Abstract
Ten-eleven translocation-2 (TET2) mutation is frequently observed in myeloid malignancies, and loss-of-function of TET2 is essential for the initiation of malignant hematopoiesis. TET2 mutation presents across disease entities and was reported in lymphoid malignancies. We investigated TET2 mutations in 27 diffuse large B-cell lymphoma (DLBCL) patients and found a frameshift mutation in 1 case (3.7%). TET2 mutation occurred in some populations of DLBCL patients and was likely involved in the pathogenesis of their malignancies.
Keywords: TET2, epigenetic modifier, DLBCL
INTRODUCTION
Diffuse large B cell lymphoma (DLBCL) is an aggressive B cell neoplasm. Gene expression profiling divided DLBCL into the following two subgroups, whose cell origins were thought to differ1: germinal center B-cell (GCB) DLBCL and activated B-cell (ABC) DLBCL. Mutations in epigenetic modifiers, such as MLL2, EZH2, CREBBP, and EP300, were frequently observed in GCB DLBCL,2 whereas gene mutations that activated nuclear factor κB (NF-κB) signaling, such as mutations in A20, CARD11, CD79B, and MYD88, were frequently observed in ABC DLBCL.3,4
Epigenetic modifiers include histone-modifying enzymes and regulators of DNA methylation. Ten-Eleven Trans-location-2 (TET2) is a regulator of DNA methylation, and plays a key role in the conversion of 5-methyl-cytosine (5-mC) to 5-hydroxymethyl cytosine (5-hmC).5 TET2 mutations, including deletions, missense, nonsense, and frameshift mutations, were shown to result in loss-of-function of TET2 and a marked reduction in global levels of 5-hmC.6,7 Somatic mutations in TET2 were first identified in myeloproliferative neoplasms (MPN) and myelodysplastic syndromes.8,9 In addition to myeloid malignancies, TET2 mutations were detected in T and B lymphomas. Of these, TET2 was most frequently mutated in angioimmunoblastic T-cell lymphomas (up to 76%) and “Th follicular (TFH)−like” peripheral T-cell lymphomas (PTCL), not otherwise specified10-12 (19–51%). TET2 mutation was also observed in approximately 10% of adult T cell leukemia/lymphoma cases.13,14 As for B-cell malignancies, 0–12% of DLBCL patients were reported to carry TET2 mutation.15-18 In this report, we examined the TET2 mutation in 27 DLBCL patients.
MATERIALS AND METHODS PATIENTS AND TUMOR SAMPLES
A series of 27 DLBCL patients with available frozen tumor cell samples was selected. The specimens were collected between 2006 and 2011. Medical records were reviewed for clinical data. This study was approved by the Research Ethics Committee of University of Miyazaki, and conducted in accordance with the Helsinki Declaration of 1975 as revised in 2008.
TET2 GENOTYPING
DNA was extracted from frozen cells using a standard protocol. The coding sequence of the TET2 gene (exons 3 through 10) was amplified by the polymerase chain reaction (PCR) method with a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The sequences of PCR primers for TET2 were designed as described in a previous report,19 and these primers were purchased from Hokkaido System Science Co., Ltd. The nucleotide sequences were determined by fluorescent dye chemistry sequencing with an ABI PRISM3000 DNA Analyzer (Applied Biosystems), and analyzed with Sequencing Analysis software (Applied Biosystems). The presence of mutations or single nucleotide polymorphisms (SNPs) was determined by referencing the assembled sequence in the Ensembl genome database.20
RESULTS
Subject characteristics are listed in Table 1. Of the 27 patients, 16 were men and 11 were women. The median age was 72 years (range, 34−81). According to the International Prognostic Index (IPI), 2 patients were classified as low risk, 4 as intermediate-1 risk, 5 as intermediate-II risk, and 15 as high risk. The surface markers of DLBCL cells were analyzed by immunohistochemistry. Based on the immunostaining of CD10, BCL6, and MUM1, 7 patients were classified as GCB DLBCL, and 9 as non-GCB DLBCL.
