Abstract
This study aimed to investigate the prevalence, serotype distribution, and antibiotic resistance, and to characterize the extended spectrum β-lactamases (ESBLs) producing Salmonella isolates from chicken and pork meats from retail markets in Guangdong province, China. A total of 903 retail meat samples (475 chicken and 428 pork meats) were obtained from six cities (Guangzhou, Shenzhen, Heyuan, Shaoguan, Foshan, and Yunfu) of Guangdong province between May 2016 and April 2017. High levels of Salmonella contamination were detected in chicken (302/475, 63.6%) and pork (313/428, 73.1%). Thirty-eight serotypes were identified in 615 detected Salmonella, and the serotypes varied greatly between chicken and pork samples. Agona (55/302, 18.2%), Corvallis (45/302, 14.9%), Kentucky (38/302, 12.6%), Mbandaka (32/302, 10.6%) was the dominant serotypes in chicken samples. However, Typhimurium (78/313, 24.9%), Rissen (67/313, 24.1%), Derby (66/313, 21.1%), and London (48, 15.3%) were the most common in pork samples. High rates of antibiotic resistance were found to sulfisoxazole (468/615, 76.1%), tetracycline (463/615, 75.3%), ampicillin (295/615, 48.0%), and ofloxacin (275/615, 44.7%). Notably, antimicrobial susceptibility tests identified resistance to polymyxin B (12/615, 2.0%) and imipenem (3/615, 0.5%). Multidrug-resistance (MDR) was detected in Salmonella isolated from chicken (245/302, 81.1%) and pork (229/313, 73.2%). The resistance rate of different Salmonella serotypes varied widely. Especially, isolates such as Typhimurium, Agona, Corvallis and Kentucky exhibited highly resistance to antibiotics. The MDR rate of Salmonella isolates from chicken was significantly higher than that from pork isolates (P < 0.05). Twenty-one Salmonella isolates were identified as ESBLs-producing, covering six Salmonella serotypes and displaying different pulse field gel electrophoresis (PFGE) genotypes. BlaOXA-1 was the dominant ESBLs gene (9/21, 42.9%), followed by blaCTX-M-55 (5/21, 23.8%). This study indicated that Salmonella was widespread in chicken and pork from retail markets in Guangdong province and the isolates showed high multidrug-resistance, especially the known multidrug-resistant Salmonella serotypes. Therefore, it is important to focus on Salmonella serotypes and strengthen the long-term monitoring of MDR Salmonella serotypes in animal-derived foods.
Keywords: Salmonella, retail meat, prevalence, antibiotic resistance, ESBLs, PFGE
Introduction
Salmonella is a foodborne pathogen that causes morbidity and mortality worldwide (Scallan et al., 2011). Until now, more than 2,600 serovars have been identified among Salmonella worldwide and almost all Salmonella can cause illness in humans and animals (Guibourdenche et al., 2010). In China, about 70–80% of foodborne disease outbreaks are caused by Salmonella, and the majority of diseases are associated with the ingestion of contaminated livestock and poultry products (Wang et al., 2007). Animal-derived foods, especially chicken and pork, are considered the major reservoirs of Salmonella dissemination (Vo et al., 2006; Jackson et al., 2013).
At the same time, antibiotic resistance of Salmonella is also one of the most important public health problems worldwide. In recent years, studies have shown that because of long-term antibiotic use during animal breeding, antibiotic resistance has markedly increased. Multidrug-resistant (MDR) Salmonella could pose a serious threat to humans through the food chain (Barza, 2002; Lai et al., 2014). In particular, extended-spectrum β-lactamases (ESBLs)-producing Salmonella have been frequently isolated from food animals in many countries, including China (Dahms et al., 2015; Ben Said et al., 2016; Ibrahim et al., 2016; Zhao et al., 2017).
For animal-derived foods, tracking the source of Salmonella infections can help to identify potential problems in food production, processing, preparation, or the sales process to prevent the introduction of other pathogens into food (Swaminathan et al., 2006; Russell et al., 2014). Guangdong is a livestock production and consumption province; furthermore, Guangdong is China’s foremost livestock production area. In 2009, laboratory-based surveillance for Salmonella infection was established in Guangdong Province, which mainly monitors the prevalence and outbreak of Salmonella in various hospitals (Deng et al., 2012). However, limited information on surveillance studies of Salmonella in animal-derived foods from retail markets in Guangdong is available, and there is a lack of epidemiological data.
The main aim of this study was to determine the prevalence, serotype distribution, antibiotic resistance of Salmonella, and the phenotype and genotype of ESBLs-producing Salmonella isolated from chicken and pork meat from retail markets in Guangdong. The results could lay the foundation for follow-up research for public health security and food safety problems caused by Salmonella.
Materials and Methods
Sample Collection
A total of 903 retail meat samples (including 475 chicken and 428 pork sample) were randomly collected from retail markets in six cities (Guangzhou, Shenzhen, Heyuan, Shaoguan, Foshan, and Yunfu; chosen from three retail markets in each district) of Guangdong province between May 2016 and April 2017. Sampling methods were as follows: In Guangzhou City, different regions were sampled monthly; in other cities, samples were taken monthly from three selected retail markets. Samples were taken of different chicken types (i.e., whole chickens and chopped chickens), pork types (i.e., dressed pork) and during different seasons (i.e., spring, summer, autumn and winter). Each sample was placed in a sterile plastic sample bag and transported to the laboratory on ice. All samples were analyzed on the day of their collection.
Salmonella Isolation and Identification
Salmonella isolation and identification was performed according to the Standard ISO-6579 (International Organization for Standardization, 2002) method, as described previously (Yang et al., 2010; Ren et al., 2016) with some modifications. Briefly, 25 g of samples was placed into a sterile plastic bag containing 225 mL of buffered peptone water and shaken for 3 min. The suspension was then cultivated at 37°C in a Shaker at 100 rpm for 6–8 h and then 1 mL of the suspension was added to 10 mL Tetrathionate Broth and Rappaport–Vassiliadis soya broth and incubated at 42°C for 18–24 h, separately. After selective enrichment, the suspensions were streaked onto xylose lysine tergitol 4 agar and CHROMagarTM incubated at 37°C over night. Isolates with typical Salmonella phenotypes were further confirmed using API 20E test strips (bioMerieux, Marcy-l’Etoile, France).
