Abstract
Ineffectiveness of cancer therapy may originate in the incompatibility of the treatment with various mutations in the cancer cells. Finding novel anticancer treatments that work efficiently for varying types of cancer cells remains challenging. Previous studies have identified that compounds in sea cucumbers are capable of inhibiting the growth of cancer cells and inducing apoptosis. However, information on the underlying mechanisms controlling cancer cell growth at a molecular level remains limited. The current study analyzed the potential of colochiroside A, ds-echinoside A, philinopside E, sphingosine and stichoposide C as inhibitors for anticancer target proteins, including mouse double minute 2 homolog (MDM2) and C-X-C chemokine receptor type 4 (CXCR4). Inhibition of MDM2 triggers apoptosis through regulation of tumor protein 53 and CXCR4 inhibition may prevent cancer cell proliferation and growth by affecting the Janus kinase 2/3 signal transducer and activator of transcription signaling pathway and protein tyrosine kinase 2. The results of a binding affinity analysis using molecular docking revealed that philinopside E and ds-echinoside A may inhibit MDM2 and CXCR4. The data suggested that these active compounds may be promising inhibitors of cell growth by binding to two targets simultaneously. Furthermore, stichoposide C and colochiroside A were predicted to inhibit CXCR4. Additional research is needed to validate the in vitro activity of the aforementioned compounds.
Keywords: sea cucumber, anticancer, apoptosis, C-X-C chemokine receptor type 4, mouse double minute 2 homolog, Janus kinase 2/3, pathway analysis, BioGRID
Introduction
The prevalence of cancer is increasing rapidly (1) and research is focusing on the exploration of novel anticancer treatments. Although the field is growing rapidly, very few cancer drugs are able to pass clinical trials (2). There are numerous types of cancer cells, which are characterized based on either the source of the cell or the development of cells due to gene mutations (3). Various types of drugs may be required for treatments based on the particular characteristics of the cancer cells. As a result, novel anticancer treatments from plants and marine invertebrates, including sea cucumbers were explored.
In general, in vitro and in vivo studies involving sea cucumbers have primarily focused on the toxicity of active compounds on cancer cells by induction of apoptosis or cell cycle arrest (4). Compounds from sea cucumbers exhibiting anticancer properties have been reported (5), including colochiroside A from Colochirus anceps (6), ds-echinoside A from Pearsonothuria graeffei (7), philinopside E from Pentacta quadrangularis (8), sphingosine from Stichopus variegatus (9) and stichoposide C from Thelenota anax (10). However, the mechanisms of action controlling cancer cell growth at a molecular level remain unclear. The current study analyzed the potential of these compounds as inhibitors of mouse double minute 2 homolog (MDM2) and C-X-C chemokine receptor type 4 (CXCR4). The inhibition of these two targets simultaneously may induce a synergistic effect, increasing treatment efficacy.
MDM2 serves a role in binding pro-apoptotic tumor protein 53 (p53) and degrading it (11). Inhibiting the activity of MDM2 may increase p53 levels in the cell, which are necessary for apoptosis (11,12). CXCR4 belongs to the G-protein-coupled receptor family that is involved in several pathways associated with cancer and serves a role in controlling cell proliferation (13). CXCR4 promotes survival of various cell types (14) and serves a critical role in tumorigenesis (15). Furthermore, it acts as receptor for the C-X-C motif chemokine ligand 12 (CXCL12) which serves a role in signal transduction for calcium uptake and enhances the activity of mitogen-activated protein kinase (MAPK)1/MAPK3 (15,16). CXCR4 was reported to be a potent inducer of apoptosis in acute myeloid leukemia cell lines (14). The protein has been a target in drug development (17) and cancer treatment (18,19), and anti-CXCR4 antibodies were demonstrated to induce apoptosis in hematologic malignancies (15). The current study describes the potential of compounds from sea cucumbers as MDM2 and CXCR4 inhibitors, aiming to reveal novel insight into the mechanisms of inhibiting cancer cell growth.