Table 1. Profiles and clinical data for each DLBCL case.
| case | age | sex | PS | LDH | stage (Ann Arbor) |
IPI | samples | Phenotype of lymphoma cells in the sample | GCB / non-GCB |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| no. | y | ECOG | IU/L | CD20 | CD10 | BCL-6 | Mum-1 | ||||||||
| 1 | 74 | M | 0 | 335 | IV | high | LNs | + | n.d. | n.d. | n.d. | n.a. | |||
| 2 | 62 | F | 1 | 882 | IV | high | Tumor | + | n.d. | n.d. | n.d. | n.a. | |||
| 3 | 34 | M | 1 | 600 | II | int1 | LNs | + | n.d. | n.d. | n.d. | n.a. | |||
| 4 | 58 | M | n.a. | n.a. | n.a. | n.a. | Bone | + | n.d. | n.d. | n.d. | n.a. | |||
| 5 | 73 | M | 3 | 1437 | IV | high | LNs | + | n.d. | - | + | n.a. | |||
| 6 | 74 | M | 2 | 397 | IV | high | Tumor | + | - | n.d. | focally + | non-GCB | |||
| 7 | 64 | F | 0 | 268 | I | int2 | LNs | + | + | + | focally + | GCB | |||
| 8 | 34 | F | 0 | 113 | II | low | LNs | + | + | - | + | non-GCB | |||
| 9 | 42 | M | 1 | 138 | IV | int1 | LNs | + | + | + | + | non-GCB | |||
| 10 | 77 | M | 2 | 254 | IV | high | Tumor | + | ± | + | + | non-GCB | |||
| 11 | 80 | M | 1 | 242 | IV | high | Tumor | + | - | - | + | non-GCB | |||
| 12 | 77 | M | 3 | 1256 | IV | high | LNs | + | - | - | + | non-GCB | |||
| 13 | 74 | M | 1 | 147 | IV | high | LNs | + | - | - | + | n.a. | |||
| 14 | 75 | M | 0 | 279 | IV | high | LNs | + | - | + | + | GCB | |||
| 15 | 67 | F | 0 | 242 | III | high | LNs | + | - | n.d. | - | n.a. | |||
| 16 | 68 | F | 2 | 326 | IV | high | LNs | + | - | - | + | non-GCB | |||
| 17 | 70 | M | 4 | 517 | IV | high | Adrenal gland | + | - | - | + | non-GCB | |||
| 18 | 79 | F | 4 | 234 | IV | high | Tumor | + | + | focally + | focally + | GCB | |||
| 19 | 60 | F | 4 | 249 | III | high | LNs | + | ± | + | + | n.a. | |||
| 20 | 34 | F | 1 | 743 | IV | int2 | Bone | + | - | + | ± | GCB | |||
| 21 | 81 | M | 1 | 210 | II | int1 | Tumor | + | - | + | + | n.a. | |||
| 22 | 72 | M | 2 | 532 | IV | high | Tumor | + | - | + | n.d | n.a. | |||
| 23 | 79 | F | 1 | 192 | IV | int2 | Tumor | + | - | - | focally + | GCB | |||
| 24 | 53 | F | 1 | 389 | III | int2 | Bone | + | ± | ± | + | non-GCB | |||
| 25 | 73 | M | 1 | 179 | III | int2 | LNs | + | + | + | + | GCB | |||
| 26 | 52 | M | 0 | 219 | II | low | LNs | + | - | + | - | GCB | |||
| 27 | 80 | F | 0 | 176 | I | int1 | Tumor | + | - | - | focally + | n.a. | |||
The results of immunohistochemical staining of tumor samples are shown as +, ±, or -, corresponding to positive, weak positive, or negative, respectively. n.a.: data not available, n.d.: not done
We examined the entire coding sequence of the TET2 gene (exons 3−10) in 27 DLBCL patients, and found a frameshift mutation in 1 patient (case 16) (Figure 1). In addition, 5 types of SNPs, as determined from referencing the base sequence in the Ensembl genome database (http://www.ensembl.org/Homo_sapiens/Transcript/Sequence_cDNA?db=core;g=ENSG00000162434;r=1:65071494-65204775;t=ENST00000342505), were found in 15 cases, including the 1 patient with a TET2 mutation (case 16).
