Tuberculosis (TB) is an ancient infectious disease of humans that has been extensively studied both clinically and experimentally. Although susceptibility to Mycobacterium tuberculosis infection is clearly influenced by factors such as nutrition, immune status, and both mycobacterial and host genetics, the variable pathogenesis of TB in infected individuals remains poorly understood.
KEYWORDS: antibiotics, microbiome, tuberculosis
ABSTRACT
Tuberculosis (TB) is an ancient infectious disease of humans that has been extensively studied both clinically and experimentally. Although susceptibility to Mycobacterium tuberculosis infection is clearly influenced by factors such as nutrition, immune status, and both mycobacterial and host genetics, the variable pathogenesis of TB in infected individuals remains poorly understood. During the past two decades, it has become clear that the microbiota—the trillion organisms that reside at mucosal surfaces within and on the body—can exert a major influence on disease outcome through its effects on host innate and adaptive immune function and metabolism. This new recognition of the potentially pleiotropic participation of the microbiome in immune responses has raised the possibility that the microbiota may influence M. tuberculosis infection and/or disease. Similarly, treatment of TB may alter the healthy steady-state composition and function of the microbiome, possibly affecting treatment outcome in addition to other host physiological parameters. Herein, we review emerging evidence for how the microbiota may influence the transition points in the life cycle of TB infection, including (i) resistance to initial infection, (ii) initial infection to latent tuberculosis (LTBI), (iii) LTBI to reactivated disease, and (iv) treatment to cure. A major goal of this review is to frame questions to guide future scientific and clinical studies in this largely unexplored but increasingly important area of TB research.
INTRODUCTION
Tuberculosis (TB) sickens more than 10 million people each year and kills 10 to 20% of them; TB is the leading cause of mortality by a single infectious agent (1) and ranks in the top 10 causes of death in the world. Mycobacterium tuberculosis is transmitted by aerosols from individuals with active TB. Although approximately one-third of the world’s population is latently infected (LTBI), only 5 to 10% develop active disease in their lifetime. Moreover, even in settings of high exposure, a sizable percentage of the population remains tuberculin skin test (TST) or interferon gamma (IFN-γ) release assay (IGRA) negative (2), suggesting that they have been able to resist infection despite likely M. tuberculosis exposure. Although drug-sensitive active TB can be cured with 6 months of daily antibiotic treatment, this long duration of therapy is frequently complicated by noncompliance, relapse, and the development of drug resistance. Often lost in the discussion of the long duration of therapy required to cure TB is the large heterogeneity in response to treatment. Six months is the shortest duration of therapy that will reliably cure >95% of subjects; however, often overlooked is the fact that >60 to 70% of subjects are cured by shorter-course regimens (3–5). Indeed, at present we have no way of predicting which patients need longer courses of treatment. Shorter regimens would be successful if we could identify biomarkers that allow those able to be cured in less than 6 months of therapy.
During the timeworn history of research on TB in humans, a number of underlying risk factors for the disease have been identified (6) that contribute to the total population attributable risk percentage (PAR%), a measure of the fraction of disease that can be accredited to a specific risk factor. Many of the risk factors directly or indirectly involve the immune system. These include HIV infection (>10 times the risk for LTBI to active disease transition) (7–9), genetic immunodeficiency (10), age (∼10% of active TB disease cases) (1, 11), indoor air pollution (>20% of TB incidence) (12), malnutrition (>25% of TB incidence) (12), metabolic syndrome (13), the use of immunosuppressive drugs (14), and substance abuse, including alcohol (15) and smoking (16, 17). These PAR estimates are imprecise due to their underlying assumptions and inability to account for overlapping exposures; nevertheless, they are an effective public health tool to understand causes of disease and prioritize potential research and health interventions (18). Mechanistically, some of these risk factors involve well-defined immunological parameters, such as the requirements for CD4 T lymphocytes and the cytokines interleukin-12 (IL-12), IFN-γ (19), and tumor necrosis factor alpha (TNF-α) (20). However, millions of people acquire LTBI or get sick with active TB disease every year with no apparent immunologic deficiency, suggesting the presence of additional, as yet unidentified, risk factors to explain the full PAR%. In this review, we summarize recent evidence implicating the composition and function of the microbiome as an additional risk factor for M. tuberculosis infection and TB disease progression.
MECHANISMS BY WHICH THE MICROBIOME MIGHT INFLUENCE TUBERCULOSIS BIOLOGY
With the advent of high-throughput sequencing technologies, the last decade has seen an explosion of microbiome research in which the composition of the microbiota has been profiled in a wide variety of diseases. In most cases, although plausible, the causal link between these commensal alterations and specific disease states (21–24) has not been established. The microbiome might contribute to tuberculosis risk and disease (i) by determining interindividual differences in immune cell subsets or function that may influence tuberculosis susceptibility or response to therapy, either remotely from the intestinal microbiota or directly in the lung, (ii) by affecting drug absorption during tuberculosis treatment, and/or (iii) by producing antimicrobial or immune activating molecules that may influence M. tuberculosis growth directly (25) (Fig. 1). Here we review the newly emerging literature addressing whether the composition of the microbiota changes with TB disease status, influences susceptibility to infection, or affects the response to therapy. We have organized the discussion around the major stages of clinical TB, namely (i) initial M. tuberculosis infection, (ii) TB disease progression, (iii) response to antibiotic treatment, and (iv) posttherapy cure and reinfection.