Salmonella Serotyping
All the Salmonella isolates were serotyped according to the White Kauffmann Le Minor scheme or National Food Safety Standard-Food microbiological examination: Salmonella (GB 4789.4-2016) by slide agglutination, using specific O and H antisera (S&A Reagents Lab Ltd., Bangkok, Thailand).
Antimicrobial Susceptibility Testing and Screening for ESBLs Strains
All Salmonella isolates were tested for antibiotic susceptibility using the Kirby-Bauer disk diffusion method, and the results were interpreted according to the standards of the Clinical and Laboratory Standards Institute (CLSI, 2013).
Agar diffusion assays were performed on Muller-Hinton agar with disks containing 18 different antibiotic agents (Oxoid, Basingstoke, United Kingdom): ampicillin 10 μg (AMP), amoxicillin-clavulanic acid 30 μg (AMC), cefotaxime 30 μg (CTX), ceftazidime 30 μg (CAZ), cefepime 5 μg (FEP), gentamicin 10 μg (GEN), streptomycin 10 μg (STR), amikacin 30 μg (AMK), sulfamethoxazole-trimethoprim 23.75/1.25 μg (SXT), sulfonamides 300 μg (S3), nalidixic acid 30 μg (NAL), ofloxacin 5 μg (OFX), ciprofloxacin 5 μg (CIP), florfenicol 30 μg (FFC), chloramphenicol 30 μg (CHL), imipenem 10 μg (IPM), polymyxin B 300 IU (PB), and tetracycline 30 μg (TET). Escherichia coli ATCC 25922 and ATCC 35218 were used as quality control organisms. Isolates exhibiting resistance to three or more antibiotic classes were defined as MDR.
The Salmonella isolates were screened for ESBLs production using cefotaxime and cefotaxime (30 μg)/clavulanic acid (10 μg) disks and ceftazidime and ceftazidime (30 μg)/clavulanic acid (10 μg) disks (Oxoid) according to the double-disk synergy test method (Emery and Weymouth, 1997). Phenotypic presence of ESBLs of the isolates was determined by detecting enhancement of the diameter of the inhibition zone of the clavulanate disk and corresponding β-lactam antibiotic disk. If there was difference of ≥5 mm between the inhibition zone of the clavulanic acid and the β-lactam antibiotic disk, the isolate confirmed as having a positive ESBLs phenotype (CLSI, 2013).
PCR Detection and DNA Sequencing
Salmonella isolates showing ESBLs phenotypes were further confirmed by PCR and DNA sequencing. ESBLs encoding genes detected included blaTEM, blaCTX-M, blaCMY, blaOXA, blaPSE, and blaSHV. The primers for the ESBLs encoding gene and the reaction conditions were as previously described (Li et al., 2013; Nguyen et al., 2016; Qiao et al., 2017). The PCR products were stained with ethidium bromide, visualized under UV light after gel electrophoresis using 1% agarose, purified, and sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing. The ESBLs encoding gene fragments were analyzed and aligned at GenBank using the online BLAST software1.
Pulse Field Gel Electrophoresis (PFGE)
Extended spectrum β-lactamases-producing Salmonella isolates were subtyped by PFGE to determine their genetic relatedness, according to the Pulse-Net protocol recommended by the CDC (Ribot et al., 2006). PFGE was preformed after digestion of the genomic DNA with the restriction enzyme XbaI, Salmonella enterica subsp. enterica serovar Braenderup (CDC no. H9812) was used as standard control strain. PFGE results were analyzed using BioNumerics Software (Version 5.1; Applied-Maths, Sint-Martens-Latem, Belgium).
Statistical Analysis
The isolation rates and antibiotic resistance rates among the different food types and isolates were analyzed using the Chi-squared test in SPSS Statistics 18.0 software (SPSS Inc., United States). A P-value of <0.05 was considered statistically significant.
Results
Prevalence and Serotypes of Salmonella
In this study, a total of 615 Salmonella isolates (615/903, 68.1%) were recovered from the meat samples. High levels of Salmonella contamination were detected: 302 (302/475, 63.6%) in chicken samples and 313 (313/428, 73.1%) in pork samples (Table 1). The isolation rate from pork was higher than that from chicken (P < 0.05).
Table 1.
Prevalence and serotypes of Salmonella isolated from chicken and pork meat at retail markets in Guangzhou, China.
| Source of samples | No. of positive samples /No. of total sample(%) | Serovar of isolates (number of samples) | Number of Serovars |
|---|---|---|---|
| Chicken | 302/475 (63.6) | Agona (55), Corvallis (45), Kentucky (38), Mbandaka (32), Braenderup (21), Enteritidis (21), Rissen (14), Indiana (12), Derby (9), London (9), Meleagridis (7), Typhimurium (6), Stanley (5), Cerro (5), Albany (4), Hadar (4), Senftenberg (3), Litchfield (3), Weltevreden (1), Anatum (1), Muenster (1), Infantis (1), Haifa (1), Rumford (1), Saint paul (1), Thompson (1), Schwarzengrund (1). | 27 |
| Pork | 313/428 (73.1) | Typhimurium (78), Rissen (67), Derby (66), London (48), Meleagridis (13), Stanley (9), Give (4), Weltevreden (3), Krefeld (3), Newport (3), Mbandaka (2), Senftenberg (2), Anatum (2), Gold coast (2), Agona (1), Corvallis (1), Muenster (1), Infantis (1), Alachua (1), Bovismorbificans (1), Brandenburg (1), Chailey (1), K é douguo (1), Kottbus (1), Reading (1). | 25 |
| Total | 615/903 (68.1) | 38 |
Thirty-eight Salmonella serotypes were identified among the 615 Salmonella isolates (Table 1). More serovars were found in chicken samples (27 serovars) than in pork samples (25 serovars). The serotypes varied greatly between the chicken and pork samples. In the chicken samples, the most common serovars were Agona (55/302, 18.2%), Corvallis (45/302, 14.9%), Kentucky (38/302,12.6%), and Mbandaka (32/302, 10.6%); and in the pork samples they were Typhimurium (78/313, 24.9%), Rissen (67/313, 24.1%), Derby (66/313, 21.1%), and London (48, 15.3%). Importantly, serovar Rumford was detected in meat samples for the first time in China.
