Abstract
Objectives: To measure the strength of agreement between the concepts and records retrieved from a computerized patient database, in response to physician-derived questions, using a semantic terminological model for clinical findings with those concepts and records excerpted clinically by manual identification. The performance of the semantic terminological model is also compared with the more established retrieval methods of free-text search, ICD-10, and hierarchic retrieval.
Design: A clinical database (Diabeta) of 106,000 patient problem record entries containing 2,625 unique concepts in an clinical academic department was used to compare semantic, free-text, ICD-10, and hierarchic data retrieval against a gold standard in response to a battery of 47 clinical questions.
Measurements: The performance of concept and record retrieval expressed as mean detection rate, positive predictive value, Yates corrected and Mantel-Haenszel chi-squared values, and Cohen kappa value, with significance estimated using the Mann-Whitney test.
Results: The semantic terminological model used to retrieve clinically useful concepts from a patient database performed well and better than other methods, with a mean detection rate of 0.86, a positive predictive value of 0.96, a Yates corrected chi-squared value of 1,537, a Mantel-Haenszel chi-squared value of 19,302, and a Cohen kappa of 0.88. Results for record retrieval were even better, with a mean record detection rate of 0.94, a positive predictive value of 0.99, a Yates corrected chi-squared value of 94,774, a Mantel-Haenszel chi-squared value of 1,550,356, and a Cohen kappa value of 0.94. The mean detection rate, Yates corrected chi-squared value, and Cohen kappa value for semantic retrieval were significantly better than for the other methods.
Conclusion: The use of a semantic terminological model in this test scenario provides an effective framework for representing clinical finding concepts and their relationships. Although currently incomplete, the model supports improved information retrieval from a patient database in response to clinically relevant questions, when compared with alternative methods of analysis.
Clinical data in computer systems have to be accurate if they are to support patient care, research, and health service management, but despite this there is little published literature devoted to measuring this in electronic health care records.1 Inaccurate data can lead to an underestimate of disease prevalence, with potentially serious consequences for monitoring the success of health care interventions and detrimental effects on decision support protocols and alert systems.2
The retrieval of meaningful information is dependent on data being entered in an organized way, and to maximize the benefit of electronic health care records, an underlying structure is required.3 If data are collected in free text, the text has to be converted to computer-understandable codes and structures to extract meaningful data.4 It is postulated that the content of the electronic health care record should be provided by a clinical terminology in which concepts and their relationships are formally expressed.5 Clinical Terms Version 3 (The Read Codes) (CTV3)6 is a clinical terminology in which concepts are represented according to their meaning (semantically) with reference to standard structured hierarchies of component values, e.g., anatomy and micro-organisms (semantic terminological model). The use of this formal semantic model for describing the intrinsic characteristics (atoms) of concepts in CTV3 has parallels in other schemes such as LOINC7 and the cross-references available in SNOMED International.8 The model employed by GALEN uses GRAIL to express sanctioned associations between primitive concepts.9 Similarly, SNOMED RT uses the KRSS description logic to formally express relationships.10 Thus, there is an emerging convergence of approaches toward the use of a concept-based clinical terminology with an underlying formal semantic terminological model (STM).
There has been little reported work on the effect of different search methods on the efficacy of data retrieval from clinical records. However, significant efforts have been invested in initiatives in the U.K. with the development of CTV3,11 in Europe with the GALEN-in-Use project,12 and in the United States with the development of SNOMED RT.10 These initiatives have confirmed that considerable resources are required to create and maintain such products, and they are all based on the assumption that a clinical terminology will bring improved data quality. There is, therefore, an urgent need to investigate whether the use of semantic-based terminologies will deliver any practical advantages over simpler existing systems.