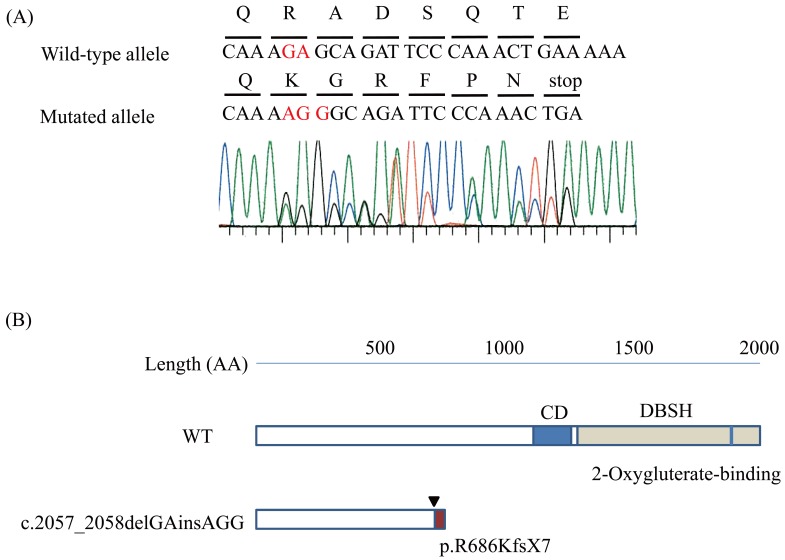
Figure 1.
TET2 mutation in DLBCL and TET2 protein
A. Sanger sequence of TET2 in case 16. Electropherogram of TET2 exon3 sequences showing the monoallelic mutation c.2057_2058delGAinsAGG (note in red). The DNA and corresponding amino acid sequences of the wild-type and mutant TET2 alleles are also shown.
B. A schematic representation of the TET2 protein. The arrowhead shows the position corresponding to the mutation. This mutation led to premature termination p.R686KfsX7. Truncated-form mutant TET2 lacks the cysteine-rich domain (CD) and double stranded b-helix (DSBH) 2OG-Fe(II)-dependent dioxygenase domain.
The frameshift mutation observed in case 16 was c.2057_2058delGAinsAGG, which led to premature termination p.R686KfsX7. This short form mutant TET2 lacks the cysteine-rich domain and double stranded b-helix (DSBH) 2OG-Fe(II)-dependent dioxygenase domain.
DISCUSSION
We found one TET2 frameshift mutation in 27 DLBCL cases. This frameshift mutation led to premature termination p.R686KfsX7, and formed truncated-type TET2 that lacks the cysteine-rich domain and DSBH 2OG-Fe(II)-dependent dioxygenase domain. Reported somatic mutations in TET2 in myeloid and lymphoid malignancies included missense, nonsense, and frameshift mutations, and thus, the TET2 mutation was thought to be loss-of-function mutation.21 As TET2 catalyzes the conversion of 5-mC to 5-hmC, loss-of-function mutations would affect the global methylation status of genes.5 Indeed, there were decreased 5-hmC levels in the DNA of myeloid malignancy patients with TET2 mutation compared with those without TET2 mutation or healthy controls.7 We and others reported that TET2-deficient hematopoietic stem cells (HSCs) exhibited increased self-renewal ability and had a competitive growth advantage over wild-type HSCs.15,22-24 This augmented self-renewal activity in HSCs may be the basis for TET2-mutated myeloid malignancies. TET2 mutation was also observed in a proportion of normal elderly individuals who exhibited clonal hematopoiesis, and one of seven such individuals subsequently developed MPN.25 The situation should be the same with DLBCL. For the development of DLBCL, several gene mutations were required.26 TET2 mutation was one of them, and DLBCL may develop with additional gene mutations.
TET2 mutation was reported in 0–12% of DLBCL patients,15-18 and in our study one of 27 DLBCL patients (3.7%) carried a TET2 mutation. As TET2 mutation was frequently observed in AITL, we examined whether case 16 harbored composite lymphoma with DLBCL and AITL.27 We carefully re-evaluated the biopsy sample from case 16, but could not find any morphological or immunohistochemical aspects of AITL. The lower incidence of TET2 mutation in our DLBCL cohort compared with previous reports may be due to racial differences or differences in methodology to detect the mutation. We analyzed TET2 mutation in Japanese patients, whereas previous reports analyzed Caucasian patients, and we detected the mutation by Sanger sequencing after PCR, whereas denaturing gradient gel electrophoresis was adopted to detect in the previous report.18
Mutations in epigenetic modifiers were dominantly found in GCB DLBCL, but our case (case 16) was classified as non-GCB DLBCL. As the cell-of-origin subtypes were not identified in reported DLBCL with TET2 mutations15-18 and we found only one non-GCB DLBCL patient with a TET2 mutation, we cannot conclude whether TET2 mutations accumulated in GCB DLBCL, as is the case with EZH2 mutation. Asmar et al. studied both a DNA methylation signature and the TET2 mutation in DLBCL, and found that TET2 mutation is associated with hyper-methylation within CpG islands, and at CpG-rich promoters of genes involved in hematopoietic differentiation and cellular development.18 They also reported that 11% of the hyper-methylated genes, which include several tumor suppressor genes, were down-regulated. These epigenetic changes may be due to TET2 mutation and its functional impairment, and may be involved in the ontogeny of DLBCL.