Antimicrobial Susceptibility Testing
The 615 isolated strains were tested for their susceptibility to 18 antibiotics (Table 2). The most resistance was to sulfisoxazole (468/615, 76.1%) and tetracycline (463/615, 75.3%), followed by ampicillin (295/615, 48.0%), ofloxacin (275/615, 44.7%), and trimethoprim/sulfamethoxazole (248/615, 40.3%). All isolates were susceptible to cefepime, ceftazidime, amikacin, and amoxicillin-clavulanic acid. Notably, the results showed very low rates of resistance to polymyxins and carbapenem antibiotics, such as polymyxin B (12/615, 2.0%) and imipenem (3/615, 0.5%).
Table 2.
Antibiotic resistance of Salmonella isolates from retail meats in Guangdong, China.
| Antibiotic | Number of resistant isolates (%) from: |
||
|---|---|---|---|
| Chicken (n = 302) | Pork (n = 313) | Total (n = 615) | |
| β-lactams | |||
| Ampicillin | 96 (31.8) | 199 (63.6) | 295 (48.0) |
| Amoxicillin/clavulanic acid | 4 (1.3) | 3 (1.0) | 7 (1.1) |
| Cefotaxime | 30 (9.9) | 10 (3.2) | 40 (6.5) |
| Ceftazidime | 10 (3.3) | 4 (1.3) | 14 (2.3) |
| Cefepime | 8 (2.6) | 9 (2.9) | 17 (2.8) |
| Aminoglycosides | |||
| Gentamicin | 34 (11.3) | 66 (21.1) | 100 (16.3) |
| Streptomycin | 84 (27.8) | 127 (40.6) | 211 (34.3) |
| Amikacin | 6 (2.0) | 2 (0.6) | 8 (1.3) |
| Sulfonamides | |||
| Trimethoprim/sulfamethoxazole | 103 (34.1) | 145 (46.3) | 248 (40.3) |
| Sulfisoxazole | 212 (70.2) | 256 (81.8) | 468 (76.1) |
| Quinolones | |||
| Nalidixic acid | 112 (37.1) | 65 (20.8) | 177 (28.8) |
| Ofloxacin | 192 (63.6) | 83 (26.5) | 275 (44.7) |
| Ciprofloxacin | 37 (12.3) | 37 (11.8) | 74 (12.0) |
| Phenicols | |||
| Florfenicol | 142 (47.0) | 96 (30.7) | 238 (38.7) |
| Chloramphenicol | 146 (48.3) | 99 (31.6) | 245 (39.8) |
| Carbapenems | |||
| Imipenem | 0 | 3 (1.0) | 3 (0.5) |
| Polymyxins | |||
| Polymyxin B | 7 (2.3) | 5 (1.6) | 12 (2.0) |
| Tetracyclines | |||
| Tetracycline | 216 (71.5) | 247 (78.9) | 463 (75.3) |
Resistance to ampicillin, tetracycline, streptomycin, sulfisoxazole, Florfenicol, Chloramphenicol, and Ofloxacin was frequently observed in the Salmonella serovars isolated (Table 3): 588 (95.6%) isolates were resistant to at least one antibiotic and 474 (77.1%) were MDR (Table 4). Resistance to 4–9 antibiotics was detected in 331 isolates (53.8%), whereas 57 isolates (9.3%) were resistant to ≥10 antibiotics. More of the serovars isolated from chicken showed MDR ≥3 (n = 245) than did the serovars isolated from pork (n = 229) (P < 0.05). The MDR rates for the Salmonella serovars are shown in Table 5. Notably, MDR was widespread serovars Kentucky, Typhimurium, Derby, Agona, and Corvallis isolated from retail meat samples.
Table 3.
Antibiotic resistance for the most common Salmonella serotypes isolated from pork and chicken meats in Guangdong.
| Antibiotic | Number (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Typhimurium (n = 84) | Rissen (n = 81) | Derby (n = 75) | London (n = 57) | Agona (n = 56) | Corvallis (n = 46) | Kentucky (n = 38) | Mbandaka (n = 34) | Others (n = 144) | |
| AMP | 70 (83.3) | 63 (77.8) | 42 (56.0) | 35 (61.4) | 9 (16.1) | 8 (17.4) | 8 (22.2) | 0 (0) | 60 (41.7) |
| AMC | 2 (2.4) | 0 (0) | 2 (2.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (2.1) |
| CTX | 8 (9.5) | 1 (1.2) | 4 (5.3) | 0 (0) | 3 (5.4) | 4 (8.7) | 4 (11.1) | 1 (2.9) | 15 (10.4) |
| CAZ | 2 (2.4) | 1 (1.2) | 0 (0) | 0 (0) | 1 (1.8) | 2 (4.3) | 0 (0) | 1 (2.9) | 7 (4.9) |
| FEP | 5 (6.0) | 0 (0) | 3 (4.0) | 0 (0) | 0 (0) | 1 (2.2) | 0 (0) | 0 (0) | 8 (5.6) |
| GEN | 13 (15.5) | 2 (2.5) | 27 (36.0) | 33 (57.9) | 0 (0) | 0 (0) | 3 (8.3) | 1 (2.9) | 21 (14.6) |
| STR | 50 (59.5) | 16 (19.8) | 27 (36.0) | 39 (68.4) | 9 (16.1) | 8 (17.4) | 5 (13.9) | 25 (73.5) | 32 (22.2) |
| AMK | 1 (1.2) | 0 (0) | 1 (1.3) | 0 (0) | 1 (1.8) | 0 (0) | 1 (2.8) | 1 (2.9) | 3 (2.1) |
| SXT | 23 (27.4) | 63 (77.8) | 32 (42.7) | 40 (70.2) | 10 (17.9) | 8 (17.4) | 26 (72.2) | 2 (5.9) | 44 (30.6) |
| S3 | 75 (89.3) | 63 (77.8) | 69 (92.0) | 44 (77.2) | 23 (41.1) | 34 (73.9) | 35 (97.2) | 34 (100.0) | 91 (63.2) |
| NAL | 25 (29.8) | 6 (7.4) | 30 (40.0) | 1 (1.8) | 11 (19.6) | 20 (43.5) | 11 (30.6) | 1 (2.9) | 72 (50.0) |
| OFX | 25 (29.8) | 7 (8.6) | 32 (42.7) | 9 (15.8) | 51 (91.1) | 40 (87.0) | 25 (69.4) | 21 (61.8) | 65 (45.1) |
| CIP | 11 (13.1) | 1 (1.2) | 25 (33.3) | 0 (0) | 1 (1.8) | 8 (17.4) | 11 (30.6) | 1 (2.9) | 16 (11.1) |
| FFC | 30 (35.7) | 5 (6.2) | 34 (45.3) | 23 (40.4) | 49 (87.5) | 25 (54.3) | 30 (83.3) | 0 (0) | 42 (29.2) |
| CHL | 34 (40.5) | 7 (8.6) | 33 (44.0) | 24 (42.1) | 50 (89.3) | 24 (52.2) | 30 (83.3) | 0 (0) | 43 (29.9) |
| IPM | 1 (1.2) | 0 (0) | 0 (0) | 2 (3.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| PB | 2 (2.4) | 1 (1.2) | 0 (0) | 1 (1.8) | 1 (1.8) | 1 (2.2) | 1 (2.8) | 1 (2.9) | 4 (2.8) |
| TET | 69 (82.1) | 77 (95.1) | 61 (81.3) | 49 (86.0) | 27 (48.2) | 37 (80.4) | 33 (91.7) | 33 (97.1) | 77 (53.5) |
Abbreviation: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; GEN, gentamicin; STR, streptomycin; AMK, amikacin; SXT, trimethoprim/sulfamethoxazole; S3, sulfisoxazole; NAL, nalidixic acid; OFX, ofloxacin; CIP, ciprofloxacin; FFC, florfenicol; CHL, chloramphenicol; IPM, imipenem; PB, polymyxin B; TET, tetracycline.
