Abstract
In recent years, there has been growing interest in optically-encoded or tagged functionalized microbeads as a solid support platform to capture proteins or nucleotides which may serve as biomarkers of various diseases. Multiplexing technologies (suspension array or planar array) based on optically encoded microspheres have made possible the observation of relatively minor changes in biomarkers related to specific diseases. The ability to identify these changes at an early stage may allow the diagnosis of serious diseases (e.g. cancer) at a time-point when curative treatment may still be possible. As the overall accuracy of current diagnostic methods for some diseases is often disappointing, multiplexed assays based on optically encoded microbeads could play an important role to detect biomarkers of diseases in a non-invasive and accurate manner. However, detection systems based on functionalized encoded microbeads are still an emerging technology, and more research needs to be done in the future. This review paper is a preliminary attempt to summarize the state-of-the-art concerning diagnostic microbeads; including microsphere composition, synthesis, encoding technology, detection systems, and applications.
Keywords: Microbeads, Optical encoding, Multiplexing technologies, Early diagnosis
1. Introduction to microbeads
Microbeads are defined as spherical polymeric particles rangingin diameter from 0.5 to 500 μm (Fig. 1) (Rödiger et al.2014). They can be functionalized with a variety of differentchemical groups and biomolecules such as antigens, antibodies,oligonucleotides etc. (Ma and Su 2013). For the first time,Singer and Plotz employed polymeric microbeads in a Blatex fixation test for the serologic diagnosis of rheumatoid arthritis in the late 1950s (Singer and Plotz 1956). Microbeads were then used in other applications such as affinity chromatography matrices for antibody purification (Staak etal. 1996), and over the succeeding years, they were more widely used in cellular immunology (Větvička and Fornůsek 1987). Up to the present microbeads have been extensively studied and now have important applications that range from their use in the biological sciences, in biomedical diagnostic fields in low-density and high-density arrays, DNA extraction, PCR product detection, micro RNA detection, gene expression analysis, single nucleotide polymorphism (SNP) discrimination, cystic fibrosis mutation analysis, identification and quantification of pathogens, drug delivery device, targeted drug delivery and so many other areas (Rödiger et al. 2014; Arshady 1993). Moreover, microbeads are also often used as carriers in purification procedures for many biomolecules extracted from natural sources (Cao et al. 2007).
Fig. 1.
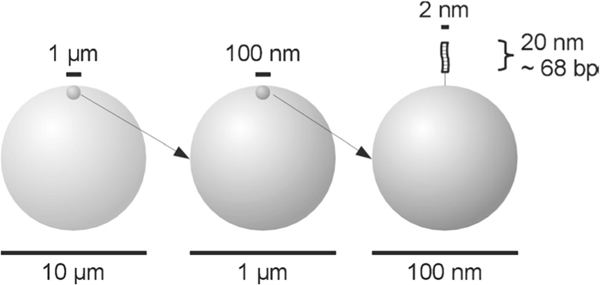
Schematic representation of the relative sizes of microbeads with a diameter of 10 μm down to a double stranded DNA molecule of 68 bp attached to a bead at a nanometer scale. The surface structures are idealized here as being smooth (Rödiger et al. 2014)
Recently developed microbead detection systems based on fluorescent-encoded nanoparticles are foundin two different formats; liquid-state suspension arrays and solid-state planar arrays. Each of these has its ownadvantages and disadvantages which will be further discussed in the following section. Flow cytometry is an ideal tool for the analysis of a large number of fluorescent microbead populations in suspension arrays (Nolan and Sklar 2002). Planar arrays on the other hand are analyzed using videoScan technology (Rödiger et al. 2014). These two procedures provide multiplexed bioassays which allows simultaneous analysis of many different targets relating to a single disease, or to different types of diseases. The advantages are, faster and more efficient analysis, lower costs, easier sample preparation, compared to the traditional single-analyte ELISA immunoassays (Leng et al. 2015). To achieve these goals, microbeads are tagged with various organic fluorescent dyes with different wavelengths and emission intensities to provide a robust and efficient platform for multiplexed assays (Jun et al. 2012).
Microbeads have been used in hundreds of different products, and one of the most important applications of these polymeric microbeads is detecting the presence, and quantifying the amount of various biological species or biomarkers in bodily fluids to detect the reference of a disease before appearing the first clinical symptoms. Using this early detection methodology, the development of more serious diseases can be prevented by detection in the first early stages, Microbeads can be useful and efficient tools for this goal whether used in suspension arrays or in a planar microarray. This review provides a general overview of these microbead-based multiplex assays, from preparation to applications.
2. Synthesis and optical encoding of microbeads
2.1. Synthesis method
Microbeads can be prepared by radical polymerization (either in suspension or in an emulsion) based on synthetic polymers including dextran, silica, melamine, polymethylmethacrylate (PMMA), poly(ethylene glycol) (PEG), hydrogels, polylactic acid (Stieber et al. 1993), and polystyrene (PS) (Rödiger et al. 2014). Overall methods for the synthesis of functionalized microbeads can be categorized into two distinct strategies. The beads can be constructed either from individual monomers assembled during the polymerization process, or else can be formed from pre-existing linear polymer chains (Freiberg and Zhu 2004).
2.1.1. Microsphere synthesis by polymerization of monomers
This method is based on starting with a defined mixture of specified monomers and an initiator to start the polymerization process. Stabilization is achieved in aqueous solutions by incorporating a suitable surfactant to preserve the microparticles in an individual microsphere form and prevent aggregation. Monomers may be soluble in an oil phase dispersed in aqueous media (oil in water emulsion, O/W) or may be dissolved in the aqueous phase dispersed in an organic medium (reverse emulsion). The most widely used techniques for the construction of microbeads are the suspension, emulsion and dispersion polymerization methods. The chief difference between these methods lies in the order of addition of the ingredients: monomer solution, initiator and surfactants (Freiberg and Zhu 2004; Saralidze et al. 2010).
In the suspension method, a mixture of monomer solution and initiator in an organic solvent is added into a non-miscible aqueous solution containing the surfactant as a stabilizing agent. Radical polymerization is triggered by heating the solution to activate the initiator. After the completion of the polymerization reaction, the microparticles are collected and the surfactants are washed away. The other method is called emulsion polymerization, in which a mixture of an aqueous solutions of initiator and emulsifier is dripped into an immiscible solution of monomer so that polymerization occurs converting micelles containing the monomers into microbeads (Fig. 2). A third method is dispersion polymerization where a blend of monomer solution, initiator and surfactant is prepared by continuous mixing, and the soluble monomers are slowly converted into an insoluble polymer and stabilized by a suitable surfactant (Saralidze et al. 2010).
Fig. 2.

Different synthetic polymeric microsphere synthesis routes. a Solvent evaporation (b) Suspension/emulsion polymerization (Saralidze et al. 2010)
Several parameters are involved in governing the final size of the microspheres, such as the concentration and molecular weight of the polymer, the type and concentration of solvent and surfactant, rate of stirring, the ratio of oil phase to water phase, the viscosity, temperature and pH of the reaction conditions etc. It is important to evaluate and optimize all these parameters to achieve microspheres with a specific and uniform size.
Suspension polymerization is usually used for microparticles in the range of 40–1000 μm. In the emulsion polymerization technique, microspheres in the size range 0.1–10 μm can be obtained, while dispersion polymerization is more suitable for the synthesis of larger particles in the range of 1 – 10 μm (Saralidze et al. 2010).
2.1.2. Microsphere synthesis from pre-formed linear polymers
This method is suitable for biodegradable aliphatic polyesters (poly (lactide), poly(glycolide), PCL and their copolymers) and for naturally occurring polymers such as chitin, chitosan, and cellulose. This class also includes proteins like collagen and albumin that nowadays are used for synthesizing spherical microparticles (Saralidze et al. 2010). The general technique that is most commonly used is the oil/water emulsion solvent extraction/evaporation method, in which a polymer solution (organic phase) is added to an aqueous phase containing the surfactant (water phase) and stirred for some time to produce an oil/water emulsion, and form the microparticles (Fig. 2). Some time is allowed before all the organic phase has evaporated or diffused into the aqueous phase so condensed solid microspheres are formed. Finally the microspheres are filtered and washed free of the emulsifier (Sinha et al. 2004).