This study explores the hypothesis that the use of an STM improves the quality of information retrieval in response to a battery of physician-derived questions from a clinical database. The Diabeta patient database,13 populated with the CTV3 terminology, has been used to compare retrieval using an STM (semantic differential retrieval) with more established approaches. The experiment measures the strength of agreement between the concepts and records retrieved from the database using the STM with those concepts and records expected clinically by manual identification. The performance of semantic differential retrieval is also compared with more traditional methods, including free-text searching, class retrieval using the CTV3 hierarchy table,14 and the framework of the International Classification of Diseases and Related Health Problems, 10th revision (ICD-10).15
Background
Clinical Terms Version 3
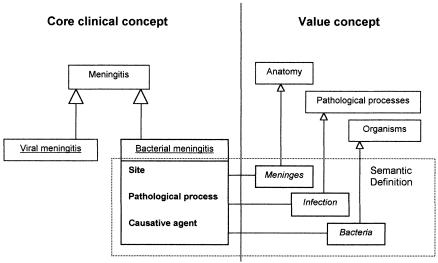
Clinical Terms Version 3 was developed during the Clinical Terms Project to provide a common terminology for electronic health care records.6,11,16,17 The structure of CTV3 provides a formal framework for representing the meaning and relationship of clinical findings and procedures, allocating each unique clinical concept a Read code.6,14 Each concept code is labeled with a unique unambiguous preferred term and, where appropriate, synonyms. Concepts are formally arranged in a hierarchy in which those of more narrow meaning appear as “types of” concepts of more general meaning (subtype hierarchy), e.g., bacterial meningitis is a subtype of meningitis (▶). Concepts are also mapped, where appropriate, to ICD-10 and the U.K. surgical procedure classification OPCS-4.18 These cross-mappings have been subject to independent quality assurance and practical evaluation in use since 1994 and are, therefore, considered to be of high quality.
Figure 1.
Concepts in Clinical Terms Version 3 are placed in a pure subtype hierarchy. The structure also allows the formal definition of concepts according their meaning (semantic definition); thus, Bacterial meningitis is represented by [Site]: Meninges; [Pathological process]: Infection; [Causative agent]: Bacteria. Note: The triangle represents a subtype relationship utilizing the notation of the Unified Modelling Language (UML), the Object Management Group (OMG) industry standard (Rational Software Corporation, Cupertino, California, 1995).
Semantic Terminology Model
The design of CTV3 employs object-attribute-value triples stored in a template file as a mechanism for defining each core clinical concept in relation to more primitive value concepts, e.g., anatomy, pathological processes, and micro-organisms.6,19 This feature allows the semantic definition of concepts according to their meaning (▶). The model describing the formal relationship between the core terminology and their constituent values (atoms) is referred to in this paper as the STM (semantic terminology model) to distinguish it from alternative models describing the structure of the record and models of health care. A detailed but provisional STM has been developed within CTV3, describing disorders.20 This has been extended in this study to provide at least partial representation of other findings, including symptoms and signs (▶).
Figure 2.
Abridged representation of the provisional semantic terminological model identifying the main characteristics of clinical findings expressed as attributes and applicable value concept hierarchies, with examples. The section mark (§) indicates that the expression of laterality is applied via anatomy.
The provisional STM defined 44.7 percent of the Diabeta concepts completely, with the 55.3 percent remaining concepts containing at least one additional nonrepresented characteristic.21 Incomplete definitions included concepts such as Osteogenesis imperfecta, Scleroderma variant, Osteopetrosis, and True hermaphrodite, whose semantic definitions are not amenable to full representation using the present STM. Characterization of these might be achievable from consideration of more detailed aspects of their embryologic, cellular, and molecular origins, but this would require considerable specialist clinical input, which might not be consistently applicable to other areas.20 The definition of other classes of concepts, such as Cicatricial junctional epidermolysis bullosa is highly dependent on the extensiveness of purely descriptive elements that do not currently exist in the supportive hierarchies.
Despite this level of incompleteness, 818 of these partially defined concepts had unique definitions that did not coincide with others. Thus, within the clinical database examined (Diabeta), the STM provides either a complete (1,175) or unique (818) semantic definition for 76 percent (1,993) of findings, with a residual 24 percent (634) of concepts having incomplete definitions that are shared with other concepts.22
Diabeta Database
Diabeta is a computerized clinical record system that has been developed and used with ongoing modification since 1973 at St. Thomas' Hospital, London (part of The Guys', King's College, and St. Thomas' Hospitals Medical and Dental School).13 It is used for supporting the management of patients attending the medical outpatient department, many of whom have diabetes mellitus.23 The original system (Diabeta 1) allowed, at every encounter between the patient and clinician, the recording of clinical findings as “problems” using a locally created list of reusable free-text term strings. The system thus allowed the collection of a large corpus of clinically relevant terms without any potential restraint of a fixed terminology. This database has been converted to a new database of patient problem records (Diabeta 3), in which every unambiguous clinical finding term string has been matched to an applicable term in CTV3. During the translation process between the existing Diabeta database and CTV3, any clinically important concept or term that was found to be absent was incorporated into the next release of CTV3 and was thus available for the analysis experiment. Consequently, the created Diabeta test database represents a valuable corpus of clinically derived concepts.