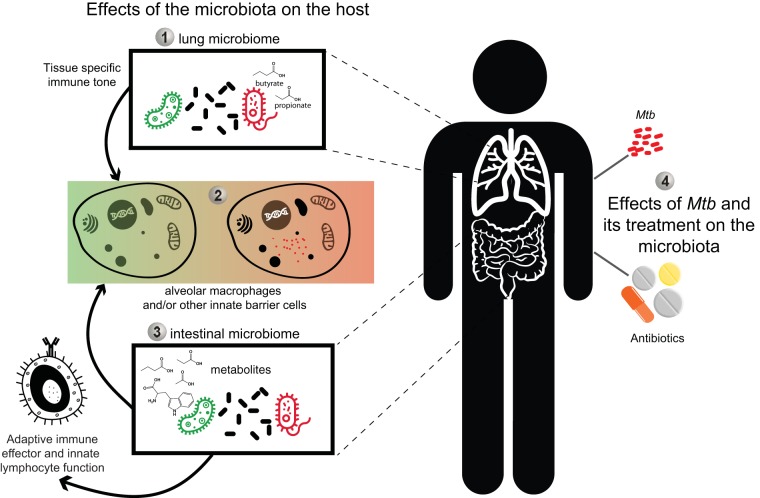
FIG 1.
Putative interactions between the intestinal and/or lung microbiome and the host that could influence the outcome of M. tuberculosis (Mtb) infection and treatment. The potential intersection points between microbiome, TB infection, and antibiotic treatment are multifold. (Point 1) Previous studies have demonstrated that specific clades of organisms (e.g., Prevotella) produce short-chain fatty acids like butyrate and propionate (44) that could set tissue-specific immune responsiveness in the lung. (Point 2) The immune state dictated in part by the interaction of the lung microbiota and innate cells such as alveolar macrophages could shape the outcome of the initial encounter of TB with the host. (Point 3) Additionally, the intestinal microbiome and its metabolites, through their previously described role in setting systemic immune tone and/or the production of antimicrobial products, may influence TB susceptibility in a related but trans fashion. (Point 4) Finally, the effects of M. tuberculosis infection and/or its treatment with antibiotics on the microbiota could influence the outcome of TB therapy and cure as well as other physiological functions.
POTENTIAL INTERACTION POINTS OF THE MICROBIOTA WITHIN THE TB LIFE CYCLE
Resistance to initial infection.
Tuberculosis is transmitted by inhalation of droplet aerosols liberated from the lungs of active tuberculosis patients through coughing. With inhalation, successful infection of the host leads to LTBI, which is detectable by TST/IGRA. It is presumed that some exposed contacts who inhale the bacterium are able to eliminate the infection before the establishment of latency and the accompanying M. tuberculosis-specific T cell responses through which LTBI is diagnosed. The explanation for why these individuals who are directly exposed remain uninfected (TST−/IGRA−) is unclear. One possibility is that differences in the microbiota could influence the clearance of initial infection through innate immune mechanisms. Comparisons of wild-type and germfree mice have revealed differences in transcriptional profiles of innate lymphoid cells (26), myeloid cell development and function (27–29), and mucus layer formation (30). Indeed, a number of studies have demonstrated the importance of the interaction between the microbiome, its metabolites, and the host innate immune system in maintaining organismal homeostasis, including immune tolerance and defense against pathogens (31, 32). In this regard, there are data indicating that certain commensal bacteria and their antimicrobial products can quantitatively influence the (initial) resistance to pathogens (e.g., vancomycin-resistant enterococcus, Clostridium difficile, and Salmonella enterica serovar Typhimurium [33–35]) via a variety of mechanisms, including niche competition (36) and bacteriolytic activity (37, 38). Nevertheless, data on the role of the microbiota in mediating initial resistance to TB infection are limited. Mice treated with broad-spectrum antibiotics were found in one study to be modestly but significantly more susceptible to aerosol M. tuberculosis challenge, particularly when bacterial burden was assessed at extrapulmonary sites (39). A more recent study utilized a mouse model of Helicobacter hepaticus in which infection with this commensal causes a defined change in the gut microbiota. The resulting dysbiosis led to an increase in bacterial burden following M. tuberculosis challenge (40). Although speculative at this time, such effects of the microbiota on initial host resistance could affect susceptibility to infection in exposed humans. This question could be approached by comparing the microbiome compositions of equivalently exposed IGRA+ and IGRA− household contacts of active TB cases and relating the differences seen to possible host protective functions.
Progression from LTBI to active TB disease.
As previously noted, numerous mechanisms have been proposed to explain the emergence of active TB in individuals with LTBI. In several studies, the microbiota has been indirectly implicated as a factor in disease progression. In one report, it was shown that LTBI patients with Helicobacter pylori in their gut flora were 50% less likely to develop active TB disease (41). This observation is consistent with other studies demonstrating similar associations between the presence of certain intestinal bacteria and pulmonary susceptibility to the manifestations of experimental respiratory syncytial virus infection and pediatric asthma (42, 43).