One issue that should always be considered is enhancing the stability of the microspheres, and prevention of the aggregation of the microbeads in the suspension medium. To decrease nonspecific binding. Stabilized microspheres will perform better in applications such as protein immobilization, cell culture and separation, affinity chromatography, drug targeting and delivery, controlled release (Ma and Su 2013). This stabilization is better with a hydrophilic coating on the surface of the outer layer of the microbeads that may be achieved by conjugating a polymer like poly (ethylene glycol) (PEG), poly (vinyl alcohol) (PVA), poly (methylmethacrylate) (PMMA) etc. Another way is the introduction of reactive groups with a specific charged functionality onto the surface of the microbeads, for instance sulfonate groups (Hermanson 2013). Microbeads should have the proper density in aqueous solution. Cross-linking agents play an important role in attaching the individual polymer chains together and improving the mechanical stability of the microspheres (Saralidze et al. 2010). However, the drawbacks of emulsion techniques for microbeads synthesizing are inhomogeneous and broad microspheres size, lack of reproducibility (Ding et al. 2005) and the use of organic solvent which may lead denaturation of protein-based drugs during processing (Yang et al. 2000).
To address these issues, an electrospraying method for producing microspheres, with average sizes ranging from 10 to 20 μm, was introduced based on the ability of an electric field to deform the interface of a liquid drop (Bock et al. 2011). The electrospraying process is a one-step technique in which a polymer solution is loaded into a syringe and infused at a constant rate using a syringe pump through a small but highly charged capillary (e.g., a 16–26-gauge needle). Once the droplets have detached from the Taylor cone, the solvent evaporates, and dense and solid particles are generated, propelled towards the collector. This method has been applied for preparation of microspheres from polymers such as polylactide (Stieber et al. 1993; Valo et al. 2009; Xu and Hanna 2006), poly(lactic-co-glycolic acid) (PLGA) (Xie et al. 2006; Hong et al. 2008), polycaprolactone (PCL) (Xie et al. 2006) and chitosan.
The challenge related to enhancing the reproducibility of the microspheres can be addressed using a microfluidic device for the microsphere synthesis. Linear emulsions can readily be made in a sealed microenvironment, using the segmented flow phenomenon. The diameter of the microspheres can be accurately tailored, with a size distributions of less than 1% coefficient of variance (Workman et al. 2007). This methodology, usually involves flow-focusing droplets of nonpolar polymers like PDMS (with a low viscosity) in an aqueous continuous phase containing a surfactant. The droplets are then treated off-chip to generate monodisperse beads that can be preserved either in the aqueous phase or as a dry powder (Jiang et al. 2012). The most important micro-fluidic configurations for droplet generation, including T-junctions, flow focusing and co-flowing (hydrodynamic method) are illustrated schematically in Fig. 3 (Basova and Foret 2015).
Fig. 3.
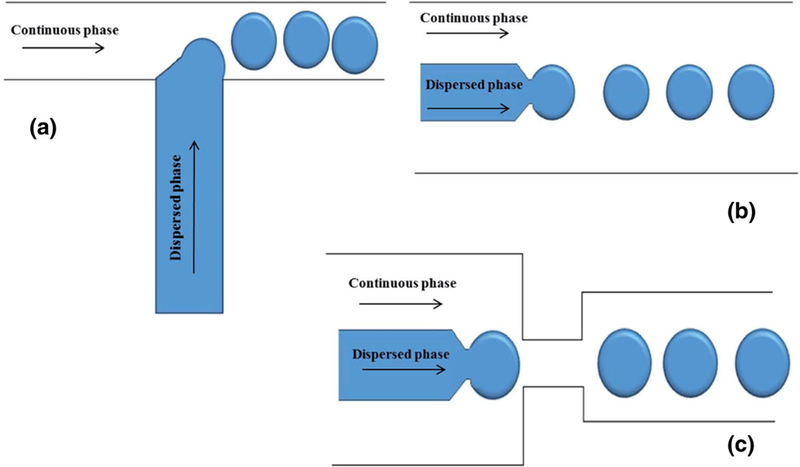
Schematics of microfluidic devices for making emulsions using (a) standard T-junction flow focusing junction; b co-flowing junction; c flow focusing junction (Basova and Foret 2015)
2.2. Polymers used for microbead synthesis
Depending on the specific application, different polymers have been utilized to form microbeads. The most common polymers are polystyrene (Guan et al. 2009; Jinhua and Guangyuan 2014), alginate (Khanna et al. 2012), gelatin (Altankov et al. 1991; Solorio et al. 2010), PMMA (Mani et al. 2013; Nord et al. 2003) etc. The microbeads must be functionalized with functional groups or “handles” In order to attach the microbeads onto the substrate in planar microarrays, and for the immobilization of antibodies, aptamers, or enzymes onto the beads (Kim et al. 2011). These handles are often incorporated by copolymerization with functionalized monomers, or by modifications carried out after polymerization. The functional groups often involve amino groups, carboxyl groups, epoxy groups, and aldehyde groups, etc. (Kim et al. 2011). If abundant carboxyl groups can be provided on the bead surface, they can be ideal handles for biomedical applications, such as nucleic acid extraction, cancer diagnosis and treatment, immunology, cell separation, biosensors, and drug delivery (Guan et al. 2009). There is a direct relationship between the bead surface chemistry (type, number, and availability of surface functional groups) and the efficiency of the beads in assays (Rödiger et al. 2014).
Some recent reports of different polymers used for preparation of microbeads include the following. Yoo et al. used silica microspheres to capture circulating tumor cells (CTCs) in whole blood. They used carboxylated silica microbeads due to their optical transparency, which facilitated the detection of CTCs by decreasing the interference caused by the microbeads themselves (Yoo et al. 2016).
Polystyrene is a common polymer employed in microbeads. Jinhua and Guangyuan (Jinhua and Guangyuan 2014) reported the synthesis of polystyrene microspheres by dispersion polymerization in a mixed solvent of ethanol and 2- methoxyethanol. Due to the poor miscibility between ethanol and 2-methoxyethanol, secondary particles were created and polydisperse polystyrene microspheres were obtained. To address this issue a small amount water (up to 2 wt%) was added into the polymerization system (Jinhua and Guangyuan 2014). Gorelik et al. applied a multiplex LabMAP system (Luminex) based on PS microbeads to identify markers in the female reproductive system in patients who either had early stage ovarian cancer or else benign pelvic tumors (Gorelik et al. 2005). Dehqanzada et al. used the same procedure as in previous research for potential analysis of a panel of serum cytokines in breast cancer patients using Luminex technology and levels of 22 different cytokines were measured (Dehqanzada et al. 2007). Brazhnik et al. described a novel bead-based (suspension) array, with each individual antigen coupled to its specific QD-encoded microbead to analyze serum samples. Polymer coated melamine resin microbeads were tagged by water soluble CdSe/ZnS QDs emitting at 585 nm (orange range), bound onto the charged surface of polystyrene latex beads (Brazhnik et al. 2015a).
Jinhua et al. successfully synthesized carboxylated magnetic polystyrene nanospheres by copolymerization of sodium 10-undecenoate together with styrene, both solubilized in a molecular bilayer on the surface of magnetic particles in the presence of aqueous magnetic fluid. In Fig. 4, SEM images of synthetized microspheres with different amounts of ethanol as solvent is illustrated (Jinhua and Guangyuan 2014).
Fig. 4.

SEM images of polystyrene microspheres synthesized with different dosages of ethanol (Jinhua and Guangyuan 2014)
In another work, Liang et al. copolymerized styrene and methacrylic acid (MAA) to prepare lanthanide-encoded p(S- MAA) particles for bead-based bioassays, using flow cytometry for sample detection applications (Liang et al. 2011). They also reported that monodisperse particles could be obtained in the presence of 2 and 4 wt% MAA. Furthermore, they compared their prepared particles to analogous particles that were copolymerized with acrylic acid (AA) as the comonomer, and concluded that with 2 wt% MAA, although the number of moles of MAA was less than AA, they produced polystyrene (PS) particles with 17 -COOH groups per nm2 on the surface compared to 6 COOH/nm2 for AA. For 4 wt% MAA, this value increased to 38 COOH/nm2 (a large number) (Liang et al. 2011).
Polycaprolactone (PCL) is semi-crystalline aliphatic polyester, which is biodegradable and easy to process (melting point at 60 °C), and possess good biocompatibility. These features make PCL an interesting substrate for biomaterials and tissue engineering. Furthermore, is appropriate for controlled drug delivery due to its high permeability to many drugs and nontoxicity (Sinha et al. 2004). In order to adjust release characteristics, microspheres can be prepared either by PCL alone, or by using copolymers with PCL or PCL blends. Gao and collaborators reported the generation of magnetic nanoparticles, composed of a PCL-PEG-PCL copolymer containing magnetite and decorated with an anti-CD40 antibody to separate cancer cells (Gao et al. 2011).