The experimental Diabeta database contained 12,696 different term strings (accounting for 106,000 “problem” record entries) mapped to 3,049 unique terms associated with 2,625 unique Read-coded concepts. Mappings to ICD-10 were available for 2,301 concepts (87.7 percent). Those concepts without an ICD-10 map fell into two groups: observations that are not specifically included in ICD-10, e.g., alcohol consumption and persistent microalbuminuria, and classes of concepts based on anatomic regions not accommodated by the axes in the classification, e.g., limb complication and limb infection.
Study Objectives
A key function of a terminology is the support it provides for the retrieval of information from a clinical system.* In practice, the interrogation of a clinical database, to answer a specified question, usually involves two main steps:
Creating a list of concepts for retrieval (in response to a posed question)
Retrieving records containing these identified concepts
The kernel of the problem is to measure how well the concepts (and records) retrieved in response to a clinical question match the expectation of the clinician who posed it. The objective of the study was to measure the strength of agreement between the concepts and records retrieved using an STM from a computerized patient database with those concepts and records expected clinically by manual identification in response to physician-derived questions. It tests the hypothesis that the use of an STM for clinical findings improves the performance of data retrieval in comparison with the more established retrieval methods of free-text search, ICD-10, and hierarchic retrieval.
Methods
Clinical Question Battery
A survey was performed to gather a battery of clinical questions relating to clinical findings that a clinician might want to ask the Diabeta clinical information system. Fifteen copies of a questionnaire were distributed to all grades of medical staff in two clinical academic departments. Eight completed forms were returned, which collectively suggested 47 unique questions relating to clinical findings (▶). These questions were then formulated into database queries using the four alternative methods of retrieval.
Table 1.
Battery of Questions Collected from a Survey of Physicians, and Their Frequency of Occurrence
| Question | Frequency |
|---|---|
| Absent foot pulses | 1 |
| Alcohol consumption | 1 |
| Amputations | 3 |
| Cataract | 1 |
| Chronic pancreatitis | 1 |
| Diabetes treated with diet alone | 1 |
| Diabetic autonomic neuropathy | 1 |
| Diabetic hand syndrome (diabetic cheiroarthropathy) | 1 |
| Diabetic ketoacidosis | 1 |
| End-stage renal failure | 1 |
| Erectile dysfunction | 1 |
| Frozen shoulder | 1 |
| Gastric paresis/autonomic bowel dyfunction | 1 |
| Hyperlipidemia | 1 |
| Hypertension | 5 |
| Impotence | 1 |
| Infection | 1 |
| Insulin-dependent diabetes mellitus | 3 |
| Insulin-treated diabetes mellitus | 2 |
| Ischemic heart disease | 2 |
| Limb amputation | 1 |
| Limb complications | 1 |
| Limb infection | 1 |
| Macrovascular complication [IHD, PVD] | 1 |
| Major limb amputation | 1 |
| Microvascular complication [retinopathy + nephropathy] | 1 |
| Myocardial infarction | 4 |
| Necrobiosis lipoidica | 2 |
| Nephropathy [diabetic nephropathy] | 3 |
| Neuropathic foot ulcer | 1 |
| Neuropathy | 4 |
| Obesity | 1 |
| Painful neuropathy | 1 |
| Peripheral vascular disease | 2 |
| Persistent microalbuminuria | 2 |
| Pregnancies | 2 |
| Pregnancy complications | 2 |
| Problematic hypoglycemia | 1 |
| Proteinuria | 1 |
| Retinopathy | 3 |
| Smokers | 1 |
| Stroke | 1 |
| Tumor of pancreas | 1 |
| Ulcer of foot | 2 |
| Ulcers [skin] | 1 |
| Unsuccessful diabetic pregnancies | 1 |
| Vascular foot ulcer | 1 |
Methods of Retrieval
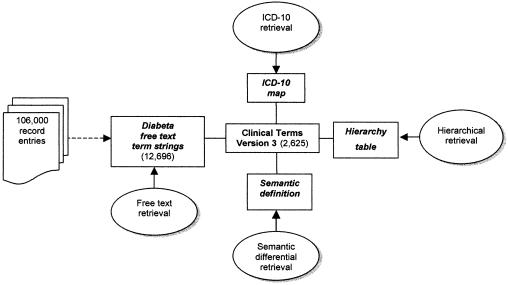
A table of the Diabeta clinical term strings mapped to CTV3 was created in an Microsoft Access database together with their frequency in records in the system and the concept (Read code) to which they had each been mapped (e.g. diabetes mellitus| 7463| C10..). A separate table was created, containing the default ICD-10 maps for each concept (Read code) using the mapping table from the October 1997 Read Codes release. Another table expressed the semantic definition of each concept with reference to the attributes expressed in the STM (▶).