Table 4.
Multidrug-resistant (MDR) Salmonella isolates from retail meats in Guangdong, China.
| No. of antibiotics | Number (%) of resistant isolates |
||
|---|---|---|---|
| Chicken (n = 302) | Pork (n = 313) | Total (n = 615) | |
| 0 | 7 (2.3) | 20 (6.4) | 27 (4.4) |
| 1–3 | 102 (33.8) | 98 (31.3) | 200 (32.5) |
| 4–6 | 130 (43.0) | 113 (36.1) | 243 (39.5) |
| 7–9 | 38 (12.6) | 50 (16.0) | 88 (14.3) |
| ≥10 | 25 (8.3) | 32 (10.2) | 57 (9.3) |
| Resistance ≥ 1 | 295 (97.7) | 293 (93.6) | 588 (95.6) |
| MDR ≥ 3 | 245 (81.1) | 229 (73.2) | 474 (77.1) |
Table 5.
Multidrug resistance (MDR) of the most prevalent Salmonella serovars isolates from retail meats.
| Serovars | Number (%) of antibiotics resistance |
||||
|---|---|---|---|---|---|
| 1–3 | 4–6 | 7–9 | ≥10 | Total (resistance ≥ 3) | |
| Typhimurium (84) | 20 (23.8) | 32 (38.1) | 16 (19.0) | 12 (14.3) | 74 (88.1) |
| Rissen (81) | 26 (32.1) | 51 (63.0) | 3 (3.7) | 1 (1.2) | 66 (81.5) |
| Derby (75) | 31 (41.3) | 12 (16.0) | 11 (14.7) | 20 (26.7) | 54 (72.0) |
| London (57) | 11 (19.3) | 22 (38.6) | 19 (33.3) | 1 (1.8) | 42 (73.7) |
| Agona (56) | 19 (34.0) | 31 (55.4) | 5 (9.0) | 1 (1.8) | 53 (94.6) |
| Corvallis (46) | 16 (34.8) | 18 (39.1) | 9 (19.6) | 2 (4.3) | 38 (82.6) |
| Kentucky (38) | 5 (13.2) | 22 (57.9) | 8 (21.1) | 3 (7.9) | 35 (92.1) |
| Mbandaka (34) | 17 (50.0) | 17 (50.0) | 0 (0) | 0 (0) | 32 (94.1) |
| Braenderup (21) | 17 (81.0) | 1 (4.8) | 1 (4.8) | 2 (9.5) | 7 (33.3) |
| Enteritidis (21) | 7 (33.3) | 13 (61.9) | 1 (4.8) | 0 (0) | 17 (81.0) |
| Meleagridis (20) | 6 (30.0) | 6 (30.0) | 3 (15.0) | 0 (0) | 9 (45.0) |
| Stanley (14) | 6 (42.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Indiana (12) | 0 (0) | 0 (0) | 0 (0) | 12 (100) | 12 (100) |
Prevalence of ESBLs-Producing Isolates and ESBLs Genes
Six serotypes were detected among 21 ESBLs-producing Salmonella isolates (Table 6). The most prevalent serotype was Indiana (7/21, 33.3%) followed by Typhimurium (5/21, 23.8%), Kentucky (3/21, 14.3%), Litchfield (3/21, 14.3%), Derby (2/21, 9.5%), Schwarzengrund (1/21, 4.8%). The resistance profiles are shown in Table 6. Eight β-lactamases and ESBLs encoding genes, including blaTEM-1, blaTEM-206, blaTEM-214, blaCTX-M-15, blaCTX-M-55, blaCTX-M-64, blaCTX-M-123, and blaOXA-1 were identified in the 21 ESBLs-producing Salmonella isolates (Table 7). Among them, 42.9% (n = 9) were found to harbor blaOXA-1, followed by blaCTX-M-55 (5/21, 23.8%), blaTEM-206 (4/21, 19.0%), blaTEM-214 (4/21, 19.0%), blaCTX-M-123 (3/21, 14.3%). Moreover, 100% of ESBLs-producing isolates carried at least one of the β-lactamases and/or ESBLs encoding genes, including blaOXA-1 (n = 9), blaCTX-M-55 (n = 5), blaCTX-M-123 (n = 3), blaTEM-206 (n = 4), and blaTEM-214 (n = 4). Seven isolates (33.3% were detected as carrying two β-lactamases and/or ESBLs encoding genes. None of the isolates were positive for blaSHV, blaPSE, or blaCMY.
Table 6.