Alginate-based materials are attractive for biomedical applications due to their hydrophilic nature, good biocompatibility, and defined physical architecture. (Khanna et al. 2012). Larson and coworkers published a protocol using a three-step process to prepare multilayered alginate microbeads (Khanna et al. 2012). Gelatin microbeads are also relatively easy to prepare. Brodvarova et al. described a novel technique for the preparation of native collagen (type I)-coated gelatin microspheres using the emulsion-polymerization technique for cell culture applications. A 20% gelatin solution was emulsified in sunflower oil and the resulting beads were cross-linked with glutaraldehyde (Altankov et al. 1991). In a more recent report, Zwolinski et al. also prepared gelatin microspheres (with diameters in the range of 2–6 μm) for controlled release of bone morphogenetic protein 2 (BMP2). These workers used an emulsification process but the small molecule genipin, was employed as a cross-linking agent (Solorio et al. 2010). Table 1 describes an overview of different polymeric microbeads formation via various technique for different application.
Table 1.
Polymeric microbeads for different usages
| Polymer | Microspheres size μm) | Method of preparation | Application |
|---|---|---|---|
| Polycaprolactone | 38.52 ±4.64 μm | emulsion solvent extraction/evaporation technique |
Drug delivery PCL beads containing Interferon-alpha (IFN-alpha) (Melo et al. 2012) |
| 10 to 20 | Electrospraying | microsphere-growth factor delivery systems (Bock et al. 2011) | |
| 452.24 | double emulsion solvent extraction/evaporation method |
endothelial cell expansion (Yuan et al. 2015) | |
| poly-L-lactide-co-glycolides (PLGA) |
40 to 330 | Emulsion technique | in vitro cell expansion of autologous chondrocytes- regenerative medicine (Gabler et al. 2007) |
| Polystyrene (PS) | 1.6 ±0.2 | Dispersion polymerization | Oligonucleotide immobilization (Zhu et al. 2013) |
| 20–27 | Membrane emulsification technique | detect prostate-specific antigen (PSA), cytokines, CA-125, CEA, CA 15–3 antigen (Han et al. 2015) | |
| Gelatin | 150–400 | emulsification process crosslinked with glutaraldehyde | Cell culture (Altankov et al. 1991) |
| 2–6 | emulsification process crosslinked with genipin | local delivery of growth factors (Solorio et al. 2010) | |
| silica | 2–5 | – | Isolation and identification of circulating tumor cells (Yoo et al. 2016) |
| Alginate | 30–60 | Microfluidic one-step synthesis | Antibody immobilization for further medical applications (Chen et al. 2013) |
| chitosan | 100–700 | solvent extraction method in co-axial microfluidic device | Bovine serum albumin (BSA) in situ encapsulation (Xu et al. 2009) |
| poly(dimethysiloxane) (PDMS) |
80 | Microfluidic system | Oxygen sensor (Jiang et al. 2012) |
In all of the above polymers used for biomedical applications, the synthesis uses one of the pre-mentioned procedures. Choosing an appropriate polymer is based on the stability, viability and biodegradability parameters which is needed to be considered. In Table 1, polymers are summarized for various applications. The most common biomaterials for microbead detection systems especially in the commercial technologies (Luminex and BD Cytometric) are PS, Silica and PMMA polymers (Dunbar 2006; Aoe et al. 2003).
2.3. Optical characteristics
Fluorescent barcodes and fluorescent dyes are commonly used as labels for microbeads. The barcodes are the key to microbead arrays applied inside or on the surface of the microbeads for multiplexing applications. After applying the barcode to a microsphere, an additional labeling step is usually required to monitor the binding of the biomolecular analyte. For data analysis, signals will arise both from the barcodes, and from labels attached to the recognized biomolecules. Ideally the microbead-based system should have three readouts: (1)the bar-coded micro-sized beads for multiplexing; (2) the labeled target molecules bound to each microparticle; (3) a background signal arising from the microbeads and biomolecules themselves, regardless of fluorescent dyes (Fig. 5). The background signals originate from Rayleigh scattering and from autofluorescence of the biomolecules which is usually more pronounced with short wavelength excitation light. Since there is little fluorescence generated when near- infrared (NIR) light interacts with biomolecules, most researchers prefer to use NIR dyes in their experiments (Leng et al. 2015; Jun et al. 2012).
Fig. 5.
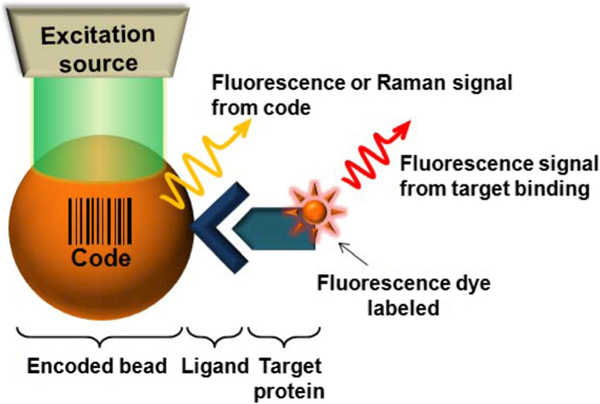
Schematic illustration of a bead-based system (Jun et al. 2012)
Fluorescent methods have the most important applications in bead encoding and labeling and can provide the highest overall levels of sensitivity. There are various fluorescent materials which have been applied in microbead based detection. These fluorophores can be chosen from different molecular systems which include small organic dyes (i.e. cyanine dyes, Alexa dyes, BODIPY dyes, fluorescein etc.), fluorophores of biological origin like fluorescent proteins (e.g. green fluorescent protein), nanocrystals with size-dependent optical properties (i.e. semiconductor quantum dots, carbon dots, and silicon nanoparticles), and size-independent fluorescent particles at the micrometer scale (lanthanides) (Resch-Genger et al. 2008).
The earliest used and the most versatile types of fluorophores are organic dyes. They have attracted much interest due to their simple applications and affordable prices. The optical properties of these compounds (excitation and emission wavelengths, absorption coefficient and quantum yield) depend on the electronic transition(s) of the molecule and can be tuned by structural modifications. Another kind of fluorophore is quantum dots (QDs) which are often used in medical diagnostic applications ranging from microarray technology to in vivo imaging. They have special features which make them a suitable option for nanoscale applications including narrow emission spectra, flexibility in excitation wavelength, tunable emission wavelengths, symmetric emission bands, high brightness, good photo-stability, and high quantum yields, thus they can overcome many drawbacks of traditional organic fluorescent dyes (Karimi et al. 2009; Alizadeh et al. 2013; Mohagheghpour et al. 2009, 2010, 2011, 2012; Mozafari et al. 2013; Tayebi et al. 2016a, b; Tayebi et al. 2016c; Yaraki et al. 2017). There is a phenomenon called the “quantum size effect” which is defined as the ability to tune the optical properties of QD by variation in the particle size which make QDs ideal candidates for the creation of diverse array of barcodes and labels (Leng etal. 2015; Junetal. 2012; Wang etal. 2005a; Kairdolf et al. 2013). Up-conversion nanoparticles (UCNPs) are lanthanide-doped nanocrystals which are more stable in comparison to QDs and organic dyes due to relying on atomic transitions instead of electronic transitions. Moreover, the Forster resonance energy transfer (FRET) effect which exist in QDs does not occur with UCNPs. Therefore, UCNPs can be considered as good choices for use as microsphere barcodes and multicolor encoding based on their multicolor emission and tunable spectra (Leng et al. 2015).
2.3.1. Encoding methods
The ability to target and detect multiple biomarkers in an analyte mixture is important in early disease detection. For these applications, parallel and simultaneous analysis is required. The basic concept in multiplexing is to develop microstructures that have a wide variety of built-in codes that can allow many different types of beads to be unambiguously detected. Fluorescent barcoded (encoded) microbeads have been widely-used for the multiplex detection of biomolecules. This approach has attractive features, such as: straight forward labeling process, easy detection of large-scale samples, and compatibility with a variety of biological chemistries, localized signals, rapid signal acquisition and high sensitivity. In the following section we provide an overview of various encoding methods (Jun et al. 2012; Resch-Genger et al. 2008; Sukhanova andNabiev 2008; Herbáth et al. 2014).