Materials and methods
Preparation of molecule structures and codes
The ligands used for docking analysis were colochiroside A, ds-echinoside A, philinopside E, sphingosine, stichoposide C, 1-(5-chloro-2-methylphenyl)-5-(3-chlorophenyl)-2-(3-methylphenyl)-1H-imidazole-4-carboxylic acid, a tetra-substituted imidazole (an MDM2 inhibitor) and chalcone-4 (a CXCR4 inhibitor). SMILES codes of the compounds were converted to 3D structures in Protein Data Bank (PDB) format using BIOVIA Discovery Studio 4.5 (20). These structures were used for ligand docking. The 3D structure for chalcone-4 was obtained from the binding database (https://www.bindingdb.org/bind/index.jsp) (21) and the 3D structure for the substituted imidazole was obtained from the PDB (PDB ID, 4OQ3). The receptor structures were retrieved from the PDB for MDM2 (PDB ID, 4OQ3) and CXCR4 (PDB ID, 3OE6). The proteins then were prepared by BIOVIA Discovery Studio.
Ligand docking studies
Interactions between receptors and ligand were analyzed by AutoDock Vina integrated in PyRx 0.8 (https://pyrx.sourceforge.io) (22,23). The docking method was used to evaluate binding affinities and to elucidate molecular mechanisms, and was performed according to previous literature (24). Docking was performed by setting receptors as rigid molecules and ligands as flexible molecules within the active site. Results of docking and bonding interactions were analyzed by BIOVIA Discovery Studio (20).
Protein-protein interactions and networks
Proteins that interact with CXCR4 were identified using BioGRID database (https://thebiogrid.org/) (25). Protein-protein interaction networks were examined using STRING (https://string-db.org/) (26).
Pathway analysis
Pathway analysis for CXCR4 was performed using Kyoto Encyclopedia Gene and Genome (KEGG; http://www.genome.jp/kegg/) (27). The role of CXCR4 proteins in various molecular pathways was identified using KEGG pathways databases with STRINGdb 10.5 software. The database covers a range of pathways that have been used as references for the determination of gene or protein function within a cell (28).
Results
Ligand docking analysis
The results of the docking between CXCR4 or MDM2 with the five compounds identified in sea cucumbers revealed that four of the compounds (Ds-echinoside A, Philinopside E, Stichoposide C and Colochiroside A, with values of −9.0, −8.5, −9.2 and −8.5 kcal/mol, respectively) exhibited higher binding affinities to CXCR4 compared with a known inhibitor (chalcone; −7.1 kcal/mol; Table I). Additionally, two compounds (Ds-echinoside A and Philinopside E) were identified to potentially bind to MDM2, with binding affinities of −7.1 and −7.5 kcal/mol, respectively. The compounds (Ds-echinoside A and Philinopside E) were predicted to inhibit MDM2 and to exhibit binding energies higher than its inhibitor, imidazole. However, the binding energies of these two molecules to MDM2 were similar to those of chalcone bound to CXCR4 (−7.1 kcal/mol), but lower when compared with protease bound to its inhibitor (−7.0 kcal/mol) (29) and to the coline receptor bound to its ligand (−6.0 kcal/mol) (30). Sphingosine exhibited lowest binding affinities for CXCR4 and MDM2 (−5.8 kcal/mol and −5.2 kcal/mol, respectively).
Table I.
Binding affinity between compounds from sea cucumbers and CXCR4 (PDB ID, 3OE6) or MDM2 (PDB ID, 4OQ3).
| Binding affinity (kcal/mol) | ||
|---|---|---|
| Ligand Name | CXCR4 | MDM2 |
| Ds-echinoside A | −9.0 | −7.1 |
| Sphingosine | −5.8 | −5.2 |
| Philinopside E | −8.5 | −7.5 |
| Stichoposide C | −9.2 | −5.7 |
| Colochiroside A | −8.5 | −7.0 |
| CXCR4 Inhibitor (Chalcone) | −7.1 | – |
| MDM2 Inhibitor (Imidazoles) | – | −9.5 |
CXCR4, C-X-C chemokine receptor type 4; MDM2, mouse double minute 2 homolog; PDB, Protein Data Base.
