulated, and MEF2, which is the core effector of the P38/MAPK signaling pathway, was upregulated with the greatest fold-change (Fig. 1a). P38/MAPK signaling as primary member of the MAPK signalings promotes myogenesis [42], but the involvement of JNK/MAPK in this process is seldom discussed. Our observation indicated that JNK/MAPK play an opposite function of P38/MAPK during muscle differentiation.
Fig. 1.
The activity of the JNK/MAPK signaling pathways is dramatically decreased, and it blocks myogenesis. a The 20-pathway assay revealed that the MAPK signaling cascade changed most during the differentiation of C2C12 myoblasts, which mimics skeletal muscle differentiation. The relative fold-change of each pathway was tested following the transfection of the pathway plasmids into C2C12 myoblasts and the maintenance of the cells in differentiation medium (DM) for 3 days and normalized to C2C12 myoblasts cultured in growth medium (D0). The data were log2-transformed. b Immunofluorescent microscopy analysis of the morphological changes and expression of p-c-Jun and myogenesis marker MHC in C2C12 cells at 48 or 72 h after the cells were treated with SP600125 or DMSO, DMSO as negative control (Ct). cMHC+ cells were calculated based on staining described in panel b. d The expression of myogenic markers (MHC, Mef2a, and MyoD) in b were detected by western blotting. e Relative expression in d were calculated. GAPDH was the internal control. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
We first explored the function of the JNK/MAPK signaling during skeletal muscle development as examined the effects of the JNK-specific inhibitor SP600125 on cells morphology [43]. After C2C12 cells were treated with SP600125 for 48 or 72 h, the cells had a higher expression of MHC, suggesting an increase in myotubes formation, compared to that of the control group treated with DMSO (Fig. 1b, c). These results were confirmed by western blot (Fig. 1d, e), showing that MHC, Mef2a and MyoD were elevated when cells were treated with SP600125. Our findings reveal that the JNK/MAPK signaling pathway inhibits muscle differentiation. Consistent with previous studies [13, 44], the JNK/MAPK signaling blocks muscle differentiation by repressing MyoD.
We further tested the messenger RNA (mRNA) and protein expression levels of key factors of the JNK/MAPK signaling. Quantitative reverse transcription PCR (RT-qPCR) analysis showed that the mRNA expression levels of key factors of the JNK/MAPK signaling were minimally changed during myogenesis (Fig. 2a). When we tested their protein expression, the western blot results showed that the total JNK protein was not changed during skeletal muscle differentiation, whereas phosphorylated-JNK (p-JNK) protein was drastically downregulated, accompanied by a reduction in its downstream factors phosphorylated-c-Jun (p-c-Jun) and total c-Jun (Fig. 2b, c). In addition, the upstream proteins MEKK1, MEKK2, phosphorylated-MKK7 (p-MKK7), total MKK7, as well as phosphorylated-MKK4 (p-MKK4), and MKK4 were all significantly downregulated during muscle differentiation (Fig. 2d–f). These results illustrated that the JNK/MAPK signaling pathway is inactivated during muscle differentiation and implied a possible mechanism of post-transcriptional regulation.
Fig. 2.
The key factors of JNK/MAPK signaling pathway are downregulated at the post-transcriptional level during muscle differentiation. a qRT-PCR assays of six key factors of the JNK/MAPK signaling (MEKK1, MEKK2, MKK7, MKK4, JNK1/2, and JUN) in C2C12 cells before and after differentiation. D0 represents cells in growth medium and D2, D4, and D6 represent cells switched into differentiation medium for 2, 4, or 6 days. b, d, e Western blotting for six JNK/MAPK signaling key factors (JNK and c-JUN, MEKK1, MEKK2, MKK7, and MKK4) and their phosphorylated forms (p-JNK and p-c-JUN, p-MKK7 and p-MKK4) in C2C12 cells before and after differentiation. c, f Relative expression in b, d and e were calculated, respectively. GAPDH was the internal control. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
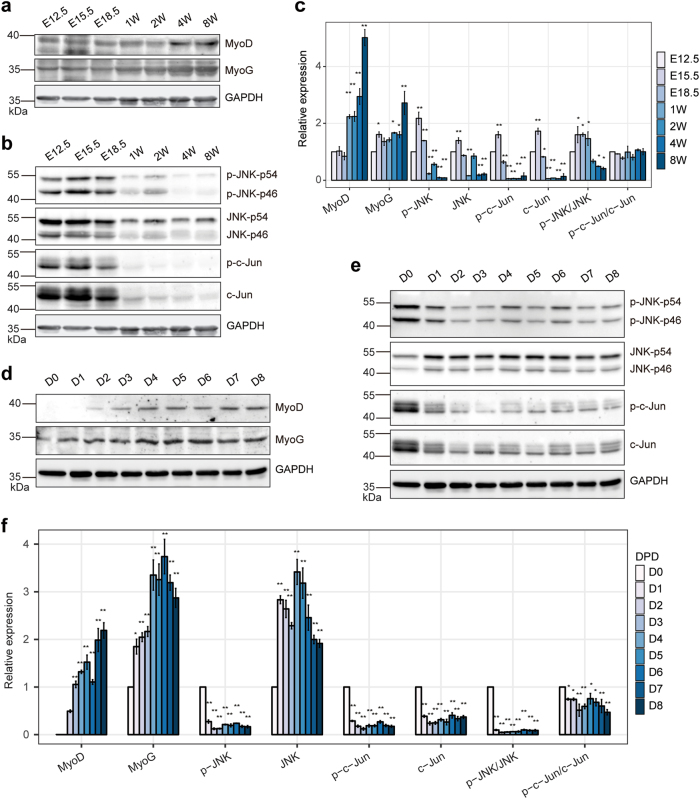
To confirm the changes of the JNK/MAPK signaling during skeletal muscle development in vivo, we investigated the JNK/MAPK factors in the hind limb muscle of developing mouse embryos. As shown in Fig. 3a–c, p-JNK and its downstream factors p-c-Jun and total c-Jun protein were drastically downregulated, negatively correlated with MyoD and MyoG. Moreover, we isolated skeletal muscle stem cells (MuSCs, VCAM1+CD31-CD45−Sca1−) from hind limb muscle in postnatal 1-week mice by fluorescence-activated cell sorting (FACS) (Supplementary Fig. S2a–c) [45]. The sorted MuSCs were activated in vitro for expansion of myogenic progenitors and subsequently to differentiation. Consistent with the results of developing mouse embryos muscle and the C2C12 cell differentiation, p-JNK and p-c-Jun expression decreased gradually during the primary skeletal muscle cell differentiation (Fig. 3d–f). Altogether, our results revealed that the JNK/MAPK signaling was significantly downregulated in the late stages of myogenesis in vivo and in vitro, suggesting that the JNK/MAPK pathway is the key player during skeletal muscle development.