Dye trapping microspheres
In this technique beads are prepared by entrapping dyes internally with different emission colors in different concentrations (Battersby et al. 2001). Traditionally, the layer-by-layer deposition method is based on alternating adsorption of oppositely charged species. So, different nanostructures can to be held together by electrostatic forces and hydrogen bonding interactions (Wang et al. 2013). Furthermore, Jun et al. used a layer-by-layer technique to prepare fluorescent microbeads and applied them for protein detection. As shown in Fig. 6, 10 forms of multi-layered fluorescent particles can be created using only two individual dyes (Jun et al. 2010, 2012).
Fig. 6.
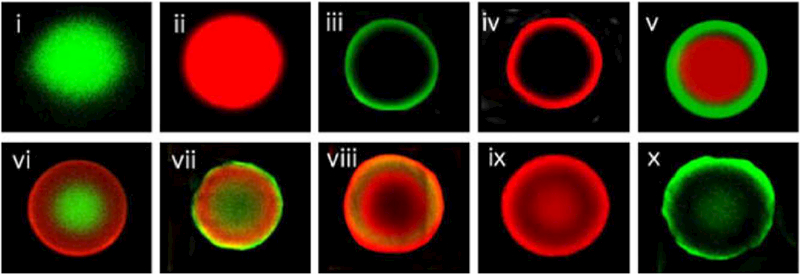
Fluorescence images of multi-layer fluorescence microbeads (Jun et al. 2010)
QDs also have promising application in encoding systems for microspheres. They have the capacity of producing emission at different wavelengths by optimization of their size. Moreover, by using QDs with different sizes and concentrations, a wide range of barcodes can be obtained, and the total number ofpossible codes increases exponentially according to the following formula:
Where C is the number of theoretically available detectable codes, N is the number of intensity levels and m is the number of colors. However, practically speaking the theoretical encoding capabilities are limited by many experimental factors like spectral overlap, variations fluorescence intensity etc. (Leng et al. 2015; Sukhanova and Nabiev 2008; Han et al. 2001).
Lifetime-encoded microspheres
Luminescence lifetime encoding can be used as a method for microbead labeling because it provides an additional independent parameter in addition to the emission wavelength and dye concentration.
In this approach, chosen dyes must have sufficiently different emission lifetimes after excitation at the same wavelength. Traditional organic dyes and QDs may therefore not be the most suitable choices owing to their relatively short lifetimes, while some metal-based materials such as lanthanides, can exhibit much longer lifetimes (Leng et al. 2015; Resch-Genger et al. 2008).
Surface-enhanced Raman scattering (SERS) encoded microspheres
Noble metal particles (gold and silver) that have nanoscale features, undergo a phenomenon known as surface plas- mon resonance (SPR) due to the excitation of electrons in the conductance band. SPR causes a tremendous enhancement in the local optical field (within a few nm of the metal nanoparticle surface). This SPR effect can be used to greatly enhance the normally weak signal from Raman spectroscopy, in “surface enhanced Raman spectroscopy” (SERS) which has many applications in diagnosis and imaging. The use of SERS as an optical tool for barcoding microbeads provides special features such as: (1) high capacity of multiplexing; (2) no photobleaching; (3) ultrasensitive detection; (4) flexible range of excitation wavelengths. For these reasons the SERS technique has gained much interest in microsphere encoding (Leng et al. 2015; Jun et al. 2012; Hoonejani et al. 2015).
2.3.2. Labeling methods
When using encoded microbeads for biodetection, a labeling step is generally required to monitor whether the molecular binding event has occurred. Direct conjugation of a fluorescent label to the individual components in the sample is the most prevalent method, but to obtain a measurable and sensitive signal, the background signal should be minimized. There should be no overlap between the barcode excitation/emission and the label emission spectra. Moreover, high densities of labeling can cause fluorescence quenching. QDs are bright fluorophores which have narrow emission peaks and large Stokes shifts; these properties make them good candidates for labeling because of minimal overlap with barcodes, reduced spectral crosstalk, and possibility for label multiplexing (as well as barcode) (Leng et al. 2015; Resch-Genger et al. 2008; Herbáth et al. 2014).
2.3.2. Label free techniques
Labeling techniques, such as: low signal intensity, high cost, time consuming and complex labeling procedures. The label- free approach is based on optical changes that occur in the microsphere structure in response to target binding. For instance, polydiacetylene (PDA) had been used as transducing material due to its special optical properties. This polymer shows a bichromic property. It has blue color visible to the unaided eye but the color alternates to red in response to environmental changes like heat, pH, ligand-receptor recognition and mechanical stress. This color transition is used for detection of different biological elements such as antibodies, proteins, bacteria, viruses, toxins etc. In immunoassay microbead detection systems blue color of the microbeads with target molecule immobilized on surface of micro bead will alternate to red after binding to the capture molecule. This result could be analyzed by microscope imaging or flow cytometry (Nie et al. 2006; Lim et al. 2011). Also there are some other label free techniques applicable in optical microbead systems such as SPR, carbon nanotubes (CNTs) and nanowires (Herbáth et al. 2014).
3. Multiplexing technologies
Multiplexing is the ability to detect various biomolecules or multiple diseases at the same time, by choosing multiple optical reporters that do not overlap and are rapid and sensitive. As mentioned above, there are two main concepts for the application of microbeads and microbead assays which vary in their potential for real-time read outs, high-output measurements; these are suspension arrays and planar arrays. Multiplexing is crucial in early diagnosis. The basic concept in multiplexing is to develop microspheres that have barcodes or labels on their surface or inside them for multi-analyte detection of various disease. Multiplexing is a useful tool for high throughput, parallel and early detection ofmultiple tumor markers (TMs), proteins, cytokines, antibodies etc., or several types of disease such as neurological, cancer and infectious disease. Accurate diagnosis is achieved by quantitative analysis of several biomarkers of interest, hence it is expected to facilitate the growth of clinical detection in the new era (Leng et al. 2015; Sukhanova and Nabiev 2008; Hsu et al. 2008).
Multiplexing is feasible through immobilization of a multitude of probe molecule onto a single bead or with mixture of a number of beads being loaded with different bio probes (Chen et al. 2013). Therefore, multiplexed bioassays increase the sensitivity and provide a more efficient, quick and precise diagnosis in a single sample volume, at the same time, in order to predict disease more efficiently than a single biomarker. Both planar and suspension array technologies can be used as a multiplexed screening tools. According to (Jin et al. 2013), multiplexing capacity of a suspension array can be enlarged to the order of about 58. Generally, these technologies have two components: the assay chemistry and the detection mechanism. Perhaps the best-publicized detection platform of recent years is the planar microarray, and now suspension arrays have emerged as a newer procedure and more articles have been published ab out them since 2006 (Nolan and Sklar 2002; Leng et al. 2015).
3.1. Planar microarrays
The use of planar microarray technology holds the potential to increase the screening throughput of biomarkers in patient samples. The planar or bead-based flat microarray contains target-specific receptors like oligonucleotides, antibodies, proteins, drug candidates etc. which are coated onto microbeads which are then deposited onto a substrate such as a chip (polymeric or glass slides) as a sensitive platform to detect their individual biological binding relating to a specific disease. These form a miniaturized analysis platform with no need for three-dimensional separation of the signals (Rödiger et al. 2014). The binding of biomarkers to the functionalized substrate can be facilitated by reverse phase arrays where lysates of tissues and cells or serum samples are attached onto the chip surface, by diverse methods such as sandwich immunoassays, or by antibody arrays to capture a mixture of fluorescent labeled antigens in samples such as cell lysates and sera (Sukhanova and Nabiev 2008; Zhu and Trau 2015).
This technology has various applications, especially in clinical diagnostics, and researchers have carried out valuable investigations and provided several routes to immobilize the microbeads onto the planar surface of the matrix. Zhu and Trau, used polyacrylamide gel-based microstructures to immobilize antibody-coated microbeads (Zhu and Trau 2012). In another approach they utilized a gel pad array chip for high-throughput and multianalyte microbead-based immunoassays (Zhu and Trau 2015). The other method to position microbeads in an array is to create a grid array of microholes, in the form of micromachined cavities on a glass substrate. This provides separation of the individual microbeads and facilitates the identification of beads using the x and y coordinates as well as by color (Han et al. 2015; Thompson 2011). The microbead loading efficiency can be enhanced by centrifugation, or by applying electric charges or magnetic fields (Manesse et al. 2013). Other techniques such as the use of affinity binding between biotin and streptavidin to functionalize the matrix and the beads to immobilize the beads (Huang et al. 2003). Surface modifications based on the composition of beads and matrix, in order to attach microbeads have been described in a patent by Margel (Margel 1997). The next step after immobilizing the beads is incubating the chip with the sample mixture, in order to couple the antigen and antibody in proteomic assays, or hybridize the cDNA sequence in the sample to specific beads on the glass slide containing its complementary sequence in genomic assay (Babu 2004). Finally the beads must be excited by a laser and the emission scanned at suitable wavelengths to detect the fluorescent dyes (Rödiger et al. 2014).