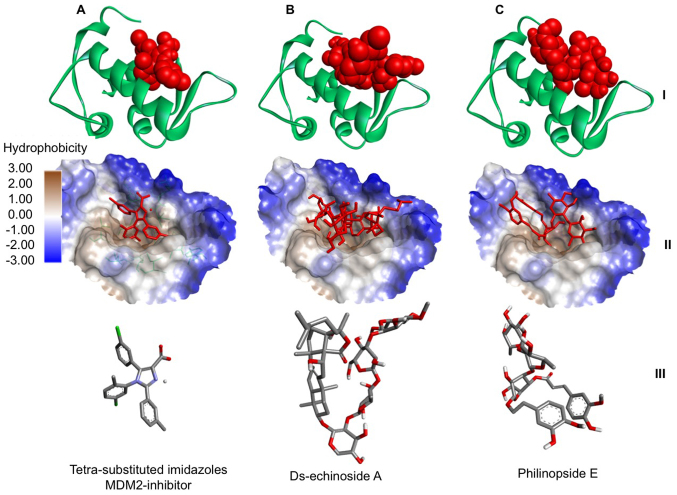
Further analysis focused on evaluating the orientation of the compounds when interacting with the active site of MDM2. This analysis describes a critical part in assessing the potential of a compound for inhibiting MDM2. It was demonstrated that two compounds (Ds-echinoside A and Philinopside E) from sea cucumbers bound to the active site of MDM2 in a similar position to the known inhibitor, a substituted imidazole (Fig. 1). The data suggested that philinopside E and ds-echinoside A, extracted from Pentacta quadrangularis and Pearsonothuria graeffei, respectively, may have potential as MDM2 inhibitors.
Figure 1.
Interaction between MDM2 and compounds from sea cucumbers. Docking of (A) a tetra-substituted imidazole, a known MDM2 inhibitor, (B) ds-echinoside A and (C) philinopside E to MDM2. At the top, MDM2 presented in green ribbon structure with ligands presented as red spheres. In the middle, hydrophobicity surface map of the active site of MDM2 with the ligands presented as red cylinders. At the bottom, cylinder representation of the ligands with carbon atoms in grey, nitrogen in blue, chlorine in green, oxygen in red and hydrogen in white, to emphasize ligand orientation. MDM2, mouse double minute 2 homolog.
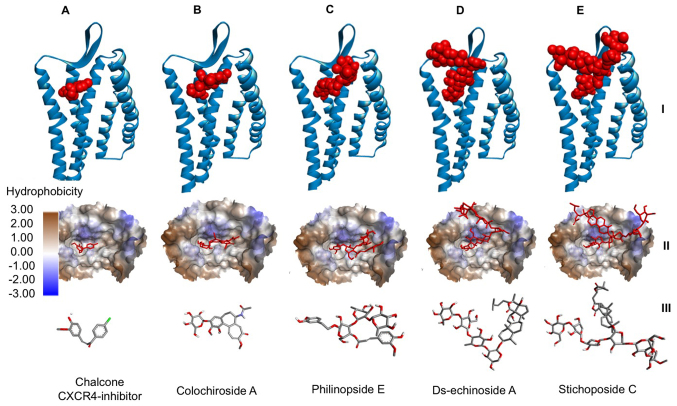
The binding of ds-echinoside A, philinopside E, stichoposide C and colochiroside A to CXCR4 were compared to the binding of chalcone-4, a known CXCR4 inhibitor, to CXCR4 (Fig. 2). It was demonstrated that all compounds bound to the active site of CXCR4 in a similar position to chalcone-4. The data indicated that these compounds may have potential as CXCR4 inhibitors.
Figure 2.
Interaction between CXCR4 and compounds from sea cucumbers. Docking of (A) chalcone-4, a known CXCR4 inhibitor, (B) colochiroside A, (C) philinopside E, (D) ds-echinoside A and (E) stichoposide C to CXCR4. At the top, CXCR4 presented in blue ribbon structure with ligands presented as red spheres. In the middle, hydrophobicity surface map of the active site of CXCR4 with the ligands presented as red cylinders. At the bottom, cylinder representation of the ligands with carbon atoms in grey, nitrogen in blue, chlorine in green, oxygen in red and hydrogen in white, to emphasize ligand orientation. CXCR4, C-X-C chemokine receptor type 4.