A recent report presented evidence for a possible link between the transition from LTBI to active disease and the composition of the lung (as opposed to intestinal) microbiota in HIV-infected South Africans (44). This study demonstrated that within a cohort of HIV-infected patients on antiretroviral therapy (ART), higher concentrations of two short-chain fatty acids (SCFAs) in serum, propionate, and butyrate were associated with increased risk for active TB. The elevation in SCFAs correlated with a corresponding increase in the abundance of a number of anaerobic bacteria, including Prevotella, a genus known to produce these lipid molecules. The same SCFAs suppressed in vitro production of IFN-γ and IL-17A by both polyclonal-stimulated T cells and TB antigen-stimulated peripheral blood mononuclear cells (PBMCs). Together, these results suggested that in some patients, ART treatment is associated with increased levels of certain pulmonary anaerobes that in turn result in increased SCFA production and suppressed T cell effector function. A broader implication of this study is that the metabolic activity of the microbiota at mucosal surfaces may be an important risk factor for the development of active TB. In this regard, indole-3-propionic acid (IPA), a metabolite produced by Clostridium sporogenes (45) and other gut commensals, was recently shown to exhibit antitubercular activity in vitro and at extrapulmonary sites in a murine experimental model (46). The mechanism underlying this antimicrobial effect remains to be elucidated.
Another plausible mechanism by which commensal metabolites may influence TB progression is through their role in stimulating innate T cell subsets through the major histocompatibility complex (MHC) class I-like proteins CD1 (47) and MR1 (48–50). These MHC-like proteins are restriction elements for the activation of invariant natural killer T cells (iNKT cells) (51), germ line-encoded mycolyl lipid-reactive (GEM) T cells (52), and mucosa-associated invariant T (MAIT) cells (48–50), all of which have been speculated to be involved in host resistance to M. tuberculosis. MAIT cells are of particular relevance since they proliferate in response to MR1-bound riboflavin biosynthetic intermediates, which can be synthesized by M. tuberculosis and are enriched at the sites of infection (53, 54). Interestingly, MAIT cells are absent in germfree mice (48), suggesting that their development and function may be influenced by the microbiota, possibly through the production of MR1-binding molecules. It is therefore plausible, although as yet unexamined, that differences in microbiome composition could influence tuberculosis progression through an effect on the abundance or function of these innate, bacterium-reactive T cell subsets.
Prospective studies that follow the development of active disease in cohorts of LTBI patients are needed to directly identify associations between microbiota composition and LTBI progression in humans. This type of analysis has been employed to identify host transcriptional profiles that correlate with, as well as predict, active TB in LTBI individuals (55). Indeed, correlations between blood transcriptional signatures and microbiota composition may exist that would be of important diagnostic as well as mechanistic significance.
Active TB versus no M. tuberculosis infection.
Two studies have examined changes in the microbiota that occur during the course of active TB in mice. Inbred specific-pathogen-free (SPF) mice, in which the intestinal microbiota is relatively uniform between animals, revealed only minor alterations in taxa as a consequence of infection with aerosol M. tuberculosis. These changes occurred largely in the relative abundance of the order Clostridiales (56, 57).
A number of microbiome studies in humans demonstrate interindividual (as well as geographic) differences in the steady-state microbiome with respect to M. tuberculosis infection or TB disease. In these reports, the microbiota was analyzed in the stool, sputum, or bronchoalveolar lavage (BAL) fluid of diseased patients. In the case of the intestinal microbiota, a recent study found increased diversity and levels of Actinobacteria and Proteobacteria in patients with recurrent TB and decreased levels of Prevotella, as well as members of the order Clostridiales, in both new and recurrent TB patients in comparison to healthy individuals (58).
Current information on changes in the lung (as opposed to intestinal) microbiome induced by active M. tuberculosis infection is based on several studies on sputum samples (59–62) and one study employing BAL fluid (63). Such analyses of the pulmonary flora are inherently more complicated than those involving the fecal microbiota due to practical limitations in obtaining material free of oral bacterial contamination, as well as the difficulty of enrolling healthy controls for BAL fluid extraction, an invasive procedure. Moreover, the lung microbiome because of its less abundant biomass and transient nature (64) makes its accurate analysis more challenging. Nevertheless, distinct changes in the diversity and composition of the lung microbiota based on sputum have been associated with new M. tuberculosis infection, recurrent TB disease, and treatment failures in humans (60), although it is difficult to discern a common pattern between the different studies, with a possible exception of increases in the common lung bacteria Streptococcus and Pseudomonas, which appear in multiple reports (65). Interestingly, although one might predict a decrease in diversity due to overcrowding of the pulmonary niche by the mycobacteria themselves, such a reduction has not been routinely observed. In addition to the above-mentioned limitations in performing human studies, our understanding of the interaction of M. tuberculosis infection and TB disease with the pulmonary microbiome has suffered from the absence of data from animal models, in part due to the lower abundance of microbiota at that tissue site. Current research on experimental M. tuberculosis infection in nonhuman primates presents an opportunity to address this shortcoming.
Effect of TB antibiotic treatment on the microbiota.
Antibiotics are a major cause of microbiota perturbation since these molecules are designed to specifically and/or broadly kill bacteria (66). Antibiotic exposure early in life is a risk factor for the development of asthma, diabetes, and weight gain later in life (67, 68). Adults taking various antibiotics can develop antibiotic-associated diarrhea (69, 70), show increased susceptibility to pathogen colonization (71, 72), and in certain cases show impaired responses to other immune-based therapies, such as checkpoint blockade for malignancy (73–75). Each of these has been linked to alterations of the intestinal microbiota, where different classes of antibiotics with distinct mechanisms of action have unique effects on the composition of the commensal flora (66).