The amount of label fluorescence emitted upon excitation relates to the amount of bound analytes (oligonucleotides or antigens) (Babu 2004). In order to analyze the interactions that have occurred on the solid platform and measure labeled target emissions, an image is captured using a CCD camera. The next steps, image processing and annotation are performed by special software (Rödiger et al. 2014). To extract the information, the resulting image must be processed. This involves three steps; identification of bound spots, determination of spot signal strength, and subtracting the background noise signal. Other steps such as normalization, filtration and data clustering are usually conducted with specialized software programs designed for microarray data analysis. The final step is, translating the extracted data into real-life biological information such as the amount of over-expression a special biomarker or groups of biomarkers related to certain disease states (Babu 2004). Fortunately, the availability of computer software, lessens the effects of human factors in the interpretation of the experimental results (Breitling 2006). Some commercial softwares are designed for data interpretation like The BioPlex 2200 Medical Decision Support Software (MDSS) which is designed for use with the BioRad BioPlex 2200 System. The mentioned software is a pattern recognition algorithm that can enhance the performance of the antinuclear antibodies (ANA) Screen by identifying associated diagnostic patterns among its multiple assay results. This system has gotten FDA approved in 2012. In this assay, the identity of the dyed beads is determined by the fluorescence of the dyes, the amount of antibody conjugated to the antigen is determined by the fluorescence of the attached phycoerythrin. Raw data is calculated in relative fluorescence intensity (RFI) and fluorescence ratio (FR) (Fig. 7) (SERVICES, D.O.H.A.H. and F.a.D. Administration 2014).
Fig. 7.
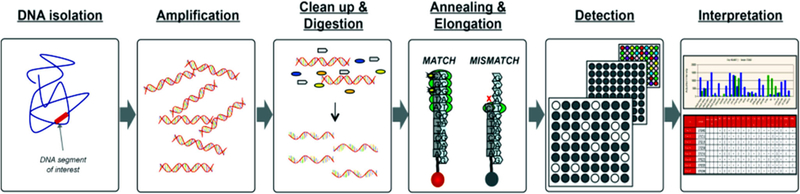
Summary of PreciseType Assay and Interpretation Steps (Thompson 2011)
Although planar microarray technology plays an important role in ultra-high-density analysis, it has limitations regarding the quality of its results, the binding rates to the chips, the decoding speed, and its overall flexibility (Leng et al. 2015). The relatively slow rates of hybridization and binding on planar arrays is due to limitations in the diffusion of analyte molecules to the surface; however the kinetics of binding to microbeads can be enhanced by more efficient mixing (Wilson et al. 2006).
3.2. Suspension microarrays
The development of multiplexed suspension arrays has currently received a great deal of attention in both research fields and in clinical diagnosis. Suspension or bead-based arrays are particularly based on a population of encoded microbeads for simultaneous high throughput detection of many analytes and specific biomarkers of many diseases in a solution. In a suspension array each individual microbead has a unique optical code for identification of the attached probes and their specific target molecules (such as, oligonucleotides, antigens, antibodies etc.) linked to the surface of the microbeads. By encoding the microbeads with different photoluminescent nanoparticles, suspension microarrays can be constructed for highly multiplexed analysis of many disease biomarkers. In contrast to conventional analytical methods, bead-based liquid phase arrays have many advantages such as a high degree of multiplexing, sensitivity, specificity, flexibility, reproducibility, reduced analysis time, low sample consumption, decreased labor and lower costs for multiple applications including early diagnosis of many disorders, identification of unknown genes and experiments for drug discovery. These advantages confirm microbeads technologies to be a useful and powerful tool in the field of personalized medicine. Personalized medicine aim at detecting personal gene expression profiles, in order to focus upon therapies that are specific for each individual since traditional symptom-diagnostics have faced failure and there are enormous various individuals’ genetic signatures. A library or database of analytical channels can be an ideal tool that can support the analysis of not tens, but thousands of distinctive molecular targets (Jin et al. 2013).
Microbeads have a high surface to volume ratio and the microbeads assist much faster reactions owing to their suspension in a homogeneous solution and the associated advantageous effects on diffusion rates (Han et al. 2015). These microparticles have a small size, so they can detect analytes down to the zeptomolar range (10(−21) M), and the reactions take place in the liquid phase resulting in faster binding kinetics. These considerations account for the increased specificity and sensitivity (Rödiger et al. 2014; Nolan and Sklar 2002; Leng et al. 2015; Sukhanova and Nabiev 2008). Optically encoded beads can be analyzed rapidly using classical flow cytometers. Flow cytometry has become accepted as a gold standard for immunophenotyping and quantitative detection of clinical biomarkers for many diseases. It can visualize hybridizations occurring on the surface of a microbead, and identify different microbeads and target analytes by measuring the optical characteristics of the photoluminescent microbeads in suspension. Flow can read the barcodes and fluorescent labels linked to their surface for a wide variety of clinical and research applications requiring detection of proteins and nucleic acids specially in multiplex microbead technology (Krishhan et al. 2009). In this technology, samples in the form of a suspension of microbeads flow through a fluidic channel system, then lasers with the required wavelengths are used to excite the fluorescent tags on both the labeled analyte and the bead- encoded barcodes to determine the identity and amount of each analyte (Wilson et al. 2006).
The first commercially available platform that used microbead-based technology, was from Luminex (Austin, TX, USA). As shown in Fig. 8, the Luminex system consists an array of 100 polystyrene microbeads, each bead has a unique optical address with immobilized specific probe molecules like antigens, antibodies and nucleic acids attached onto the surface of the microbeads. By incubating microbeads with serum samples containing a fluorescently labeled mixture of analytes, the target molecules couple to the beads. Microbeads are encoded with different red-to-infrared ratio fluorochromes, then two lasers excite fluorescence, a red laser (635-nm) excites the red and NIR fluorochromes inside the microbeads which allows identification of the individual beads and a green laser (532-nm) excites fluorochrome such as R-phycoerythrin and Alexa 532 that have been used to label the analytes to detect hybridization that has taken place on the surface of the microbeads. The green fluorophore can be linked to the probe molecule through streptavidin-biotin interactions. Thousands of different reactions that have taken place on the surface the microbeads can be analyzed in just a few seconds per sample in a single reaction vessel (Dunbar 2006).
Fig. 8.
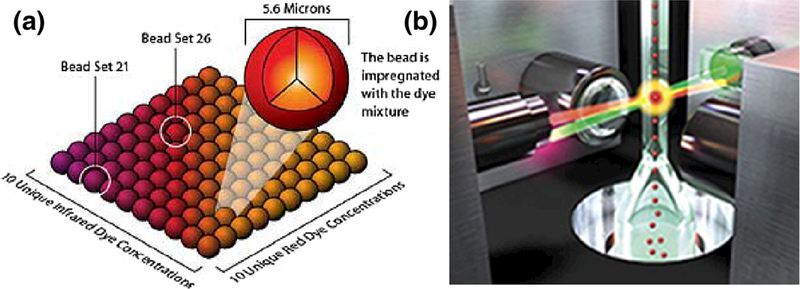
Suspension bead-based array. Luminex xMAP system consists a panel of 100 distinct 5.6-μm polystyrene microspheres, each microsphere having a unique ratio of red to infrared fluorescent dyes b. The microspheres are identified individually in a rapidly flowing fluid stream that passes by two laser beams: one reveals the color code of the bead, and one quantifies the biomolecular reaction by measuring the fluorescence intensity of the reporter. (Courtesy of Luminex Corporation)
The differences between planar and suspension arrays can be summarized as follows:
In suspension arrays all biomolecular interactions happen in solution which is three dimensional, but in planar array, two dimensional slides used to immobilize microbeads.