In conclusion, TET2 mutation was observed in one of 27 DLBCL patients (3.7%), and was likely to be involved in DLBCL ontogeny.
ACKNOWLEDGMENT
This study was supported by a Grant-in-Aid for Clinical Research from University of Miyazaki Hospital.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest
REFERENCES
- 1.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. : Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503-511, 2000. 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 2.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, et al. : Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189-195, 2011. 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, et al. : Oncogenically active MYD88 mutations in human lymphoma. Nature 470: 115-119, 2011. 10.1038/nature09671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young RM, Staudt LM: Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov 12: 229-243, 2013. 10.1038/nrd3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, et al. : Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 108: 14566-14571, 2011. 10.1073/pnas.1112317108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, et al. : Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200-213, 2011. 10.1016/j.stem.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, et al. : Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature, 2010. 10.1038/nature09586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, et al. : Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289-2301, 2009. 10.1056/NEJMoa0810069 [DOI] [PubMed] [Google Scholar]
- 9.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, et al. : Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838-842, 2009. 10.1038/ng.391 [DOI] [PubMed] [Google Scholar]
- 10.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, et al. : Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 46: 171-175, 2014. 10.1038/ng.2872 [DOI] [PubMed] [Google Scholar]
- 11.Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, et al. : A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 123: 1293-1296, 2014. 10.1182/blood-2013-10-531509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, et al. : Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120: 1466-1469, 2012. 10.1182/blood-2012-02-408542 [DOI] [PubMed] [Google Scholar]
- 13.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, et al. : Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 47: 1304-1315, 2015. 10.1038/ng.3415 [DOI] [PubMed] [Google Scholar]
- 14.Shimoda K, Shide K, Kameda T, Hidaka T, Kubuki Y, et al. : TET2 Mutation in Adult T-Cell Leukemia/Lymphoma. J Clin Exp Hematop 55: 145-149, 2015. 10.3960/jslrt.55.145 [DOI] [PubMed] [Google Scholar]
- 15.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, et al. : TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25-38, 2011. 10.1016/j.ccr.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 16.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, et al. : Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298-303, 2011. 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, et al. : Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA 109: 3879-3884, 2012. 10.1073/pnas.1121343109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, et al. : Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica 98: 1912-1920, 2013. 10.3324/haematol.2013.088740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiel A, Beier M, Ingenhag D, Servan K, Hein M, et al. : Comprehensive array CGH of normal karyotype myelodysplastic syndromes reveals hidden recurrent and individual genomic copy number alterations with prognostic relevance. Leukemia 25: 387-399, 2011. 10.1038/leu.2010.293 [DOI] [PubMed] [Google Scholar]
- 20.Kameda T, Shide K, Shimoda HK, Hidaka T, Kubuki Y, et al. : Absence of gain-of-function JAK1 and JAK3 mutations in adult T cell leukemia/lymphoma. Int J Hematol 92: 320-325, 2010. 10.1007/s12185-010-0653-2 [DOI] [PubMed] [Google Scholar]
- 21.Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W: The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia 28: 485-496, 2014. 10.1038/leu.2013.337 [DOI] [PubMed] [Google Scholar]
- 22.Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, et al. : TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia: official journal of the Leukemia Society of America. Leukemia Research Fund, UK 26: 2216-2223, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, et al. : Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11-24, 2011. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Cai X, Cai CL, Wang J, Zhang W, et al. : Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509-4518, 2011. 10.1182/blood-2010-12-325241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, et al. : Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44: 1179-1181, 2012. 10.1038/ng.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW: Cancer genome landscapes. Science 339: 1546-1558, 2013. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuppers R, Duhrsen U, Hansmann ML: Pathogenesis, diagnosis, and treatment of composite lymphomas. Lancet Oncol 15: e435-e446, 2014. 10.1016/S1470-2045(14)70153-6 [DOI] [PubMed] [Google Scholar]