Abstract
Background
The rhesus macaque (RM, Macaca mulatta) is the most important nonhuman primate model in biomedical research. We present the first genomic survey of wild RMs, sequencing 81 geo-referenced individuals of five subspecies from 17 locations in China, a large fraction of the species’ natural distribution.
Results
Populations were structured into five genetic lineages on the mainland and Hainan Island, recapitulating current subspecies designations. These subspecies are estimated to have diverged 125.8 to 51.3 thousand years ago, but feature recent gene flow. Consistent with the expectation of a larger body size in colder climates and smaller body size in warmer climates (Bergman's rule), the northernmost RM lineage (M. m. tcheliensis), possessing the largest body size of all Chinese RMs, and the southernmost lineage (M. m. brevicaudus), with the smallest body size of all Chinese RMs, feature positively selected genes responsible for skeletal development. Further, two candidate selected genes (Fbp1, Fbp2) found in M. m. tcheliensis are involved in gluconeogenesis, potentially maintaining stable blood glucose levels during starvation when food resources are scarce in winter. The tropical subspecies M. m. brevicaudus showed positively selected genes related to cardiovascular function and response to temperature stimuli, potentially involved in tropical adaptation. We found 118 single-nucleotide polymorphisms matching human disease-causing variants with 82 being subspecies specific.
Conclusions
These data provide a resource for selection of RMs in biomedical experiments. The demographic history of Chinese RMs and their history of local adaption offer new insights into their evolution and provide valuable baseline information for biomedical investigation.
Keywords: Macaca mulatta, population genomics, adaptive selection, biomedical model
Introduction
Understanding how species evolve and adapt to their environments is an essential question in evolutionary biology. Rhesus macaques (RMs, Macaca mulatta) are, after humans, the world's most widely distributed primates [1–5], occupying a vast geographic distribution spanning from Afghanistan to the Chinese shore of the Pacific Ocean and south into Myanmar, Thailand, Laos, and Vietnam [5]. As the most widely distributed nonhuman primate species, RMs occupy diverse ecological landscapes and habitats, making them an interesting model to address questions about how species evolve and adapt to local environmental variation, including characterizing the genomic architecture of adaptation to habitat, climate, and other biotic and abiotic factors. Yet, despite much work on primate comparative genomics, very few population genomic studies have been carried out on wild RMs [6, 7]. Importantly, as RMs are widely used as a primate model in physiological, psychological, and cognitive studies [8–10], knowledge about their genomic architecture could improve and refine biomedical research [10] as the genomic composition of experimental animals can have a considerable influence on the outcome of experiments [11, 12]. Therefore, information on the genomic diversity of captive and wild RMs that could become a genomic resource for future utilization in medical research is essential.
In biomedical research, two main RM populations (Indian and Chinese) are recognized [6, 13]. They diverged from each other ∼162 thousand years ago (kya) and are characterized by extensive differences in morphology, behavior, ecology, physiology, reproduction, and disease progression [6, 13–19]. In 1978 India banned all RM exports to breeding centers across the world, thus curtailing the availability of wild Indian RMs and subsequently increasing the demand for Chinese RMs in biomedical research, thereby making a detailed characterization of genetic variants from Chinese RMs crucial for biomedical usage of this species.
Until recently, the genomes of 133 captive RMs from eight colonies have been sequenced; however, 124 of them are of Indian origin and only 9 individuals were presumed to be of Chinese origin [6]. In addition, Zhong et al. [7] reported genomic variation in 26 Chinese captive RMs identifying ∼46 million (M) single-nucleotide polymorphisms (SNPs). Nevertheless, most of the RM genetic variation known to date is limited to captive populations that may contain composite genotypes due of admixture among animals of different and unclear origin [20]. Here, we present the first attempt to survey the geo-referenced genomic diversity in wild Chinese RM populations, which is the largest extant population of the species. The current effective population size of Chinese and Indian RMs was estimated to be approximately 240,000 and 17,000, respectively, indicating that the Chinese RMs are likely to harbor substantially more genomic diversity compared to their Indian conspecifics [13]. Therefore, this population genomic survey of 81 RMs originating from 17 wild locations across China, including phylogenetic and demographic analyses as well as genome-wide selection scans, corresponds to the most comprehensive characterization of RM genetic diversity to date. The aim in this survey is to characterize the processes that lead to the extant patterns of variability as well as to identify the potential implications for the use of these populations in biomedical research.
Results and Discussion
Genetic diversity, phylogeny, and population structure
Blood and tissue samples from 79 wild-born RMs, representing five subspecies [21, 22], were collected at 17 sites in China (M. m. tcheliensis: TH; M. m. littoralis: AH, FJ, HB, GX, GZ; M. m. brevicaudus: HN; M. m. lasiotis: SX, SC1, SC2, SC3, SC4; M. m. mulatta: YN1, YN2, YN3, YN4, YN5; Fig. 1a and Supplementary Table S1). Genome sequences of two additional Chinese RMs (CR1 and CR2) were retrieved from the National Center for Biotechnology Information (NCBI) [9, 23, 24]. Resequencing was at a high average depth of 28.06 ± 5.08 × for 10 individuals and a moderate average depth of 9.98 ± 1.05 × for the remainder (n = 71), with an overall average genome coverage of 93.77% of the RM reference (Mmul_8.0.1; Supplementary Table S1). A total of 52,534,348 autosome SNPs were identified in these 81 wild Chinese RMs (Supplementary Table S2), and the nucleotide diversity measured by segregating sites (Watterson's θ, θW) and mean pairwise differences (θπ) were 0.00375 and 0.00247, respectively (Table 1). The number of SNPs (all positions with differences to the genome reference) per individual ranged from 7.0 to 9.2 M (mean of 8.50 M; Supplementary Fig. S1 and Supplementary Table S3). Among all detected SNPs, 8,171,139 were shared among all subspecies and 22,768,395 were shared by at least two subspecies, with the remaining SNPs confined to a single subspecies (Supplementary Fig. S2a). For each subspecies, the subspecies-specific SNPs (ssSNPs) ranged from 702,099 to 7,736,924 and the nonsynonymous ssSNPs ranged from 3,056 to 25,960 (Supplementary Fig. S2a, S2b). Among Chinese RM subspecies, M. m. mulatta had the highest heterozygosity (2.29 × 10−3 ± 3.24 × 10−5), followed by M. m. lasiotis (2.04 × 10−3± 1.40 × 10−4) and M. m. littoralis (2.00 × 10−3 ± 1.18 × 10−4). The lowest heterozygosity rates were found in M. m. brevicaudus (1.82 × 10−3 ± 1.28 × 10−4) and M. m. tcheliensis (1.46 × 10−3 ± 2.65 × 10−4) (Supplementary Fig. S3).