Fig. 3.
The phosphorylation level of JNK/MAPK signaling pathway are downregulated during developing mouse embryos muscle and primary skeletal muscle cell differentiation. a The protein expression levels of myogenic markers (MyoD and MyoG) in developing mouse embryos muscle. Mouse hind limb muscles were isolated from seven time points: E12.5, E15.5, E18.5, postnatal 1 week, postnatal 2 weeks, postnatal 4 weeks, and postnatal 8 weeks (adult). b Western blotting for phosphorylation level of JNK/MAPK signaling: JNK, c-JUN, and their phosphorylated forms p-JNK and p-c-JUN in developing mouse embryos muscle. c Relative expression in a and b were calculated. Data in the same group were compared to E12.5. GAPDH was the internal control. d The protein expression levels of myogenic markers (MyoD and MyoG) during primary skeletal muscle cell differentiation. MuSCs were isolated from hind limb muscles in postnatal 1-week mice, and then the cells were activated in vitro for expansion of myogenic progenitors (D0) and subsequently to differentiation (D1 to D8). e Western blotting for phosphorylation level of JNK/MAPK signaling: JNK, c-JUN, and their phosphorylated forms p-JNK and p-c-JUN during primary skeletal muscle cell differentiation. f Relative expression in d and e were calculated. Data in the same group were compared to D0 (except for MyoD was compared to D2). GAPDH was the internal control. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
Genome-wide identification of miRNA-target interaction in undifferentiated and differentiated C2C12 cells by PAR-CLIP
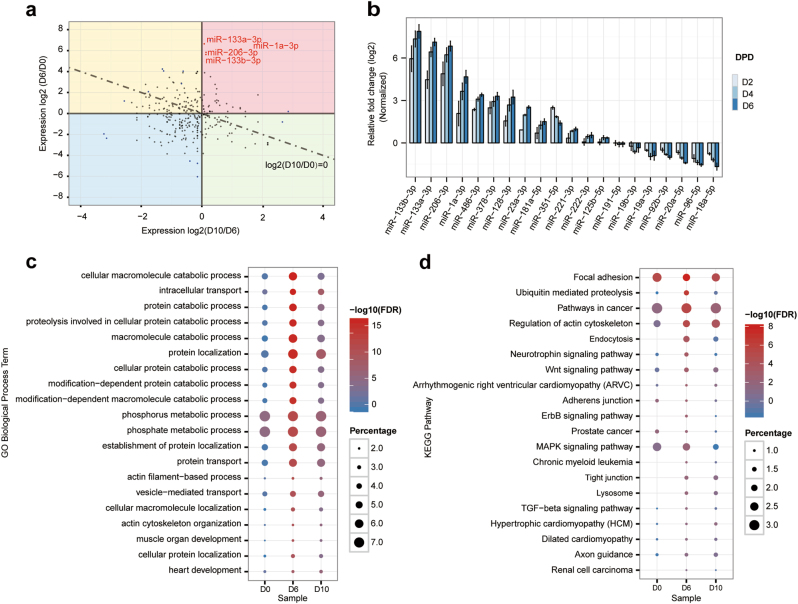
Given the recent observations that various miRNAs are a large group of post-transcriptional factors that participate in many biological processes, we reasoned that elevated miRNA targeting of the JNK/MAPK signaling pathway might account for its reduced activity during muscle differentiation. To explore these possibilities, we first performed miRNA expression profiling during C2C12 differentiation. Small RNA-sequencing analysis data showed that a group of skeletal muscle-specific or enriched miRNAs was dramatically elevated during muscle differentiation, including those that have been reported previously, such as miR-1a-3p (1a), 133a-3p (133a), 133b-3p (133b), 206-3p (206), and 486-3p, as well as some other miRNAs, such as miR-128-3p (128) and miR-351-5p (351) (Fig. 4a, Supplementary Fig. S3a, and Supplementary Table S1). Additionally, dozens of miRNAs were analyzed for miRNA expression by RT-qPCR, and the results were consistent with our small RNA-sequencing data (Fig. 4b, and Supplementary Fig. S3b).
Fig. 4.
miRNA expression profile and mapping of Ago2–RNA interactions by PAR-CLIP during C2C12 differentiation. a Small RNA sequencing revealed that myomiRs were remarkably upregulated remarkable during C2C12 cell differentiation. myomiRs are indicated in red color. C2C12 myoblasts cultured in growth medium (D0) and in C2C12 myotubes maintained in differentiation medium (DM) for 6 or 10 days. b The relative expression of partial miRNAs that had >2-folds changes in small RNA sequencing were analyzed by qRT-PCR. c, d Functional enrichment of miRNA target genes found by Ago2-PAR-CLIP before and after C2C12 cell differentiation. c Top 20 enriched Gene ontology (GO) biological process terms. d Top 20 enriched KEGG pathways. FDR false discovery rate; Percentage, number of genes involved in one particular terms divided by total genes
Because of the robust functional roles of myomiRs (miR-1a, 133a, 133b, and 206) in promoting skeletal muscle myogenesis, we examined whether these myomiRs correlate with the activity of the JNK/MAPK signaling pathway. To determine the relationship between miRNAs and their targets, we used a PAR-CLIP sequencing approach. Firstly, the protein expression of the Argonaute2 (Ago2) was increased during C2C12 differentiation (Supplementary Fig. S4a). And the RNA-immunoprecipitation (RIP) [46] results shown that two Ago2-specific antibodies, but not control IgG, could pull down the Ago2 protein and capture it’s binding RNAs efficiently (Supplementary Fig. S4b–d). Therefore, the miRNA machinery (miRNA–Argonaute–target triple complex) could be captured in undifferentiated and differentiated C2C12 cells. And then, we performed PAR-CLIP to decode the miRNA-target interaction. Three representative differentiation stages were selected: D0 (myoblasts, undifferentiated state), D6, and D10 (myotubes, differentiated for 6 and 10 days, respectively) (Supplementary Fig. S5). We yielded over 10 million uniquely mapped tags from each PAR-CLIP-sequencing library (Supplementary Table S2), and most tags were located on the 3'UTR or CDS of genes, which was consistent with the miRNA targeting signatures (Supplementary Fig. S6).