Salmonella carrying ESBLs encoding genes and corresponding antibiotic resistance profile.
| Number of isolates | Sample types | Serotypes | Resistance phenotype | ESBLs genes |
|---|---|---|---|---|
| 7 | Pork | Derby | AMP+CTX+FEP+NAL+TET | blaCTX-M-55 |
| Pork | Derby | AMP+CTX+FEP+NAL+TET | blaCTX-M-55 | |
| Pork | Typhimurium | AMP+CTX+CAZ+FEP+STR+S3+OFX+TET | blaTEM-206 | |
| Pork | Typhimurium | AMP+CTX+GEN+STR+SXT+S3+FFC+CHL+TET | blaTEM-214 | |
| Pork | Typhimurium | AMP+CTX+FEP+STR+SXT+S3+OFX+FFC+CHL+TET | blaTEM-206, blaCTX-M-55 | |
| Pork | Typhimurium | AMP+CTX+STR+S3+TET | blaTEM-214, blaCTX-M-55 | |
| Pork | Typhimurium | AMP+CTX+FEP+STR+S3+NAL+OFX+CHL+TET | blaCTX-M-55 | |
| 14 | Chicken | Indiana | AMP+CTX+GEN+STR+SXT+S3+NAL+OFX+CIP+FFC+CHL | blaTEM-214, blaOXA-1 |
| Chicken | Indiana | AMP+CTX+FEP+GEN+STR+AMK+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaCTX-M-123 | |
| Chicken | Indiana | AMP+CTX+CAZ+FEP+GEN+AMK+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaOXA-1 | |
| Chicken | Indiana | AMP+CTX+FEP+GEN+STR+AMK+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaTEM-1, blaOXA-1 | |
| Chicken | Kentucky | AMP+CTX+GEN+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaTEM-214 | |
| Chicken | Indiana | AMP+CTX+GEN+STR+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaOXA-1 | |
| Chicken | Litchfield | AMP+CTX+CAZ+FEP+GEN+STR+SXT+S3 | blaCTX-M-123 | |
| Chicken | Schwarzengrun | AMP+CTX+GEN+S3+NAL+OFX+CIP+FFC+CHL+TET | blaOXA-1 | |
| Chicken | Indiana | AMP+CTX+CAZ+FEP+GEN+STR+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaCTX-M-64, blaOXA-1 | |
| Chicken | Indiana | AMP+CTX+CAZ+FEP+GEN+SXT+S3+NAL+OFX+CIP+FFC+CHL+TET | blaCTX-M-15, blaOXA-1 | |
| Chicken | Litchfield | AMP+CTX+CAZ+FEP+GEN+STR+SXT+OFX+FFC+CHL+TET | blaCTX-M-123, blaTEM-206 | |
| Chicken | Litchfield | AMP+CTX+GEN+STR+SXT+S3+OFX+FFC+CHL+TET | blaTEM-206 | |
| Chicken | Kentucky | AMP+CTX+S3+OFX+FFC+CHL | blaOXA-1 | |
| Chicken | Kentucky | AMP+CTX+GEN+S3+NAL+OFX+CIP+FFC+CHL | blaOXA-1 |
Abbreviation: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; GEN, gentamicin; STR, streptomycin; AMK, amikacin; SXT, trimethoprim/sulfamethoxazole; S3, sulfisoxazole; NAL, nalidixic acid; OFX, ofloxacin; CIP, ciprofloxacin; FFC, florfenicol; CHL, chloramphenicol; IPM, imipenem; PB, polymyxin B; TET, tetracycline.
Table 7.
Prevalence of Salmonella carrying β-lactamase and ESBLs encoding genes (n = 21).
| Genes | Number of isolates | Percentage of isolates | |
|---|---|---|---|
| blaTEM | blaTEM-1 | 1 | 4.8 |
| blaTEM-206 | 4 | 19.0 | |
| blaTEM-214 | 4 | 19.0 | |
| blaCTX | blaCTX-M-15 | 1 | 4.8 |
| blaCTX-M-55 | 5 | 23.8 | |
| blaCTX-M-64 | 1 | 4.8 | |
| blaCTX-M-123 | 3 | 14.3 | |
| blaOXA | blaOXA-1 | 9 | 42.9 |
PFGE
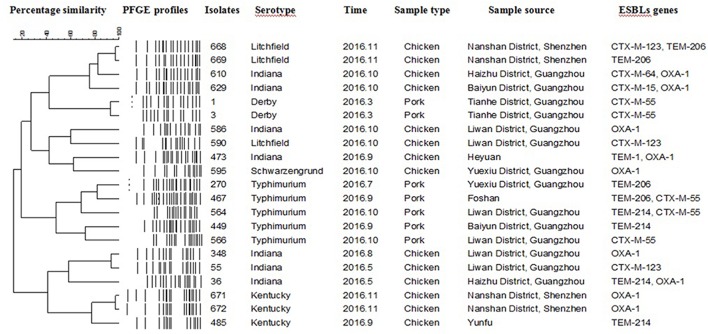
Twenty-one PFGE patterns were identified among the 21 ESBLs-producing isolates (Figure 1). Seven ESBLs-producing Indiana isolates, five Typhimurium isolates, three Kentucky isolates, three Litchfield isolates, two Derby isolates and, one Schwarzengrund isolate were analyzed via PFGE with the enzyme XbaI. The PFGE profile analysis demonstrated that Salmonella of the same serotype isolated from different times and locations have markedly different genotypes. Similarly, serovars isolated from the same time and place also showed diverse genotypes.
FIGURE 1.
Dendrogram of pulse field gel electrophoresis profiles of 21 extended spectrum β-lactamases (ESBLs)-producing Salmonella isolates with the respective ESBL genes from chicken and pork meat.
