Upper respiratory tract infections are the number one reason for a child to visit the emergency department, and otitis media (middle ear infection) ranks third overall. Biofilms contribute significantly to the chronic nature of bacterial respiratory tract infections, including otitis media, and make these diseases particularly difficult to treat. Several mucosa-associated human pathogens utilize a mechanism of rapid adaptation termed the phasevarion, or phase variable regulon, to resist environmental and host immune pressures. In this study, we assessed the role of the phasevarion in regulation of biofilm formation by nontypeable Haemophilus influenzae (NTHI), which causes numerous respiratory tract diseases. We found that the NTHI phasevarion regulates biofilm structure and critical biofilm matrix components under disease-specific conditions. The findings of this work could be significant in the design of improved strategies against NTHI infections, as well as diseases due to other pathogens that utilize a phasevarion.
KEYWORDS: DNABII, HU, NTHI, eDNA, otitis media, phasevarion
ABSTRACT
Biofilms play a critical role in the colonization, persistence, and pathogenesis of many human pathogens. Multiple mucosa-associated pathogens have evolved a mechanism of rapid adaptation, termed the phasevarion, which facilitates a coordinated regulation of numerous genes throughout the bacterial genome. This epigenetic regulation occurs via phase variation of a DNA methyltransferase, Mod. The phasevarion of nontypeable Haemophilus influenzae (NTHI) significantly affects the severity of experimental otitis media and regulates several disease-related processes. However, the role of the NTHI phasevarion in biofilm formation is unclear. The present study shows that the phasevarions of multiple NTHI clinical isolates regulate in vitro biofilm formation under disease-specific microenvironmental conditions. The impact of phasevarion regulation was greatest under alkaline conditions that mimic those known to occur in the middle ear during disease. Under alkaline conditions, NTHI strains that express the ModA2 methyltransferase formed biofilms with significantly greater biomass and less distinct architecture than those formed by a ModA2-deficient population. The biofilms formed by NTHI strains that express ModA2 also contained less extracellular DNA (eDNA) and significantly less extracellular HU, a DNABII DNA-binding protein critical for biofilm structural stability. Stable biofilm structure is critical for bacterial pathogenesis and persistence in multiple experimental models of disease. These results identify a role for the phasevarion in regulation of biofilm formation, a process integral to the chronic nature of many infections. Understanding the role of the phasevarion in biofilm formation is critical to the development of prevention and treatment strategies for these chronic diseases.
INTRODUCTION
Nontypeable Haemophilus influenzae (NTHI) naturally resides asymptomatically within the human nasopharynx (NP). However, changes in NTHI or the host can result in increased virulence and subsequent disease. NTHI causes multiple human respiratory tract diseases, which include otitis media (OM), sinusitis, conjunctivitis, and pneumonia (1). NTHI is the predominant pathogen in the establishment of chronic and recurrent OM (2), diseases that result in conductive hearing loss and often require costly surgical intervention (3–5).
To date, phasevarions have been identified in Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, Helicobacter pylori, Kingella kingae, and Neisseria species (6–11). The phasevarion mechanism simultaneously regulates the expression of multiple genes throughout the bacterial chromosome via phase variation of the DNA methyltransferase, Mod. Expression of Mod results in site-specific chromosomal methylation, which alters the expression of genes within the regulon. This epigenetic regulatory mechanism results in two genetically identical, yet phenotypically distinct, subpopulations of bacteria referred to as “mod OFF” when the Mod protein is not expressed and “mod ON” when the Mod protein is expressed. The phasevarions of multiple species have been shown to regulate disease-related phenotypes in vitro and virulence in experimental models of disease. In NTHI, ModA has been identified as the phase variable methyltransferase, of which over 21 distinct alleles have been identified. However, two-thirds of NTHI strains isolated from the NP or middle ear (ME) of children with chronic and/or recurrent OM have one of just five alleles (modA2, -4, -5, -9, or -10) (12). Phasevarions controlled by these five predominant alleles regulate expression of virulence determinants, which include potential vaccine antigens. ModA2 of NTHI strain 723 has also been shown to significantly impact disease severity in a chinchilla model of OM (13).
During the course of OM, NTHI cells in the NP ascend the eustachian tube and gain access into the ME, initially resulting in acute otitis media (AOM) that can later evolve to chronic and/or recurrent OM. The microenvironments of the ME, sinuses, and lungs are very different from those of the NP and one another. In healthy individuals, the NP is at 34°C, and association with the mucosal membranes provides bacteria with nutrients and a pH neutral surface (14). In contrast, the ME has varied nutrient availability and is warmer at 37°C, and ME effusions increase in pH as disease progresses (15–17). It has been demonstrated that Haemophilus influenzae responds to changes in pH by regulating mechanisms of pH homeostasis or induction of biofilm formation in a strain-dependent manner (18). The formation of a biofilm contributes significantly to the chronic and recurrent nature of diseases, such as OM.
Bacteria within biofilms are more resistant to environmental stresses, antimicrobial treatments, and host immunity due to multiple factors. The protective nature of the extracellular polymeric substance (EPS), or biofilm matrix, provides structural integrity to the biofilm and acts a physical barrier to separate cells within the biofilm from the extracellular milieu (19–23). Biofilm architecture and structure play a key role in the exchange of nutrients and waste within the biofilm (24, 25). Metabolic quiescence, induced by the stringent response, and changes in the growth rate of bacteria within a biofilm significantly contribute to increased resistance to antibiotics. A shift in bacterial metabolism within Pseudomonas biofilms, due to nutrient or oxygen limitation, has been shown to confer increased resistance to antibiotics (26, 27). NTHI biofilms are known to be more resistant to antibiotic killing than planktonic bacteria, yet the mechanism behind this observation is not fully understood. Proteomic analysis by Post et al. revealed decreased energy metabolism and protein synthesis by NTHI within biofilms compared to planktonic NTHI (28), which may contribute to increased antibiotic resistance.
The NTHI phasevarions potentially regulate numerous genes and gene products involved in multiple aspects of biofilm formation. Here we assess the role of the five most predominant modA alleles in biofilm formation under conditions specifically designed to mimic the microenvironments of the human nasopharynx, middle ear, and middle ear during disease.
RESULTS
Multiple ModA phasevarions regulate biofilm formation under diverse environmental conditions.
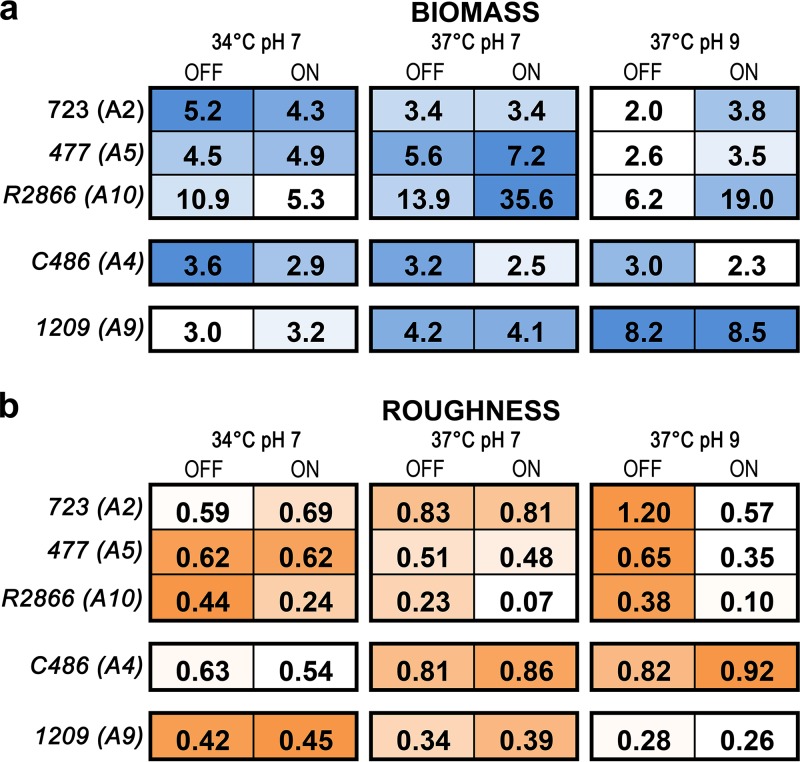
Representative strain pairs (modA ON and modA OFF) for each of the 5 most common alleles, modA2, -4, -5, -9, and -10, were tested for their ability to form biofilms within the wells of a chambered cover glass. NTHI strains 723, C486, 477, and 1209 were all isolated from the middle ears of children with otitis media. Strain R2866 is a blood isolate collected from a child with bacterial meningitis. For all of the strains tested in Fig. 1, the modA allele was free to phase vary or switch, but populations were predominantly (>90%) in the indicated modA status at the time of inoculation. Biofilms were formed at 34°C and a neutral pH (pH 7), 37°C with a neutral pH (pH 7), or 37°C with an alkaline pH (pH 9). Biofilms were started in brain heart infusion supplemented with hemin and NAD (sBHI) buffered to the indicated pH (7 or 9). Due to factors such as acidifying metabolic by-products, the pH of sBHI adjusted to a pH of 9 dropped and stabilized near a pH of 8 (see Fig. S1 in the supplemental material). As the pH of middle ear fluids collected from children with OM ranges from pH 8 to 9, we selected medium with an initial pH of 9 (16). The heat map in Fig. 1 represents the average biomass (μm3/μm2) and roughness (Ra) of biofilms formed by each strain under these three representative conditions. ModA status significantly affected biofilm formation by 3 of the 5 strains (Fig. 1; see Table S1 in the supplemental material). Under conditions that mimic those of the ME with chronic effusions (37°C, pH 9), NTHI strains 723 (modA2), 477 (modA5), and R2866 (modA10) produced biofilms that were larger in biomass and thickness and significantly less rough when ModA was expressed (ON status) compared to when ModA was not expressed (OFF status) (Fig. 1, P < 0.01; Table S1). Biofilm roughness is a measure of structural heterogeneity, and increased roughness indicates more variation in the biofilm surface. ModA expression did not affect biofilm formation by NTHI strain C486 (modA4) or NTHI strain 1209 (modA9), with no differences observed between variants in either the ON or OFF status under the conditions tested; however, alkaline conditions did significantly affect biofilm formation by strain 1209 irrespective of modA9 status (Fig. 1; α < 0.01; Table S1). These results indicated that phasevarion status significantly affects biofilm formation by multiple NTHI clinical isolates and that microenvironmental changes significantly impact how NTHI strains form biofilms in both phasevarion-dependent and -independent manners.