Comparing the miRNA reads of the CLIP data, we found that the miRNA expression profiles were altered greatly, which agreed with the small RNA-sequencing results (Supplementary Fig. S7). By comparing the top 20 elevated miRNAs from these two sets of sequencing data, we found that five miRNAs, miR-133a-3p, 1a-3p, 206-3p, 101a-3p, and 133b-3p, are in both groups (Supplementary Fig. S8), indicating that myomiRs might exert dominating effects during muscle differentiation. Investigations of the targets of myomiRs (Supplementary Table S3), showed that the distribution of target sites of each miRNA differed among the three libraries (Supplementary Fig. S9a). Additionally, the target sites in D6 were richer than those of the other two libraries (Supplementary Fig. S9b). Moreover, motif enrichment analysis of the tags using MEME [47] showed that the top 20 resulting motifs matched the seed sequence of miRNAs, which including miR-133abc and miR-1ab/206/613 (Supplementary Fig. S10a, b). And the seed centers were downstream of the corresponding cluster centers (Supplementary Fig. S10c), this characteristic was consistent with the miRNA functional feature.
We next combined tags to generate an Ago2 interaction map and analyzed its function. Gene ontology (GO) analysis of the genes in which the tags resided in showed enrichments in the cellular macromolecule catabolic process, protein localization, the phosphorus metabolic process, and the phosphate metabolic process during C2C12 differentiation (Fig. 4c). This result implied that the phosphorylation reaction is a major physiologic process during skeletal muscle differentiation. KEGG pathway analysis of the miRNA targetome revealed enrichments in multiple essential pathways, and among those pathways was the MAPK signaling pathway (Fig. 4d). These data indicated that phosphorylation of the MAPK signaling pathway might be the predominant phosphorylation reaction in these processes. Combining the three libraries of miRNA-target interactions from PAR-CLIP data, we found a group of miRNAs that could target the key factors of the JNK/MAPK signaling pathway (Supplementary Table S4). Then, we narrowed down to 12 candidates that are upregulated with more than twofold changes both in the small RNA-sequencing and RT-qPCR data (Supplementary Table S5). Notably, no miRNA in our candidates targets JNK itself.
MamiRs convergently and cooperatively target the JNK/MAPK signaling pathway
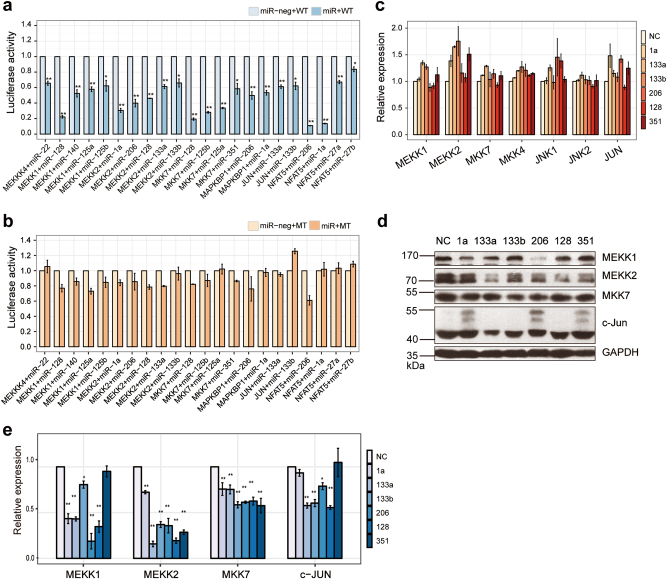
To confirm that these miRNAs could target those JNK factors functionally, we examined the effects of each miRNA on the expression of its putative targets using a heterologous luciferase reporter assay. The 3′UTRs of mouse MEKKK4, MEKK1, MEKK2, MKK7, MAPKBP1, Jun, and NFAT5 were inserted downstream of the luciferase gene, co-transfected with the corresponding miRNAs, and then assayed in C2C12 cells. Interestingly, all the reporter assay activity levels of the putative targets were repressed by its corresponding miRNAs individually (Fig. 5a), and mutating the miRNA complementary sequences on each target abrogated the miRNA-mediated regulation (Fig. 5b). The mapped miRNA-target sites in all the targets fulfilled the established base-pairing rule for miRNA targeting (Supplementary Fig. S11). These results revealed a bona fide targeting relationship between the key factors of the JNK/MAPK signaling and the elevated miRNAs during muscle differentiation.
Fig. 5.
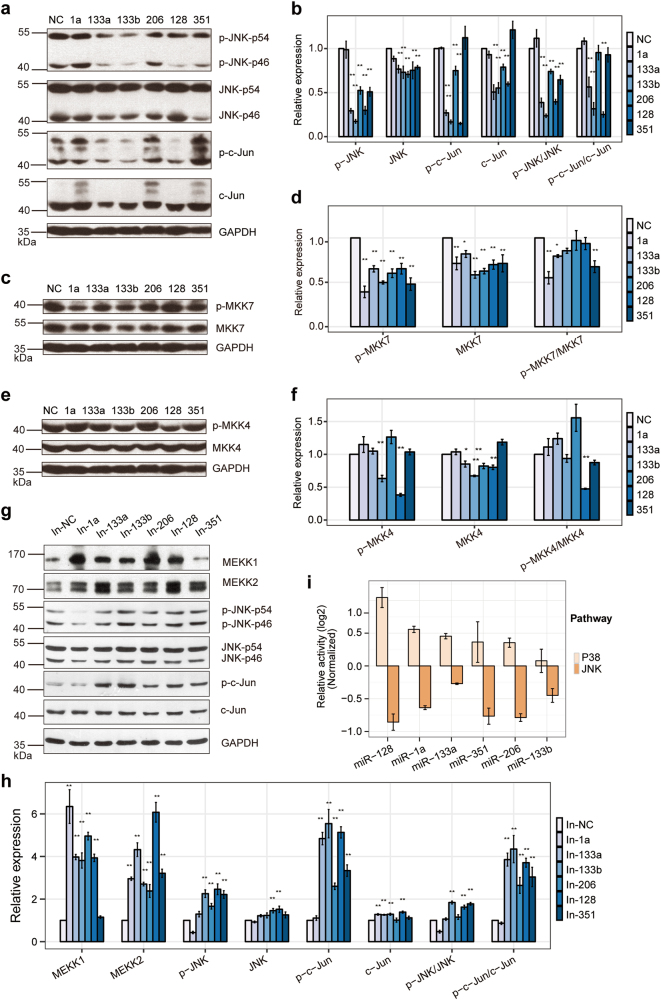
Multi-miRNAs target JNK/MAPK signaling. a, b Luciferase assays show the effects of multi-miRNAs on reporter constructs containing the wild-type (a) and mutant (b) target fragments of each candidate target gene. MT mutant, WT wild-type. The target sites constructs were co-transfected into C2C12 cells with either the corresponding miRNA or a miR-neg expression vector. The pcDNA6.2-miR-negplasmid (miR-neg) was used as a negative control. The relative luciferase activities are the ratio of renilla/firefly luciferase normalized to the negative control for each target group. c The relative expression of mRNA of the six JNK/MAPK signaling key factors (MEKK1, MEKK2, MKK7, MKK4, JNK1/2, and JUN) in C2C12 cells transfected with mimics of mamiRs or with a miRNA NC by qRT-PCR. d Western blotting showed the effect of the overexpression of mamiRs (1a, 133a, 133b, 206, 128, and 351) on the protein expression of targets of JNK/MAPK signaling (MEKK1, MEKK2, MKK7, and c-Jun). C2C12 cells were transfected with mamiR mimics or with NC. e Relative expression in d were calculated. GAPDH was the internal control. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
Further network analysis of the targets of the candidate miRNAs displayed convergent and cooperative functions of the six miRNAs, namely miR-1a, miR-133a, miR-133b, miR-206, miR-128, and miR-351, which we hereafter refer to as “mamiRs” (Supplementary Fig. S12). To provide decisive evidence, we transfected mamiRs into C2C12 cells and then tested the expressions of key factors of the JNK/MAPK signaling pathway using RT-qPCR and western blot. The results showed that mamiRs downregulated their protein but not mRNA expression levels (Fig. 5c–e). Furthermore, we tested the protein expression of p-JNK, JNK, p-MKK7, p-MKK4, and MKK4, and the western blot results were consistent with the PAR-CLIP data and the pathway reporter array results mostly(Fig. 6a–f), we hypothesized that there are some non-classical or indirect regulatory mechanism. Remarkably, the p-JNK protein was drastically downregulated without changed in total JNK protein. Thus, mamiRs can functionally target the JNK effectors and reduce the phosphorylation of the JNK/MAPK signaling pathway. Additionally, down-expression of each miRNA by inhibitor elevated the protein expression of MEKK1, MEKK2, p-JNK, and p-c-Jun, as well as JNK and c-Jun with a very slight increase (Fig. 6g, h), supporting the findings that mamiRs target the JNK/MAPK cascade convergently.