Figure 3.
Relationship between the four methods of retrieval, the original Diabeta clinical finding term string and the Clinical Terms Version 3 concepts (with their mapping to ICD-10, hierarchical position represented in the hierarchy table, and semantic definition) in the experimental database.
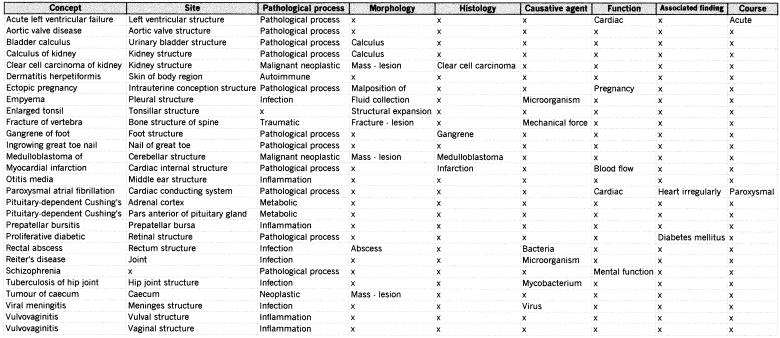
The semantic definition table contained an entry for each concept with a separate column for every attribute. The applicable values (atoms) for each concept were entered in the appropriate column field (▶). The database design dealt with the uncommon circumstance in which a concept had an attribute containing more than one value by replicating the line for each value (e.g., Vulvovaginitis has a separate row for the attribute [Site]: Vulval structure and Vaginal structure).
Figure 4.
An extract of the semantic definition table from the experimental database, illustrating its use of a separate column for each attribute. The atoms for each concept are indicated by an entry of the appropriate value in the applicable attribute field. (Only a subset of attributes is shown.)
These resource tables were then used to retrieve concepts and records in response to each of the 47 clinical questions. The principle adopted was that a user would want to retrieve the chosen clinical concept and any subtypes of that clinical concept, e.g., Gallstone chronic pancreatitis was retrieved when searching for cases of Chronic pancreatitis. The concepts and records of the experimental Diabeta database were identified and flagged for each question and stored in 47 separate tables using each of the following four approaches:
Free-text Retrieval
Free-text searching was performed using standard Access query methods to find phrases containing the required string, which has previously been found to be effective.24 For example, record entries of Frozen shoulder were retrieved by searching for all strings containing <*froz*> (where * is a wildcard representing any characters). Multiple searches were allowed to identify alternative expressions of the same concept; for example, both <*IHD*> and <*ischaemic heart*> were used to identify records of Ischaemic heart disease.
ICD-10 Retrieval
The ICD-10 categories required for retrieval for each question were identified with reference to the ICD-10 (volume 3) index. A single ICD-10 code or list of codes was constructed and its appropriateness to the clinical question validated by an independent clinician. The ICD-10 code (or codes) were then used to retrieve a unique list of Read-coded concepts that were relevant to the question, to identify the concepts with applicable maps, e.g., L92.1 for cases of Necrobiosis lipoidica.
Hierarchic Retrieval
The October Read Version 3 browser25 was used to identify the Read-code node of the hierarchy required for each question. For example, Myocardial infarction (Read code X200E) was identified as the superordinate node marking the hierarchy of concepts required, in response to the question “Find all types of Myocardial infarction.” A unique descent from this identified node was then performed using the “descent” functionality of the Version 3 browser. Occasionally, more than one Read code descent was needed to retrieve the concepts required to answer the clinical question, e.g., Ischaemic heart disease and Peripheral vascular disease.