FIG 1.
Biomass and roughness of biofilms formed by NTHI clinical isolates that represent the 5 most common modA alleles. (a and b) Biofilms were formed by NTHI clinical isolates that represent the most common modA alleles: strain 723 (modA2), strain 477 (modA5), strain R2866 (modA10), strain C486 (modA4), and strain 1209 (modA9). Biofilms were grown for 24 h under conditions that mimic the human NP (34°C, pH 7), the healthy ME (37°C, pH 7), or the ME during chronic disease (37°C, pH 9). COMSTAT2 analysis was performed to determine (a) biomass and (b) roughness. Individual values are colored to indicate biomass (blue) or roughness (orange) for each strain. Darker colors indicate greater values. Strains were grouped based on biofilm formation at 37°C, pH 9. Values are an average of duplicate biofilms from 3 independent experiments.
Effect of NTHI growth on the pH of buffered sBHI. NTHI cells were inoculated into sBHI supplemented with 0.1 M HEPES or 0.1 M TAPS and adjusted to a pH of 6, 7, 8, or 9. Medium was collected after 6 or 12 h, and the pH was measured. sBHI buffered to a starting pH of 9 dropped to an approximate pH of 8. sBHI buffered to a starting pH of 8 dropped to an approximate pH of 7. sBHI buffered to a starting pH of 7 dropped to an approximate pH of 6.4. sBHI buffered to a starting pH of 6 dropped to an approximate pH of 5.2. Starting pHs of 7 and 9 were selected to represent neutral and alkaline conditions, respectively. Download FIG S1, TIF file, 0.1 MB (62.6KB, tif) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis of biomass and roughness of biofilms formed by NTHI clinical isolates that represent the 5 most common modA alleles. Student’s t test was performed to compare modA OFF versus modA ON populations under each of the conditions tested, as indicated. One-way ANOVA with multiple comparisons (Holms-Sidak) was used to compare the effect of microenvironmental conditions among members of a single modA subpopulation. Download Table S1, XLSX file, 0.1 MB (14.1KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
ModA2 regulates biofilm structure at alkaline pH.
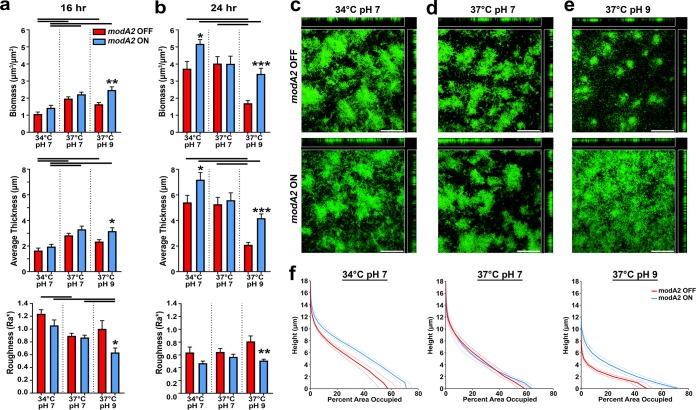
To identify key phasevarion-regulated factors that contributed to the observed differences in biofilm structure, we first investigated biofilm formation by NTHI strain 723. NTHI strain 723 carries the modA2 allele and has been studied in vitro as well as in the chinchilla model of OM (12, 13, 29). Locked versions of NTHI strain 723, in which modA2 is unable to phase vary, were utilized, and biofilm formation was assessed after both 16 and 24 h (29). As was observed with the enriched populations that were free to phase vary (Fig. 1), biofilms formed for 16 or 24 h by strain 723 modA2 locked ON and modA2 locked OFF were of similar total biomasses at 37°C and a neutral pH (Fig. 2a and b). In contrast, biofilms formed for 16 or 24 h by modA2 locked ON at an alkaline pH had significantly greater biomass than those formed by modA2 locked OFF (Fig. 2a and b; P < 0.001). Additionally, biofilms formed by modA2 locked ON bacteria and modA2 locked OFF bacteria exhibited major differences in biofilm architecture and bacterial distribution under alkaline conditions (Fig. 2e). At neutral pH, regardless of modA2 status, biofilms had dense tower-like structures (viewed as green) with interspersed water channels (viewed as black), characteristic of biofilms formed by other NTHI strains under standard laboratory conditions (Fig. 2c and d) (30, 31). modA2 locked OFF bacteria formed biofilms with similar tower-like structures under alkaline conditions, but with significantly less biomass than those formed at a neutral pH (Fig. 2b and e; P < 0.005). In contrast, modA2 locked ON bacteria formed biofilms with a distinctive architecture that lacked tower-like structures and formed a mat-like biofilm that was significantly denser at the base of the biofilm compared to modA2 OFF biofilms (compare Fig. 2e top and bottom images; P < 0.01, unpaired t test). To confirm that these observations were not due to differences in growth rate, planktonic growth in sBHI buffered to pH 6, 7, 8, or 9 was assessed. There were no differences in growth rates between the ON and OFF subpopulations at any of the pHs tested (see Fig. S2 in the supplemental material).
FIG 2.
Biofilm formation by NTHI strain 723. (a and b) Biomass, average thickness, and roughness of modA2 locked ON and modA2 locked OFF biofilms grown for (a) 16 h or (b) 24 h. Biofilms were analyzed by COMSTAT2, and values are shown as mean ± standard error of the mean. Lines over graphs indicate significance due to growth condition by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons (α < 0.05). Asterisks denote significance between subpopulations under each condition: *, P < 0.05, **, P < 0.01, and ***, P < 0.001, by Student's t test. (c to e) Representative orthogonal image renderings of modA2 locked ON and modA2 locked OFF biofilms grown at (c) 34°C at pH 7, (d) 37°C at pH 7, and (e) 37°C at pH 9. Scale bars, 100 μm. (f) Average percentage of area occupied by bacteria at each 1-μm optical section (“layer”). Dashed lines indicate standard error of the mean.