Fig. 6.
MamiRs target JNK/MAPK signaling biologically and attenuated its activity. a, c, e Western blotting analysis shows the expression of JNK/MAPK signaling protein (JNK, c-Jun, MKK7, and MKK4) and their phosphorylated forms (p-JNK, p-c-JUN, p-MKK7, and p-MKK4) in C2C12 cells that were transfected with mamiR mimics or NC. b, d, f Relative expression in a, c, and e were calculated, respectively. GAPDH was the internal control. g Western blotting analysis shows the protein expression of key factors of JNK/MAPK signaling (MEKK1, MEKK2, p-JNK, JNK, p-c-Jun, and c-Jun) in C2C12 cells that were transfected with mamiR inhibitors or NC inhibitor. h Relative expression in g was calculated. eThe pathway assay revealed mamiRs affected the activity of JNK/MAPK and P38/MAPK signaling. The results are normalized to those of with NC. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
We proceeded to ask whether these mamiRs regulate the MAPK signaling pathway. Transfection of any one of the mamiRs repressed the activity of the JNK/MAPK signaling, while the P38/MAPK signaling activity changed reversely (Fig. 6i). To verify the role of P38/MAPK signaling during muscle differentiation, we inhibited the P38/MAPK pathway with its specific inhibitor SB203580 [48]. The differentiation of C2C12 cells treated with SB203580 was largely blocked and was accompanied by attenuated myotubes fusion (Supplementary Fig. S13a), as well as decreased endogenous myogenic biomarkers expression (Supplementary Fig. S13b, c). These data agreed with the existing research results and provided strong biochemical evidence for the function of the MAPK signaling pathway during skeletal muscle development. Collectively, mamiRs promoted muscle differentiation via repressing the JNK/MAPK signaling pathway and the activating P38/MAPK signaling pathway.
MamiRs promote muscle differentiation by regulating the MAPK signaling pathway
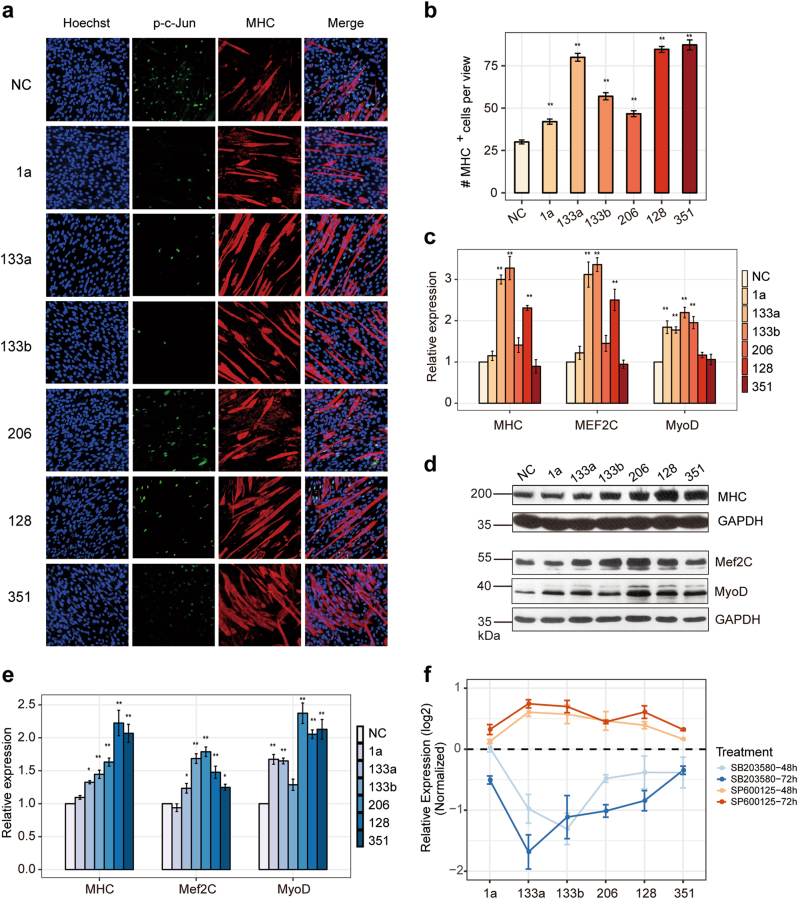
Previous studies have shown that miR-1a and miR-206 can promote myogenesis by targeting various genes, but the function of miR-133a and miR-133b is a controversial issue [28, 49–51]. Some studies have shown that miR-133a and miR-133b function as proliferation factors that can target SRF [25]. To stretch their function a little further, we performed immunofluorescence as showed that miR-133a, miR-133b, as well as miR-1, miR-206, miR-128, and miR-351 can promote muscle differentiation (Fig. 7a, b). Additionally, these mamiRs partially increased the mRNA expression levels of MHC, MEF2C, and MyoD (Fig. 7c), so did their protein expression (Fig. 7d, e). These results are consistent with the data of JNK-specific inhibitor, strongly support the fact that JNK/MAPK signaling inhibits skeletal muscle differentiation by negatively regulating MyoD. Notably, miR-128 and miR-351, two seldom-discussed miRNAs, had the apparent effect of promoting the myogenesis marker expression, suggesting that there are some undiscovered mechanism during skeletal muscle development.