As noted above, treatment of drug-susceptible TB requires multiple daily administrations of oral antibiotics for a duration of at least 6 months according to World Health Organization guidelines (76). Millions of people receive these drugs every year, making TB chemotherapy one of the most widely administered treatment interventions, as well as one of the longest-duration antibiotic regimens utilized globally. Of the four first-line antibiotics used in TB treatment, only rifampin (R/RIF) has a broad-spectrum activity against a wide range of Gram-positive and Gram-negative bacteria. Isoniazid (I/INH), pyrazinamide (Z/PZA), and ethambutol (E/EMB) specifically target mycobacterial species, with isoniazid and pyrazinamide being prodrugs that need to be activated by mycobacterium-specific enzymes, which then inhibit or require for function mycobacterium-specific targets (77–80). Due to this mycobacterial specificity, the effects of antituberculosis treatment on intestinal or pulmonary microbiome composition are not predictable and, until recently, were unknown.
Acute effects of TB treatment on the microbiota.
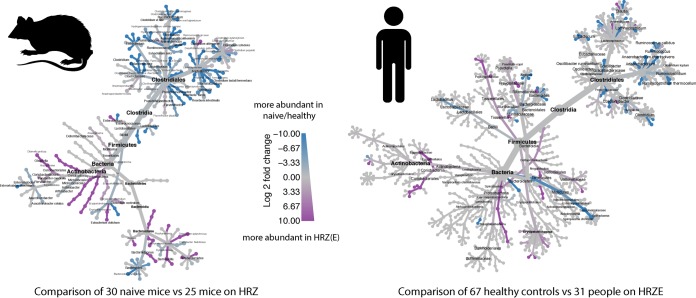
Recent studies in mice and TB patients have examined the effects of TB treatment on the microbiome (Table 1). These studies have shown that conventional TB antibiotic therapy causes a defined and persistent dysbiosis in the intestinal microbiota, which somewhat unexpectedly has marked similarities in terms of the taxa altered in the two host species (57, 81) (Fig. 2).
TABLE 1.
Summary of antituberculosis treatment-induced alterations in the microbiota
| Antibiotic(s)a | Effect on intestinal microbiota | Reference |
|---|---|---|
| HRZ (mice) | Decreases in Acetivibrio, Robinsoniella, Alkaliphilus, Stomatobaculum, Butyricicoccus, Acetanaerobacterium, Tyzzerella, Ruminococcus, and Peptococcus and increase in Erysipelatoclostridium |
57 |
| Post-HRZ (mice) | Decrease in Lactobacillus and increase in Barnesiella, Porphyromonas, Paraprevotella, Parasutterella, and Desulfovibrio and Actinobacteria genera |
57 |
| HRZE (humans) | Decrease in Lactobacillus, Coprococcus, Ruminococcus, and Bifidobacterium
and increase in Erysipelatoclostridium, Fusobacterium, and Prevotella |
81 |
| HRZE (humans) | Decrease in Prevotella and Lachnospira | 58 |
| Post-HRZE (humans) | Decrease in Bacteroides and increase in Faecalibacterium, Eubacterium, and Ruminococcus |
81 |
| H alone | Alterations in Barnesiella and certain Clostridium species | 57 |
| R alone | Decrease in diversity and a number of Clostridium species | 57 |
| Z alone | Alterations in Anaeroplasma and certain Clostridium species | 57 |
Abbreviations: H, isoniazid; R, rifampin; Z, pyrazinamide; E, ethambutol.
FIG 2.
Cladograms depicting the parallel effects of TB antibiotic treatment on the intestinal microbiomes of mice and humans. Two recent studies reported the effects of antituberculosis therapy, HRZ(E), on the gut flora of mice (57) and humans (81). A combined comparison of data from naive/healthy mice/humans versus mice administered HRZ or humans taking HRZE is shown based on published (57, 81) as well as additional unpublished data. In both host species, in comparison to corresponding healthy untreated controls, members of the order Clostridiales were depleted following treatment, whereas certain taxa of the order Erysipelotrichales and phylum Actinobacteria were enriched. Cladograms were generated using Metacoder (92).
The first surprising observation is that isoniazid-rifampin-pyrazinamide (ethambutol) [HRZ(E)] treatment has minimal effects on the diversity of the intestinal microbiome, a unitary metric of species number (richness) and distribution (evenness). In mice, there is only a transient decrease in diversity (57), while in humans who have been in treatment for at least 3 months, no significant change in diversity was observed (81). Thus, diversity during chronic HRZ(E) treatment may not be a good metric of mycobacterial drug effects.
In terms of the specific taxa affected by HRZ(E) treatment, both mice and humans have shown strikingly similar effects on the order Clostridiales of the phylum Firmicutes. Clostridia are important players of gut homeostasis, barrier function, and metabolism, particularly via their production of SCFAs (82). Among the members of the Bacteroidetes phylum, Bacteroides species are decreased with TB antibiotic treatment, whereas Prevotellaceae are increased—a shift that has also been associated with a protein versus carbohydrate-enriched diet (83). Among the prominent increases in microbiota observed following antituberculosis treatment were those affecting Erysipelotrichaceae, species of which have been associated with immune function (84) and metabolism (85). In mice, where the effect of monotherapy was analyzed, it was shown that RIF is the major driver of taxonomic alterations (57). Interestingly, in the same experiments, the mycobacterium-specific antibiotics INH and PZA, as well as EMB (S. Namasivayam, unpublished data), also independently affected the microbiota, and these changes were distinct from those observed during combination therapy. Since in wealthier countries isoniazid prophylaxis (IPT) is used frequently in adults with LTBI to prevent conversion to active disease, the effects of INH administration observed in the mouse model suggest that this group of antibiotic-treated individuals may also exhibit an intestinal dysbiosis, a hypothesis that has yet to be formally investigated.