Biological interactions have a faster and more probable binding in suspension arrays compared to planar flat arrays
Detection analysis system in suspension arrays is flow cytometry. Modern flow cytometers can read fluorescent labels at extremely high speeds (50 million events per day) and it only requires low consumption of sample. In planar arrays a CCD camera is used to track fluorescent signals. Planar microarrays can provide ultra-high-density analysis, but with more sample consumption.
Suspension arrays have higher quality, such as better reproducibility and higher sensitivity. In planar microarray technology, the number of spots being analyzed at the same time is limited. This causes relatively poor productivity and detection sensitivity.
4. Microbead assay advantages and applications
As mentioned above, multiplexed microbead arrays have been successfully used to detect several different types of analytes such as proteins, antibodies, autoantibodies, nucleic acids, unknown genes, cytokines etc. Accordingly, there have been several published papers concerned with detection of multiple disease biomarkers in clinical samples from patients such as different types of cancers like breast and ovarian cancer, prostate cancer, lung cancer, infectious and viral diseases, neurological conditions such as Alzheimer’s disease and multiple sclerosis (Brazhnik et al. 2015a, b; Hsu et al. 2008; Krishhan et al. 2009; Liu et al. 2016; Kang et al. 2012; Bystrom et al. 2014; Yuetal. 2011).
Development of multiplexed microbead arrays based on encoded microbeads has emerged as an advanced alternative platform compared to conventional techniques like enzyme linked immunoassays (ELISA) for clinical diagnosis and especially early detection of disease. ELISA methodologies are time-consuming, costly and can usually measure only one analyte at a time, while microbead technologies have a multiplex capability that can quantify several analytes simultaneously which gives them greater power for speedy and robust analyses. In microbead arrays probe and target biomolecules link on spherical microbeads in suspension, while ELISAs generally depend on flat surfaces in 96 well microtiter plates. Microbead immunoassays have proven to be reliable, easy to perform, accurate, cost and time-effective, reproducible, highly sensitive and require lower sample consumption. It is likely that 1 day multiplex bead array assays will replace older technologies (Fritzler and Fritzler 2009; Elshal and McCoy 2006).
The measurement of multiple cytokines in serum and plasma is becoming crucial in the study and detection of diseases related to inflammation, cancer and immunology. Several of these studies have also examined expression pattern of different cytokines (interferons, interleukins and growth factors) which are small, soluble proteins that are analyzed for determination of the condition of the immune system. These immune mediators have been also validated by ELISA assays but these are problematic due to only one single cytokine at a time, and the need for large sample volumes, but microbead systems are gaining prominence for the analysis of a panel of cytokines and commercial vendors for many multiplex analyses are now available.
Luminex® xMAP™ Technology can analyze up to 100 multiple cytokines in a single reaction vessel with advantages of higher throughput, lower cost and smaller sample volume. Wang et al. evaluated ninety-six specimens related to reproductive immunology in women for cytokine concentrations such as IL-1|3, IL-4, IL-5, IL-6, IL-10, IFN y, and TNF a, IL-12 p70 and IL-13, and the results demonstrated significant accuracy and sensitivity for the Luminex technology in contrast to traditional ELSA methods (Wang et al. 2005b). The Bio-Plex® suspension array system applies xMAP technology, licensed from Luminex Corp., for multiplexed detection of 100 different assays within a single sample. The Bio-Plex MAGPIX system utilizes low-cost lightemitting diodes (LEDs) and a charge-coupled device (CCD) imager to illuminate and screen magnetic microbeads (Houser 2012).
Another company namely BD Biosciences (San Jose, CA, USA) produces the “Cytometric Bead Array” (CBA) system that accomplishes several assays and offers CBA kits for the multiplexed diagnosis of human cytokines (IL-2, IL-4, IL-5, IL-10, IFN-g, and TNF-a) and data has been published for these kits. Singulex®, Inc., also provides a key tool in the researcher’s arsenal to aid them move novel biology forward, fueling the discovery and development of new therapeutics. SMC™ technology facilitates the detection of low-abundance biomarkers, such as proteins and nucleic acids, with incomparable sensitivity and accuracy, capturing concentrations as low as femtogram/mL level. Make possible detecting, and monitoring changes in, extremely low levels of established disease biomarkers such as cardiac troponin I and cytokines.
As a result, there is a growing demand for accurate, rapid and cost-effective management of such cytokines in both clinical and research studies (Aoe et al. 2003; Soldan et al. 2004).
5. Challenges and limitations
As discussed above, researchers have invested great energy in order to improve the performance of microbead-based array technology, including multiplexing ability, sensitivity, robustness, portability, and high throughput capability. Despite all of these advantages, there are still some challenges that need to be resolved.
Considering the coding materials, an unlimited number of coding possibilities has not been fully achieved. Fluorescence-based beads can be limited in their number and their possible toxicity needs to be taken into consideration. Different signal intensity and signal complexity sometimes limit the coding number capacity.
Better resolution is required to address the limitations presented by the use of antibodies, such as cross-reactivity (non-specificity), and the number of matched antibody pairs available is limited. Label-free detection methods require to be further investigated. This is still at the beginning stage for protein binding detection, but currently there is a necessity for the progression of more smart, fast, sensitive and multiplexing capacities
The major benefit of optically-encoded beads originates in well-developed decoding and sorting systems. Up to now, decoding and sorting with flow cytometry has provided best performance and can be immediately applicable and could be more promising in the future.
There is a strong demand for developing “point-of-care” (POC) diagnostic platforms, which are economical and can be managed by untrained personnel. These are expected to have more efficiency and much higher throughput and be incorporated in faster automated biosystems (Leng et al. 2015; Jun et al. 2012).
6. Conclusions
In this review article, we have discussed of the state-of-the-art in microbead-based clinical analysis, focusing on different types of multiplexing technologies, optical characteristics of microbeads, and the advantages which make them prominent in clinical diagnosis are discussed. Microbead-based assays use a wide range of optical codes for labeling and detection of different biomolecules for constructing affordable devices to have an accurate and valid detection read-out. Optical nanoparticles used either as labels or barcodes are vital to detect the identity of microbead and receive signals from hybridizations occur on the surface of microbead. Early diagnosis of biomarkers is crucial to prevent disease progression and achieve successful treatment. In recent years, there has been a great progress in both clinical and research diagnostic approaches, two common approaches for multiplexing detections are liquid-based suspension arrays or solid-state planar arrays, each of them has its own advantages and disadvantages. It is anticipated that multiplexed technologies based on optically encoded microbeads could replace conventional singleanalyte ELISA methods because of their high multiplexing capacity and sensitivity with a low cost and sample consumption. Thus, microbead technologies enable precise and efficient quantification of multiple biomolecules and biomarkers for medical diagnostic applications.