Semantic Differential Retrieval
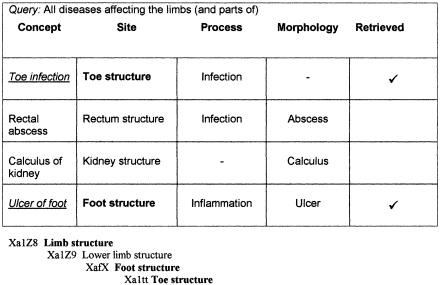
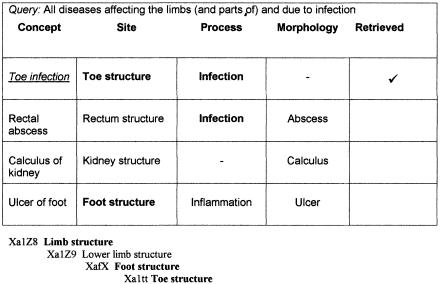
Semantic retrieval was performed by exploiting the “atoms” of concepts in the STM and the formal structuring of the underlying primitive values that have a strict subtype arrangement.22 For example, to find all “disorders affecting the limbs,” a list of concepts was created that had a semantic definition containing [Site]: Limb structure, or a subtype of Limb structure (▶). In the example shown, this would result in the retrieval of two concepts, Toe infection ([Site]: Toe structure) and Ulcer of foot ([Site]: Foot structure). The described STM of clinical findings contains a large number of attributes, and a more complex query might involve the differential retrieval of concepts that have more than one characteristic in common (semantic differential retrieval). For example, to retrieve all “limb infections,” a list of concepts was created that had a semantic definition containing [Site]: Limb structure or a subtype and [Process]: Infection (▶). In the example shown, this would result in the retrieval of a single concept, Toe infection, but not Ulcer of foot (as its semantic definition does not contain the [Process]: Infection).
Figure 5.
The specification of a query to retrieve all disorders affecting the limb includes those disorder concepts having a semantic definition [Site]: Limb structure or part of limb structure. (An extract from the anatomy value limb structure hierarchy is illustrated.)
Figure 6.
The specification of a query to retrieve all disorders caused by infection and affecting the limbs includes those concepts having both a semantic definition of [Pathological process]: Infection and [Site]: Limb structure or part of limb structure. (An extract from the anatomy value limb structure hierarchy is illustrated.)
Gold Standard
To evaluate the retrieval performance of a terminology, comparison against a gold standard is needed. The creation of such gold standards in medical informatics is recognized as problematic.26 Ideally, such a standard should represent the (perfect) “truth” about the population against which the performance of the information resource can be compared. In the evaluation of the performance of a terminology, the gold standard is the complete subset of concepts from the terminology that all clinicians (with perfect knowledge) would expect to retrieve in response to a particular clinical question. Thus, a separate gold standard is required for each discrete clinical question (and relating to the version of the terminology at the time of the query).
An initial flagged list of concepts was created that one would expect to be retrieved from the total 2,625 concepts in the database, in response to each question posed. The quality of these was then independently assured by a second clinician, who had a good knowledge of the contents of the original Diabeta database. A 10 percent sample was then further validated by a third clinician, to create the final gold standard list of concepts expected to be retrieved from the database for each of the 47 questions.
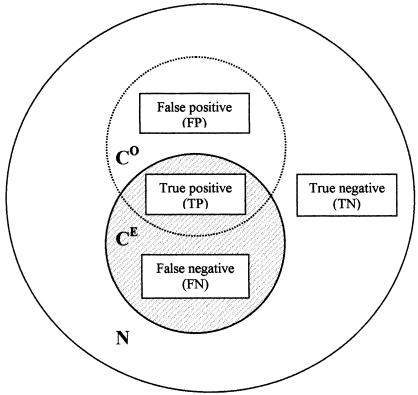
A table was constructed for each question and search method for all concept database tables (141 fields) and record database tables (168 fields). These tables contained those concepts and records that were expected for each question and those that were actually observed for each method (▶).
Figure 7.
Venn diagram showing the relationship between the population of concepts observed by data retrieval (CO), with respect to the actual (gold standard) expected population (CE), and the total population (N); and the true positive, false positive, false negative and true negative populations forming the 2 × 2 contingency tables for each question in the experimental database.
Statistical Analysis
The choice of the statistical method for the evaluation of the retrieval performance of a clinical terminology requires careful consideration.27 The retrieved concepts and records are nominal data, dictating the use of 2 × 2 continency tables as the most appropriate method to compare the observed retrieval with that of the gold standard.17 Individual derived measures and means of performance across all 47 questions were calculated and expressed by the mean detection rate (TP/(TP + FN)) and mean positive predictive value (TP/(TP + FP)). The likelihood of the association was estimated using the Yates continuity-corrected chi-squared test. This test was chosen because it provides a more robust estimate of the exact probability (compared with the chi-squared test) where the expected and observed numbers are relatively small, e.g., when the clinical questions are specific and generate only small frequencies of retrieved concepts (e.g., Frozen shoulder).