Protein interaction and pathway analysis
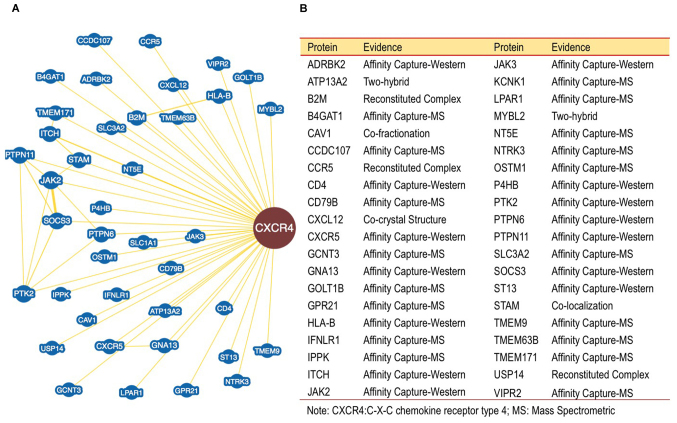
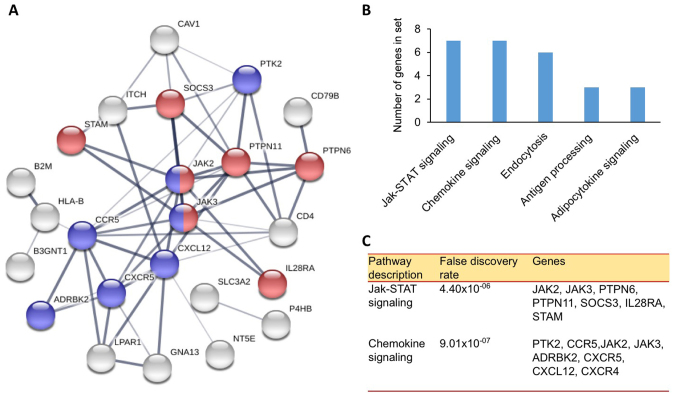
Furthermore, the binding of proteins to CXCR4 were investigated. The data obtained using BioGRID revealed over 40 proteins interacting with CXCR4 (Fig. 3). This analysis is essential to map and resolve functions of proteins that interact with CXCR4. The data may be used in further pathway analysis to help understand the role of CXCR4 in the mechanism of cancer cell growth regulation. The results of the analysis are summarized as a map of proteins interacting with CXCR4. Identified proteins may serve a role in the pathways involved in the pathomechanism of cancer. The proteins are involved in two pathways associated with cancer cell signaling, including the chemokine and Janus kinase (JAK) signal transducer and activator of transcription (STAT) signaling pathway (Fig. 4). A minimum of 15 proteins participates in these pathways, including protein tyrosine kinase 2 (PTK2), C-C chemokine receptor type 5, JAK2, JAK3, β-adrenergic receptor kinase 2, CXCR5, CXCL12, CXCR4, protein tyrosine phosphatase (PTP)6, PTPN11, suppressor of cytokine signaling 3, interleukin-28 α receptor and signal transducing adapter molecule 1. These pathways serve a role in cell proliferation, angiogenesis, cell growth, and metastasis (31). Inhibition of these two pathways may inhibit cancer cell growth and induce apoptosis.
Figure 3.
CXCR4 binding proteins. (A) CXCR4 protein interaction obtained from the BioGrids Database. (B) List of proteins which interact with CXCR4 based on BioGrids Database. CXCR4, C-X-C chemokine receptor type 4.
Figure 4.
CXCR4-protein interaction network. (A) Network of CXCR4 interactions based on the String Database which involve the Jak-STAT (red) and Chemokine signaling pathways (violet). (B) Calculated number of genes involved in various pathways connected to the CXCR4 network. (C) Genes identified to participate in the Jak-STAT and chemokine signaling pathways. The false discovery rates were also determined. CXCR4, C-X-C chemokine receptor type 4; Jak-STAT, Janus kinase signal transducer and activator of transcription.
The results of the current study indicated that two out of the five chosen compounds, philinopside E and ds-echinoside A, may inhibit MDM2 and CXCR4 (Fig. 5). Two other compounds, stichoposide C and colochiroside A, were predicted to inhibit CXCR4. The data suggested that philinopside E and ds-echinoside A may exhibit higher efficiencies due to inhibiting two targets simultaneously. Inhibition of MDM2 may trigger apoptosis through p53 activation (32) and inhibition of CXCR4 may affect cell proliferation and growth through JAK2/3-STAT and PTK2 signalling pathways.