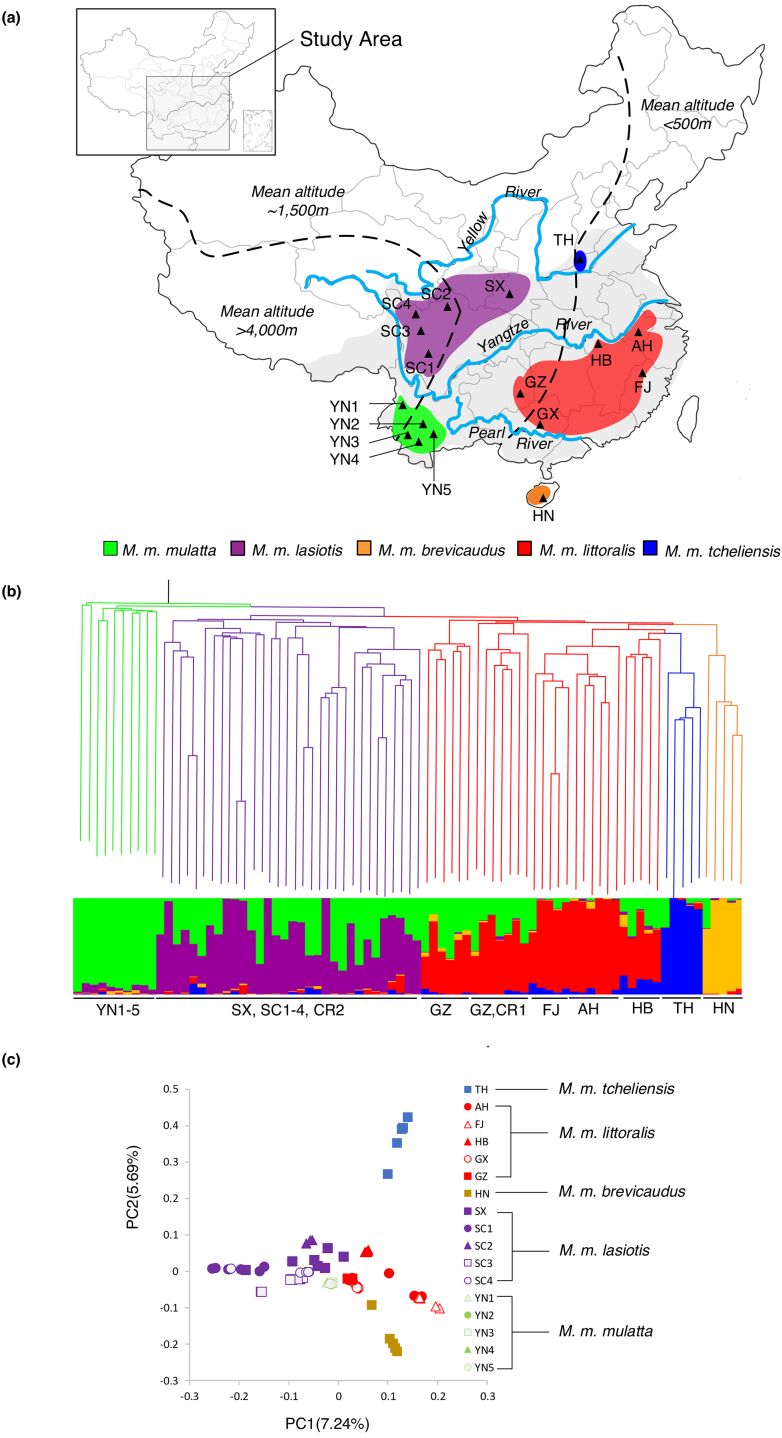
Figure 1:
Phylogeny and population genetic structure of 81 wild Chinese RMs. (a) Geographic distribution of RMs in China (gray shadow) and the 17 sampling sites along with their subspecies assignment. (b) Neighbor-joining (NJ) tree and clustering solution inferred using STRUCTURE and displaying five populations (inferred with Evanno's ∆K method; Supplementary Fig. S5). (c) Principal component analysis plots depicting the first two components (variance explained by PC1 = 7.24% and PC2 = 5.69%).
Table 1:
Genetic diversity (θ) and effective population size (Ne) in Chinese rhesus macaques based on segregating sites (θW) and nucleotide diversity (θπ)
| θW | θπ | |||||
|---|---|---|---|---|---|---|
| Sample size (n) | θ | N e | θ | N e | ||
| Chinese rhesus macaques (all samples) | 81 | 0.00375 | 93 750 | 0.00247 | 61 750 | |
| Subspecies | M. m. littoralis | 29 | 0.00313 | 78 250 | 0.00240 | 60 000 |
| M. m. tcheliensis | 5 | 0.00215 | 53 750 | 0.00230 | 57 500 | |
| M. m. brevicaudus | 5 | 0.00203 | 50 750 | 0.00207 | 51 750 | |
| M. m. lasiotis | 32 | 0.00298 | 74 500 | 0.00239 | 59 750 | |
| M. m. mulatta | 10 | 0.00303 | 75 750 | 0.00245 | 61 250 | |
We reconstructed a neighbor-joining (NJ) tree for Chinese RMs based on autosomal SNPs, using Indian RMs and M. sylvanus as outgroups (Fig. 1b and Supplementary Fig. S4). Individuals from M. m. lasiotis, M. m. brevicaudus, and M. m. tcheliensis form monophyletic lineages, respectively, while M. m. mulatta and M. m. littoralis are paraphyletic. Next, we performed a population structure analysis using STRUCTURE (version 2.3.4) [25], which estimates individual ancestry and admixture proportions assuming K ancestral populations. Plots of ΔK generated from STRUCTURE results indicated five genetic clusters present in the full dataset (Fig. 1b and Supplementary Fig. S5). A principal component analysis (PCA) corroborated the division of Chinese RMs into five groups. The first eigenvector separated M. m. mulatta and M. m. lasiotis from M. m. tcheliensis, M. m. littoralis, and M. m. brevicaudus (variance explained = 7.24%, Tracy-Widom P = 4.78 × 10−44), and the second eigenvector further separated M. m. tcheliensis, M. m. littoralis, and M. m. brevicaudus (variance explained = 5.69%, Tracy-Widom P = 4.21 × 10−27) (Fig. 1c, Supplementary Table S4). The division of Chinese RMs into five geographic lineages supports the former taxonomic division of Chinese RMs into five subspecies [21, 22]. M. m. mulatta (YN1–5) and M. m. lasiotis (SC1–4, SX) form the pan-western populations of Chinese RMs, with both subspecies inhabiting the montane Tibetan Plateau regions with an altitude ≥1500 meters above sea level in western China and separated from each other by the Yangtze River. M. m. littoralis (AH, FJ, HB, GX, GZ), M. m. tcheliensis (TH), and M. m. brevicaudus (HN) occur in the eastern coastal lowland of China and form the pan-eastern population. M. m. tcheliensis from the Taihang Mountains area is the northernmost (34°54’–35°16’ N; 112°02’–112°52’ E), while M. m. brevicaudus, restricted to Hainan Island, is the most southern Chinese RM subspecies.
Demographic and phylogeographic history
The estimated effective population sizes, based on θW and θπ, are approximately 93,750 and 61,750 for Chinese RMs (Table 1). In order to infer the ancient demographic history of Chinese RMs, we applied a pairwise sequential Markovian coalescent (PSMC) [26] analysis using 10 RM individuals with an average sequencing coverage depth higher than 20 × (one individual of M. m. tcheliensis and one of M. m. brevicaudus, two of M. m. lasiotis, three of M. m. littoralis, as well as three individuals of M. m. mulatta). The inferred PSMC trajectories were very similar for all analyzed individuals throughout most of the species’ history, reflecting the species’ cohesiveness (Fig. 2a). The ancient demographic history of RMs is marked by population fluctuations following the glacial periods during the Pleistocene [27]. Approximately 1200–800 kya all Chinese RMs experienced a population reduction at the time of the Xixiabangma Glaciation, followed by an expansion during the Mid-Pleistocene inter-glaciation (800–200 kya). This expansion was then interrupted by the Penultimate Glaciation (200–130 kya) when suitable habitat might have been lost leading to a population decline [27]. PSMC analyses also suggested that all the Chinese RMs had a population expansion during the last interglacial (around 100 kya) and a subsequent bottleneck during the Last Glaciation (LG; 70–10 kya) (Fig. 2a). Interestingly, the demographic inference by Xue et al. [6], derived from genomic data of a single Chinese RM (CH_37 945) from AH (M. m. littoralis), qualitatively resembled the demographic trajectory of M. m. littoralis presented herein.