The Cohen kappa (κ) has been used as an index of the strength of agreement (between the observed retrieval and the gold standard) against that which might be expected by chance.26,28 A value of +1 indicates perfect agreement, and some authorities consider a kappa value above 0.4 as evidence of useful agreement, but this threshold obviously depends on the clinical application and may need to be set at a level higher than 0.8 for the evaluation of retrieval performance.27
To express the collective results for all retrievals and produce a summary index of the overall performance of a terminology, means of the derived indexes from the 2 × 2 contingency were used. In addition, the Mantel-Haenszel chi-squared test was quoted, which also pools the results of the individual subsets using the following formula:
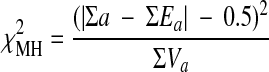 |
where a is the observed value, Ea is the expected value of a, and Va is the variance of a.
The Mann-Whitney test was used as a measure of the significance of association because it provides a more conservative estimate, as the assumption that the data always come from a normal distribution may not always be true. For example, the very fact that a question may (by design) retrieve concepts sharing one or more particular characteristics suggests that it is safer to assume that the data are nonparametric.
Results
A 2 × 2 contingency table was constructed for each of the 47 questions for all four methods relating to both concept and record retrieval (although free-text retrieval related only to records). These individual contingency tables related the actual numbers of concepts and records observed by each method compared with those expected (by the gold standard). The number of concepts expected to be retrieved ranged from 1 to 219 (from a total of 2,625 concepts); the number of records expected ranged from 0† to 4,231 (from a total of 106,000 records).
The mean detection rate of semantic differential retrieval was significantly better for both concept (0.86) and record (0.94) retrieval (▶). The use of the ICD-10 framework was slightly better than hierarchic retrieval, with free-text retrieval having the lowest detection rate (0.61). The detection rate is a useful indicator in assessing the ability of the retrieval method to identify all relevant concepts (avoiding false negatives), in contrast to the positive predictive value, which is a valuable indicator of the retrieval of false positives. This latter index again shows that semantic differential retrieval performs better than ICD-10, hierarchic, or free-text searching, although this does not reach significance in comparison with hierarchic retrieval.
Table 2.
Statistical Values for Free-text, ICD-10, Hierarchy, and Semantic Analysis for Concept and Record Retrieval in Response to the Battery of Clinical Questions
| Free Text | ICD | Hierarchy | Semantic | |
|---|---|---|---|---|
| Concept retrieval (N = 2,625): | ||||
| Mean detection rate (SD) | NA | 0.68 (0.35)** | 0.66 (0.32)* | 0.86 (0.26) |
| Mean positive predictive value (SD) | NA | 0.79 (0.27)* | 0.93 (0.19) | 0.96 (0.08) |
| Mean Yates value (SD) | NA | 955 (695)* | 1034 (752)** | 1537 (783) |
| Mantel-Haenszel chi-squared value | NA | 11,467 | 10,013 | 19,302 |
| Mean Cohen kappa value (SD) | NA | 0.64 (0.30)* | 0.73 (0.30)** | 0.88 (0.21) |
| Record retrieval (N = 106,000) | ||||
| Mean detection rate (SD) | 0.61 (0.34)* | 0.81 (0.32)* | 0.79 (0.32)* | 0.94 (0.20) |
| Mean positive predictive value (SD) | 0.82 (0.32)* | 0.83 (0.29)* | 0.95 (0.20) | 0.99 (0.03) |
| Mean Yates value (SD) | 57,969 (36,932)* | 73,454 (33,154)* | 79,520 (34,870)** | 97,774 (23,962) |
| Mantel-Haenszel chi-squared value | 987,190 | 1,243,364 | 1,277,119 | 1,550,356 |
|
Mean Cohen kappa value (SD)
|
0.65 (0.33)* |
0.75 (0.32)* |
0.83 (0.30)* |
0.94 (0.19)
|
| Note: Comparison of semantic differential retrieval is included with other methods. NA indicates not applicable. The performance of semantic retrieval with other methods is calculated using the Mann-Whitney test with P values expressed as P < 0.001 (indicated by a single asterisk) and P < 0.01 (indicated by a double asterisk). | ||||
The large mean Yates corrected chi-squared and Mantel-Haenszel chi-squared values confirm the expectation of a strong association between the focused set of concepts and records retrieved compared with the gold standard, in the context of a large total population. The strength of association indicated by the mean Yates chi-squared is significantly greater with semantic differential retrieval for both concepts and records, with ICD-10 and hierarchic retrieval being of intermediary status and free-text retrieval again fairing least well.