These initial studies of the short-term effects of TB treatment on the microbiota suggest that delineation of how future TB drugs in the development pipeline affect the diversity, taxonomic structure, and metabolism of the microbiota may yield important information relevant to understanding their efficacy and potentially in predicting variations in treatment outcome between individuals.
Chronic effects of M. tuberculosis treatment.
An important aspect of the TB antibiotic-induced dysbiosis is its long-lasting nature. In mice treated with HRZ, this dysbiosis lasted at least 3 months after cessation of therapy (57). In humans, active TB subjects who completed the 6-month standard course of HRZE treatment and were clinically cured for an average of at least 1.2 years displayed altered intestinal microbiome composition compared to healthy LTBI controls (81). Biomarkers of dysbiosis in treated mice were Barnesiella, Porphyromonas, Paraprevotella, Parasutterella, Desulfovibrio, and interestingly some Actinobacteria, the phylum to which mycobacteria belong. In humans, Bacteroides, Faecalibacterium, Eubacterium, and Ruminococcus were all biomarkers of dysbiosis when measured more than 1 year posttreatment.
While TB antibiotic treatment has well-defined long-term effects on the microbiome, far less is known about the possible consequences of these alterations on human health. Interestingly, after an individual is cured by TB antibiotics, their risk for reinfection is increased (86, 87)—up to 4-fold in one study, suggesting a possible link between the posttreatment effects of chemotherapy on the microbiome and TB recurrence. A recent study employed an IFN-γ enzyme-linked immunospot assay to compare immune responses in patients who were treated for TB with those who were LTBI/IGRA+ but had never had active disease or received TB treatment (88). The authors found that a defined subset of M. tuberculosis T cell epitopes was recognized poorly by PBMCs from patients who were treated less than 6 years previously compared with PBMCs from untreated LTBI individuals. Interestingly, many of the M. tuberculosis epitopes in this subset had sequence homology with bacterial peptides from the Human Microbiome Project data set. In contrast, a second pool of M. tuberculosis epitopes with relatively weaker homology to microbiome peptides stimulated indistinguishable T cell responses in the treated TB and untreated LTBI groups. Based on these observations, the authors suggested that the cross-reactivity between certain microbiota and M. tuberculosis epitopes is important in maintaining long-term host resistance to TB and that the effects of TB antibiotics on these commensal taxa result in increased susceptibility to reinfection and disease. These interesting observations suggest a mechanism by which the dysbiosis induced by TB antibiotic therapy could be a risk factor for M. tuberculosis reinfection in cured individuals. Future analyses that incorporate matched immune readouts and microbiome measurements from the same patient to allow for direct correlations are required to validate this hypothesis.
The intestinal dysbiosis observed both during and after antituberculosis therapy may have additional consequences. For example, it may affect the absorption or metabolism of the antimycobacterial drugs themselves during the prolonged treatment regimen. Such effects could hinder or perhaps even promote the efficacy of the individual antibiotics against TB infection. Although as yet to be documented in patients, TB antibiotic treatment through the dysbiosis it induces may influence the host resistance to other diseases. Studies employing the murine TB chemotherapy model can be used to specifically address these questions.
CONCLUSIONS
Our understanding of the interrelationship of the microbiome and TB infection and treatment is still at an early stage, and how microbiome composition and function influence M. tuberculosis infection and TB disease risk is far from being precisely delineated. It is becoming clear, however, that active M. tuberculosis infection in both mice and humans causes alterations in the microbiota, although these changes are variable between studies and in many cases of minor magnitude. In contrast, M. tuberculosis antibiotic treatment induces both profound and long-lasting changes in the intestinal microbiome that are largely shared between mice and humans. In mice, these changes occur within 2 weeks of treatment, the time in which M. tuberculosis is being rapidly cleared from the sputum in infected humans. Given these findings, critical questions that should be addressed in future studies include:
Does the early alteration in microbiome composition that occurs during TB treatment, which is both substantial and variable between individuals, correlate with the efficacy of TB treatment? This could be approached through prospective studies in which microbiome composition, TB bacterial load, and immune function are measured in the same subjects.
Are there differences in the microbiome, and immune correlates of those differences, that identify individuals resistant to initial infection or who will control latent infection?
Does the posttreatment dysbiosis that occurs following cessation of TB therapy have consequences for susceptibility to reinfection by M. tuberculosis or other pathogens, and if so, what mechanism(s) underlie this effect?