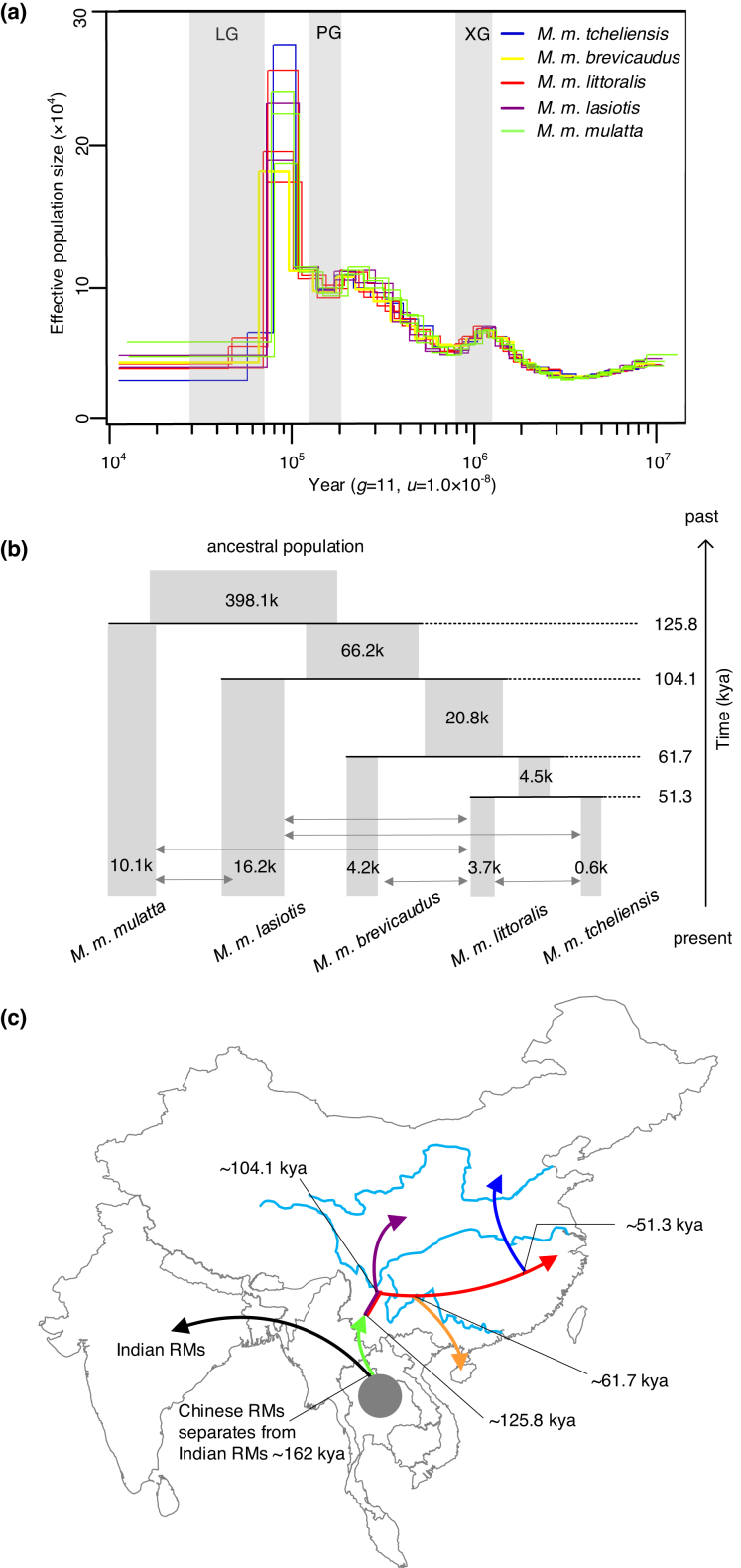
Figure 2:
Demographic history and differentiation scenarios of Chinese RMs. (a) Historical changes in effective population size reconstructed using the PSMC applied on individual whole genomes for each of the five subspecies. The generation length (g) and the neutral mutation rate per generation (μ) were assumed to be 11 years and 1.08 × 10–8, respectively. The Xixiabangma Glaciation (1200–800 kya), Penultimate Glaciation (200–130 kya), and Last Glaciation (70–10 kya) are shaded in gray. (b) Demographic history inferred by fastsimcoal2. The width of the gray bars and numbers on them indicate the estimated effective population size (all effective population sizes were converted to individuals). The arrows indicate migration rate between different subspecies. The detailed migration rates are listed in Supplementary Table S5. Numbers at the right show the divergence times between subspecies (all times were converted to years assuming a generation time of 11 years). (c) Biogeographic scenario for RMs. Chinese RMs separated from Indian RMs ∼162 kya [13], followed by further migration into China by the different RM subspecies indicated with arrows colored following the color key in Fig. 1a.
To further describe the divergence process among the five Chinese RM subspecies, we also employed the SVDquartes approach [28–31] that takes incomplete lineage sorting into account. The obtained phylogenetic tree suggests a “step-by-step” divergence of the five subspecies. Accordingly, the M. m. mulatta lineage diverged from that of the remaining Chinese RMs first, and then the M. m. lasiotis diverged from the ancestral lineage of pan-eastern RMs (M. m. tcheliensis, M. m. littoralis, and M. m. brevicaudus). Subsequently, M. m. brevicaudus diverged from the ancestor of M. m. tcheliensis and M. m. littoralis, the divergence of which occurred last (Supplementary Fig. S6). Under this step-by-step divergence scenario, we performed the joint site frequency spectrum (SFS)-based approach implemented in fastsimcoal2 [32] to model demographic fluctuations, respective divergence times, and gene flow events among the five RM subspecies. Following the divergence between the ancestral lineages of Indian and Chinese RMs (∼162 kya), the ancestor of M. m. mulatta diverged from the remaining Chinese RMs ∼125.8 kya (95% confidence interval [CI]: 92.0–162.1 kya) (Fig. 2b) [6, 13]. Subsequently, M. m. lasiotis diverged from the ancestral lineage of pan-eastern RMs ∼104.1 kya (95% CI: 50.2–154.5 kya) near the end of the last interglacial. The divergence time between M. m. brevicaudus and the ancestor of M. m. tcheliensis and M. m. littoralis was estimated at ∼61.7 kya (95% CI: 43.6–115.1 kya), while the divergence between the latter two occurred ∼51.3 kya (95% CI: 7.2–55.4 kya) during the last glacial maximum [33, 34]. Interestingly, the coalescence analysis revealed a large ancestral population size of the Chinese RMs 125.8 kya (95% CI: 92.0–162.1 kya) and a subsequent population decline and divergence among the five subspecies (Fig. 2b), which coincided with the population expansion during the last interglacial (around 100 kya) and the subsequent bottleneck of Chinese RMs during the LG (70–10 kya) revealed by PSMC analyses. Our results indicate that substantial gene flow occurred between all five extant lineages of Chinese RMs (Fig. 2b, Supplementary Table S5, and Supplementary Fig. S7).
A previous study of mitochondrial DNA identified two major haplogroups dividing Chinese RMs into a western and an eastern clade. Modern Chinese RMs were thought to have undergone a northward expansion while entering China via two possible routes: the first into the western mountains and the second following the eastern coast [35]. Our evolutionary model, however, suggests a step-by-step colonization process of RMs in China (Fig 2c). After the divergence from the Indian population (∼162 kya) [6, 13], the ancestor of Chinese RMs colonized the Tibetan Plateau from southwestern China and then experienced a range expansion north and eastward. The pan-western population (M. m. mulatta and M. m. lasiotis) inhabited the western montane region in China, while the pan-eastern population (M. m. tcheliensis, M. m. littoralis, and M. m. brevicaudus) entered the eastern coastal region. These five subspecies further diverged from each other during the bottleneck caused by the LG. Additionally, barriers such as the Yellow, Yangtze, and Pearl rivers and open sea (Fig. 1a) led to further differentiation by limiting gene flow among them. Water bodies and mountains could therefore be described as driving the formation of a habitat “lattice,” with the different subspecies of RMs occupying different grids in the lattice.
Signatures of selection and local adaptation
The wide distribution of Chinese RMs and their respective contrasting habitat types, as well as their wide use in biomedical studies, makes them an important case study for the analysis of signatures of local adaptation to divergent selective pressures [36–38]. We identified putative targets of selection by carrying out pairwise comparisons between RM subspecies inhabiting the most different environments to increase the chance of finding selection signatures, i.e., M. m. tcheliensis that occurs in the northernmost range of the species under cold conditions and M. m. brevicaudus that inhabits the southernmost range of the species, a tropical island. For each analysis, we compared the target subspecies (i.e. M. m. tcheliensis or M. m. brevicaudus) to the each of the other subspecies using the fixation index (FST) and genetic diversity (θπ), calculated on 50-kb-long sliding windows (Fig. 3 and Supplementary Figs. S8–S13). The top 5% of the windows with the largest FST and θπ ratios (θπ2/θπ1) in each pairwise comparison were considered to be potentially under positive selection. For each target subspecies, we identified potential selective-sweep regions as the intersection between the top 5% outliers in all pairwise comparisons (four pairwise comparisons in each case) (Supplementary Fig. S8). We used these consistent selective-sweep regions for further analyses, as they represent robust putative positively selected regions. The sizes of candidate selective-sweep regions ranged from 0.100 Mb to 11.075 Mb, and the number of genes located in these regions, which are expected to represent targets of selection for each subspecies, varied from 6 to 176 in different subspecies (Supplementary Table S6).