Discussion
We found the high levels of Salmonella contamination rate in chicken (63.6%) and pork (73.1%) collected from retail markets in Guangdong Province, China. These prevalences were significantly higher than those reported in similar studies in other districts in China, such as Hebei (Yan et al., 2010), Sichuan (Li et al., 2013), Henan (Li et al., 2014), Beijing (Wang et al., 2014), Jiangsu (Bai et al., 2015), and Shandong (Cui et al., 2016). Meanwhile, the Salmonella contamination rate was higher than that of other parts of Asia, such as South Korea (Choi et al., 2015), Tokyo (Katoh et al., 2015), and Northern Vietnam (Thai et al., 2012), and higher than that reported in Egypt (Gharieb et al., 2015). This indicated that contamination by Salmonella in animal-derived foods in Guangdong Province is more serious, which greatly increases the risk of humans being infected. The results suggested that Salmonella in livestock and poultry products should be a priority area for supervision in Guangdong Province. It is also a reminder that animal-derived foods in the tropics should be paid more attention and reflected the need for strengthening of the monitoring of Salmonella the animal-origin food supply; for example, improving environmental sanitation in food processing and health awareness among employees should be enhanced, and the disinfection management of processing equipment and tableware containing food should be improved.
Thirty-eight Salmonella serovars were found in retail meat samples, among which, serovars Typhimurium, Rissen, Derby (pork), and Agona, Corvallis, and Kentucky (chicken) were the most common serotypes, respectively. The serotypes found in the pork samples were the same as those identified in previous studies in China and abroad (Liang et al., 2015). The distribution of the serotypes in the chicken samples was very different from those in other regions. It was reported that serovars Enteritidis and Typhimurium were two predominant serotypes in chickens from other regions (Li et al., 2017). Interestingly, Agona, Corvallis, and Kentucky were reported less frequently in previous studies (Arnedo-Pena et al., 2016), whereas they were the dominant isolates in the current investigation. Additionally, serovar Agona is one of the top 10 serotypes responsible for nosocomial infections in China (Zhang et al., 2014); serovar Corvallis has been reported in Brazil, Germany, and Turkey (Kocabiyik et al., 2006; Fischer et al., 2013; Yamatogi et al., 2015); and serovar Kentucky was the most common serovar in the United States (van Duijkeren et al., 2002; Galanis et al., 2006). The results suggest that Agona, Corvallis, and Kentucky may cause public health concerns in Guangdong. Importantly, serovar Rumford was detected in animal-derived food for the first time in China. The detection of this strain not only fills the blank of the bacterial species library in China, but also provides a new basis for diagnostic tests and epidemiological investigation of this strain. Notably, Typhimurium is the most frequently isolated serovar from infants in Guangdong (Deng et al., 2012). Therefore, further study is required to determine the source of this serovar (Liang et al., 2015; O’Leary et al., 2015). Our results also showed that an association might exist between Salmonella-contaminated food and the sample source. Guangdong is mainly involved in yellow broiler production and its breeding, production mode, and climatic conditions are different from those in other regions, which might explain the different serotype distribution. Thus, China should strengthen yellow chicken monitoring and pay attention to changes in serotypes in different regions.
The susceptibility results showed more than 75% of the Salmonella isolates were resistant to tetracycline and sulfisoxazole. These high resistance rates reflect their widespread use in animal feed and are consistent with other reports (Bai et al., 2015). In addition, resistance to florfenicol and chloramphenicol in 47.0% and 48.3% of chicken isolates, in 30.7 and 31.6% of the pork isolates deserves our attention because resistance to these antibiotics may cause human resistance. In this study, all of the Salmonella isolates were susceptible to amoxicillin/clavulanic acid and amikacin, possibly because these antibiotics are not used for therapeutic purposes in clinical veterinary medicine; this result was consistent with previous reports (Sinwat et al., 2015). Different serotypes of Salmonella showed different resistance rates. In the present study, we observed a high prevalence of MDR Salmonella isolates among those isolated from chicken and pork. Serovars Agona, Corvallis, and Kentucky had higher resistance rates to the third-generation cephalosporins (CTX, CAZ) and fluoroquinolones than did the serovars Rissen and Derby. Serovars Agona, Corvallis, and Kentucky also exhibited higher resistance rates to ofloxacin. They also provided more information for further study related to cephalosporin or fluoroquinolones-resistant Salmonella. The increasing prevalence of MDR Salmonella involving multiple resistances to various antibiotics could cause Salmonella to evolve toward becoming a super bacteria (Campioni et al., 2014). The association between antibiotic resistance and specific serotypes should that Kentucky, Typhimurium, Corvallis, and Agona were strongly associated with MDR phenotypes. In addition, resistant strains were found for first line of drug used in human medicine, such as Carbapenems and Polymyxins, which showed that Salmonella had a tendency toward drug resistance. Hence, there is a need to strengthen epidemiological investigations of Salmonella infections and further studies are required to improve our knowledge concerning the development of MDR.
In the present study, the most commonly detected ESBLs encoding genes were OXAs (42.9%), followed by CTX-Ms (23.8%). This was similar to the results of previous studies in which CTX-M- types were the main family of ESBLs (Nguyen et al., 2016; Qiao et al., 2017). Furthermore, reports indicated that CTX-Ms have been disseminating rapidly among populations of gram-negative bacteria in clinical settings in recent years (Shahid et al., 2009). The 21 ESBLs-producing isolates identified in this study were subjected to PFGE analysis. The resultant dendrograms of isolate patterns showed clusters with a high level of diversity. This indicated that the Salmonella prevailing in Guangdong’s retail market during this period not comprised diverse serotypes, but also strains of the same serotype have more complex genotypes and exhibit diverse features. Thus, we should pay attention to this easily overlooked repository of MDR genes, and to the food safety issues they invoke in terms of consumers’ health. Moreover, more research into the characteristics of the dissemination of Salmonella and antibiotic resistance genes is warranted.
Conclusion
Our results indicated the diversity of Salmonella serotypes and the high prevalence and antibiotic resistance that existed among Salmonella recovered from chicken and pork meat at retail markets. In particular, MDR Salmonella serotypes Agona, Corvallis, and Kentucky may indicate the potential risk of resistant Salmonella foodborne infections in Guangdong, China. Thus, this study suggests that epidemic trends information about the Salmonella serovars in Guangdong province should be further monitored and studied.
Author Contributions
ML and LZ contributed to the conception of the study. JZ and LZ contributed significantly to analysis and manuscript preparation. LZ performed the data analyses and wrote the manuscript. YF, ZX, and YM helped to perform the analysis with constructive discussions. YW, XQ, and HZ collected the samples and conducted the experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the Special Fund for Agro-Scientific Research in the Public Interest (201403054), National Key R&D Program of China (2017YFC1600101 and 2018YFD0500500), Project Supported by Guangdong Province Universities and Colleges, Pearl River Scholar Funded Scheme (2018), Pearl River S&T Nova Program of Guangzhou (201806010183), National Natural Science Foundation of China (31402193), Walmart Foundation (SA1703162), Province Science and Technology of Guangdong Research Project (2017A020208055), and Earmarked Fund for China Agriculture Research System (CARS-41-G16).