Fig. 7.
mamiRs promote myogenesis and form an autoregulatory loop with JNK/MAPK and P38/MAPK signaling. a Immunofluorescent microscopy analysis of the morphological changes and expression of p-c-Jun and myogenesis markers in C2C12 cells transfected with mamiR mimics or NC. bMHC+ cells were calculated based on staining described in panel a. c qRT-PCR assays of myogenic markers (MHC, MEF2C, and MyoD) in C2C12 cells transfected with mamiR mimics or NC. d Western blotting for myogenic markers (MHC, MEF2C, and MyoD) in C2C12 cells transfected with mamiR mimics or NC. e Relative expression in d were calculated. GAPDH was the internal control. f qRT-PCR assays of mamiR expression in C2C12 cells treated with SP600125, SB203580, or DMSO. The results are normalized with to those of cells treated with DMSO. Values are means ± SEM. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01
Inhibition of the MAPK signaling pathway alter the expression of mamiRs
Based on the regulation between the activity of the JNK/P38 signaling pathways and mamiRs, we supposed that the inhibition of the MAPK signaling pathway during C2C12 differentiation could affect mamiRs expression. Therefore, we inhibited the activity of JNK/MAPK pathway and found that the expression profiles of these mamiRs showed a gradually increasing pattern. In agreement with this finding, knockdown of P38/MAPK signaling resulted in a gradual decrease of the mamiRs (Fig. 7f), confirming the presence of a regulatory balance between the JNK/MAPK and P38/MAPK signaling pathways. These results revealed that the inhibition of the JNK/MAPK signaling pathway can promote C2C12 cell differentiation accompanied by the upregulation of mamiRs expression, which further induced myoblasts differentiation, whereas inhibiting the P38/MAPK pathway showed the opposite effects.
Discussion
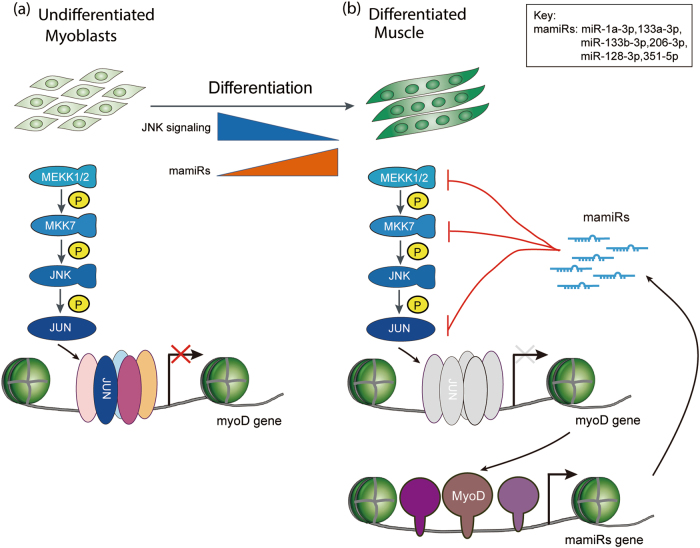
Extracellular-regulated kinases and mitogen-activated protein kinases are activated upon signals via different routes to alter the stability and translation of various cytokine mRNAs, enabling cells to respond promptly. Using a global approach, we described a novel regulatory network controlling muscle differentiation through the cooperation of multiple myogenesis-associated miRNAs, which we termed mamiRs, (i) were upregulated in C2C12 myoblast-mimic skeletal muscle differentiation, (ii) functional targeted the upstream and downstream core factors of the JNK/MAPK pathway, including MEKK1, MEKK2, MKK7, and c-Jun, but did not target JNK itself, resulting in the repression of JNK/MAPK pathway activity and promotion of muscle differentiation, and (iii) stimulated the P38/MAPK signaling pathway, which also contributed to promote myogenesis. Furthermore, we demonstrated that these mamiRs were, in turn, regulated by the JNK/MAPK and P38/MAPK pathways. Overall, our results, combined with the fact that JNK-induced activation of c-Jun could repress miRNAs via the attenuated expression of MyoD, established a novel double-negative feedback regulatory network among six convergent and cooperative mamiRs and the JNK/MAPK signaling pathway (Fig. 8). Additionally, these mamiRs could stimulate the P38/MAPK signaling pathway, thus resulting in a shift in the balance between the JNK/MAPK and P38/MAPK signaling pathways.
Fig. 8.
A model of mamiRs and JNK/MAPK signaling forming a double-negative feedback regulatory network in skeletal muscle development. a In undifferentiated myoblasts, when JNK/MAPK signaling is activated, it phosphorylates its downstream effector c-Jun. P-c-Jun enters into the nucleus, and together with Fos or ATF family proteins forms a demeric compound that called AP-1 that can could inhibit MyoD transcription, therefore, blocking differentiation. b In differentiated muscle, the activity of JNK/MAPK signaling is attenuated because its key factors are targeted by mamiRs (miR-1a-3p, 133a-3p, 133b-3p, 206-3p, 128-3p, and 351-5p), c-Jun is rarely phosphorylated, so forms less AP-1 compound in nucleus and turns on MyoD transcription, activating the transcription of mamiR genes. In turn, mature mamiRs further repress the JNK/MAPK signaling, forming a double-negative feedback regulatory network in skeletal muscle development
Among the mamiRs, miR-1a and miR-206 have been confirmed to promote muscle differentiation, while the exact roles of miR-133a and miR-133b in controlling skeletal muscle proliferation are controversial [25, 26, 51]. Here, by miRNA expression profiling and miRNA-target identification via PAR-CLIP, we confirmed that miR-133a and miR-133b can promote muscle differentiation by targeting the major effectors of the JNK/MAPK signaling pathway. Recently, a study has showed that miR-133 and miR-135 were competed by linc-MD1, resulting in regulating the expression of MAML1 and MEF2C, revealed a new regulatory circuitry in which RNAs can crosstalk with each other by competing for shared miRNAs [52]. Additionally, our data expanded the roles of miR-1a, miR-206, miR-128, and miR-351 in promoting muscle differentiation, and suggested that the role of these mamiRs is to coordinate regulatory networks during muscle differentiation.