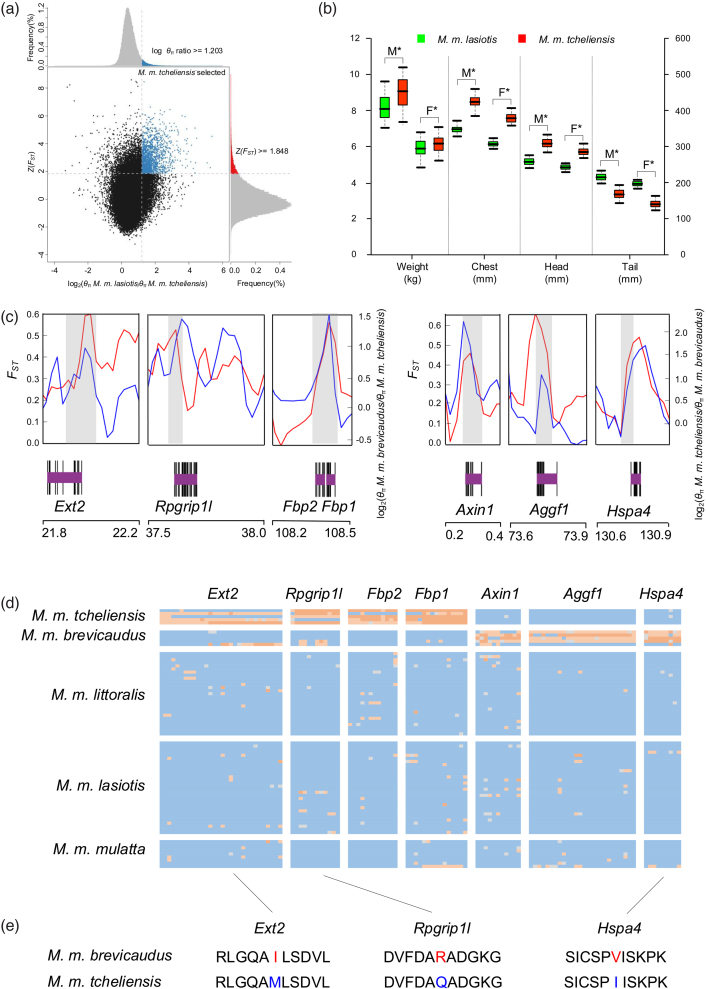
Figure 3:
Genomic regions with selection-sweep signals in RM. (a) Distribution of log2 (θπM. m. lasiotis/θπM. m. tcheliensis) and Z (FST) of 50-kb windows with 25-kb steps. Blue dots located in the selected regions requirement (corresponding to Z test P< 0.05, where Z (FST) ≥ 1.848 and θπ log-ratio ≥ 1.203) represent selected windows for M. m. tcheliensis. (b) Morphological comparison between M. m. tcheliensis and M. m. lasiotis. M and F represent males and females. (c) Example of genes with selection sweep signals. Ext2, Rpgrip1l, Fbp2, and Fbp1 in M. m. tcheliensis and Axin1, Aggf1, and Hspa4 in M. m. brevicaudus. FST and θπ log-ratios between the two subspecies are represented in red and blue, respectively. All values in Fig. 3c are plotted using 50-kb windows with half steps. Genome annotations are show at the bottom (black bar, coding sequences (purple bar, genes). (d) SNP genotypes in putative selective sweeps containing Ext2, Rpgrip1l, Fbp2, Fbp1, Axin1, Aggf1, and Hspa4. (e) Nonsynonymous variants in gene Ext2, Rpgrip1l and Hspa4.
M. m. tcheliensis from the Taihang Mountains area is the northernmost population of the species. The Taihang Mountains are characterized by a continental monsoon climate, and conditions for RMs are harsh during winter and early spring, with extreme cold temperatures of –14°C [39]. Food resources are limited and consist mainly of barks, twigs, roots of crops, and withered grass, thus, all sources are high in fiber but low in energy and nutritional value [40, 41]. Therefore, M. m. tcheliensis suffers from starvation due to food shortages during winter and early spring. In starvation, blood glucose levels are maintained by gluconeogenesis through which glucose is converted from other molecules, such as amino acids and lactic acid [42]. For M. m. tcheliensis, the positive-selection genes are enriched in the gene ontology (GO) term “fructose 1, 6-bisphosphate 1-phosphatase activity” with two genes (Fbp1, Fbp2; modified Fisher exact P = 1.90E-02; Fig. 3c, 3d; Supplementary Table S7). These two genes encode for fructose-1, 6-bisphosphatase 1 and fructose-1, 6-bisphosphatase isozyme 2, which catalyze the hydrolysis of fructose 1, 6-bisphosphate and play a rate-limiting role in gluconeogenesis. Furthermore, in starved zebrafish it was shown that the expression of Fbp1 was significantly upregulated in brain and liver tissues [43]. The positive-selection genes are also enriched in other terms and pathways related to gluconeogenesis, including the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway “fructose and mannose metabolism” (modified Fisher exact P = 4.35E-02) and GO terms “hexose biosynthetic process,” “monosaccharide biosynthetic process,” and “cellular carbohydrate biosynthetic process” (modified Fisher exact P = 3.36E-02, P = 4.64E-02, and P = 2.65E-02, respectively; Supplementary Table S7). Our findings suggest that the regulation of gluconeogenesis might be a mechanism of M. m. tcheliensis to adapt to food shortages in winter.
According to Bergman's rule, animals living in cold climates tend to have larger body sizes compared to their relatives in warm climates (i.e., they have a lower surface area to volume ratio), so they radiate less body heat per unit of mass [44]. Consistent with this expectation, among all RM subspecies, M. m. tcheliensis exhibits the largest body size and mass and the largest head and chest circumference (Fig. 3b and Supplementary Table S8) [40, 45]. Among the consistent signatures of positive selection identified in M. m. tcheliensis (176 genes), we found signatures of selective sweeps in eight genes linked to limb morphogenesis or skeletal system development (Supplementary Table S6). Among these genes, Fto and Rpgripl1 play an essential role in post-natal growth of mammals [46]. Mice lacking Fto completely display immediate post-natal growth retardation, with shorter body length, lower body weight, and lower bone mineral density, than control animals [47]. Furthermore, Sox5 and Sox6 (Fig. 3c, 3d) play an essential role in synovial joint morphogenesis via promoting both growth plate and articular chondrocyte differentiation [48]. Mutations in Atp6v0a4 could cause developmental delay and delayed closure of the anterior fontanelle in human [49], while expression of Ext2 enhances the bone formation in mice [50]. These genes, which are involved in the growth and development of the skeletal system and appendages, are likely contributors to the larger body size of M. m. tcheliensis and represent an undescribed adaptive pathway for primates living in colder climates.
In contrast, M. m. brevicaudus inhabits the tropical island of Hainan where it copes with a mean annual temperature of 24°C. M. m. brevicaudus has the smallest body size and the smallest body mass among RM subspecies [45]. As described above, they radiate more body heat per unit of mass (Bergman's rule) [44]. We found 127 putatively selected genes in M. m. brevicaudus (Supplementary Table S6), four of which were found to be enriched in GO term “bone morphogenetic protein (BMP) signaling pathway” (modified Fisher exact P = 4.65E-02; Supplementary Table S9), and two genes were found to be enriched in GO term “I-SMAD (inhibitory small mothers against decapentaplegic) binding" (modified Fisher exact P = 4.65E-02; Supplementary Table S9). BMP and I-SMAD signaling pathways are involved in the development of bones and the skeleton [51, 52]. Mutations in Axin1, a gene of the I-SMAD pathway, cause kinked tails in mice [53]. In M. m. brevicaudus, we found two nonsynonymous mutations in this gene (A674G, T656I) (Supplementary Fig. S14 and Supplementary Tables S10 and S11).
Additionally, putatively selected genes in M. m. brevicaudus (Fig. 3c, 3d and Supplementary Table S6) were also involved in GO terms related to cardiovascular system and blood circulation, e.g., Aggf1 related to GO term “blood vessel morphogenesis” and Ctnna3 related to GO term “regulation of heart rate by cardiac conduction.” The upregulated Aggf1 expression is capable of increasing blood flow in mouse hindlimb [54]. In addition, Hspa4, heat shock 70kDa protein 4, is directly involved in GO term “response to temperature stimulus.” We thus hypothesize that the cardiovascular system of M. m. brevicaudus might play an important role in stabilizing body temperature, assisted by blood flow through different body parts requiring good fluidity and vascular permeability to transfer heat out of the body [55]. Testing these hypotheses needs further functional assays; however, these genes, together with the positively selected genes identified in M. m. tcheliensis, are known to be relevant to human physical function and thus are likely of importance in the adaptation of Chinese RMs to different climate conditions.