Another critical need is for improved techniques and experimental models to delineate the cross talk between M. tuberculosis and the lung microbiome. As discussed above, major problems exist in both the sampling of the lung flora free of oral contamination and the accurate classification of taxa due to the lower bacterial biomass in this tissue site. Interestingly, while the effects of TB antibiotics on the intestinal flora are now well documented, to the best of our knowledge, there is no information on how these antibiotics affect the lung microbiota either directly or indirectly through their effect on the removal of M. tuberculosis from that niche. Although TB is primarily a pulmonary disease, it is important to note that the intestinal microbiota may be equally as important as the lung microbiome in influencing pulmonary TB given its abundant size and well-known effects on systemic immunity. Indeed, studies using the mouse influenza model have shown that a homeostatic gut microbiome is critical for mounting an optimal immune response to respiratory tract flu (89) and that influenza infection can in turn affect the intestinal microbiota in a type I interferon-dependent manner (90). That a similar gut-lung axis (Fig. 1) occurs in M. tuberculosis infection was suggested in one report (39); however, this, as well as the above studies all employ antibiotics which in addition to affecting the intestinal microbiota could potentially alter the pulmonary flora (91). Therefore, future experimental as well as prospective clinical studies aimed at deciphering the role of the microbiome in the different stages of the TB life cycle, whenever possible, should attempt to sample both anatomical sites. Finally, given the well-documented association of host nutritional and metabolic status in TB, it is not unreasonable to predict that the microbiome, which strongly influences these physiological parameters, will also be shown to be an important factor in determining the outcome of M. tuberculosis infection, progression to TB disease, and risk of reinfection.
ACKNOWLEDGMENTS
This work was supported by the Tri-I TBRU supported by National Institutes of Health TBRU-Network (U19 AI-111143) and P30 CA008748. M.F.W. acknowledges support from the National Center for Advancing Translational Sciences (grant TL1TR002386-01). S.N. and A.S. are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Footnotes
Citation Namasivayam S, Sher A, Glickman MS, Wipperman MF. 2018. The microbiome and tuberculosis: early evidence for cross talk. mBio 9:e01420-18. https://doi.org/10.1128/mBio.01420-18.
REFERENCES
- 1.WHO. 2016. Global tuberculosis report. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, Doherty TM, Hanekom WA, Eley B, Jaïs J-P, Boland-Auge A, van Helden P, Casanova J-L, Abel L, Hoal EG, Schurr E, Alcaïs A. 2009. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 206:2583–2591. doi: 10.1084/jem.20090892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous 1981. Controlled clinical trial of five short-course (4-month) chemotherapy regimens in pulmonary tuberculosis. Second report of the 4th study. East African/British Medical Research Councils Study. Am Rev Respir Dis 123:165–170. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie SH, Crook AM, McHugh TD, Mendel CM, Meredith SK, Murray SR, Pappas F, Phillips PP, Nunn AJ, REMox TB Consortium. 2014. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N Engl J Med 371:1577–1587. doi: 10.1056/NEJMoa1407426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous 1979. Sputum-smear-negative pulmonary tuberculosis: controlled trial of 3-month and 2-month regimens of chemotherapy. Lancet i:1361–1363. [PubMed] [Google Scholar]
- 6.Narasimhan P, Wood J, Macintyre CR, Mathai D. 2013. Risk factors for tuberculosis. Pulm Med 2013:828939. doi: 10.1155/2013/828939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havlir DV, Getahun H, Sanne I, Nunn P. 2008. Opportunities and challenges for HIV care in overlapping HIV and TB epidemics. JAMA 300:423–430. doi: 10.1001/jama.300.4.423. [DOI] [PubMed] [Google Scholar]
- 8.Ford N, Shubber Z, Meintjes G, Grinsztejn B, Eholie S, Mills EJ, Davies MA, Vitoria M, Penazzato M, Nsanzimana S, Frigati L, O'Brien D, Ellman T, Ajose O, Calmy A, Doherty M. 2015. Causes of hospital admission among people living with HIV worldwide: a systematic review and meta-analysis. Lancet HIV 2:e438–e444. doi: 10.1016/S2352-3018(15)00137-X. [DOI] [PubMed] [Google Scholar]
- 9.Horsburgh CR Jr, Rubin EJ. 2011. Latent tuberculosis infection in the United States. N Engl J Med 364:1441–1448. doi: 10.1056/NEJMcp1005750. [DOI] [PubMed] [Google Scholar]
- 10.Grant AV, Sabri A, Abid A, Abderrahmani Rhorfi I, Benkirane M, Souhi H, Naji Amrani H, Alaoui-Tahiri K, Gharbaoui Y, Lazrak F, Sentissi I, Manessouri M, Belkheiri S, Zaid S, Bouraqadi A, El Amraoui N, Hakam M, Belkadi A, Orlova M, Boland A, Deswarte C, Amar L, Bustamante J, Boisson-Dupuis S, Casanova JL, Schurr E, El Baghdadi J, Abel L. 2016. A genome-wide association study of pulmonary tuberculosis in Morocco. Hum Genet 135:299–307. doi: 10.1007/s00439-016-1633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant AV, El Baghdadi J, Sabri A, El Azbaoui S, Alaoui-Tahiri K, Abderrahmani Rhorfi I, Gharbaoui Y, Abid A, Benkirane M, Raharimanga V, Richard V, Orlova M, Boland A, Migaud M, Okada S, Nolan DK, Bustamante J, Barreiro LB, Schurr E, Boisson-Dupuis S, Rasolofo V, Casanova JL, Abel L. 2013. Age-dependent association between pulmonary tuberculosis and common TOX variants in the 8q12-13 linkage region. Am J Hum Genet 92:407–414. doi: 10.1016/j.ajhg.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonnroth K, Castro KG, Chakaya JM, Chauhan LS, Floyd K, Glaziou P, Raviglione MC. 2010. Tuberculosis control and elimination 2010–50: cure, care, and social development. Lancet 375:1814–1829. doi: 10.1016/S0140-6736(10)60483-7. [DOI] [PubMed] [Google Scholar]