References
- Arnedo-Pena A., Sabater-Vidal S., Herrera-Leon S., Bellido-Blasco J. B., Silvestre-Silvestre E., Meseguer-Ferrer N., et al. (2016). An outbreak of monophasic and biphasic Salmonella Typhimurium, and Salmonella Derby associated with the consumption of dried pork sausage in Castellon (Spain). Enferm. Infecc. Microbiol. Clin. 34 544–550. 10.1016/j.eimc.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Bai L., Lan R. T., Zhang X. L., Cui S. H., Xu J., Guo Y. C., et al. (2015). Prevalence of Salmonella Isolates from chicken and pig slaughterhouses and emergence of ciprofloxacin and cefotaxime Co-resistant S. enterica serovar Indiana in Henan, China. PLoS One 10:e0144532. 10.1371/journal.pone.0144532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barza M. (2002). Potential mechanisms of increased disease in humans from antimicrobial resistance in food animals. Clin. Infect. Dis. 34 S123–S125. 10.1086/340249 [DOI] [PubMed] [Google Scholar]
- Ben Said L., Jouini A., Alonso C. A., Klibi N., Dziri R., Boudabous A., et al. (2016). Characteristics of extended-spectrum beta-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total Environ. 550 1103–1109. 10.1016/j.scitotenv.2016.01.042 [DOI] [PubMed] [Google Scholar]
- Campioni F., Zoldan M. M., Falcao J. P. (2014). Characterization of Salmonella Enteritidis strains isolated from poultry and farm environments in Brazil. Epidemiol. Infect. 142 1403–1410. 10.1017/S0950268814000491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Chon J. W., Kim H. S., Kim D. H., Lim J. S., Yim J. H., et al. (2015). Incidence, antimicrobial resistance, and molecular characteristics of nontyphoidal Salmonella including extended-spectrum beta-lactamase producers in retail chicken meat. J. Food Prot. 78 1932–1937. 10.4315/0362-028X.JFP-15-145 [DOI] [PubMed] [Google Scholar]
- Cui M. Q., Xie M. Y., Qu Z. N., Zhao S. J., Wang J. W., Wang Y., et al. (2016). Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao. China. Food Control 62 270–276. 10.1016/j.foodcont.2015.10.036 [DOI] [Google Scholar]
- Dahms C., Hubner N. O., Kossow A., Mellmann A., Dittmann K., Kramer A. (2015). Occurrence of ESBL-producing Escherichia coli in livestock and farm workers in mecklenburg-western pomerania, Germany. PLoS One 10:e0143326. 10.1371/journal.pone.0143326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. L., Ran L., Wu S. Y., Ke B. X., He D. M., Yang X. F., et al. (2012). Laboratory-based surveillance of non-typhoidal Salmonella infections in guangdong province. China. Foodborne Pathog. Dis. 9 305–312. 10.1089/fpd.2011.1008 [DOI] [PubMed] [Google Scholar]
- Emery C. L., Weymouth L. A. (1997). Detection and clinical significance of extended-spectrum beta-lactamases in a tertiary-care medical center. J. Clin. Microbiol. 35 2061–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J., Schmoger S., Jahn S., Helmuth R., Guerra B. (2013). NDM-1 carbapenemase-producing Salmonella enterica subsp enterica serovar Corvallis isolated from a wild bird in Germany. J. Antimicrobial. Chemother. 68 2954–2956. 10.1093/jac/dkt260 [DOI] [PubMed] [Google Scholar]
- Galanis E., Wong D. M. A. L. F., Patrick M. E., Binsztein N., Cieslik A., Chalermchaikit T., et al. (2006). Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12 381–388. 10.3201/eid1203.050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharieb R. M., Tartor Y. H., Khedr M. H. E. (2015). Non-Typhoidal Salmonella in poultry meat and diarrhoeic patients: prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 7:34. 10.1186/s13099-015-0081-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibourdenche M., Roggentin P., Mikoleit M., Fields P. I., Bockemuhl J., Grimont P. A. D., et al. (2010). Supplement 2003-2007 (No. 47) to the white-kauffmann-le minor scheme. Res. Microbiol. 161 26–29. 10.1016/j.resmic.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Ibrahim D. R., Dodd C. E. R., Stekel D. J., Ramsden S. J., Hobman J. L. (2016). Multidrug resistant, extended spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol. Ecol. 92:fiw013. 10.1093/femsec/fiw013 [DOI] [PubMed] [Google Scholar]
- Jackson B. R., Griffin P. M., Cole D., Walsh K. A., Chai S. J. (2013). Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg. Infect. Dis. 19 1239–1244. 10.3201/eid1908.121511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh R., Matsushita S., Shimojima Y., Ishitsuka R., Sadamasu K., Kai A. (2015). Serovars and Drug-resistance of Salmonella strains isolated from domestic chicken meat in Tokyo (1992-2012). Kansenshogaku Zasshi 89 46–52. [DOI] [PubMed] [Google Scholar]
- Kocabiyik A. L., Cetin C., Dedicova D. (2006). Detection of Salmonella spp. in stray dogs in Bursa province, Turkey: first isolation of Salmonella corvallis from dogs. J. Vet. Med. B Infect. Dis. Vet. Public Health 53 194–196. 10.1111/j.1439-0450.2006.00932.x [DOI] [PubMed] [Google Scholar]
- Lai J., Wu C. M., Wu C. B., Qi J., Wang Y., Wang H. Y., et al. (2014). Serotype distribution and antibiotic resistance of Salmonella in food-producing animals in shandong province of China, 2009 and 2012. Int. J. Food Microbiol. 180 30–38. 10.1016/j.ijfoodmicro.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Li R. C., Lai J., Wang Y., Liu S. L., Li Y., Liu K. Y., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province. China. Int. J. Food Microbiol. 163 14–18. 10.1016/j.ijfoodmicro.2013.01.020 [DOI] [PubMed] [Google Scholar]