Acknowledgments
Funding The work was supported by the Faculty of Biomedical Engineering, Amirkabir University of Technology. Michael R Hamblin was supported by US National Institute of Health (NIH) under Grants R01AI050875 and R21AI121700.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
References
- Alizadeh M, Salimi R, Sameie H, Sarabi AA, Sabbagh Alvani AA, Tahriri M, The wet-chemical synthesis of functionalized Zn1- xOMnx quantum dots utilizable in optical biosensors. Mater. Technol. 47(2), 235–237 (2013) [Google Scholar]
- Altankov G, Brodvarova I, Rashkov I, Synthesis of protein-coated gelatin microspheres and their use as microcarriers for cell culture. Part I Derivatization with native collagen. J. Biomater. Sci. Polym. Ed. 2(2), 81–89 (1991) [DOI] [PubMed] [Google Scholar]
- Aoe K et al. , Relative abundance and patterns of correlation among six cytokines in pleural fluid measured by cytometric bead array. Int. J. Mol. Med. 12(2), 193–198 (2003) [PubMed] [Google Scholar]
- Arshady R, Microspheres for biomedical applications: Preparation of reactive and labelled microspheres. Biomaterials 14(1), 5–15 (1993) [DOI] [PubMed] [Google Scholar]
- Babu MM, Introduction to microarray data analysis. Computational Genomics: Theory and Application, 225–249 (2004) [Google Scholar]
- Basova EY, Foret F, Droplet microfluidics in (bio) chemical analysis. Analyst 140(1), 22–38 (2015) [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Lawrie GA, Trau M, Optical encoding ofmicrobeads for gene screening: Alternatives to microarrays. Drug Discov. Today 6, 19–26 (2001)11165168 [Google Scholar]
- Bock N et al. , Electrospraying, a reproducible method for production of polymeric microspheres for biomedical applications. Polymers 3(1), 131–149(2011) [Google Scholar]
- Brazhnik K et al. , Quantum dot-based lab-on-a-bead system for multiplexed detection of free and total prostate-specific antigens in clinical human serum samples. Nanomedicine 11(5), 1065–1075 (2015a) [DOI] [PubMed] [Google Scholar]
- Brazhnik K et al. , Multiplexed analysis of serum breast and ovarian cancer markers by means of suspension bead-quantum dot microarrays. Phys. Procedia 73, 235–240 (2015b) [Google Scholar]
- Breitling R, Biological microarray interpretation: the rules of engagement. BBA-Gen Subjects 1759(7), 319–327 (2006) [DOI] [PubMed] [Google Scholar]
- Bystrom S et al. , Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J. Proteome Res. 13(11), 4607–4619 (2014) [DOI] [PubMed] [Google Scholar]
- Cao Y et al. , Immobilization staphylococcal protein A on magnetic cellulose microspheres for IgG affinity purification. Artif. Cell. Blood Sub. 35(5), 467–480 (2007) [DOI] [PubMed] [Google Scholar]
- Chen W et al. , Microfluidic one-step synthesis of alginate microspheres immobilized with antibodies. J. R. Soc. Interface 10(88), 20130566 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehqanzada ZA et al. , Assessing serum cytokine profiles in breast cancer patients receiving a HER2/neu vaccine using Luminex® technology. Oncol. Rep. 17(3), 687–694 (2007) [PubMed] [Google Scholar]
- Ding L, Lee T, Wang C-H, Fabrication of monodispersed Taxol-loaded particles using electrohydrodynamic atomization. J. Control. Release 102(2), 395–413 (2005) [DOI] [PubMed] [Google Scholar]
- Dunbar SA, Applications of Luminex® xMAP™ technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363(1), 71–82 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshal MF, McCoy JP, Multiplex bead array assays: Performance evaluation and comparison of sensitivity to ELISA. Methods 38(4), 317–323 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberg S, Zhu X, Polymer microspheres for controlled drug release. Int. J. Pharm. 282(1), 1–18 (2004) [DOI] [PubMed] [Google Scholar]
- Fritzler MJ, Fritzler ML, Microbead-based technologies in diagnostic autoantibody detection. Expert Opin. Med. Diagn. 3(1), 81–89 (2009) [DOI] [PubMed] [Google Scholar]
- Gabler E et al. , Emulsion-based synthesis of PLGA-microspheres for the in vitro expansion of porcine chondrocytes. Biomol. Eng. 24(5), 515–520 (2007) [DOI] [PubMed] [Google Scholar]
- Gao X et al. , Preparation of anti-CD40 antibody modified magnetic PCL-PEG-PCL microspheres. J. Biomed. Nanotechnol 7(2), 285–291 (2011) [DOI] [PubMed] [Google Scholar]
- Gorelik E et al. , Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol. Biomark. Prev. 14(4), 981–987 (2005) [DOI] [PubMed] [Google Scholar]
- Guan N et al. , A facile method to synthesize carboxyl-functionalized magnetic polystyrene nanospheres. Colloids Surf. A Physicochem. Eng. Asp. 335(1), 174–180 (2009) [Google Scholar]
- Han M et al. , Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 19(7), 631–635 (2001) [DOI] [PubMed] [Google Scholar]
- Han SW, Jang E, Koh W-G, Microfluidic-based multiplex immunoassay system integrated with an array of QD-encoded microbeads. Sensors Actuators B Chem. 209, 242–251 (2015) [Google Scholar]
- Herbath M et al. , Exploiting fluorescence for multiplex immunoassays on protein microarrays. Methods and Applications in Fluorescence 2(3), 032001 (2014) [DOI] [PubMed] [Google Scholar]
- Hermanson ET, Bioconjugate techniques (Academic Press, Cambridge, 2013) [Google Scholar]
- Hong Y et al. , Electrohydrodynamic atomization of quasi-monodisperse drug-loaded spherical/wrinkled microparticles. J. Aerosol Sci. 39(6), 525–536 (2008) [Google Scholar]
- Hoonejani M et al. , Quantitative multiplexed simulated-cell identification by SERS in microfluidic devices. Nanoscale 7(40), 16834-–16840. (2015) [DOI] [PubMed] [Google Scholar]
- Houser B, Bio-Rad’s bio-Plex® suspension array system, xMAP technology overview. Arch. Physiol. Biochem. 118(4), 192–196 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H-Y et al. , Suspension microarrays for the identification of the response patterns in hyperinflammatory diseases. Med. Eng. Phys. 30(8), 976–983 (2008) [DOI] [PubMed] [Google Scholar]
- Huang TT et al. , Micro-assembly of functionalized particulate monolayer on C18-derivatized SiO2 surfaces. Biotechnol. Bioeng. 83(4), 416–427 (2003) [DOI] [PubMed] [Google Scholar]
- Jiang K et al. , Microfluidic synthesis of monodisperse PDMS microbeads as discrete oxygen sensors. Soft Matter 8(4), 923–926 (2012) [Google Scholar]
- Jin D, Lu Y, Zhao J, Multiplex suspension assay/array using lifetime coding. Google Patents (2013) [Google Scholar]
- Jinhua L, Guangyuan Z, Polystyrene microbeads by dispersion polymerization: Effect of solvent on particle morphology. Int. J. Polym. Sci. 2014, 1 (2014) [Google Scholar]
- Jun B-H et al. , Multilayer fluorescence optically encoded beads for protein detection. Anal. Biochem. 396(2), 313–315 (2010) [DOI] [PubMed] [Google Scholar]
- Jun B-H et al. , Fluorescence-based multiplex protein detection using optically encoded microbeads. Molecules 17(3), 2474–2490 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kairdolf BA et al. , Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu Rev Anal Chem (Palo Alto, Calif) 6(1), 143 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-H et al. , Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer’s disease. Methods 56(4), 484–493 (2012) [DOI] [PubMed] [Google Scholar]
- Karimi M et al. , Ammonia-free method for synthesis of CdS nanocrystalline thin films through chemical bath deposition technique. Solid State Commun. 149(41), 1765–1768 (2009) [Google Scholar]
- Khanna O et al. , Generation of alginate microspheres for biomedical applications. JoVE (Journal of Visualized Experiments) 66, e3388–e3388(2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-H et al. , Encoding peptide sequences with surface-enhanced Raman spectroscopic nanoparticles. Chem. Commun. 47(8), 2306–2308 (2011) [DOI] [PubMed] [Google Scholar]
- Krishhan V, Khan IH, Luciw PA, Multiplexed microbead immunoassays by flow cytometry for molecular profiling: Basic concepts and proteomics applications. Crit. Rev. Biotechnol. 29(1), 29–43 (2009) [DOI] [PubMed] [Google Scholar]
- Leng Y et al. , Suspension arrays based on nanoparticle-encoded microspheres for high-throughput multiplexed detection. Chem. Soc. Rev. 44(15), 5552–5595 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y et al. , The synthesis and characterization of lanthanide-encoded poly (styrene-co-methacrylic acid) microspheres. Polymer 52(22), 5040–5052 (2011) [Google Scholar]
- Lim M-C et al. , Microbead-assisted PDA sensor for the detection of genetically modified organisms. Anal. Bioanal. Chem. 400(3), 777–785 (2011) [DOI] [PubMed] [Google Scholar]