- 13.Jeon CY, Murray MB. 2008. Diabetes mellitus increases the risk of active tuberculosis: a systematic review of 13 observational studies. PLoS Med 5:e152. doi: 10.1371/journal.pmed.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CH, Kim K, Hyun MK, Jang EJ, Lee NR, Yim JJ. 2013. Use of inhaled corticosteroids and the risk of tuberculosis. Thorax 68:1105–1113. doi: 10.1136/thoraxjnl-2012-203175. [DOI] [PubMed] [Google Scholar]
- 15.Rehm J, Samokhvalov AV, Neuman MG, Room R, Parry C, Lonnroth K, Patra J, Poznyak V, Popova S. 2009. The association between alcohol use, alcohol use disorders and tuberculosis (TB). A systematic review. BMC Public Health 9:450. doi: 10.1186/1471-2458-9-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates MN, Khalakdina A, Pai M, Chang L, Lessa F, Smith KR. 2007. Risk of tuberculosis from exposure to tobacco smoke: a systematic review and meta-analysis. Arch Intern Med 167:335–342. doi: 10.1001/archinte.167.4.335. [DOI] [PubMed] [Google Scholar]
- 17.Glickman MS, Schluger N. 2016. Adding insult to injury: exacerbating TB risk with smoking. Cell Host Microbe 19:432–433. doi: 10.1016/j.chom.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Northridge ME. 1995. Public health methods—attributable risk as a link between causality and public health action. Am J Public Health 85:1202–1204. doi: 10.2105/AJPH.85.9.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. 1993. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med 178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. 1995. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 21.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat Rev Immunol 17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 22.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada N, Seo SU, Chen GY, Nunez G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC. 2015. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26:26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooks MG, Garrett WS. 2016. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, Dori-Bachash M, Pevsner-Fischer M, Lorenzo-Vivas E, Keren-Shaul H, Paul F, Harmelin A, Eberl G, Itzkovitz S, Tanay A, Di Santo JP, Elinav E, Amit I. 2016. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166:1231–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 27.Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F, Koo LY, Brenchley JM, Fraser ID, Belkaid Y. 2013. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat Med 19:713–721. doi: 10.1038/nm.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, Arike L, Wising C, Svensson F, Bäckhed F, Hansson GC. 2015. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubeda C, Djukovic A, Isaac S. 2017. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunol 6:e128. doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 33.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, Gobourne A, Ling L, Pamer EG. 2015. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J Infect Dis 212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Nunez G. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN. 2016. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol 7:529. doi: 10.3389/fimmu.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majlessi L, Sayes F, Bureau JF, Pawlik A, Michel V, Jouvion G, Huerre M, Severgnini M, Consolandi C, Peano C, Brosch R, Touati E, Leclerc C. 2017. Colonization with Helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of Mycobacterium tuberculosis. Mucosal Immunol 10:1178–1189. doi: 10.1038/mi.2016.140. [DOI] [PubMed] [Google Scholar]
- 41.Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, Hansen LM, Talat N, Hill PC, Hussain R, Adegbola RA, Flynn J, Canfield D, Parsonnet J. 2010. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One 5:e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, Boushey HA, Zoratti E, Ownby D, Lukacs NW, Lynch SV. 2014. House dust exposure mediates gut microbiome lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A 111:805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, CHILD Study Investigators, Mohn WW, Turvey SE, Finlay BB. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 44.Segal LN, Clemente JC, Li Y, Ruan C, Cao J, Danckers M, Morris A, Tapyrik S, Wu BG, Diaz P, Calligaro G, Dawson R, van Zyl-Smit RN, Dheda K, Rom WN, Weiden MD. 2017. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe 21:530–537.e4. doi: 10.1016/j.chom.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. 2017. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Negatu DA, Liu JJJ, Zimmerman M, Kaya F, Dartois V, Aldrich CC, Gengenbacher M, Dick T. 2018. Whole-cell screen of fragment library identifies gut microbiota metabolite indole propionic acid as antitubercular. Antimicrob Agents Chemother 62:e01571-17. doi: 10.1128/AAC.01571-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA. 2003. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun 71:3076–3087. doi: 10.1128/IAI.71.6.3076-3087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 49.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YY, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, Levy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, Riteau B, Duban L, Robert D, Huang S, Rottman M, Soudais C, Lantz O. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol 11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 51.Chackerian A, Alt J, Perera V, Behar SM. 2002. Activation of NKT cells protects mice from tuberculosis. Infect Immun 70:6302–6309. doi: 10.1128/IAI.70.11.6302-6309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, Rossjohn J, Moody DB. 2013. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol 14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dias J, Leeansyah E, Sandberg JK. 2017. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci U S A 114:E5434–E5443. doi: 10.1073/pnas.1705759114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O'Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 55.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA, ACS and GC6-74 Cancer Cohort Study Groups. 