Figure 5.
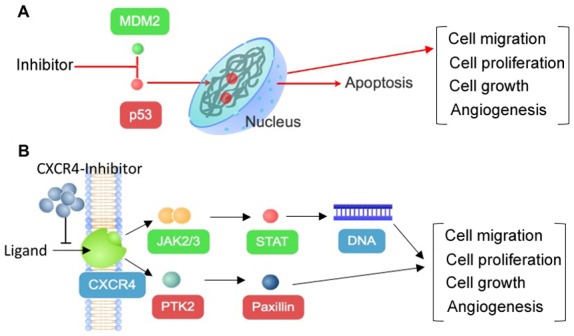
Anticancer mechanism of compounds from sea cucumbers through inhibition of (A) MDM2 or (B) CXCR4. Inhibition of both proteins may lead to decreased cell migration, cell proliferation, cell growth and angiogenesis, which may lead to apoptosis. MDM2, mouse double minute 2 homolog; CXCR4, C-X-C chemokine receptor type 4; p53, tumor protein 53; CXCL12, C-X-C motif chemokine 12; JAK, Janus kinase; STAT, signal transducer and activator of transcription; PTK2, protein tyrosine kinase 2.
Discussion
Previous studies have demonstrated that sea cucumbers contain compounds which exhibit anticancer properties and are described as beneficial agents for human health (33). However, mechanisms of action explaining the anticancer properties remain unclear. The current study analyzed the anticancer mechanisms of compounds from five species of sea cucumber (Table II) using an in silico approach. These compounds included colochiroside A, ds-echinoside A, philinopside E, sphingosine and stichoposide C. Molecular docking was conducted to examine the binding affinity between these compounds and MDM2 or CXCR4. MDM2 has been used as a target in cancer therapy (32). It serves a role in the degradation of p53, a pro-apoptotic protein (32). CXCR4 has also been described as a target in the development of cancer treatments (13). The protein is a receptor that regulates cell cycle, cell proliferation, metastasis and angiogenesis (34).
Table II.
Compounds from sea cucumbers exhibiting anticancer activities.
| Author, year | Organism | Compound | Cell effect | (Refs.) |
|---|---|---|---|---|
| Zhang and Yi, 2011 | Colochirus anceps | Colochiroside A | Antitumor activity | (6) |
| Zhao et al, 2011 | Pearsonothuria graeffei | Ds-echinoside A | Antimetastatic, angiogenesis, apoptosis | (7) |
| Tian et al, 2007 | Pentacta quadrangularis | Philinopside E | Antimetastatic, angiogenesis, apoptosis | (8) |
| Sugawara et al, 2006 | Stichopus variegatus | Sphingosine | Apoptosis | (9) |
| Yun et al, 2012 | Thelenota anax | Stichoposide C | Apoptosis, growth inhibition | (10) |
Details on mechanisms of action for compounds from sea cucumbers as anticancer treatments remain limited. An in-depth docking analysis was conducted to evaluate the potential of compounds from sea cucumbers as anticancer treatments through inhibition of MDM2 and CXCR4. The findings indicated that four out of the five chosen compounds from sea cucumbers are predicted to inhibit CXCR4 and two of these further inhibit MDM2. However, based on the in silico analysis performed in the current study, sphingosine may not be a suitable inhibitor for CXCR4 or MDM2.
The data obtained from pathway analysis suggested that the studied compounds may inhibit cancer cell growth through the chemokine and JAK-STAT signaling pathway or p53 pathway (4). These three pathways serve a central role in the process of controlling cell cycle, migration and apoptosis (13,31,32). Therefore, the data indicated that the anticancer mechanism of the active compound of sea cucumber occurs through inference with the JAK-STAT and Chemokine signaling pathways.
CXCR4 is a receptor located on the cell surface, functioning as a communicator between cells and their environment (14). The receptor binds chemokines and other growth factors, which then transmit signals into cells through multiple pathways, including JAK2/3 and PTK2, which regulate cell division (35). By interrupting the signal transmission through CXCR4, inhibition of cancer cell growth may occur. CXCR4 is known to bind CXCL12, which influences calcium uptake and enhances MAPK1/MAPK3 (15,16) and may be a suitable target in cancer treatment (17,18).