Both coding and noncoding changes could contribute to local adaptations of organisms [56]. To further investigate the adaptive mechanism of M. m. tcheliensis and M. m. brevicaudus to the opposite climates (cold vs hot), we focused on SNPs in the gene regions of above-described candidate genes. A total of 5,817 SNPs were found with significant differences at the 5% level in the distributions of genotypes between these two subspecies, and 10 SNPs were nonsynonymous variants (Supplementary Tables S10 and S11). In M. m. tcheliensis, nonsynonymous mutations were found in the coding regions of Atp6v0a4 (R667Q), Ext2 (I363M), Fto (N10S), and Rpgrip1l (R1281Q) (Supplementary Table S11 and Supplementary Fig. S14), implying that selection might have acted on protein sequence changes. No nonsynonymous changes were detected in Fbp1, Fbp2, Sox5, and Sox6. However, SNPs are located in the 1 kb up/downstream, 5’ and 3’ UTR, and intronic regions of these genes (Supplementary Table S10), indicating selection on noncoding regulatory variants. In addition, nonsynonymous mutations in Aggf1 (H343Y), Axin1 (A674G, T656I), Hspa4 (I782V), and Ctnna3 (V551I, T577M) were revealed for M. m. brevicaudus (Supplementary Table S11 and Supplementary Fig. S14).
In addition to the genes related to the adaptation to various climate conditions, we also found signatures of positive selection in genes related to the nervous system. In M. m. tcheliensis, the 176 identified candidate genes are enriched in GO term “synapse” (modified Fisher exact P = 4.28E-02; Supplementary Table S7) with eight genes; two of these gene, Gabra2 and Chrm2, are associated with alcohol dependence [57]. For M. m. brevicaudus, 18 putatively selected genes related to nervous system development were found. For example, Dcc is reported to be required for long-term potentiation and memory [58]. Auts2, one of the eight putatively selected genes in M. m. lasiotis, has been shown to regulate neuronal migration, and mutations in this gene cause mental dysfunction in human [59] (Supplementary Table S6). Our findings suggest that RM subspecies have experienced different adaptive processes in the nervous system. Respective genomic differences should be taken into account when animals are selected for neurobiological research.
Disease-causing variants and implication for biomedical research
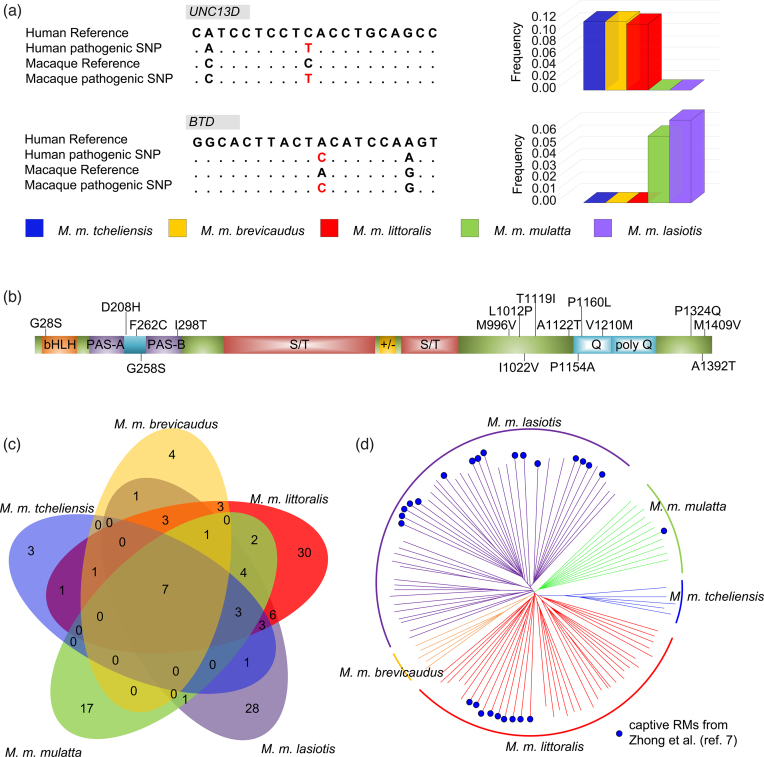
Given the large evolutionary similarity between macaques and humans, human diseases are better modeled in RMs than in many other animals. Thus, variants in RMs that match to orthologous human variants annotated as “pathogenic” are of particular interest. We examined presumed homologous Chinese RM SNPs in the human genome. A total of 34,850,330 RM SNPs analyzed in this study were successfully identified in the human genome (hg19). Among these SNPs, 118 [60] variants matched human variants with the accordant reference alleles, and alternative alleles were annotated as “disease causing” in HGMD or pathogenic in ClinVar. These 118 RM SNPs affect genes that cause specific human diseases including acromesomelic dysplasia maroteaux type, anonychia, atransferrinemia, blau syndrome, carcinoma of the colon, Charcot-Marie-Tooth disease, deafness, early infantile epileptic encephalopathy 7, glycogen storage disease, and others (Supplementary Table S12). Among these 118 SNPs, only 7 pathogenic SNPs are shared by all five subspecies, while 82 are subspecies specific (Fig. 4c, Supplementary Table S12). For example, the SNP rs116229331 in the gene Unc13d (human Chr17: 73836585C>T), known to cause juvenile idiopathic arthritis in humans [60], has an RM homologue (RM Chr16: 69559126 C>T, Fig. 4a) that is present in M. m. tcheliensis, M. m. brevicaudus, and M. m. littoralis but absent in M. m. lasiotis and M. m. mulatta. Another pathogenic variant (rs397514345, human Chr3: 15686724 A>C) in the Btd gene is involved in biotinidase deficiency [61]. Its homologous RM variant (RM Chr2: 172277927 A>C, Fig. 4a) is found only in M. m. lasiotis and M. m. mulatta. In addition, we also identified 16 nonsynonymous SNPs in the Noca3 gene, which encodes a protein that modulates the replication and transcriptional reactivation of human immunodeficiency virus type 1 (HIV-1) during virus latency [62] (Fig. 4b). Ten of these 16 nonsynonymous SNPs are private to one subspecies (Supplementary Table S13). The effects of these variants on HIV-1 replication and reactivation are unknown and need further investigation, but the high number of mutations suggests a complex response of the host to the virus.
Figure 4:
Population study of putative pathogenic SNPs in Chinese RM subspecies. (a) The site and frequency of pathogenic SNPs located in Unc13d and Btd genes. (b) Scheme of the Ncoa3 gene in RM. The positions of nonsynonymous polymorphisms (black) and three amino acid deletions (in red) are marked. (c) Private and shared pathogenic SNPs in Chinese RM subspecies (blue: M. m. tcheliensis; orange: M. m. brevicaudus; red: M. m. littoralis; green: M. m. mulatta; purple: M. m. lasiotis). The sizes of the areas are not proportional to the magnitude of the numbers. (d) NJ tree including the 81 Chinese RMs derived from this study. The 26 captive Chinese RMs from Zhong et al. [7] are indicated by blue dot.
Overall, these findings suggest that the genomic architecture of Chinese RMs used in biomedical research and their geographic origin could strongly influence the outcome of biomedical experiments and should be taken into account when using Chinese RMs in clinical and neurobiological research. Unfortunately, genome-wide screening of RMs used in biomedical research is, to date, only rarely conducted, and uncharacterized animals are most often used. Importantly, individuals from all five Chinese RM subspecies are used in biomedical research [63, 64]. Combined with our data, 9 of the 26 captive Chinese RMs reported by Zhong et al. [7] were found to cluster with M. m. littoralis, 16 with M. m. lasiotis, and 1 with M. m. mulatta (Fig. 4d). Thus, the data and results presented here provide the basis to trace the origin of captive RMs and to allow for the selection of appropriate animal models when testing for particular diseases and are thus a significant contribution to the “3 Rs” principle, which aims to reduce, refine, and replace experimental animals [65].
Conclusion
We present the first description of the evolutionary history and genomic variation of geo-referenced wild RMs throughout China, including scenarios on potential functions of this variation in adaptation to local environments. This genomic resource represents a valuable contribution to the understanding of the biology and evolution of a highly successful and important biomedical research species. In particular, it is important to note that due to the difference in evolutionary history of the subspecies identified here, it can be expected that animals originating from different regions may react differently to experimental tests, and thus their background needs to be assessed beforehand [10]. Our results highlight the importance that genome typing can play in biomedical research where animal origins are uncertain, and the resources generated here provide a baseline for genomic assessment of biomedical research populations and genetic resource conservation and for refined usage of RMs in future research.
Materials and Methods
Ethics statement
The methods were carried out in accordance with the approved guidelines of the Good Experimental Practices adopted by the Institute of Zoology, Chinese Academy of Sciences (CAS). All experimental procedures and animal collection were conducted under the supervision of the Committee for Animal Experiments of the Institute of Zoology (CAS).