2016. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winglee K, Eloe-Fadrosh E, Gupta S, Guo H, Fraser C, Bishai W. 2014. Aerosol Mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS One 9:e97048. doi: 10.1371/journal.pone.0097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Namasivayam S, Maiga M, Yuan W, Thovarai V, Costa DL, Mittereder LR, Wipperman MF, Glickman MS, Dzutsev A, Trinchieri G, Sher A. 2017. Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome 5:71. doi: 10.1186/s40168-017-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo M, Liu Y, Wu P, Luo DX, Sun Q, Zheng H, Hu R, Pandol SJ, Li QF, Han YP, Zeng Y. 2017. Alternation of gut microbiota in patients with pulmonary tuberculosis. Front Physiol 8:822. doi: 10.3389/fphys.2017.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung MK, Lam WY, Fung WY, Law PT, Au CH, Nong W, Kam KM, Kwan HS, Tsui SK. 2013. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One 8:e54574. doi: 10.1371/journal.pone.0054574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Liu W, He L, Huang F, Chen J, Cui P, Shen Y, Zhao J, Wang W, Zhang Y, Zhu M, Zhang W, Zhang Y. 2013. Sputum microbiota associated with new, recurrent and treatment failure tuberculosis. PLoS One 8:e83445. doi: 10.1371/journal.pone.0083445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishna P, Jain A, Bisen PS. 2016. Microbiome diversity in the sputum of patients with pulmonary tuberculosis. Eur J Clin Microbiol Infect Dis 35:1205–1210. doi: 10.1007/s10096-016-2654-4. [DOI] [PubMed] [Google Scholar]
- 62.Cui Z, Zhou Y, Li H, Zhang Y, Zhang S, Tang S, Guo X. 2012. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol 12:276. doi: 10.1186/1471-2180-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Lin F, Cui Z, Zhang X, Hu C, Shen T, Chen C, Zhang X, Guo X. 2015. Correlation between either Cupriavidus or Porphyromonas and primary pulmonary tuberculosis found by analysing the microbiota in patients' bronchoalveolar lavage fluid. PLoS One 10:e0124194. doi: 10.1371/journal.pone.0124194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huffnagle GB, Dickson RP, Lukacs NW. 2017. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol 10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickson RP, Erb-Downward JR, Huffnagle GB. 2013. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med 7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozyrskyj AL, Ernst P, Becker AB. 2007. Increased risk of childhood asthma from antibiotic use in early life. Chest 131:1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 69.Beaugerie L, Flahault A, Barbut F, Atlan P, Lalande V, Cousin P, Cadilhac M, Petit JC, Study Group. 2003. Antibiotic-associated diarrhoea and Clostridium difficile in the community. Aliment Pharmacol Ther 17:905–912. doi: 10.1046/j.1365-2036.2003.01531.x. [DOI] [PubMed] [Google Scholar]
- 70.Kelly CP, Pothoulakis C, LaMont JT. 1994. Clostridium difficile colitis. N Engl J Med 330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 71.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bohnhoff M, Drake BL, Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med 86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 73.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Berard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L. 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zitvogel L, Galluzzi L, Viaud S, Vetizou M, Daillere R, Merad M, Kroemer G. 2015. Cancer and the gut microbiota: an unexpected link. Sci Transl Med 7:271ps1. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. 2013. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.WHO. 2010. Treatment of tuberculosis: guidelines, 4th ed WHO, Geneva, Switzerland. [Google Scholar]
- 77.Brennan PJ, Young DB. 2008. Isoniazid. Tuberculosis 88:112–116. doi: 10.1016/S1472-9792(08)70011-8. [DOI] [PubMed] [Google Scholar]
- 78.Brennan PJ, Young DB. 2008. Pyrazinamide. Tuberculosis 88:141–144. doi: 10.1016/S1472-9792(08)70021-0. [DOI] [PubMed] [Google Scholar]
- 79.Brennan PJ, Young DB. 2008. Rifampin. Tuberculosis 88:151–154. doi: 10.1016/S1472-9792(08)70024-6. [DOI] [PubMed] [Google Scholar]
- 80.Brennan PJ, Young DB. 2008. Ethambutol. Tuberculosis 88:102–105. doi: 10.1016/S1472-9792(08)70008-8. [DOI] [PubMed] [Google Scholar]
- 81.Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A, Bean JM, Bucci V, Glickman MS. 2017. Antibiotic treatment for tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep 7:10767. doi: 10.1038/s41598-017-10346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Rie A, Warren R, Richardson M, Victor TC, Gie RP, Enarson DA, Beyers N, van Helden PD. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med 341:1174–1179. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 87.Verver S, Warren RM, Beyers N, Richardson M, van der Spuy GD, Borgdorff MW, Enarson DA, Behr MA, van Helden PD. 2005. Rate of reinfection tuberculosis after successful treatment is higher than rate of new tuberculosis. Am J Respir Crit Care Med 171:1430–1435. doi: 10.1164/rccm.200409-1200OC. [DOI] [PubMed] [Google Scholar]
- 88.Scriba TJ, Carpenter C, Pro SC, Sidney J, Musvosvi M, Rozot V, Seumois G, Rosales SL, Vijayanand P, Goletti D, Makgotlho E, Hanekom W, Hatherill M, Peters B, Sette A, Arlehamn CSL. 2017. Differential recognition of Mycobacterium tuberculosis-specific epitopes as a function of tuberculosis disease history. Am J Respir Crit Care Med 196:772–781. doi: 10.1164/rccm.201706-1208OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, Rozengurt N, Shi W, Cheng G. 2016. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog 12:e1005572. doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. 2018. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med 198:497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Foster ZS, Sharpton TJ, Grunwald NJ. 2017. Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol 13:e1005404. doi: 10.1371/journal.pcbi.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]