- Li S., Zhou Y. F., Miao Z. M. (2017). Prevalence and antibiotic resistance of non-typhoidal salmonella isolated from raw chicken carcasses of commercial broilers and spent hens in Tai’an, China. Front. Microbiol. 8:2106 10.3389/Fmicb.2017.02106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. C., Pan Z. M., Kang X. L., Geng S. Z., Liu Z. Y., Cai Y. Q., et al. (2014). Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu Province, Eastern China. J. Food Prot. 77 236–245. 10.4315/0362-028X.JFP-13-269 [DOI] [PubMed] [Google Scholar]
- Liang Z. M., Ke B. X., Deng X. L., Liang J. H., Ran L., Lu L. L., et al. (2015). Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009-2012. BMC Infect. Dis. 15:53. 10.1186/s12879-015-0784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. T. A., Kanki M., Do Nguyen P., Le H. T., Ngo P. T., Tran D. N. M., et al. (2016). Prevalence, antibiotic resistance, and extended-spectrum and AmpC beta-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City. Vietnam. Int. J. Food Microbiol. 236 115–122. 10.1016/j.ijfoodmicro.2016.07.017 [DOI] [PubMed] [Google Scholar]
- O’Leary D., McCabe E. M., McCusker M. P., Martins M., Fanning S., Duffy G. (2015). Acid environments affect biofilm formation and gene expression in isolates of Salmonella enterica Typhimurium DT104. Int. J. Food Microbiol. 206 7–16. 10.1016/j.ijfoodmicro.2015.03.030 [DOI] [PubMed] [Google Scholar]
- Qiao J., Zhang Q., Alali W. Q., Wang J. W., Meng L. Y., Xiao Y. P., et al. (2017). Characterization of extended-spectrum beta-lactamases (ESBLs)-producing Salmonella in retail raw chicken carcasses. Int. J. Food Microbiol. 248 72–81. 10.1016/j.ijfoodmicro.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Ren X., Li M., Xu C., Cui K., Feng Z., Fu Y., et al. (2016). Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 144 2989–2999. 10.1017/S0950268816001515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot E. M., Fair M. A., Gautom R., Cameron D. N., Hunter S. B., Swaminathan B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157 : H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3 59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- Russell D. A., Bowman C. A., Hatfull G. F. (2014). Genome sequence of Salmonella enterica subsp. enterica strain durban. Genome Announc 2:e00399-14.. 10.1128/genomeA.00399-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Sobia E., Singh A., Malik A., Khan H. M., Jonas D., et al. (2009). Beta-lactams and Beta-lactamase-inhibitors in current- or potential-clinical practice: a comprehensive update. Crit. Rev. Microbiol. 35 81–108. 10.1080/10408410902733979 [DOI] [PubMed] [Google Scholar]
- Sinwat N., Angkittitrakul S., Chuanchuen R. (2015). Characterization of antimicrobial resistance in Salmonella enterica isolated from pork, chicken meat, and humans in Northeastern Thailand. Foodborne Pathog. Dis. 12 759–765. 10.1089/fpd.2015.1946 [DOI] [PubMed] [Google Scholar]
- Swaminathan B., Barrett T. J., Fields P. (2006). Surveillance for human Salmonella infections in the United States. J. AOAC Int. 89 553–559. [PubMed] [Google Scholar]
- Thai T. H., Hirai T., Lan N. T., Yamaguchi R. (2012). Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. 156 147–151. 10.1016/j.ijfoodmicro.2012.03.016 [DOI] [PubMed] [Google Scholar]
- van Duijkeren E., Wannet W. J. B., Heck M. E. O. C., van Pelt W., van Oldruitenborgh-Oosterbaan M. M. S., Smit J. A. H., et al. (2002). Sero types, phage types and antibiotic susceptibilities of Salmonella strains isolated from horses in The Netherlands from 1993 to 2000. Vet. Microbiol. 86 203–212. 10.1016/S0378-1135(02)00007-X [DOI] [PubMed] [Google Scholar]
- Vo A. T. T., van Duijkeren E., Fluit A. C., Heck M. E. O. C., Verbruggen A., Maas H. M. E., et al. (2006). Distribution of Salmonella enterica serovars from humans, livestock and meat in vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet. Microbiol. 113 153–158. 10.1016/j.vetmic.2005.10.034 [DOI] [PubMed] [Google Scholar]
- Wang S. J., Duan H. L., Zhang W., Li J. W. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51 8–13. 10.1111/j.1574-695X.2007.00305.x [DOI] [PubMed] [Google Scholar]
- Wang Y. R., Chen Q., Cui S. H., Xu X., Zhu J. H., Luo H. P., et al. (2014). Enumeration and characterization of Salmonella isolates from retail chicken carcasses in Beijing, China. Foodborne Pathog. Dis. 11 126–132. 10.1089/fpd.2013.1586 [DOI] [PubMed] [Google Scholar]
- Yamatogi R. S., Oliveira H. C., Camargo C. H., Fernandes S. A., Hernandes R. T., Pinto J. P., et al. (2015). Clonal relatedness and resistance patterns of Salmonella Corvallis from poultry carcasses in a Brazilian slaughterhouse. J. Infect. Dev. Ctries 9 1161–1165. 10.3855/jidc.5634 [DOI] [PubMed] [Google Scholar]
- Yan H., Li L., Alam M. J., Shinoda S., Miyoshi S., Shi L. (2010). Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int. J. Food Microbiol. 143 230–234. 10.1016/j.ijfoodmicro.2010.07.034 [DOI] [PubMed] [Google Scholar]
- Yang B. W., Qu D., Zhang X. L., Shen J. L., Cui S. H., Shi Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 141 63–72. 10.1016/j.ijfoodmicro.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Zhang J., Jin H., Hu J., Yuan Z., Shi W., Ran L., et al. (2014). Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006-2010. Epidemiol. Infect. 142 826–832. 10.1017/S0950268813001659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Ye C., Chang W., Sun S. (2017). Serotype distribution, antimicrobial resistance, and class 1 integrons profiles of Salmonella from animals in slaughterhouses in Shandong Province, China. Front. Microbiol. 8:1049. 10.3389/fmicb.2017.01049 [DOI] [PMC free article] [PubMed] [Google Scholar]