Sample collection and sequencing
Samples from 79 individuals with information about geographic origin were collected from 17 local wildlife rescue centers, which covered most of the species’ range in China. Muscle samples were collected from deceased individuals, and the blood samples were taken during routine physical examinations. Total genomic DNA was extracted from blood or tissue samples using standard phenol/chloroform methods. For each individual, ∼3 μg DNA was sheared into fragments of 500 bp with the Covaris system. DNA fragments were then processed and sequenced using the Illumina HiSeq 2000 and 2500 platforms. Furthermore, published genomic data for two individuals were downloaded from NCBI [9, 23] and filtered using the same conditions. Raw reads were first filtered with the following criteria: reads with unidentified nucleotides (N) exceeded 10% were discarded and reads with the proportion of low-quality base (phred quality ≤ 5) larger than 50% were discarded. After quality control, 3,095.6 Gb of high-quality sequences with 22.53 billion pair-end reads (100 or 125 bp) were generated.
Sequence data preprocessing and variant calling
High-quality sequence reads were mapped to the macaque reference genome, Mmul_8.0.1 [66], using the Burrows–Wheeler Aligner 0.7.10-r789 (BWA, RRID:SCR_010910) [67]. Sequence Alignment/Map (SAM) format files were imported to SAMtools v0.1.19 (SAMtools, RRID:SCR_002105) [68] for sorting and then imported to Picard, version 1.118 (Picard, RRID:SCR_006525) [69] for removing duplicated reads. To improve the quality of sites reported, we performed SNP calling following GATK's best practice, version 3.3–0 (GATK, RRID:SCR_001876) on autosomal sites only [70]. We got the GVCF file for each individual using the “HaplotypeCaller” method in GATK and then using the GenotypeGVCFs-based method with the “-includeNonVariantSites” flag to get the population VCF file, including all the confident sites. After that, we applied the “SelectVariants” to exclude the Indel and split the variant and nonvariant sites. Then, we applied the hard filter command “VariantFiltration” to exclude potential false-positive variant calls with the following criteria: “-filterExpression QD < 5.0 ‖ FS > 60.0 ‖ MQ < 40.0 ‖ ReadPosRankSum< -8.0 || MQRankSum < -12.5” and “–genotypeFilterExpression DP < 4.0”. Additionally, the sites are filtered out if there is an “N” in the reference sequence or the site is fixed difference to the reference genome or the site including more than 20% missing genotypes. For nonvariant sites, we filtered the sites if there is an “N” in the reference sequence or if the site is including more than 20% missing genotypes. All the SNPs were annotated by ANNOVAR v2013–06-21 (ANNOVAR, RRID:SCR_012821) [71] (Supplementary Table S2). For each individual, the heterozygosity was calculated as the heterozygous SNP rate across the whole genome based on the whole number of sites that are callable (Supplementary Table S3).
Genetic diversity and structure analysis
An NJ tree was constructed for the 81 individuals based on the autosomal genome data using the software TreeBeST. The bootstrap was set to 1,000 times to assess branch support, with the genome information of Indian RMs and M. sylvanus as outgroups. FigTree ([72], v1.4.0) was used to visualize the phylogenetic tree (Fig. 1b and Supplementary Fig. S4). Population structure analysis was performed using the software STRUCTURE 2.3.4 [25], which estimates individual ancestry and admixture proportions assuming K ancestral populations. We ran STRUCTURE five times to assess convergence and tested the number of genetic clusters (K) from 2–9 (Supplementary Fig. S5). We also carried out a PCA using the smartPCA program from the Eigensoft package, v5.0 (Eigensoft, RRID:SCR_004965) [73]. To determine the significance level of principal components, a Tracy-Widom test was done after the PCA (Supplementary Table S4). Decay of linkage disequilibrium against physical distance for the different populations was calculated using the Haploview software [74], with the maxdistance set as 500 kb (Supplementary Fig. S15).
Demographic and divergence inference using PSMC and fastsimcoal2
We called the consensus sequences using Samtools mpileup [68] by applying: “samtools mpileup -q 1 -C 50 -S -D -m 2 -F 0.002 -u -f *.fa(genome) *.bam | bcftools view -c—| vcfutils.pl vcf2fq -d 10 -D 100 -Q 20 - > *.psmc.fq” and “fq2psmcfa -q10 -s 100 *.psmc.fq >*.psmc.fa.” To ensure the quality of consensus sequences, we used data of 10 individuals with an average coverage >20 × (22.20–34.32 ×). The PSMC model [26] was used to estimate the population histories from the individual genomes (sex chromosomes excluded) with the following parameters: –N30 –t15 –r5 –p “4+25 × 2+4+6.” We chose a generation length of 11 years and a mutation rate per generation (μ) of 1.0 × 10−8 (for the rationale to use these values, see [6, 75]).
We used PAUP* 4.0a142 (PAUP, RRID:SCR_014931) [30] to run SVDquartets to estimate the branching pattern among the five subspecies with the following command: SVDQuartets SpeciesTree = yes bootstrap evalQuartets = all seed = 0 nthreads = 40. The joint SFS approach implemented in fastsimcoal2 [32] was performed to model more recent demographic fluctuations and respective divergence times based on the species tree estimation by SVDquartets. A VCF file containing callable variant sites was converted to fastsimcoal style folded SFS. To mitigate the effect of linkage disequilibrium, we filtered out the SNPs located within 10 kb from genes and then we took one SNP every 10 kb randomly. The multidimensional folded SFS for all five subspecies was generated with easySFS [76]. During the likelihood calculation, a conditional maximization algorithm (ECM) is used to maximize the likelihood of each parameter while keeping the others stabilized. This ECM procedure runs through 40 cycles where each composite likelihood was calculated using 100,000 coalescent simulations. Additionally, in order to avoid likelihood estimates that oversample parameter values at local maxima across the composite likelihood surface, we ran 50 replicates with each starting from different initial conditions. We chose the replicate with the highest estimated maximum likelihood score to estimate confidence intervals using parametric bootstrapping. The SFS used in the bootstrap was simulated with the parameter values from the highest likelihood model and then new parameter values were re-estimated from the simulated SFS. We ran 100 parametric bootstraps (Supplementary Fig. S7).
Positive selection
To identify genomic regions that may have been subject to selection for each subspecies in different habitats, we scanned the genome using one-to-one pairwise comparisons between all five subspecies. We calculated the genome-wide distribution of FST values [77] and θπ ratios for each pairwise comparison among five RM subspecies. We calculated θπ for each population and the FST between the two populations in each comparison using VCFtools (VCFtools, RRID:SCR_001235) [78] with a genome-wide sliding window strategy (50 kb in length with 25-kb step). The FST values were Z-transformed, and the log value of θπ ratio (θπ2/θπ1) was estimated. Candidate regions under positive selection were extracted based on the top 5% of log-odds ratios for both Z (FST) and log (θπ –ratio). Finally, for each subspecies, we used the intersection of putatively selected regions generated by all the pairwise comparisons with other subspecies as the candidate regions under positive selection (i.e., consistent signatures of selective sweeps). Genes located in these regions are expected to represent targets of selection. Functional classification and enrichment analysis of GO categories and KEGG pathways for these candidate genes were performed using DAVID v6.8 (DAVID, RRID:SCR_001881) [79]. The modified Fisher exact P value cutoff was 0.05. Chi-square and P values for the allele frequencies in M. m. tcheliensis vs M. m. brevicaudus for the resequenced SNPs from the candidate genes were assessed with the Haploview program [74].
Genomic divergence and implication for biomedical research
A total of 118 of 52,534,348 RM SNPs analyzed in this study were successfully mapped to human reference sequence version hg19 (GRCh37) using liftOver [80] and were annotated as “disease causing” in HGMD (version 2015.1) or pathogenic in ClinVar (downloaded 25/02/2018) (Supplementary Table S12).
Availability of supporting data
All data generated from this study have been submitted to the NCBI Sequence Read Archive under BioProject PRJNA345528. The datasets supporting the results of this article are available in the GigaScience GigaDB repository [81].
Additional files
Supplementary Fig. S1. Variant number for the 81 individuals in this study.
Supplementary Fig. S2. Private and shared SNPs per Chinese RM subspecies.
Supplementary Fig. S3. Heterozygosity per base-pair for five subspecies of Chinese RMs.
Supplementary Fig. S4. Neighbor-joining tree derived from 1000 bootstrap replicates.