In conclusion, the current study suggested that several compounds from sea cucumbers may have potential as MDM2 or CXCR4 inhibitors. Philinopside E and ds-echinoside A were predicted as MDM2 and CXCR4 inhibitors, while stichoposide C and colochiroside A were predicted as CXCR4 inhibitors. The compounds may be able to inhibit MDM2 and CXCR4 and induce apoptosis in cancer cells. Further research should be conducted in vitro to validate the activity of the studied compounds.
Acknowledgements
The authors would like to thank the Ministry of Research, Technology and Higher Education for supporting the present study.
Funding
The present study was funded by the Ministry of Research, Technology and Higher Education, Republic of Indonesia (grant no. 1598/K4/KM/2017).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SP and NW designed the study. TWL and NW conducted the research and prepared the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Khazaei S, Salehiniya H, Mohammadian-Hafshejani A. Some facts about cancer in the world using registered cancer in 2012. Iran J Public Health. 2015;44:1559–1560. [PMC free article] [PubMed] [Google Scholar]
- 2.Chary KV, Pandian K. Accelerated approval of drugs: Ethics versus efficacy. Indian J Med Ethics. 2017;2:244–247. doi: 10.20529/IJME.2017.062. [DOI] [PubMed] [Google Scholar]
- 3.Pilié PG, Johnson AM, Hanson KL, Dayno ME, Kapron AL, Stoffel EM, Cooney KA. Germline genetic variants in men with prostate cancer and one or more additional cancers. Cancer. 2017;123:3925–3932. doi: 10.1002/cncr.30817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wargasetia TL, Permana S, Widodo The role of sea cucumber active compound and its derivative as an anti-cancer agent. Curr Pharmacol Rep. 2018;4:27–32. doi: 10.1007/s40495-018-0121-x. [DOI] [Google Scholar]
- 5.Janakiram NB, Mohammed A, Rao CV. Sea cucumbers metabolites as potent anti-cancer agents. Mar Drugs. 2015;13:2909–2923. doi: 10.3390/md13052909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Yi Y. Studies on antitumor activities of triterpene glycoside colochiroside A from sea cucumber Colochirus anceps. Zhongguo Zhong Yao Za Zhi. 2011;36:504–507. (In Chinese) [PubMed] [Google Scholar]
- 7.Zhao Q, Liu Z, Xue Y, Wang JF, Li H, Tang QJ, Wang YM, Dong P, Xue CH. Ds-echinoside A, a new triterpene glycoside derived from sea cucumber, exhibits antimetastatic activity via the inhibition of NF-κB-dependent MMP-9 and VEGF expressions. J Zhejiang Univ Sci B. 2011;12:534–544. doi: 10.1631/jzus.B1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian F, Zhu CH, Zhang XW, Xie X, Xin XL, Yi YH, Lin LP, Geng MY, Ding J. Philinopside E, a new sulfated saponin from sea cucumber, blocks the interaction between kinase insert domain-containing receptor (KDR) and alphavbeta3 integrin via binding to the extracellular domain of KDR. Mol Pharmacol. 2007;72:545–552. doi: 10.1124/mol.107.036350. [DOI] [PubMed] [Google Scholar]
- 9.Sugawara T, Zaima N, Yamamoto A, Sakai S, Noguchi R, Hirata T. Isolation of sphingoid bases of sea cucumber cerebrosides and their cytotoxicity against human colon cancer cells. Biosci Biotechnol Biochem. 2006;70:2906–2912. doi: 10.1271/bbb.60318. [DOI] [PubMed] [Google Scholar]
- 10.Yun SH, Park ES, Shin SW, Na YW, Han JY, Jeong JS, Shastina VV, Stonik VA, Park JI, Kwak JY. Stichoposide C induces apoptosis through the generation of ceramide in leukemia and colorectal cancer cells and shows in vivo antitumor activity. Clin Cancer Res. 2012;18:5934–5948. doi: 10.1158/1078-0432.CCR-12-0655. [DOI] [PubMed] [Google Scholar]