The calculated means of the Cohen kappa for all methods of retrieval were greater than 0.4 (empirically stated as evidence of useful agreement).26,28 Cohen's kappa value for concept (0.88) and record (0.94) semantic retrieval were significantly better than for the other approaches and were above the benchmark of 0.81 (considered by some as almost perfect29), indicating that high levels of retrieval accuracy from clinical records are achievable.
Discussion
The evaluation of data retrieval can be tested against technical markers such as speed, but from the clinical perspective, although these usability issues are important, it is more vital to evaluate whether an information system can accurately answer the spectrum of questions that might be posed. The lack of reported work on the effect of different search methods on the efficacy of data retrieval from clinical records1 is surprising, considering the large investment in terminology development in the United Kingdom and abroad10,11,12 and the assumptions that data storage in computers will deliver sufficiently accurate information to underpin health service planning and decision support.30 The effort required by terminology developers to semantically define concepts is considerable and has been quoted as varying between 4.6 and 20.5 minutes per concept.22 The recent commencement of the cooperative development of SNOMED Clinical Terms (SNOMED CT)31 heralds yet another massive effort embracing the principle of an underlying STM to provide reference functionality. This study provides the first practical evidence of the utility of such an approach in providing enhanced and accurate data retrieval from clinical records.
The clinical questions required retrieval of information from multiple perspectives. The multi-axial directed acyclic graph structure of CTV36 offered a moderately good mechanism for retrieval. This finding mirrors experiments using the cross-references of SNOMED International, which enabled the attainment of good detection rates and positive predictive values.32 The performance of hierarchic retrieval in the study, however, was dependent on the expressed relationships between concepts in CTV3. Theoretically, if all possible relationships were expressed within the hierarchy, retrieval using this approach would be as good as any other method. The lack of some hierarchies representing a particular clinical perspective (e.g., “limb disorders”) led to a number of false negatives, resulting in reduced performance (indicated by poor detection rates) in response to some clinical questions, e.g., retrieval of “limb complications.” Creating an exhaustive list of all potential clinical perspectives could rectify hierarchy omissions, but these may be difficult to predict and would eventually cause an unwieldy “explosion” of large numbers of relationships. The performance measures indicate that, although the semantic method had significantly better detection rates compared with hierarchic retrieval rates, the positive predictive values were more comparable. This result reflects the formality of the pure subtype hierarchy and its success in providing a robust mechanism to avoid false positives.
While the disorder hierarchy present in CTV3 was unable to support retrieval of “limb disorders,” the semantic method was able to exploit the more complete value hierarchy. The anatomy chapter of CTV3 utilizes the notion of “structure,” which encompasses both the whole and “part of” descriptions of that site to ensure that the subtype structure is maintained.33 This anatomy hierarchy contains the concept “limb structure” with applicable class members, allowing the creation of a semantic differential retrieval of all concepts having a definition [Site]: Limb structure or a type of limb structure (▶). Thus, although the complete classification of “core” disorder concepts may not be possible (or desirable), the complete multiple classification of the underlying primitive values is essential. It is only with the exhaustive polyhierarchic arrangement (complete multiple classification) of primitive values that retrieval from the multiple perspectives that are clinically required is assured.
Evidence from examination of the raw free-text entries in Diabeta 1 suggests that clinicians may not always record the concepts they require with sufficient semantic pedantry. For example, the term “Paget's disease” may have a clear meaning in the context of a patient's record juxtaposed to an entry of Mastectomy, but following retrieval of the concept, outside this environment, it acquires ambiguity as to whether it is Paget's disease of the breast or Paget's disease of the bone. The study also revealed some discrepancy in the interpretation of the semantics of the question posed between the standard setter and the retrieval performer. This discrepancy was the main reason for the suboptimal performance of semantic differential retrieval in 13 of the 47 questions posed. For example, the meaning of the question “retrieve all cases of diabetic cataract” might be interpreted to include all cases of “cataract” (and its class) associated with diabetes; or only cases where the disorder of “diabetic cataract” was explicitly recorded. The rules of data entry can thus affect the result of the retrieval; e.g., if all cataracts in patients with diabetes mellitus are recorded as Cataract but retrieved as Diabetic cataract, no cases would be found. This experience identifies the important association between of the semantics of data entry and data retrieval and suggests that caution may be necessary before wholesale adoption of the principle of separation between the interface and reference features of terminologies that has been suggested.