Supplementary Fig. S5. Plots of ΔK generated from STRUCTURE results.
Supplementary Fig. S6. The species tree based on SVDquartets+PAUP*.
Supplementary Fig. S7. Confidence Intervals from 100 parametric bootstraps for inferred demographic parameters.
Supplementary Fig. S8. Positive selection flow chart for each subspecies.
Supplementary Fig. S9. Distribution of Z(FST) and θπ log ratio of 50kb windows with 25kb sliding window for M. m. tcheliensis.
Supplementary Fig. S10. Distribution of Z(FST) and θπ log ratio of 50kb windows with 25kb sliding window for M. m. brevicaudus.
Supplementary Fig. S11. Distribution of Z(FST) and θπ log ratio of 50kb windows with 25kb sliding window for M. m. littoralis.
Supplementary Fig. S12. Distribution of Z(FST) and θπ log ratio of 50kb windows with 25kb sliding window for M. m. lasiotis.
Supplementary Fig. S13. Distribution of Z(FST) and θπ log ratio of 50kb windows with 25kb sliding window for M. m. mulatta.
Supplementary Fig. S14. Non-synonymous variants in putatively selected genes.
Supplementary Fig. S15. Linkage disequilibrium pattern of the five Chinese RM subspecies.
Supplementary Table S1. Overview of sample information and sequencing statistics.
Supplementary Table S2. Distribution of autosomal SNPs within various genomic regions of RM.
Supplementary Table S3. Identified SNPs and heterozygosity for 81 individuals from 17 sampling locations.
Supplementary Table S4. Tracy-Widom (TW) statistics and P values for the ten first eigenvalues in PCA.
Supplementary Table S5. Inferred demographic parameters with 95% confidence intervals for fastsimcoal2 model.
Supplementary Table S6. List of positively selected genes in the five Chinese RM subspecies.
Supplementary Table S7. Enrichment of genes under selective sweep in M. m. tcheliensis.
Supplementary Table S8. Morphological differences between the five investigated Chinese RM subspecies.
Supplementary Table S9. Enrichment of the genes under selective sweep in M. m. brevicaudus.
Supplementary Table S10. Distribution of SNPs in the selected genes described in the part of “Signatures of selection and local adaptation”.
Supplementary Table S11. Non-synonymous SNPs with significant differences at the 5% level in the distributions of genotypes between M. m. tcheliensis and M. m. brevicaudus.
Supplementary Table S12. List of RM variants scored by HGMD and ClinVar as “disease causing” or “pathogenic”.
Supplementary Table S13. Population study for Ncoa3 reveals multiple genotypes.
Abbreviations
BMP: bone morphogenetic protein CI: confidence interval GO: Gene Ontology HIV-1: human immunodeficiency virus type 1 I-SMAD: inhibitory small mothers against decapentaplegic KEGG: Kyoto Encyclopedia of Genes and Genomes kya: thousand years ago LG: Last Glaciation M: million NCBI: National Center for Biotechnology Information NJ: neighbor-joining PCA: principal component analysis PSMC: pairwise sequential Markovian coalescent RM: rhesus macaque SAM: Sequence Alignment/Map SFS: site frequency spectrum SNP: single-nucleotide polymorphism ssSNP: subspecies-specific single-nucleotide polymorphism
Funding
This project was sponsored by grants to the following: M.L. (National Natural Science Foundation of China, 31530068 and 31821001; Strategic Priority Research Program of the Chinese Academy of Sciences, XDB31000000 and XDA19050202; National Key R&D Program of China, 2016YFC0503200; and Science & Technology Department of Sichuan Province, 2018JZ0008); Z.L. (Natural Science Foundation of China, 31471989).
Competing interests
The authors declare that they have no competing interests.
Author contributions
M.L., Z.L., and M.B. conceived the study and designed the project. Z.L., X.T., P.O., X.Z., L.Z., and S.T. managed the project, performed the analyses, and wrote the manuscript. Z.L., B.S., and H.X. prepared samples. Z.L., X.T., and P.O. performed genetic analyses. Z.L., X.T., P.O., B.R., L.Z., G.L., Z.Y., Z.P., Z.X., C.R., M.B., and M.L. discussed the data. Z.L. and X.T. wrote the manuscript with contributions from P.O., B.W., H.X., W.Z., C.R., M.B., and M.L.. All authors contributed to data interpretation.
Supplementary Material
12/27/2017 Reviewed
04/19/2018 Reviewed
01/04/2018 Reviewed
04/15/2018 Reviewed
07/12/2018 Reviewed
ACKNOWLEDGEMENTS
The authors thank Baoguo Li, Meng Yao, Songtao Guo, Jiqi Lu, Zhenlong Wang, Xuelong Jiang, Tao Meng and Qihai Zhou for their help in sampling and Daniel Pitt, Quan Kang, Qi Wu, Chuanyun Li and Qi Pan for their assistance in data analysis.
References
- 1. Moreno-Estrada A, Gignoux CR, Fernández-López JC et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2013;344:1280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allentoft ME, Sikora M, Sjögren KG et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–72. [DOI] [PubMed] [Google Scholar]
- 3. Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2504 human genomes. Nature. 2015;526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maestripieri D. Macachiavellian Intelligence: How Rhesus Macaques and Humans Have Conquered the World. Chicago: The University of Chicago Press; 2007. [Google Scholar]
- 5. Zinner D, Fickenscher GH, Roos C. Family cercopithecidae (Old World Monkeys). In: Mittermeier RA, Rylands AB, Wilson DE, eds. Handbook of the Mammals of the World. Vol. 3 Primates. Barcelona: Lynx Edicions; 2013. pp. 550–753. [Google Scholar]
- 6. Xue C, Raveendran M, Harris RA, et al. The population genomics of rhesus macaques (Macaca mulatta) based on whole genome sequences. Genome Res. 2016;26:1651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong X, Peng J, Shen QS et al. RhesusBase PopGateway: genome-wide population genetics atlas in rhesus Macaque. Mol Biol Evol. 2016;33:1370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fawcett GL, Raveendran M, Deiros DR et al. Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta). BMC Genomics. 2011;12:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan G, Zhang G, Fang X et al. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nature Biot. 2011;29:1019–23. [DOI] [PubMed] [Google Scholar]
- 10. Haus T, Ferguson B, Rogers J, et al. Genome typing of nonhuman primate models: implications for biomedical research. Trends Genet. 2014;30:482–7. [DOI] [PubMed] [Google Scholar]
- 11. Flynn S, Satkoski J, Lerche N et al. Genetic variation at the TNF-alpha promotor and malaria susceptibility in rhesus (Macaca mulatta) and long-tailed (Macaca fascicularis) macaques. Infect Genet Evol. 2009;9:769–77. [DOI] [PubMed] [Google Scholar]
- 12. de Groot NG, Heijmans CMC, Koopman G et al. TRIM5 allelic polymorphism in macaque species/populations of different geographic origins: its impact on SIV vaccine studies. Tissue Antigens. 2011;78:256–62. [DOI] [PubMed] [Google Scholar]
- 13. Hernandez RD, Hubisz MJ, Wheeler DA, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–3. [DOI] [PubMed] [Google Scholar]
- 14. Champoux M, Higley JD, Suomi SJ. Behavioral and physiological characteristics of Indian and Chinese-Indian hybrid rhesus macaque infants. Dev Psychobiol. 1997;31:49–63. [PubMed] [Google Scholar]
- 15. Trichel AM, Rajakumar PA, Murphey-Corb M. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J Med Primatol. 2002;31:171–8. [DOI] [PubMed] [Google Scholar]
- 16. Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–35. [DOI] [PubMed] [Google Scholar]
- 17. Smith DG. Genetic characterization of Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta). Comp Med. 2005;55:227–30. [PubMed] [Google Scholar]
- 18. Ferguson B, Street SL, Wright H et al. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta). BMC Genomics. 2007;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubisch HM, Falkenstein KP, Deroche CB, et al. Reproductive efficiency of captive Chinese- and Indian-origin rhesus macaque (Macaca mulatta) females. Am J Primatol. 2012;74:174–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanthaswamy S, Johnson Z, Trask JS et al. Development and validation of a SNP-based assay for inferring the genetic ancestry of rhesus macaques (Macaca mulatta). Am J Primatol. 2014;76:1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fooden J. Systematic review of the rhesus macaque, Macaca mulatta (Zimmermann, 1780). Field Zool. 2000;96:1–180. [Google Scholar]
- 22. Jiang X, Wang Y, Ma S. Taxonomic revision and distribution of subspecies of rhesus monkey (Macaca mulatta) in China. Zool Res. 1991;12:241–7. [Google Scholar]