In this study, we provided a series of evidence for the MAPK signaling pathway acting as the primary functional pathway during skeletal muscle differentiation. Initially, the activity of the JNK/MAPK signaling pathway was significantly downregulated not only during C2C12 cell differentiation, but also in developing mouse embryos muscle in vivo and primary skeletal muscle cell differentiation. And conversely, the activity of the P38/MAPK signaling pathway was elevated. Moreover, clustering the function of miRNA targets upon C2C12 cell differentiation showed enrichments in the phosphorus metabolic process and in the phosphate metabolic process, which is consistent with the fact that MAPK signaling is the main phosphorus pathway in physiology and pathology conditions. Importantly, our data revealed a dramatic downregulation in phosphorylated JNK protein but not in total JNK protein levels during this process. Mechanically, we illustrated that the core factors of the JNK/MAPK signaling were all targeted by mamiRs except for JNK itself. Furthermore, we demonstrated a mamiRs-MAPK-mamiRs autoregulatory loop that maintains the myogenic program. Therefore, we report a newly elucidated role of the miRNA complex in regulating signaling pathways, suggesting a new miRNA-signaling pathway regulatory pattern. Published studies have shown that JNK/MAPK and P38/MAPK signaling might have two regulatory patterns. On one hand, they may have cooperative effect on some conditions [53]. On the other hand, they may be entangled in an ever-lasting arms race during certain biological processes [7, 54]. Here, we presented antagonism of the two pathways, where the JNK/MAPK signaling repressed muscle differentiation, while the P38/MAPK signaling facilitated this process. And the balance between these two pathways was regulated by the mamiRs favors myogenesis (Supplementary Fig. S14). This pivotal regulatory mechanism could also be validated in the differentiation of rat myoblast L6 cells (Supplementary Fig. S15). Therefore, the JNK/MAPK and P38/MAPK signaling pathways might play crucial roles in other developmental and cellular systems, potentially extending the regulatory repertoire of miRNAs beyond skeletal muscle development.
Overall, we explored a mamiRs-MAPK axis that is responsible for skeletal muscle development. We believe our study provides a better understanding of the integration of genes regulation by multiple miRNAs during skeletal muscle differentiation, and reveals that convergent miRNAs control cell fate by controlling kinases cascades and influencing multiple phosphorylation processes cooperatively. Our study will hold promising potential for rationales of small RNAs-mediated signaling pathway activity for such development.
Materials and methods
Mice and animal care
C57/BL6 mice were purchased form the Guangdong Medical Laboratory Animal Center, Guangdong, China. Mice were housed in the animal facility and had free access to water and standard rodent chow. Fetal and postnatal hind limb muscles were isolated.
MuSCs sorting
MuSCs were FACS-sorted from hind limb muscles in postnatal 1-week mice according to the reported protocol [45]. Briefly, mechanical and enzymatic dissociation of cells, released resident mononucleated cells, then followed by antibody staining and FACS isolation.
Mononuclear cells were incubated with antibodies against CD45 (PE/Cy7-CD45, eBioscience, 25-0451-82), CD31 (FITC-CD31, BioLegend, 102506), Sca1 (V450-Sca1, BD, 560653) and VCAM1 (efluor660-VCAM1, eBioscience, 50-1061-80). Immunostained cells were briefly washed, passed through a 70-μM nylon mesh (Falcon) and suspended at a concentration of 106–107 cells per mL. Cells were further separated with the Beckman Coulter MoFlo XDP system. Sorting gates were strictly defined on the basis of control cells stained with single antibodies as well as the patterns of forward scatter and side scatter of MuSCs in preliminary tests. Collection of the cells that were positive for VCAM1 expression and negative for CD31, CD45, and Sca1.
Myoblast cell culture and differentiation
Mouse skeletal muscle C2C12 cells, rat muscle L6 cells [55, 56] were maintained in growth medium (GM), which consisted of Dulbecco’s minimal essential medium (DMEM; Life Technology) supplemented with 10% FBS (Life Technology). The sorted MuSCs were cultured in GM supplemented with 1 × penicillin–streptomycin and 2.5 ng/mL bFGF. To induce C2C12, L6 cells and MuSCs differentiation, the cells were first grown to 80–90% confluency, and the GM was subsequently replaced with differentiation medium (DM, which was DMEM plus 2% horse serum). Phenotypic differentiation was typically observed during 3 days of culturing the cell in DM. More significant myotube formation was observed after 5 days. To inhibit the activity of the JNK/MAPK or P38/MAPK signaling pathways, C2C12 cells were treated with 10 μM SP600125 or SB203580 (Sigma-Aldrich), respectively, and DMSO (Sigma-Aldrich) was used as the negative control.
For transfection, the cells were seeded in 6-well plates and were transfected using Lipofectamine 2000 (Life Technology) after the cells reached 30–40% confluency, according to manufacturer’s instructions. Briefly, RNA and Lipofectamine 2000 were diluted in Opti-MEM and incubated for 20 min. During this interval, the cells were seeded in 6-well plates, and the transfection mixture was added to the culture medium. After 72 h, the cells were harvested with Trizol (Life Technology) for RT-PCR or RIPA protein lysis buffer for western blot.
RNA extraction and qRT-PCR assays
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. qRT-PCR assays were performed as previously described [57]. U6 and GAPDH were employed as endogenous controls for miRNA and mRNA, respectively. The comparative Ct Method (ΔΔCT Method) was used to determine the expression levels of genes. The primers used for qRT-PCR are shown in Supplementary Table S6a.
Mimics
mirVanaTM miR-1a-3p, 133a-3p, 133b-3p, 206-3p, 128-3p, 351-5p Mimics (# 4464066), and mirVanaTM Inhibitors (# 4464084), as well as corresponding negative control mimic (# 4464058) and inhibitor (# 4464076) were obtained from Ambion. The sequence of RNA oligonucleotides are shown in Supplementary Table S7.