Growth of NTHI strain 723 modA2 ON and modA2 OFF subpopulations. Bacteria were diluted into buffered sBHI adjusted to a starting pH of 6, 7, 8, or 9 and seeded into wells of a 96-well flat-bottom plate. Plates were incubated shaking at either 34 or 37°C with 5% atmospheric CO2. Absorbance at 490 nm was recorded every 15 min for 12 h. No significant differences in planktonic growth rates occurred between the modA2 ON (circles) or OFF (squares) populations at any of the temperatures or pHs tested. Download FIG S2, TIF file, 0.5 MB (510.8KB, tif) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
In order to compare these architectural differences in greater detail, we determined the area occupied by layer for each population. The area occupied by layer is a calculation of the amount (or percentage) of bacterial biomass that is present within each 1-μm optical section of the biofilm taken from the base of the biofilm to the top. These data were plotted wherein the layer closest to the glass surface is at the bottom of the y axis and the top of the biofilm (farthest from the surface) is at the top of the y axis. At 34°C at pH 7, modA2 locked ON biofilms were denser near the base of the biofilm compared to those formed by modA2 locked OFF bacteria, but were of similar densities and heights near the top of the biofilm (Fig. 2f). There were no significant differences between biofilms grown at 37°C at pH 7. Under alkaline conditions, modA2 locked ON biofilms were composed of significantly greater biomass throughout the biofilm compared to modA2 locked OFF biofilms. modA2 locked ON biofilms were significantly denser at the bottom of the biofilm and occupied up to 75% of the glass surface compared to 50% by modA2 locked OFF biofilms (Fig. 2f; P < 0.05).
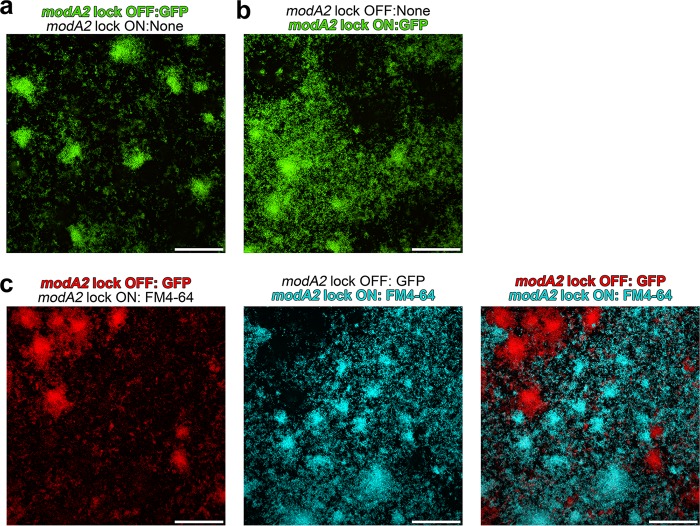
In nature, NTHI strains naturally exist as a mixture of both modA2 ON and modA2 OFF subpopulations. To verify that subpopulations maintained their unique biofilm phenotype when grown together, biofilms were formed with an equal 1:1 mixture of modA2 locked OFF bacteria and modA2 locked ON bacteria, in which the modA2 locked OFF or modA2 locked ON population had been engineered to constitutively express green fluorescent protein (GFP). The bacteria were incubated at 37°C at pH 9 to mimic the conditions of the ME during disease, and biofilms were allowed to form for 24 h. Each subpopulation maintained its distinct phenotype in which modA2 locked OFF bacteria formed discrete tower-like structures (Fig. 3a) and modA2 locked ON bacteria formed a more mat-like biofilm (Fig. 3b). When mixed biofilms were counterstained with the membrane stain FM4-64, to visualize the nonfluorescent bacteria, a mat-like biofilm formed by modA2 locked ON bacteria (pseudocolored blue) surrounded dense tower-like structures formed by modA2 locked OFF bacteria (pseudocolored red) (Fig. 3c). Taken together, these data suggested that the ModA2 phasevarion regulates biofilm formation under alkaline conditions, such as those in the ME during chronic disease, with both modA2 ON and modA2 OFF subpopulations contributing substantially to the large biofilms we have previously observed formed by populations that contain a mixture of modA2 ON bacteria and modA2 OFF bacteria (13). As disease progresses, selection for a predominant modA status, such as the shift to modA2 ON status within the chinchilla middle ear (12), will influence the resultant character of the biofilm within each specific niche.
FIG 3.
Distribution of individual subpopulations within an NTHI strain 723 mixed biofilm. Biofilms were formed by a 1:1 mixture of NTHI strain 723 modA2 locked OFF and modA2 locked ON bacteria. Biofilm cultures were incubated for 24 h at 37°C at pH 9. (a) Representative biofilm formed by a 1:1 mixture of modA2 locked OFF bacteria that constitutively expresses GFP and nonfluorescent modA2 locked ON. The GFP fluorescent modA2 OFF population formed dense tower-like structures, shown in green. (b) Representative biofilm formed by a 1:1 mixture of nonfluorescent modA2 locked OFF bacteria and modA2 locked ON bacteria that constitutively expresses GFP. The GFP fluorescent modA2 ON population formed a more mat-like biofilm, shown in green. (c) Representative biofilm formed by a 1:1 mixture of modA2 locked OFF bacteria that constitutively expresses GFP and nonfluorescent modA2 locked ON bacteria. Biofilms were counterstained with the bacterial outer membrane stain FM4-64 to visualize the modA2 ON population, which does not express GFP. modA2 locked OFF (pseudocolored red) is shown in the left panel, and modA2 locked ON (pseudocolored blue) is shown in the middle panel. The localization and distribution of both subpopulations are shown as a merged image in the panel on the right. Scale bars, 100 μm.
ModA5 regulates biofilm formation in response to alkaline conditions.
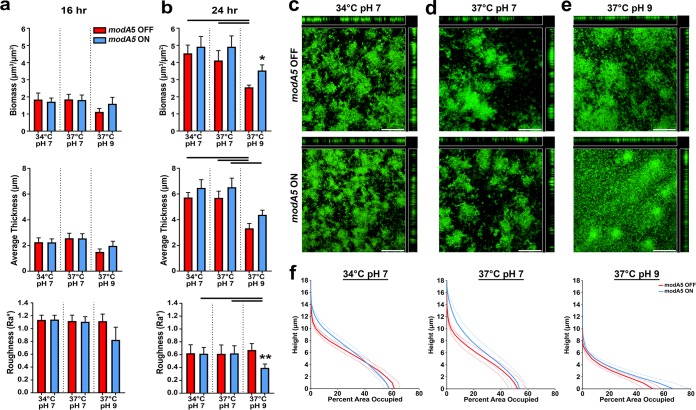
To determine if the above observations were restricted to strains that carry the modA2 allele, we selected a second clinical isolate, NTHI strain 477, to assess biofilm formation by a strain that contained a different modA allele than NTHI strain 723. NTHI strain 477 carries a modA5 allele that is free to phase vary or switch modA status. ModA5 recognizes and methylates a different DNA sequence than other ModA methyltransferases and thus regulates a unique set of genes (12). Biofilm formation was studied with modA5 ON and modA5 OFF populations that were greater than 90% ON or OFF at time of inoculation, but free to switch (12). As such, biofilm formation by strain 477 was expected to be more heterogeneous than those formed by pure strain 723 locked populations that were unable to phase vary. After 16 h of biofilm growth, there were no significant differences between the biofilms as assessed by COMSTAT (Fig. 4a). However, after 24 h there were significant differences in biofilm formation between the modA5 subpopulations when grown at 37°C at pH 9, like what was observed with strain 723 (compare Fig. 4b and Fig. 2b). At an alkaline pH, modA5 ON bacteria formed biofilms that were more mat-like in architecture with significantly greater biomass than those formed by modA5 OFF bacteria (Fig. 4b and e; P = 0.01, unpaired t test). modA5 ON bacteria also formed biofilms with greater biomass at the base of the biofilm, compared to modA5 OFF biofilms; these data are presented as area occupied by layer (Fig. 4f). At a neutral pH, both strain 477 subpopulations formed biofilms with towers and intervening water channels, as was observed with strain 723 at a neutral pH (Fig. 2c and d). To ensure that the biofilm phenotypes observed were due to the indicated modA5 status and not switching, we assayed for this and found that the relative modA5 status for each of the populations (>90% modA5 ON or >90% modA5 OFF at time of inoculation) had not significantly shifted after 24 h of biofilm formation under any condition tested (see Table S2 in the supplemental material). Thereby, even when modA5 was free to phase vary, biofilm formation was significantly different between modA5 ON and modA5 OFF subpopulations when grown at an alkaline pH.
FIG 4.
Biofilm formation by NTHI strain 477. (a and b) Biomass and average thickness and roughness of modA5 ON and modA5 OFF biofilms grown for (a) 16 h or (b) 24 h. Biofilms were analyzed by COMSTAT2, and values are shown as mean ± standard error of the mean. Lines over graphs indicate significance due to growth condition by one-way ANOVA with Tukey’s multiple comparisons (α < 0.05). Asterisks denote significance between subpopulations under each condition: *, P < 0.01, and **, P < 0.001, Student's t test. (c to e) Representative orthogonal image renderings of modA5 ON and modA5 OFF biofilms grown at (c) 34°C at pH 7, (d) 37°C at pH 7, and (e) 37°C at pH 9. Scale bars, 100 μm. (f) Average percentage of area occupied by bacteria at each individual 1-μm optical section (“layer”). Dashed lines indicate standard error of the mean.