- 11.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bally C, Adès L, Renneville A, Sebert M, Eclache V, Preudhomme C, Mozziconacci MJ, de The H, Lehmann-Che J, Fenaux P. Prognostic value of TP53 gene mutations in myelodysplastic syndromes and acute myeloid leukemia treated with azacitidine. Leuk Res. 2014;38:751–755. doi: 10.1016/j.leukres.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Des Devel Ther. 2015;9:4953–4964. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremer KN, Peterson KL, Schneider PA, Meng XW, Dai H, Hess AD, Smith BD, Rodriguez-Ramirez C, Karp JE, Kaufmann SH, Hedin KE. CXCR4 chemokine receptor signaling induces apoptosis in acute myeloid leukemia cells via regulation of the Bcl-2 family members Bcl-XL, Noxa, and Bak. J Biol Chem. 2013;288:22899–22914. doi: 10.1074/jbc.M113.449926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng SB, Zhang X, Paul D, Kays LM, Ye M, Vaillancourt P, Dowless M, Stancato LF, Stewart J, Uhlik MT, et al. Inhibition of CXCR4 by LY2624587, a fully humanized anti-CXCR4 antibody induces apoptosis of hematologic malignancies. PLoS One. 2016;11:e150585. doi: 10.1371/journal.pone.0150585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng T, Chou J, Zhang F, Liu Y, Ni H, Li X, Zheng L, Tang T, Jin L, Xi T. CXCR4 3′UTR functions as a ceRNA in promoting metastasis, proliferation and survival of MCF-7 cells by regulating miR-146a activity. Eur J Cell Biol. 2015;94:458–469. doi: 10.1016/j.ejcb.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Narumi T, Tanaka T, Hashimoto C, Nomura W, Aikawa H, Sohma A, Itotani K, Kawamata M, Murakami T, Yamamoto N, Tamamura H. Pharmacophore-based small molecule CXCR4 ligands. Bioorg Med Chem Lett. 2012;22:4169–4172. doi: 10.1016/j.bmcl.2012.04.032. [DOI] [PubMed] [Google Scholar]
- 18.Fontanella R, Pelagalli A, Nardelli A, D'Alterio C, Ieranò C, Cerchia L, Lucarelli E, Scala S, Zannetti A. A novel antagonist of CXCR4 prevents bone marrow-derived mesenchymal stem cell-mediated osteosarcoma and hepatocellular carcinoma cell migration and invasion. Cancer Lett. 2016;370:100–107. doi: 10.1016/j.canlet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Jiang K, Li J, Yin J, Ma Q, Yan B, Zhang X, Wang L, Wang L, Liu T, Zhang Y, et al. Targeted delivery of CXCR4-siRNA by scFv for HER2(+) breast cancer therapy. Biomaterial. 2015;59:77–87. doi: 10.1016/j.biomaterials.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 20.Dassault Systèmes BIOVIA, Discovery studio modeling environment, Version 4.5. San Diego: Dassault Systèmes; 2015. [Google Scholar]
- 21.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 23.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widodo, Wisnasari S, Saifur Rohman M, et al. Alu insertion/deletion of ACE gene polymorphism might not affect significantly the serum bradykinin level in hypertensive patients taking ACE inhibitors. Egypt J Med Hum Genet. 2017;18:187–191. doi: 10.1016/j.ejmhg.2016.08.001. [DOI] [Google Scholar]
- 25.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O'Donnell L, Oster S, Theesfeld C, Sellam A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. J Chem Inf Model. 2007;47:1258–1262. doi: 10.1021/ci700044s. [DOI] [PubMed] [Google Scholar]
- 30.Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinforma Chem. 2014;7:23–36. doi: 10.2147/AABC.S63749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groner B, von Manstein V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14. doi: 10.1016/j.mce.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Zeng SX, Lu H. Targeting p53-MDM2-MDMX loop for cancer therapy. Subcell Biochem. 2014;85:281–319. doi: 10.1007/978-94-017-9211-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordbar S, Anwar F, Saari N. High-Value components and bioactives from sea cucumbers for functional Foods-A review. Mar Drugs. 2011;9:1761–1805. doi: 10.3390/md9101761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi WT, Yang Y, Xu Y, An J. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Curr Top Med Chem. 2014;14:1574–1589. doi: 10.2174/1568026614666140827143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curnock AP, Logan MK, Ward SG. Chemokine signalling: Pivoting around multiple phosphoinositide 3-kinases. Immunology. 2002;105:125–136. doi: 10.1046/j.1365-2567.2002.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.