- 23. Fang X, Zhang Y, Zhang R, et al. Genome sequence and global sequence variation map with 5.5 million SNPs in Chinese rhesus macaque. Genome Biol. 2011;12:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prado-Martinez J, Sudmant PH, Kidd JM, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. [DOI] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng B, Xu Q, Shen Y. The relationship between climate change and Quaternary glacial cycles on the Qinghai–Tibetan Plateau: review and speculation. Quatern Int. 2002;97:93–101. [Google Scholar]
- 28. Chifman J, Kubatko L. Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. J Theor Biol. 2014;374:35–47. [DOI] [PubMed] [Google Scholar]
- 29. Chifman J, Kubatko L. Quartet inference from SNP data under the coalescent model. Bioinformatics. 2014;30:3317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swofford D, et al. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- 31. Song S, Liu L, Edwards SV et al. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci U S A. 2012;109:14942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Excoffier L, Dupanloup I, Huerta-Sánchez E, et al. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Owen LA, Finkel RC, Caffee MW. A note on the extent of glaciation throughout the Himalaya during the global Last Glacial Maximum. Quaternary Sci Rev. 2002;21:147–57. [Google Scholar]
- 34. Owen LA. Latest Pleistocene and Holoene glacier fluctuations in the Himalaya and Tibet. Quaternary Sci Rev. 2009;28:2150–64. [Google Scholar]
- 35. Wu S, Luo J, Li Q et al. Ecological genetics of Chinese rhesus macaque in response to mountain building: all things are not equal. PLoS One. 2013;8:e55315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yi X, Liang Y, Huerta-Sanchez E, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhatia G, Patterson N, Pasaniuc B et al. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011;89:368–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao SC, Zheng PP, Dong SS et al. Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet. 2013;45:67–71. [DOI] [PubMed] [Google Scholar]
- 39. Tian JD, Wang ZL, Lu JQ, et al. Reproductive parameters of female Macaca mulatta tcheliensis in the temperate forest of mount Taihangshan, Jiyuan, China. Am J Primatol. 2013;75:605–12. [DOI] [PubMed] [Google Scholar]
- 40. Zhao X, Zhang H, Lv X et al. Survey and research of morphological characters of monkeys (Macaca mulatta) in the Taihang Mountains. J Henan Nor Uni. 1989;62:120–5. [Google Scholar]
- 41. Lu JQ, Hou JH, Wang HF et al. Current status of Macaca mulatta in Taihangshan Mountains Area, Jiyuan, Henan, China Int J Primatol. 2007;28:1085–91. [Google Scholar]
- 42. Sadava DE, Heller HC, Orians GH, et al. Life: The Science of Biology, 8th edn New York: Macmillan; 2008. [Google Scholar]
- 43. Drew RE, Rodnick KJ, Settles M, et al. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio). Physiol Genomics. 2008;35:283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bergmann C. Über die Verhältnisse der Wärmeökonomie der Thiere zu ihrer Grösse. Göttinger Studien. 1847;3:595–708. [Google Scholar]
- 45. Zhang P, Lyu MY, Wu CF, et al. Variation in body mass and morphological characters in Macaca mulatta brevicaudus from Hainan, China. Am J Primatol. 2016;78:679–98. [DOI] [PubMed] [Google Scholar]
- 46. Jevsinek Skok D, Kunej T, Kovac M et al. FTO gene variants are associated with growth and carcass traits in cattle. Animal Genetics. 2016;47:219–22. [DOI] [PubMed] [Google Scholar]
- 47. Gao X, Shin YH, Li M, et al. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dy P, Smits P, Silvester A et al. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev Biol. 2010;341:346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greally MT, Kalis NN, Agab W et al. Autosomal recessive cutis laxa type 2A (ARCL2A) mimicking Ehlers‐Danlos syndrome by its dermatological manifestations: report of three affected patients. Am J Med Genet A. 2014;164A:1245–53. [DOI] [PubMed] [Google Scholar]
- 50. Morimoto K, Shimizu T, Furukawa K, et al. Transgenic expression of the EXT2 gene in developing chondrocytes enhances the synthesis of heparan sulfate and bone formation in mice. Biochem Biophys Res Commun. 2002;292:999–1009. [DOI] [PubMed] [Google Scholar]
- 51. Salazar VS, Gamer LW, Rosen V. BMP signaling in skeletal development, disease and repair. Nat Rev Endocrinology. 2016;12:203–21. [DOI] [PubMed] [Google Scholar]
- 52. Bragdon B, Moseychuk O, Saldanha S et al. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609–20. [DOI] [PubMed] [Google Scholar]
- 53. Ruvinsky A, Flood WD, Costantini F. Developmental mosaicism may explain spontaneous reappearance of the AxinFu mutation in mice. Genesis. 2001;29:49–57. [DOI] [PubMed] [Google Scholar]
- 54. Lu QL, Yao YH, Yao YF et al. Angiogenic factor AGGF1 promotes therapeutic angiogenesis in a mouse limb ischemia model. PLoS One. 2012;7:e46998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. González-Alonso J. Human thermoregulation and the cardiovascular system. Exp Physiol. 2012;97:340–6. [DOI] [PubMed] [Google Scholar]
- 56. Meadows JRS, Lindblad-Toh K. Dissecting evolution and disease using comparative vertebrate genomics. Nat Rev Genet. 2017;18:624–36. [DOI] [PubMed] [Google Scholar]
- 57. Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151–7. [DOI] [PubMed] [Google Scholar]
- 58. Horn KE, Glasgow SD, Gobert D et al. DCC expression by neurons regulates synaptic plasticity in the adult brain. Cell Rep. 2010;31:173–85. [DOI] [PubMed] [Google Scholar]
- 59. Hori K, Hoshino M. Neuronal migration and AUTS2 syndrome. Brain Sci. 2017;7:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hazen MM, Woodward AL, Hofmann I, et al. Mutations of the hemophagocytic lymphohistiocytosis-associated gene UNC13D in a patient with systemic juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:567–70. [DOI] [PubMed] [Google Scholar]
- 61. Procter M, Wolf B, Mao R. Forty-eight novel mutations causing biotinidase deficiency. Mol Genet Metab. 2016;117:369–72. [DOI] [PubMed] [Google Scholar]
- 62. Munier S, Delcroix-Genete D, Carthagena L, et al. Characterization of two candidate genes, NCoA3 and IRF8, potentially involved in the control of HIV-1 latency. Retrovirology. 2005;2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fan ZY, Song YL. Chinese primate status and primate captive breeding for biomedical research in China. In: Institute for Laboratory Animal Research, National Research Council. International Perspectives: The Future of Nonhuman Primate Resources. Washington, DC: National Academy Press; 2003. [Google Scholar]
- 64. Xin H. Monkey research in China: developing a natural resource. Cell. 2007;129:1033–6. [DOI] [PubMed] [Google Scholar]
- 65. Zhou Q. Balancing the welfare: the use of non-human primates in research. Trends Genet. 2014;30:476–8. [DOI] [PubMed] [Google Scholar]
- 66. Gradnigo JS, Majumdar A, Norgren RB Jr, et al. Advantages of an improved rhesus macaque genome for evolutionary analyses. PLoS One. 2016;11:e0167376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li H, Handsaker B, Wysoker A et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. http://broadinstitute.github.io/picard/ [Google Scholar]
- 70. Mckenna A, Hanna M, Banks E et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164, http://annovar.openbioinformatics.org/en/latest/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Figtree http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- 73. Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 75. Ségurel L, Wyman M J, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. [DOI] [PubMed] [Google Scholar]
- 76.EasySFS https://github.com/isaacovercast/easySFS#easysfs [Google Scholar]
- 77. Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70. [DOI] [PubMed] [Google Scholar]
- 78. Danecek P, Auton A, Abecasis G et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 80.hgLiftOver https://genome.ucsc.edu/cgi-bin/hgLiftOver [Google Scholar]
- 81. Liu Z, Tan X, Orozco-terWengel P et al. Supporting data for “Population genomics of wild Chinese rhesus macaques reveals a dynamic demographic history and local adaptation, with implications for biomedical research.”. GigaScience Database. 2018. 10.5524/100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
12/27/2017 Reviewed
04/19/2018 Reviewed
01/04/2018 Reviewed
04/15/2018 Reviewed
07/12/2018 Reviewed