Ratio of modA5 OFF to modA5 ON in 24-h biofilms. Biofilms were formed by NTHI strain 477 populations in the predominantly (>90%) modA5 OFF or modA5 ON status at time of seed (inoculation into well of chambered coverglass). Planktonic bacteria as well as adherent biofilm bacteria were collected after 24 h at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9. Fragment length analysis was performed as described previously (12) to identify any shift in population status. No significant shift was observed for any of the samples or conditions tested. Download Table S2, XLSX file, 0.1 MB (13.1KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ModA2 phasevarion regulates proteins required for biofilm formation and structural stability.
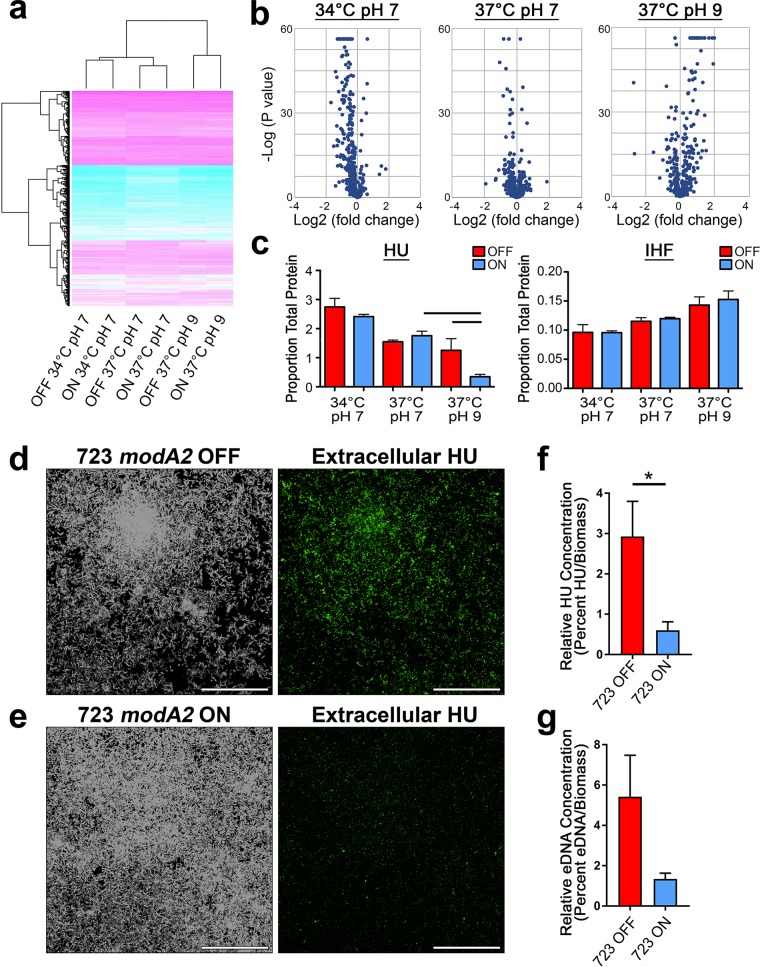
To now elucidate specific differences between the proteomes of biofilms formed under alkaline conditions, proteomic analysis was performed to determine relative protein abundances within biofilms formed by each modA2 subpopulation. Sequential window acquisition of all theoretical mass spectra (SWATH-MS) (32), which is a data-independent acquisition (DIA) mass spectrometry (MS) approach, was used to determine relative quantification of every detectable peptide within the biofilm sample. Biofilms were formed by NTHI strain 723 modA2 locked populations for 16 h under conditions designed to mimic the microenvironments of the human nasopharynx (34°C, pH 7), healthy middle ear (37°C, pH 7), or middle ear during chronic OM (37°C, pH 9). The 16-h time point was selected because significant differences in biofilm structure were observed (Fig. 2) and to avoid potential effects of replacing the growth medium. The biofilms were rinsed to remove any nonadherent bacteria, and then the entire biofilm mass was collected in order to analyze the total protein content of the biofilm; this included total bacterial proteins as well as proteins integrated into the biofilm extracellular matrix. SWATH-MS proteomic analysis identified the abundance of a total of 644 proteins in each sample. The most important environmental determinant of protein expression in these samples was pH, followed by temperature, as depicted in the clustered heat map in Fig. 5a. Significant differences in protein abundance between the modA2 locked ON and modA2 locked OFF populations at each condition are represented as volcano plots (Fig. 5b). The x axis indicates relative fold difference in protein abundance in modA2 ON biofilms compared to modA2 OFF biofilms; the y axis indicates statistical significance. The greatest differences in ModA2-dependent protein expression occurred at 37°C at pH 9. Proteins with significant changes in abundance (log2 fold change [FC] of ≥1 or ≤−1, P < 0.05) between modA2 locked ON biofilms and modA2 locked OFF biofilms are presented in Table S3 in the supplemental material.
FIG 5.
Relative protein abundance within NTHI strain 723 biofilms. (a) Clustered heat map of protein abundance within biofilms formed by NTHI strain 723 at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9. (b) Volcano plots of protein abundance between modA2 locked ON biofilms versus modA2 locked OFF biofilms. (c) Relative protein abundance of the DNABII proteins HU and IHF as percentage of proportion of total protein. Lines over the graph indicate adjusted P value of <1.00E−12. (d to e) Laser-scanning confocal images of (d) modA2 locked OFF biofilms and (e) modA2 locked ON biofilms imaged for total biomass (left [white]) and extracellular HU (right [green]). Scale bars, 30 μm. (f) Relative HU abundance within the biofilms normalized to total biomass. *, P < 0.05, unpaired t test. (g) Relative eDNA abundance within the biofilms normalized to total biomass.
NTHI strain 723 SWATH results. Relative protein abundance in biofilms formed by NTHI strain 723 modA2 locked ON and modA2 locked OFF populations at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9. Log2 fold change of ≥1. Download Table S3, XLSX file, 0.1 MB (15.7KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
At 34°C at pH 7, we observed a 2-fold or greater reduction (log2 FC of ≤−1) in relative abundance of 15 proteins within biofilms formed by modA2 locked OFF bacteria compared to those formed by modA2 locked ON bacteria. The abundance of only one protein changed greater than 2-fold at 37°C at pH 7. This protein, an alkylhydroperoxidase (723_00583), was more abundant in modA2 locked ON biofilms compared to modA2 locked OFF biofilms (log2 FC of −1.05). Under conditions that mimic those within the ME during disease (37°C, pH 9), there was a 6.6-fold increase (log2 FC of −2.72) of a putative Fe2+-trafficking protein (723_00852) in the modA2 locked ON biofilms compared to the modA2 locked OFF biofilms. A total of 19 proteins were 2-fold or less (log2 FC of ≤−1) abundant in the modA2 locked ON biofilms, and 5 of these proteins were ribosomal proteins (Table S3). There was also a significant decrease in abundance of the DNABII DNA-binding protein HU (723_01259) within biofilms formed by modA2 locked ON bacteria compared to those formed by modA2 locked OFF bacteria (log2 FC = 1.70, P < 1E−15). HU abundance was only significantly decreased within biofilms formed by modA2 locked ON bacteria at pH 9, the same conditions under which the modA2 locked ON subpopulation formed the distinct mat-like biofilm (Fig. 5c). There were no significant differences in the abundance of the DNABII DNA-binding protein IHF (723_00064) within these early biofilms (Fig. 5c). These results indicated the role of the phasevarion in regulation of numerous proteins, but more intriguing was the fact that the greatest changes in protein abundance occurred under physiologically relevant conditions, such as the pHs and temperatures that mimic those within the nasopharynx and diseased middle ear.
To confirm the decreased abundance of HU within biofilms formed by modA2 locked ON bacteria at pH 9, we performed immunofluorescence. Biofilms were formed by the modA2 locked OFF and modA2 locked ON subpopulations for 16 h at 37°C at pH 9 and then labeled with a polyclonal antibody directed against NTHI HU in order to determine the relative abundance and distribution of extracellular HU within these biofilms. Significantly more relative HU was present within biofilms formed by modA2 locked OFF bacteria than in biofilms formed by modA2 locked ON bacteria (compare Fig. 5d and e and see Fig. 5f; P = 0.03, unpaired t test) confirming the SWATH-MS results.