- Liu L et al. , Bead-based microarray immunoassay for lung cancer biomarkers using quantum dots as labels. Biosens. Bioelectron. 80, 300–306 (2016) [DOI] [PubMed] [Google Scholar]
- Ma G, Su Z-G, Microspheres and microcapsules in biotechnology: design, preparation and applications (CRC Press, Boca Raton, 2013) [Google Scholar]
- Manesse M et al. , Dynamic microbead arrays for biosensing applications. Lab Chip 13(11), 2153–2160 (2013) [DOI] [PubMed] [Google Scholar]
- Mani N et al. , Novel use of polymethyl methacrylate (PMMA) microspheres in the treatment of infraorbital rhytids. J. Cosmet. Dermatol 12(4), 275–280 (2013) [DOI] [PubMed] [Google Scholar]
- Margel S, Method for attaching microspheres to a substrate. Google Patents (1997)
- Melo C. d.S. et al. , Poly-ε-caprolactone microspheres containing interfer on alpha as alternative formulations for the treatment of chronic hepatitis C. Braz. J. Pharm. Sci. 48(1), 51–59 (2012) [Google Scholar]
- Mohagheghpour D et al. , Controllable synthesis, characterization and optical properties of ZnS: Mn nanoparticles as a novel biosensor. Mater. Sci. Eng. C 29(6), 1842–1848 (2009) [Google Scholar]
- Mohagheghpour E et al. , Effect of manganese (Mn) doping on the optical properties of zinc sulfide (ZnS) semiconductor nanocrystals. J. Ceram. Process. Res. 11(2), 144 (2010) [Google Scholar]
- Mohagheghpour E, et al. , A new optical bio-sensor: Wet-chemical synthesis and surface treatment of nanocrystalline Zn1-xS: Mn+2×. In Optical Sensors. Optical Society of America. (2011) [Google Scholar]
- Mohagheghpour E et al. , Micro-emulsion synthesis, surface modification, and photophysical properties of nanocrystals for biomolecular recognition. IEEE Trans. Nanobioscience 11(4), 317–323 (2012) [DOI] [PubMed] [Google Scholar]
- Mozafari M, Moztarzadeh F, Tahriri M, Green synthesis and characterisation of spherical PbS luminescent micro-and nanoparticles via wet chemical technique. Adv. Appl. Ceram. 110(1), 30–34 (2013) [Google Scholar]
- Nie Q et al. , Immobilization of polydiacetylene onto silica microbeads for colorimetric detection. J. Mater. Chem. 16(6), 546–549 (2006) [Google Scholar]
- Nolan JP, Sklar LA, Suspension array technology: Evolution of the flat-array paradigm. Trends Biotechnol. 20(1), 9–12 (2002) [DOI] [PubMed] [Google Scholar]
- Nord O, Uhlen M, Nygren P-A, Microbead display of proteins by cell-free expression of anchored DNA. J. Biotechnol. 106(1), 1–13 (2003) [DOI] [PubMed] [Google Scholar]
- Resch-Genger U et al. , Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 5(9), 763–775 (2008) [DOI] [PubMed] [Google Scholar]
- Rödiger S et al. , Nucleic acid detection based on the use of microbeads: A review. Microchim. Acta 181(11–12), 1151–1168 (2014) [Google Scholar]
- Saralidze K, Koole LH, Knetsch ML, Polymeric microspheres for medical applications. Materials 3(6), 3537–3564 (2010) [Google Scholar]
- SERVICES, D.O.H.A.H. and F.a.D. Administration, Immucor PreciseType™ Human Erythrocyte Antigen Molecular BeadChip Test. FDA; MARyland: (2014) [Google Scholar]
- Singer JM, Plotz CM, The latex fixation test: I. Application to the serologic diagnosis of rheumatoid arthritis. Am. J. Med. 21(6), 888–892 (1956) [PubMed] [Google Scholar]
- Sinha V et al. , Poly-e-caprolactone microspheres and nanospheres: An overview. Int. J. Pharm. 278(1), 1–23 (2004) [DOI] [PubMed] [Google Scholar]
- Soldan SS et al. , Dysregulation of 1L-10 and lL-12p40 in secondary progressive multiple sclerosis. J. Neuroimmunol. 146(1), 209–215 (2004) [DOI] [PubMed] [Google Scholar]
- Solorio L et al. , Gelatin microspheres crosslinked with genipin for local delivery of growth factors. J. Tissue Eng. Regen. Med. 4(7), 514–523 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staak C et al. , Polystyrene as an affinity chromatography matrix for the purification of antibodies. J. Immunol. Methods 194(2), 141–146 (1996) [DOI] [PubMed] [Google Scholar]
- Stieber P et al. , CYFRA 21–1: A new marker in lung cancer. Cancer 72(3), 707–713 (1993) [DOI] [PubMed] [Google Scholar]
- Sukhanova A, 1. Nabiev, Fluorescent nanocrystal-encoded microbeads for multiplexed cancer imaging and diagnosis. Crit. Rev. Oncol. Hematol. 68(1), 39–59 (2008) [DOI] [PubMed] [Google Scholar]
- Tayebi M et al. , Synthesis, surface modification and optical properties of thioglycolic acid-capped ZnS quantum dots for starch recognition at ultralow concentration. J. Electron. Mater, 1–8 (2016a) [Google Scholar]
- Tayebi M et al. , Determination of total aflatoxin using cysteamine- capped CdS quantum dots as a fluorescence probe. Colloid Polym. Sci. 294(9), 1453–1462 (2016b) [Google Scholar]
- Tayebi M et al. , Thioglycolic acid-capped CdS quantum dots conjugated to α-amylase as a fluorescence probe for determination of starch at low concentration. J. Fluoresc. 26(5), 1787–1794 (2016c) [DOI] [PubMed] [Google Scholar]
- Thompson JA, Microbead-based biosensing in microfluidic devices (2011) [Google Scholar]
- Valo E et al. , Electrospray encapsulation of hydrophilic and hydrophobic drugs in poly (L-lactic acid) nanoparticles. Small 5(15), 1791–1798 (2009) [DOI] [PubMed] [Google Scholar]
- Větvička V, Fornůsek L, Polymer microbeads in immunology. Biomaterials 8(5), 341–345 (1987) [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang Z, Kotov NA, Bioapplication of nanosemiconductors. Mater. Today 8(5), 20–31 (2005a) [Google Scholar]
- Wang K et al. , Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: Determinations of a panel of nine cytokines in clinical sample culture supernatants. J. Reprod. 1mmunol. 66(2), 175–191 (2005b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X et al. , NIR-emitting quantum dot-encoded microbeads through membrane emulsification for multiplexed immunoassays. Small 9(19), 3327–3335 (2013) [DOI] [PubMed] [Google Scholar]
- Wilson R, Cossins AR, Spiller DG, Encoded microcarriers for high-throughput multiplexed detection. Angew. Chem. 1nt. Ed. 45(37), 6104–6117 (2006) [DOI] [PubMed] [Google Scholar]
- Workman VL et al. , Microfluidic chip-based synthesis of alginate microspheres for encapsulation of immortalized human cells. Biomicrofluidics 1(1), 014105 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J et al. , Electrohydrodynamic atomization for biodegradable polymeric particle production. J. Colloid Interface Sci. 302(1), 103–112 (2006) [DOI] [PubMed] [Google Scholar]
- Xu Y, Hanna MA, Electrospray encapsulation of water-soluble protein with polylactide: Effects of formulations on morphology, encapsulation efficiency and release profile of particles. Int. J. Pharm. 320(1), 30–36 (2006) [DOI] [PubMed] [Google Scholar]
- Xu J et al. , Preparation of monodispersed chitosan microspheres and in situ encapsulation of BSA in a co-axial microfluidic device. Biomed. Microdevices 11(1), 243–249 (2009) [DOI] [PubMed] [Google Scholar]
- Yang Y-Y et al. , Effect of preparation conditions on morphology and release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion method. Chem. Eng. Sci. 55(12), 2223–2236 (2000) [Google Scholar]
- Yaraki MT et al. , Synthesis and optical properties of cysteamine-capped ZnS quantum dots for aflatoxin quantification. J. Alloys Compd. 690, 749–758 (2017) [Google Scholar]
- Yoo BE et al. , Highly dense, optically inactive silica microbeads for the isolation and identification of circulating tumor cells. Biomaterials 75, 271–278 (2016) [DOI] [PubMed] [Google Scholar]
- Yu B et al. , Rapid detection of common viruses using multi-analyte suspension arrays. J. Virol. Methods 177(1), 64–70 (2011) [DOI] [PubMed] [Google Scholar]
- Yuan S et al. , PCL microspheres tailored with carboxylated poly (glyc-idyl methacrylate)-REDV conjugates as conducive microcarriers for endothelial cell expansion. J. Mater. Chem. B 3(44), 8670–8683 (2015) [DOI] [PubMed] [Google Scholar]
- Zhu Q, Trau D, Multiplex detection platform for tumor markers and glucose in serum based on a microfluidic microparticle array. Anal. Chim. Acta 751, 146–154 (2012) [DOI] [PubMed] [Google Scholar]
- Zhu Q, Trau D, Gel pad array chip for high throughput and multi-analyte microbead-based immunoassays. Biosens. Bioelectron. 66, 370–378 (2015) [DOI] [PubMed] [Google Scholar]
- Zhu S, Panne U, Rurack K, A rapid method for the assessment of the surface group density of carboxylic acid-functionalized polystyrene microparticles. Analyst 138(10), 2924–2930 (2013) [DOI] [PubMed] [Google Scholar]