Western blot and antibodies
Protein samples were extracted from muscles or cells by using RIPA protein lysis buffer with protease inhibitor cocktail (Roche, 4693132001) and phosphatase inhibitor (Roche, 4906837001). The concentration of protein samples were determined by the BCA Protein Assay Kit (KeyGEN, China). Equal amounts of protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then were transferred to polyvinylidene difluoride membrane. After that, membranes were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature. Primary antibodies were incubated and a horseradish peroxidase-conjugated secondary antibody was followed. The primary antibodies used were as following: antibodies for phospho-JNK (Thr183/Tyr185, # 4671), JNK (# 9258), phospho-c-Jun (Ser63, # 9261), c-Jun (# 9165), phospho-MKK4 (Ser257/Thr261, # 9156), MKK4 (# 9152), MKK7 (# 4172), MEF2A (# 9736), MEF2C (# 5030), and GAPDH (# 2118) were obtained from Cell Signaling Technology. Antibodies for phospho-MKK7 (Ser271/Thr275, ab4762), MEKK1 (ab69533), MEKK2 (ab40805) were obtained from Abcam. Antibodies for MHC (MAB4470), MyoD (sc-760), and MyoG (sc-12732) were obtained from R&D, SantaCruz, and SantaCruz, respectively. Two Ago2 antibodies were obtained from Wako (018-22021) and Abnova (H00027161-M01). Normal mouse IgG was obtained from Abcam (ab13830). All proteins were visualized by Chemiluminescent HRP Substrate (Millipore, WBKLS0500) and chemical luminescence of membranes was detected by BioRad luminescent imaging system. Gray value of each image was quantified by gel-pro analyzer. Results were presented as mean ± SEM. An unpaired, two-tailed Student’s t-test was used for comparisons between two groups. The statistical significance of difference between two means was calculated with the t-test, *p < 0.05; **p < 0.01.
Vectors construction
To screen for the miRNAs that target MAPK signaling, psiCHECK2-Target-binding site luciferase reporter vectors were constructed. Briefly, we synthesized 58 nt DNA oligos containing the putative binding site for the chosen miRNAs and their mutant sequences, and then, these oligos were annealed and cloned into the psiCHECK2 vector (Promega). The genomic fragments encoding the miRNA precursors (~200 bp) were cloned into pcDNA6.2 (K4936-00, Invitrogen). The primers used for primers for vectors construction are shown in Supplementary Table S6b.
The Cignal 20-pathway reporter arrays, which including JNK/MAPK and P38/MAPK pathway reporters that measure the activity of the corresponding pathways, as well as standard negative control, were purchased from Qiagen (Qiagen, Hilden, Germany).
Lucierase reporter assays
For the analysis of the miRNA targets reporter assay and the Cignal 20-pathway reporter arrays, cells were transfected 24 h after seeding using the lipofectamine 2000 reagent (Invitrogen), then cultured for 2 days in growth medium for D0, another 96-well plate subsequently replaced with differentiation medium for 3 days for D3, cells were lysed in passive lysis buffer and luciferase activities were measured with a Dual Luciferase Assay Kit (Promega). When assaying the pathway activities during C2C12 and L6 cell differentiation, pathway reporters and standard negative control were transfected at a 50 ng/well dose in 96-well plates, For testing the influence of mamiRs on pathway activities, mamiRs plasmids were separately transfected C2C12 and L6 cells together with pathway reporters. MamiRs plasmids and pathway reporters at the dose of 100 ng/well and 100 ng/well in 96-well plates, respectively. All luciferase assays were repeated at least three times and done in triplicate each time.
Immunofluorescence
Cells cultured on coverslips were fixed with 4% paraformaldehyde and incubated with antibodies against MHC (R&D, MAB4470, 1:50) and p-c-Jun (CST, #9261, 1:100). Then, the cells were stained with secondary antibodies (Alexa Fluor® 594, donkey anti-mouse, Invitrogen, A-21203, 1:400) and nuclear counter-stained with Hoechst 33342 (Life Technologies, H1399). Fluorescent images were examined using a Zeiss 7 DUO NLO microscope (Leica Microsystems, Bannockburn, IL, USA).
Analysis of miRNA expression profiling by small RNA sequencing
Total RNA were extracted from C2C12 cells from three time point, proliferation (D0) and differentiation for 6 and 10 days (D6 and D10), and used for small RNA sequencing as previously described. Briefly, small RNA (sRNA) libraries were constructed using the Ion Total RNA-Seq Kit v2 (Life Technology), followed by sRNA-seq on an Ion Personal Genome MachineTM (PGMTM) System. Small RNAs were annotated by directly aligning to genome and various known RNAs using bowtie.
Ago2-PAR-CLIP and data analysis
The detailed CLIP experiments were performed according to the Argonaute HIST-CLIP method [33], but 4-thiouridine (4SU, 100 μM) was added and crosslinked under 365 nm UV light (0.15 J/cm2), as described [37]. There were some modifications to the original HITS-CLIP method as follows. Briefly, cells from four 155 cm2 dishes were used for a CLIP reaction with two Ago2 antibodies (15 μg Ab, 300 μL Protein G Dynabeads for each reaction), respectively. After alkaline phosphatase (Fermentas, EF0651) treatment, a 3′ adapter (IDT miRNA Cloning Linker 1) was ligated on-bead using truncated T4 RNA ligase 2 (NEB, M0242). After PNK treatment (with or without 32P-γ-ATP), the samples were run on a Novex 10% NuPAGE gel and then transferred onto a Protran™ nitrocellulose membrane (GE Healthcare, 0.2 µm). Following autorad exposure, 115–200 kDa regions were excised from the membrane for RNA isolation (samples without radiolabel were cut out according to the pre-stained molecular weight, Fermentas, SM0671). Next, 5′ RNA adapters (IDT, with different barcodes) were added to the recovered RNAs, and then the RNAs were reverse transcribed into cDNAs. After determining the best amplification cycles for each sample by qPCR, the final libraries were generated, recovered and sent to high-throughput sequencing. Bioinformatics analysis was performed as described [58]. Briefly, Ago-PAR-CLIP raw data were preprocessed with the FASTX-Toolkit v0.0.13 and mapped to the mouse genome using the Bowtie program. Overlapping reads were grouped to identify clusters that contained at least five reads and one T to C conversion location. The Ago-binding clusters were scanned to find miRNAs-target sites using the seed sequences of all known mouse miRNAs (mirBase v 20). Only miRNA-target sites with at least one T to C mutation were kept. All miRNA-target sites were further annotated using mouse transcripts (mm9). Functional enrichment analysis was performed using DAVID functional annotation tools [59]. The linkers and primers used for AGO-PAR-CLIP experiment are shown in Supplementary Table S8.
Electronic supplementary material
Acknowledgements
We thank Wen-Li Xu and Zi-Rui Liang for performing small RNA sequencing experiments. We thank Qiao-Juan Huang, Xiao-Hong Chen, and Yan Xu for their technical assistances. This work was supported by the National Natural Science Foundation of China (No. 31230042, 31471223, 81070589, and 31701116), the project of Science and Technology of Guangzhou (No. 201504010022) and the National Key R&D Program of China (No. 2017YFA0504400) from the Ministry of Science and Technology of China.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0063-1) contains supplementary material, which is available to authorized users.