HU binds to DNA within the extracellular matrix of biofilms. Therefore, we utilized immunofluorescence to determine potential differences in extracellular DNA (eDNA) within biofilms formed under alkaline conditions (37°C, pH 9). Sixteen-hour biofilms formed by the 723 modA2 locked OFF or 723 modA2 locked ON populations were labeled for the presence of eDNA with commercial antibody directed against double-stranded DNA (dsDNA). Biofilms formed by the modA2 OFF population contained more relative eDNA; however, considerable variation was observed with the OFF population due to the heterogeneous nature of the biofilms, and thereby, this difference was not statistically significant (Fig. 5g; P = 0.062, unpaired t test). Multiple factors can contribute to the integration of eDNA and DNA-binding proteins into the biofilm matrix; however, lower abundance of these components is consistent with the observed decrease in overall structure of biofilms formed by the modA2 ON population. Increased adherence by modA2 ON bacteria, as suggested by increased bacterial density near the base of the biofilms, likely contributes to the greater overall biomass observed under alkaline conditions.
To determine if HU expression was regulated at the transcriptional level, we performed RNA sequencing to compare transcriptional profiles of NTHI within biofilms formed under alkaline conditions. RNA was isolated from 16-h biofilms formed by strain 723 modA2 OFF and strain 723 modA2 ON populations at 37°C at pH 9. Surprisingly, there was no significant difference in relative expression of hupA (HU) between modA2 OFF and modA2 ON populations (see Table S4 in the supplemental material). The hupA expression result was confirmed by quantitative reverse transcription-PCR (RT-PCR). As hupA transcription was not directly regulated by ModA2, alternative mechanisms, such as translational regulation or posttranslation modification may occur. Differences in eDNA abundance or composition may also affect HU integration into the biofilm matrix.
NTHI strain 723 complete RNA sequencing results. Relative transcript abundance in biofilms formed by NTHI strain 723 modA2 ON and modA2 OFF populations at 37°C at pH 9. Differential gene expression was determined with the Bioconductor package EdgeR. Download Table S4, XLSX file, 0.2 MB (209.9KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Based on RNA sequencing results, 9 genes were significantly upregulated and 8 genes were significantly downregulated more than 2-fold in biofilms formed by modA2 ON bacteria compared to biofilms formed by modA2 OFF bacteria (Table 1). The gene that encodes the high-molecular-weight (HMW) adhesin was significantly upregulated by the modA2 ON population compared to the modA2 OFF population during alkaline biofilm formation. Members of the HMW family of adhesins are involved in attachment to epithelial surfaces and in colonization (33, 34). Several genes that encode putative phage proteins were also overexpressed in the modA2 ON biofilm samples. In contrast, the dimethyl sulfoxide (DMSO) reductase operon, which is involved in anaerobic respiration, was significantly upregulated in modA2 OFF biofilms compared to modA2 ON biofilms. Components of the nitrate and nitrite reductases were also significantly upregulated, as were several genes involved in cytochrome c maturation (Table S4), which suggests a shift to a more anaerobic metabolism within biofilms formed by the modA2 OFF population.
TABLE 1.
Significant differential gene expression in NTHI strain 723 biofilms formed by modA2 locked ON bacteria compared to modA2 locked OFF bacteria
| Gene | Protein description | Log FC | FC | P value |
|---|---|---|---|---|
| Greater expression in modA2 ON | ||||
| NTHI723_01859 | Hypothetical protein (phage) | −2.25 | 4.76 | 1.09E−04 |
| NTHI723_01862 | Phage minor tail protein U | −1.83 | 3.56 | 1.12E−04 |
| NTHI723_01861 | Prophage minor tail protein Z (GPZ) | −1.68 | 3.21 | 3.54E−05 |
| hxuA_1 | HMW adhesin | −1.67 | 3.17 | 3.54E−05 |
| NTHI723_01863 | Hypothetical protein (phage) | −1.56 | 2.95 | 1.09E−04 |
| fis | Hin recombinational enhancer-binding protein | −1.27 | 2.40 | 6.14E−03 |
| glxK | Glycerate kinase | −1.22 | 2.33 | 9.08E−04 |
| gsiA_5 | Glutathione import ATP-binding protein GsiA | −1.13 | 2.19 | 2.04E−05 |
| clpP2 | ATP-dependent Clp protease proteolytic subunit 2 | −1.10 | 2.15 | 1.09E−04 |
| Greater expression in modA2 OFF | ||||
| yhjE_1 | Inner membrane metabolite transport protein | 1.25 | 2.37 | 8.55E−03 |
| NTHI723_00144 | Hypothetical protein | 1.21 | 2.32 | 4.82E−02 |
| dmsD | Twin-arginine leader-binding protein DmsD | 1.17 | 2.24 | 3.44E−04 |
| thiE | Thiamine-phosphate synthase | 1.15 | 2.21 | 3.64E−02 |
| ccmE | Heme chaperone CcmE | 1.09 | 2.14 | 4.25E−04 |
| dmsA | DMSO reductase DmsA precursor | 1.05 | 2.07 | 4.17E−04 |
| dmsC | DMSO reductase anchor subunit | 1.03 | 2.04 | 9.80E−05 |
| clpB | Heat shock protein F84.1 | 1.00 | 2.00 | 1.07E−02 |
DISCUSSION
In this study, we demonstrated how the phasevarions of multiple NTHI clinical isolates influenced biofilm formation under host-relevant microenvironmental conditions. The most significant differences occurred at the pH and temperature found within the ME during chronic disease. We also found that multiple phasevarions (modA2, -A5, and -A10), which each recognize a unique DNA sequence, regulated similar patterns of biofilm formation under specific conditions that mimic those in the ME during chronic OM (37°C, pH 9). Yet, under neutral conditions at the temperature of the nasopharynx or middle ear, biofilm formation was diverse among these strains. Furthermore, other phasevarions, such as ModA9 of NTHI strain 1209, did not affect biofilm formation under any of the conditions tested. NTHI phasevarions appeared to regulate biofilm formation in a microenvironment-dependent fashion.
Phasevarions have been identified in multiple human mucosa-associated pathogens, all of which are known to form biofilms during pathogenesis and disease. The phasevarions of several species have been shown to regulate disease-relevant phenotypes such as the ability to acquire nutrients and resistance to oxidative stress (29). Here we show for the first time that the phasevarions of multiple NTHI clinical isolates regulated biofilm formation under host-specific microenvironmental conditions, yet others, such as the ModA9 phasevarion of strain 1209, had no effect on biofilm formation. While ModA9 did not impact biofilm formation by NTHI strain 1209 under the conditions tested here, ModA9 does significantly affect other phenotypes, such as expression of the outer membrane protein OMP P6 (12). The conservation of the relationship between multiple phasevarions and biofilm formation under the conditions of chronic ME disease is remarkable and signifies the importance of phasevarion-influenced biofilm formation within this microenvironment. Laser-scanning confocal microscopy of biofilms formed at an alkaline pH revealed unique differences in biofilm architecture formed between the ON and OFF subpopulations of the modA2 and modA5 representative strains. Under alkaline conditions, the modA2 ON and modA5 ON populations formed mat-like biofilms with significantly less discrete towers and water channels than those formed by the modA2 OFF and modA5 OFF populations at pH 9 or biofilms formed by either subpopulation at a neutral pH. Several models of biofilm formation suggest that more heterogeneous biofilms with tower-like structures and intervening water channels may allow for better exchange of nutrients and oxygen into the biofilm, as well as removal of waste from the biofilm (35, 36). Biofilm formation and maturation are a dynamic process with ongoing changes in gene and protein expression over time (26, 37). As such, the impact of phasevarion-induced differences in biofilm architecture may become more important as the biofilms mature. Future studies that assess biofilms formed for greater than 24 h may reveal additional long-term benefits of a particular phasevarion status or switching on the persistence of NTHI within biofilms.
While the effect of changes in NTHI biofilm architecture in vivo is not yet known, loss of biofilm structural stability has been shown to increase bacterial dispersion in a chinchilla model of OM (38). Biofilm formation is a major factor in bacterial recalcitrance and persistence within the middle ear. Increased resistance to antimicrobials is attributed to changes in bacterial metabolism and decreased growth rates (27). Regulatory mechanisms that balance the need for nutrient acquisition and growth with the benefits of biofilm quiescence will facilitate effective microniche adaptation and survival. While NTHI biofilm formation is important in colonization of the nasopharynx, NTHI can utilize mechanisms such as the phasevarion to also regulate biofilm formation in response to stress or microenvironmental changes, such as the alkaline condition found in the ME during chronic disease.
Under alkaline conditions, the relative abundances of biofilm-associated proteins were different between biofilms formed by the modA2 locked OFF and modA2 locked ON populations. Differences in nutrient availability and stress could contribute to the observed changes in iron acquisition, oxidative stress, and metabolic proteins within these biofilms. In addition, differences in protein expression and biofilm formation on mucosal surfaces diminish host response and recognition and can alter the effectiveness of innate immune defenses (39–41).
Biofilms formed by modA2 ON bacteria contained significantly less of the bacterial DNABII DNA-binding protein HU. HU and the related IHF play a critical role in biofilm structure by stabilization of extracellular DNA within the biofilm matrix (30, 42). Strategies directed against these DNA-binding proteins are highly effective at disrupting biofilms formed in vitro and in vivo and present very promising targets for biofilm prevention and treatment (38, 43). We also found that biofilms formed by modA2 ON bacteria at an alkaline pH had relatively less eDNA than comparable biofilms formed by modA2 OFF bacteria. Decreased extracellular DNA in biofilms formed by the modA2 ON population corresponded with the reduced abundance of HU integrated into the adherent biomass. As eDNA and DNABII proteins are required to build and maintain the stability of the biofilm matrix, loss of these critical structural components would be expected to result in biofilms similar to the less-structured and more mat-like biofilms formed by modA2 ON bacteria under alkaline conditions. Future work is necessary to fully understand the integration of eDNA and DNA binding proteins into the biofilm matrix under alkaline conditions and the role of the phasevarion in this process.
The most significant differences in biofilm formation occurred at an alkaline pH, and pH was the strongest driver overall for changes in the biofilm proteomes. Middle-ear effusions of children with chronic OM and chinchillas during experimental OM are alkaline in nature and can reach a pH of 9 or greater (16). Such alkaline conditions are uncommon elsewhere in the human body. The results of this study highlight the need to understand the influence of this alkaline environment on bacteria within the ME during infection. While clear differences in biofilm formation by NTHI strain 723 were obvious when grown under alkaline conditions that mimic the diseased ME, there was no difference in biofilm formation by this strain when grown under standard laboratory conditions at 37°C and at a neutral pH. Biofilm formation was affected by phasevarion status in NTHI strains 477, R2866, and C486, which carry the modA5, modA10, and modA4 alleles, respectively. Interestingly, strain R2866, which formed biofilms with the greatest biomass and thickness under all conditions tested, is the only blood isolate assessed and also the only strain that expresses the Hia adhesin (44, 45). In strain R2866, Hia expression can be regulated via ModA10-independent phase variation of the poly(T) tract in the promoter region (12). Additional work is necessary to determine the specific role of Hia in biofilm formation by strain R2866. All other strains tested express the high-molecular-weight (HWM) adhesins. In biofilms formed at 37°C at pH 9, there was greater expression of the HWM gene by modA2 ON compared to modA2 OFF. Increased expression of adhesins such as HMW adhesin may contribute to greater attachment to the coverslip, and as such, the unique mat-like architecture observed with the modA2 ON population at an alkaline pH. Regulation of the different adhesins expressed by each of these strains likely plays an important role in the biofilm phenotypes observed, as well as adherence and colonization, and will require further investigation (46).
The presence of phasevarions in pathogenic bacteria adds an extra level of complexity to understanding bacterial responses to changing conditions and microenvironments, and it is critical to understand the implication of these mechanisms for disease. Highly effective treatment and prevention strategies will require a more complete understanding of not only which virulence factors are being regulated, but also when and where they are expressed. It is also important to consider the phase of bacterial lifestyle (planktonic versus biofilm versus newly released from a biofilm) in the development of vaccines, as expression of bacterial antigens can vary significantly (47). Here, identification of genes and proteins regulated by the NTHI phasevarion during biofilm formation at disease-relevant pHs and temperatures will facilitate continued research to better understand the role of these proteins in biofilm formation over the course of disease. Furthermore, interventions that eradicate only a single modA subpopulation may lead to a shift in modA status and result in more severe disease (13). Therefore, an increased understanding of the role of the phasevarion in disease processes, such as biofilm formation, will facilitate the development of new combinatorial strategies that synergistically target the entire NTHI population regardless of modA status, to not only prevent but also treat chronic diseases due to nontypeable Haemophilus influenzae.
MATERIALS AND METHODS
Bacterial strains and culture medium.
NTHI strains 723, 477, and 1209 were received from the Finnish Otitis Media study group (48). NTHI strain C486 was isolated from a child with otitis media (49), and NTHI strain R2866 was isolated from the blood of a child with meningitis (50). NTHI cells were routinely cultured in brain heart infusion broth supplemented with hemin (2 µg/ml) and NAD (2 µg/ml) (sBHI) or on chocolate agar and grown at 37°C with 5% CO2.
Biofilm formation.
Biofilms were formed by NTHI cultured within chambers of eight-well-chambered coverglass slides (Thermo Scientific, Waltham, MA) as described previously (51). Briefly, mid-log-phase cultures of NTHI strains were diluted with buffered sBHI that contained 100 mM HEPES (Fisher BioReagents), adjusted to pH 7 or 100 mM TAPS (Sigma-Aldrich), adjusted to pH 9. NTHI cells were inoculated at 4 × 104 CFU in a 200-μl total volume per well and slides were incubated at 34 or 37°C, as indicated, with 5% atmospheric CO2. Biofilms were grown for either 16 or 24 h, with the growth medium replaced after 16 h. To visualize, biofilms were stained with LIVE/DEAD BacLight stain (Life Technologies) and fixed overnight in fixative (1.6% paraformaldehyde, 2.5% glutaraldehyde, and 4% acetic acid in 0.1 M phosphate buffer, pH 7.4). Fixative was replaced with saline before imaging with a Zeiss 510 Meta-laser scanning confocal microscope. Images were rendered with Zeiss Zen software.
Analysis of biofilm formation and architecture.
z-stack images acquired at 63× with a Zeiss 510 Meta-laser scanning confocal microscope were analyzed by COMSTAT2 to determine biomass (μm3/μm2), average thickness (μm), roughness (Ra) and percentage of area occupied by layers (52–54). Area occupied by layer was plotted as percentage of bacterial biomass coverage per 1-μm optical section from the base of the biofilm. The standard error of the mean for replicate biofilms was calculated for each individual layer with GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA).
Formation of mixed biofilms with NTHI strain 723.
Biofilms were formed as indicated above in sBHI buffered with 100 mM TAPS and adjusted to a pH of 9. NTHI were inoculated into the chambers of an eight-well-chambered cover glass as a 1:1 mixture of modA2 locked OFF bacteria that constitutively expressed green fluorescent protein and nonfluorescent modA2 locked ON bacteria. A total of 4 × 104 CFU of NTHI (2 × 104 cells of each subpopulation) were seeded in a total volume of 200 μl per well. Cultures were incubated at 37°C with 5% atmospheric CO2 for 24 h. The medium was replaced with fresh medium after 16 h. Biofilms were rinsed with Dulbecco’s phosphate-buffered saline (DPBS) and then counterstained with the fluorescent bacterial outer membrane stain FM4-64 (Thermo Fisher, Waltham, MA) to visualize the modA2 locked ON subpopulation. The biofilms were fixed overnight in fixative (1.6% paraformaldehyde, 2.5% glutaraldehyde, and 4% acetic acid in 0.1 M phosphate buffer, pH 7.4) and the fixative was replaced with saline before imaging with a Zeiss 510 Meta-laser scanning confocal microscope. Images were rendered with Zeiss Zen software.
SWATH-MS analysis.
NTHI cells were inoculated into the wells of 96-well flat-bottom microtiter plates (Corning, Corning, NY) and incubated for 16 h at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9 with 5% atmospheric CO2 in buffered sBHI, as described above. Biofilms were rinsed once with DPBS, and then the total adherent biomass was suspended in DPBS with aggressive scraping. The biomass was collected by centrifugation and extracted with TRIzol reagent (ThermoFisher, Waltham, MA) at 107 CFU per 750 μl. Isopropanol was added to the phenol-ethanol supernatant of the TRIzol-extracted samples. Proteins were pelleted by centrifugation at 12,000 × g for 10 min at 4°C, washed three times with 95% ethanol, and precipitated with 100% ethanol. The protein pellets were resuspended in 200 μl 6 M guanidinium-HCl–50 mM Tris-HCl (pH 8). Proteins were then reduced and alkylated by 10 mM dithiothreitol (DTT) and 25 mM acrylamide, respectively. The resultant proteins were precipitated with 1 ml of 1:1 methanol/acetone and then suspended in 50 μl of 50 mM Tris-HCl (pH 8) with 1 μg of trypsin and incubated overnight at 37°C. Tryptic-digested peptides were cleaned with C18 ZipTips (Millipore, Burlington, MA). SWATH-MS analysis was performed as described previously (55).