References
- 1.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4:a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008;9:632–46. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- 3.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–38. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Brand-Saberi B. Genetic and epigenetic control of skeletal muscle development. Ann Anat. 2005;187:199–207. doi: 10.1016/j.aanat.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–52. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 7.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 8.Pearson G. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–83. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 9.Edmunds JW, Mahadevan LC. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J Cell Sci. 2004;117:3715–23. doi: 10.1242/jcs.01346. [DOI] [PubMed] [Google Scholar]
- 10.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Genet Dev. 2002;12:14–21. doi: 10.1016/S0959-437X(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 11.Jochen H. PAaMS-K. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 12.Andreucci JJ, Grant D, Cox DM, Tomc LK, Prywes R, Goldhamer DJ, et al. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J Biol Chem. 2002;277:16426–32. doi: 10.1074/jbc.M110891200. [DOI] [PubMed] [Google Scholar]
- 13.Bengal E, Raphael L, Scharfmann R, Dwarki VJ, T SJ, Weintraub H, Verma lM. Functional antagonism between c-Jun and MyoD proteins A direct physical association. Cell. 1992;68:507–19. doi: 10.1016/0092-8674(92)90187-H. [DOI] [PubMed] [Google Scholar]
- 14.Trouche D, Grigoriev M, Lenormand JL, Robin P, Leibovitch SA, Sassone-Corsi P, Harel-Bellan A. Repression of c-fos promoter by MyoD on muscle cell differentiation. Nature. 1993;363:79–82. doi: 10.1038/363079a0. [DOI] [PubMed] [Google Scholar]
- 15.Pedraza-Alva G, Zingg JM, Jost JP. AP-1 binds to a putative CAMP response element of the MyoD1 promoter and negatively modulates MyoD1 expression in dividing myoblasts. J Biol Chem. 1994;269(March 4):6978–85. [PubMed] [Google Scholar]
- 16.Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–79. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabrizchi A. jun N-terminal kinase mediates activation of skeletal muscle glycogen synthase by insulin in vivo. J Biol Chem. 1996;271:30765–73. doi: 10.1074/jbc.271.48.30765. [DOI] [PubMed] [Google Scholar]
- 18.Sabio G, Davis RJ. cJun NH2-terminal kinase 1 (JNK1): roles in metabolic regulation of insulin resistance. Trends Biochem Sci. 2010;35:490–6. doi: 10.1016/j.tibs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henstridge DC, Bruce CR, Pang CP, Lancaster GI, Allen TL, Estevez E, et al. Skeletal muscle-specific overproduction of constitutively activated c-Jun N-terminal kinase (JNK) induces insulin resistance in mice. Diabetologia. 2012;55:2769–78. doi: 10.1007/s00125-012-2652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Chen D, Zhang K, Yu B, Chen X, Meng J. Regulation of myostatin signaling by c-Jun N-terminal kinase in C2C12 cells. Cell Signal. 2007;19:2286–95. doi: 10.1016/j.cellsig.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–48. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–76. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–42. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 24.Xie S, Zhang Y, Qu L, Xu H. A Helm model for microRNA regulation in cell fate decision and conversion. Sci China Life Sci. 2013;56:897–906. doi: 10.1007/s11427-013-4547-4. [DOI] [PubMed] [Google Scholar]
- 25.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–52. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Et Biophys Acta. 2008;1779:682–91. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang MB, Xu H, Xie SJ, Zhou H, Qu LH. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE. 2011;6:e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–79. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, et al. A novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS ONE. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moresi V, Marroncelli N, Coletti D, Adamo S. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim Et Biophys Acta. 2015;1849:309–16. doi: 10.1016/j.bbagrm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–44. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–86. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–19. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley interdisciplinary reviews. RNA. 2012;3:159–77. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Xie S, Xu H, Qu L. CLIP: viewing the RNA world from an RNA-protein interactome perspective. Sci China Life Sci. 2015;58:75–88. doi: 10.1007/s11427-014-4764-5. [DOI] [PubMed] [Google Scholar]
- 41.Burattini S, Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development. Eur J Histochem. 2004;48:223–34. [PubMed] [Google Scholar]
- 42.Lluis F, Perdiguero E, Nebreda AR, Munoz-Canoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006;16:36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Bennett BL. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–6. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lassar AB, Thayer MJ, Overell RW, Weintraub H. Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell. 1989;58:659–67. doi: 10.1016/0092-8674(89)90101-3. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Cheung TH, Charville GW, Rando TA. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat Protoc. 2015;10:1612–24. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan LP, Seinen E, Duns G, de Jong D, Sibon OC, Poppema S, et al. A high throughput experimental approach to identify miRNA targets in human cells. Nucl Acids Res. 2009;37:e137. doi: 10.1093/nar/gkp715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucl Acids Res. 2006;34:W369–W73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bu HQ, Liu DL, Wei WT, Chen L, Huang H, Li Y, et al. Oridonin induces apoptosis in SW1990 pancreatic cancer cells via p53- and caspase-dependent induction of p38 MAPK. Oncol Rep. 2014;31:975–82. doi: 10.3892/or.2013.2888. [DOI] [PubMed] [Google Scholar]
- 49.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–54. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491–9. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 51.Feng Y, Niu LL, Wei W, Zhang WY, Li XY, Cao JH, et al. A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis. 2013;4:e934. doi: 10.1038/cddis.2013.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamoto E, Koshinaka K, Yoshimura T, Masuda H, Kawanaka K. Immobilization rapidly induces muscle insulin resistance together with the activation of MAPKs (JNK andp38) and impairment of AS160 phosphorylation. Physiol Rep. 2016;4:e12876. doi: 10.14814/phy2.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Chen L, Ko CI, Zhang L, Puga A, Xia Y. Distinct signaling properties of mitogen-activated protein kinase kinases 4 (MKK4) and 7 (MKK7) in embryonic stem cell (ESC) differentiation. J Biol Chem. 2012;287:2787–97. doi: 10.1074/jbc.M111.281915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iovino S, Oriente F, Botta G, Cabaro S, Iovane V, Paciello O, et al. PED/PEA-15 induces autophagy and mediates TGF-beta1 effect on muscle cell differentiation. Cell Death Differ. 2012;19:1127–38. doi: 10.1038/cdd.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar N, Shaw P, Uhm HS, Choi EH, Attri P. Influence of nitric oxide generated through microwave plasma on L6 skeletal muscle cell myogenesis via oxidative signaling pathways. Sci Rep. 2017;7:542. doi: 10.1038/s41598-017-00154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou AD, Diao LT, Xu H, Xiao ZD, Li JH, Zhou H, et al. beta-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/beta-catenin-signaling pathway. Oncogene. 2012;31:2968–78. doi: 10.1038/onc.2011.461. [DOI] [PubMed] [Google Scholar]
- 58.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBasev2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucl Acids Res. 2014;42(database issue):D92–D7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.