Detection of HU and eDNA within biofilms.
NTHI strain 723 modA2 locked ON and modA2 locked OFF populations that were engineered to constitutively expressed GFP (29) were inoculated into the chambers of an eight-well-chambered cover glass, as described above. After 16 h, biofilms were rinsed twice with DPBS to remove nonadherent bacteria and then incubated with anti-NTHI HU polyclonal rabbit sera (1:200) for 30 min or commercially available murine antibody against dsDNA (AbCam) (1:200) for 1 h. Biofilms were then rinsed and incubated for 15 min with goat anti-rabbit or goat anti-mouse IgG secondary antibody conjugated to Alexa Fluor 647 (Thermo Fisher, Waltham, MA). Biofilms were rinsed with DPBS and imaged with a Zeiss 700 Meta-laser scanning confocal microscope. Relative HU and eDNA abundance was calculated as total fluorescence in the Alexa Fluor 647 channel divided by total biomass in the GFP channel, multiplied by 100.
RNA sequencing.
NTHI were inoculated into the wells of 96-well flat-bottom microtiter plates (Corning, Corning, NY) and incubated for 16 h at 37°C with 5% atmospheric CO2 in sBHI buffered to pH 9, as described above. Biofilms were rinsed once with DPBS, and then the total adherent biomass was collected. Total bacterial RNA was collected by acid-phenol-chloroform extraction and purified with the RNAeasy kit (Qiagen, Hilden, Germany), according to the manufacturer’s directions. Library preparation and sequencing were performed as described previously (56). Differential gene expression was determined with the Bioconductor package EdgeR (57).
Statistical analysis.
All statistical analyses were performed with GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA). The statistical tests used and P values are indicated within the text and figure legends. All experiments were performed a minimum of three times on separate days in duplicate.
ACKNOWLEDGMENTS
We thank Amy Wetzel, James Fitch, and Peter White in the Institute for Genomic Medicine at Nationwide Children’s Hospital for assistance with RNA sequencing. We acknowledge the assistance of The University of Queensland School of Chemistry and Molecular Biosciences mass spectrometry facility. We thank Jennifer Neelans for manuscript preparation.
This work was funded by NIH/NIDCD R01DC003915 to L.O.B. and NIH/NIDCD R01DC015688 to L.O.B. and M.P.J. and NHMRC Fellowship 1138466 to M.P.J.
K.L.B., M.P.J., and L.O.B., conceived the research project and designed the experiments. K.L.B., P.N.A., M.T.B., J.M.A., B.L.S., and F.E.J. performed the experiments and analyzed the experimental results. K.L.B. and P.N.A. prepared the manuscript. J.M.A., B.L.S., F.E.J., M.P.J., and L.O.B. edited the manuscript. All authors reviewed the manuscript.
Footnotes
Citation Brockman KL, Azzari PN, Branstool MT, Atack JM, Schulz BL, Jen FE-C, Jennings MP, Bakaletz LO. 2018. Epigenetic regulation alters biofilm architecture and composition in multiple clinical isolates of nontypeable Haemophilus influenzae. mBio 9:e01682-18. https://doi.org/10.1128/mBio.01682-18.
REFERENCES
- 1.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J 28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- 2.Ruohola A, Meurman O, Nikkari S, Skottman T, Salmi A, Waris M, Osterback R, Eerola E, Allander T, Niesters H, Heikkinen T, Ruuskanen O. 2006. Microbiology of acute otitis media in children with tympanostomy tubes: prevalences of bacteria and viruses. Clin Infect Dis 43:1417–1422. doi: 10.1086/509332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter LL, Margolis RH, Giebink GS. 1994. Identification of hearing loss in children with otitis media. Ann Otol Rhinol Laryngol Suppl 163:59–61. [DOI] [PubMed] [Google Scholar]
- 4.Paradise JL, Feldman HM, Campbell TF, Dollaghan CA, Rockette HE, Pitcairn DL, Smith CG, Colborn DK, Bernard BS, Kurs-Lasky M, Janosky JE, Sabo DL, O'Connor RE, Pelham WE Jr.. 2007. Tympanostomy tubes and developmental outcomes at 9 to 11 years of age. N Engl J Med 356:248–261. doi: 10.1056/NEJMoa062980. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld RM, Schwartz SR, Pynnonen MA, Tunkel DE, Hussey HM, Fichera JS, Grimes AM, Hackell JM, Harrison MF, Haskell H, Haynes DS, Kim TW, Lafreniere DC, LeBlanc K, Mackey WL, Netterville JL, Pipan ME, Raol NP, Schellhase KG. 2013. Clinical practice guideline: tympanostomy tubes in children. Otolaryngol Head Neck Surg 149:S1–35. doi: 10.1177/0194599813487302. [DOI] [PubMed] [Google Scholar]
- 6.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A 102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, Fox KL, Seib KL, Maguire TL, Wang AH, Maiden MC, Grimmond SM, Apicella MA, Jennings MP. 2009. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog 5:e1000400. doi: 10.1371/journal.ppat.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP. 2011. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One 6:e27569. doi: 10.1371/journal.pone.0027569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, Seib KL. 2014. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J 28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 11.Srikhanta YN, Fung KY, Pollock GL, Bennett-Wood V, Howden BP, Hartland EL. 2017. Phasevarion-regulated virulence in the emerging pediatric pathogen Kingella kingae. Infect Immun 85:e00319-17. doi: 10.1128/IAI.00319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, Grimmond SM, Smith AL, Barenkamp SJ, Korlach J, Bakaletz LO, Jennings MP. 2015. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun 6:7828. doi: 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockman KL, Jurcisek JA, Atack JM, Srikhanta YN, Jennings MP, Bakaletz LO. 2016. ModA2 phasevarion switching in nontypeable Haemophilus influenzae increases the severity of experimental otitis media. J Infect Dis 214:817–824. doi: 10.1093/infdis/jiw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb P. 1951. Air temperatures in respiratory tracts of resting subjects in cold. J Appl Physiol 4:378–382. doi: 10.1152/jappl.1951.4.5.378. [DOI] [PubMed] [Google Scholar]
- 15.Nuutinen J, Torkkeli T, Penttila I. 1993. The pH of secretions in sinusitis and otitis media. J Otolaryngol 22:79–82. [PubMed] [Google Scholar]
- 16.Wezyk MT, Makowski A. 2000. pH of fluid collected from middle ear in the course of otitis media in children. Otolaryngol Pol 54:131–133. [PubMed] [Google Scholar]
- 17.Chamberlain JM, Terndrup TE, Alexander DT, Silverstone FA, Wolf-Klein G, O'Donnell R, Grandner J. 1995. Determination of normal ear temperature with an infrared emission detection thermometer. Ann Emerg Med 25:15–20. doi: 10.1016/S0196-0644(95)70349-7. [DOI] [PubMed] [Google Scholar]
- 18.Ishak N, Tikhomirova A, Bent SJ, Ehrlich GD, Hu FZ, Kidd SP. 2014. There is a specific response to pH by isolates of Haemophilus influenzae and this has a direct influence on biofilm formation. BMC Microbiol 14:47. doi: 10.1186/1471-2180-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slinger R, Chan F, Ferris W, Yeung SW, St Denis M, Gaboury I, Aaron SD. 2006. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis 56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB Jr.. 2006. Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med 174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swords WE. 2012. Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front Cell Infect Microbiol 2:97. doi: 10.3389/fcimb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, Daigle BJ, Ehrlich MD, Post JC. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710–1715. doi: 10.1001/jama.287.13.1710. [DOI] [PubMed] [Google Scholar]
- 24.de Beer D, Stoodley P, Roe F, Lewandowski Z. 1994. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng 43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 25.Stewart PS. 2012. Mini-review: convection around biofilms. Biofouling 28:187–198. doi: 10.1080/08927014.2012.662641. [DOI] [PubMed] [Google Scholar]
- 26.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Post DM, Held JM, Ketterer MR, Phillips NJ, Sahu A, Apicella MA, Gibson BW. 2014. Comparative analyses of proteins from Haemophilus influenzae biofilm and planktonic populations using metabolic labeling and mass spectrometry. BMC Microbiol 14:329. doi: 10.1186/s12866-014-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brockman KL, Branstool MT, Atack JM, Robledo-Avila F, Partida-Sanchez S, Jennings MP, Bakaletz LO. 2017. The ModA2 phasevarion of nontypeable Haemophilus influenzae regulates resistance to oxidative stress and killing by human neutrophils. Sci Rep 7:3161. doi: 10.1038/s41598-017-03552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. 2011. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol 4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 31.Novotny LA, Jurcisek JA, Ward MO Jr, Jordan ZB, Goodman SD, Bakaletz LO. 2015. Antibodies against the majority subunit of type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol 96:276–292. doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St Geme JW III, Falkow S, Barenkamp SJ. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci U S A 90:2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buscher AZ, Burmeister K, Barenkamp SJ, St Geme JW III.. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J Bacteriol 186:4209–4217. doi: 10.1128/JB.186.13.4209-4217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das J, Mokrzan E, Lakhani V, Rosas L, Jurcisek JA, Ray WC, Bakaletz LO. 2017. Extracellular DNA and type IV pilus expression regulate the structure and kinetics of biofilm formation by nontypeable Haemophilus influenzae. mBio 8:e01466-17. doi: 10.1128/mBio.01466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davit Y, Byrne H, Osborne J, Pitt-Francis J, Gavaghan D, Quintard M. 2013. Hydrodynamic dispersion within porous biofilms. Phys Rev E Stat Nonlin Soft Matter Phys 87:012718. doi: 10.1103/PhysRevE.87.012718. [DOI] [PubMed] [Google Scholar]
- 37.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novotny LA, Jurcisek JA, Goodman SD, Bakaletz LO. 2016. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine 10:33–44. doi: 10.1016/j.ebiom.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahearn CP, Gallo MC, Murphy TF. 2017. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis 75:ftx042. doi: 10.1093/femspd/ftx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones EA, McGillivary G, Bakaletz LO. 2013. Extracellular DNA within a nontypeable Haemophilus influenzae-induced biofilm binds human beta defensin-3 and reduces its antimicrobial activity. J Innate Immun 5:24–38. doi: 10.1159/000339961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong W, Juneau RA, Pang B, Swords WE. 2009. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J Innate Immun 1:215–224. doi: 10.1159/000205937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurcisek JA, Brockman KL, Novotny LA, Goodman SD, Bakaletz LO. 2017. Nontypeable Haemophilus influenzae releases DNA and DNABII proteins via a T4SS-like complex and ComE of the type IV pilus machinery. Proc Natl Acad Sci U S A 114:E6632–E6641. doi: 10.1073/pnas.1705508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rocco CJ, Davey ME, Bakaletz LO, Goodman SD. 2017. Natural antigenic differences in the functionally equivalent extracellular DNABII proteins of bacterial biofilms provide a means for targeted biofilm therapeutics. Mol Oral Microbiol 32:118–130. doi: 10.1111/omi.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ecevit IZ, McCrea KW, Pettigrew MM, Sen A, Marrs CF, Gilsdorf JR. 2004. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J Clin Microbiol 42:3065–3072. doi: 10.1128/JCM.42.7.3065-3072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, Unrath WC, Campos J, Crook DW, Farley MM, Henderson FW, Jacobs RF, Muhlemann K, Satola SW, van Alphen L, Golomb M, Smith AL. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect Immun 73:5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atack JM, Day CJ, Poole J, Brockman KL, Bakaletz LO, Barenkamp SJ, Jennings MP. 2018. The HMW2 adhesin of non-typeable Haemophilus influenzae is a human-adapted lectin that mediates high-affinity binding to 2-6 linked N-acetylneuraminic acid glycans. Biochem Biophys Res Commun 503:1103–1107. doi: 10.1016/j.bbrc.2018.06.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harro JM, Peters BM, O'May GA, Archer N, Kerns P, Prabhakara R, Shirtliff ME. 2010. Vaccine development in Staphylococcus aureus: taking the biofilm phenotype into consideration. FEMS Immunol Med Microbiol 59:306–323. doi: 10.1111/j.1574-695X.2010.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cody AJ, Field D, Feil EJ, Stringer S, Deadman ME, Tsolaki AG, Gratz B, Bouchet V, Goldstein R, Hood DW, Moxon ER. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect Genet Evol 3:57–66. doi: 10.1016/S1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nizet V, Colina KF, Almquist JR, Rubens CE, Smith AL. 1996. A virulent nonencapsulated Haemophilus influenzae. J Infect Dis 173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- 51.Jurcisek JA, Dickson AC, Bruggeman ME, Bakaletz LO. 2011. In vitro biofilm formation in an 8-well chamber slide. J Vis Exp doi: 10.3791/2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 53.Anonymous Comstat www.comstat.dk.
- 54.Vorregaard M. 2008. Comstat2—a modern 3D image analysis environment for biofilms. Thesis. Informatics and Mathematical Modelling, Technical University of Denmark, Kongens Lyngby, Denmark. [Google Scholar]
- 55.Peak IR, Chen A, Jen FE, Jennings C, Schulz BL, Saunders NJ, Khan A, Seifert HS, Jennings MP. 2016. Neisseria meningitidis lacking the major porins PorA and PorB is viable and modulates apoptosis and the oxidative burst of neutrophils. J Proteome Res 15:2356–2365. doi: 10.1021/acs.jproteome.5b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santana EA, Harrison A, Zhang X, Baker BD, Kelly BJ, White P, Liu Y, Munson RS Jr.. 2014. HrrF is the fur-regulated small RNA in nontypeable Haemophilus influenzae. PLoS One 9:e105644. doi: 10.1371/journal.pone.0105644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikolayeva O, Robinson MD. 2014. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol 1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of NTHI growth on the pH of buffered sBHI. NTHI cells were inoculated into sBHI supplemented with 0.1 M HEPES or 0.1 M TAPS and adjusted to a pH of 6, 7, 8, or 9. Medium was collected after 6 or 12 h, and the pH was measured. sBHI buffered to a starting pH of 9 dropped to an approximate pH of 8. sBHI buffered to a starting pH of 8 dropped to an approximate pH of 7. sBHI buffered to a starting pH of 7 dropped to an approximate pH of 6.4. sBHI buffered to a starting pH of 6 dropped to an approximate pH of 5.2. Starting pHs of 7 and 9 were selected to represent neutral and alkaline conditions, respectively. Download FIG S1, TIF file, 0.1 MB (62.6KB, tif) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis of biomass and roughness of biofilms formed by NTHI clinical isolates that represent the 5 most common modA alleles. Student’s t test was performed to compare modA OFF versus modA ON populations under each of the conditions tested, as indicated. One-way ANOVA with multiple comparisons (Holms-Sidak) was used to compare the effect of microenvironmental conditions among members of a single modA subpopulation. Download Table S1, XLSX file, 0.1 MB (14.1KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of NTHI strain 723 modA2 ON and modA2 OFF subpopulations. Bacteria were diluted into buffered sBHI adjusted to a starting pH of 6, 7, 8, or 9 and seeded into wells of a 96-well flat-bottom plate. Plates were incubated shaking at either 34 or 37°C with 5% atmospheric CO2. Absorbance at 490 nm was recorded every 15 min for 12 h. No significant differences in planktonic growth rates occurred between the modA2 ON (circles) or OFF (squares) populations at any of the temperatures or pHs tested. Download FIG S2, TIF file, 0.5 MB (510.8KB, tif) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ratio of modA5 OFF to modA5 ON in 24-h biofilms. Biofilms were formed by NTHI strain 477 populations in the predominantly (>90%) modA5 OFF or modA5 ON status at time of seed (inoculation into well of chambered coverglass). Planktonic bacteria as well as adherent biofilm bacteria were collected after 24 h at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9. Fragment length analysis was performed as described previously (12) to identify any shift in population status. No significant shift was observed for any of the samples or conditions tested. Download Table S2, XLSX file, 0.1 MB (13.1KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
NTHI strain 723 SWATH results. Relative protein abundance in biofilms formed by NTHI strain 723 modA2 locked ON and modA2 locked OFF populations at 34°C at pH 7, 37°C at pH 7, or 37°C at pH 9. Log2 fold change of ≥1. Download Table S3, XLSX file, 0.1 MB (15.7KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.
NTHI strain 723 complete RNA sequencing results. Relative transcript abundance in biofilms formed by NTHI strain 723 modA2 ON and modA2 OFF populations at 37°C at pH 9. Differential gene expression was determined with the Bioconductor package EdgeR. Download Table S4, XLSX file, 0.2 MB (209.9KB, xlsx) .
Copyright © 2018 Brockman et al.
This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.