The use of ICD-10 as a retrieval tool was comparatively effective as a method for retrieving data, a finding recently supported by an investigation of the coding and retrieval of stroke patients.34 Retrieval with ICD-10 performed less well when questions fell outside its scope, e.g., levels of alcohol consumption, or required detail not present in the restricted number of categories available, e.g., absent foot pulses. Overall, the performance of ICD-10 as a retrieval tool was good when it was used within its scope. The method of free-text retrieval fared less well, despite this approach having previously been shown to be effective.24 The performance may have been improved with the use of more sophisticated natural language processing tools,35,36,37 and there have been reports of better detection rates in limited studies.38
The collected battery contained clinical questions from a diversity of perspectives, and the STM supported semantic differential retrieval from these various viewpoints significantly better than the other methods. The indications of the experiments on the Diabeta database are that good detection rates and Cohen's kappa values are achievable (despite the STM being incomplete), and the success supports further investment in exploring this approach. The success of the STM for retrieval was achieved by the flexibility of approach this afforded; however, this was at the expense of its being more complex to use. The method required the analyst to have a good understanding of relational databases and the resource tables (such as the hierarchy table) in CTV3. These operational issues could be improved by the fashioning of clinically intuitive human-computer interface designs.
Conclusion
The investigation has demonstrated a number of key points (see sidebar). It has shown that the use of an STM significantly improves information retrieval from a clinical database in response to physician-derived questions, in comparison with free-text, hierarchic, and ICD-10 approaches. This indicates that an STM provides a useful framework for the representation of clinical findings and that there is merit in the current approach of defining health care concepts semantically. The study has also indicated that alternative methods of analysis will give different results, depending on the purpose for which they have been designed. Greater understanding of the design and scope of ICD may help with the appreciation of its merits as well as its restrictions as a statistical tool for data analysis.▶
Figure 8.
The experiment used a clinical database that was oriented toward the collection of data from patients with diabetes. These patients, with their diversity of complications and associated comorbidities, make the Diabeta database a valuable test bed for retrieval experiments. It is likely that the principles identified in this study are applicable to data represented by a terminology in other health care environments, but this will need confirmation.
The corpus of concepts in the disorder chapter of CTV3 is large, and the extent to which these have been completely semantically represented has been shown to be dependent on the specialist area.20 For example, concepts describing mental health, neurologic, and dermatologic conditions are less amenable to complete characterization. Further effort is required to model these areas and to test whether such specialist extensions can be developed and be mutually compatible with a global STM for clinical findings. Finally, although the use of an STM as the basis for retrieval appears to be valuable technically, further investigation is required into the development of intuitive user-friendly interfaces and the practicability of its use by clinicians.
Acknowledgments
The authors thank Steve Carey at St. Thomas' Hospital for technical support in extracting data from the Diabeta database, the clinicians who contributed to the formulation of the battery of clinical questions, and Henri Brown for additional quality assurance. They are also indebted to Philip Young, Department of Mathematics, University of York, for statistical advice.
Some of the results of this work were presented at the AMIA Annual Symposium, Washington, DC, November 6-10, 1999.
Footnotes
A system in this context refers to the hardware, software and terminology populating the database.
Zero records were retrieved for the question concerning the concept Diabetes treated with diet alone as the status of the patient whose record contained this entry changed during the interval between the original extraction of the Diabeta database and the subsequent retrieval experiments.
References
- 1.Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc. 1997;4: 342-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson N, Mant D, Jones L, Randall T. Use of computerised general practice data for population surveillance: comparative study of influenza data. BMJ. 1991;302: 763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rector AL, Nowlan WA, Kay S. Foundations for an electronic medical record. Methods Inf Med. 1991;30: 179-86. [PubMed] [Google Scholar]
- 4.McDonald CJ. The barriers to electronic medical record systems and how to overcome them. J Am Med Inform Assoc. 1997;4: 213-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans DA, Cimino JJ, Hersh WR, Huff SM, Bell DS, for the CANON Group. Toward a medical-concept representation language. J Am Med Inform Assoc. 1994;1: 207-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Neil M, Payne C, Read JD. Read Codes version 3: a userled terminology. Methods Inf Med. 1995;34: 187-92. [PubMed] [Google Scholar]
- 7.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observations Identifiers, Names, and Codes (LOINC) vocabulary. J Am Med Inform Assoc. 1998;5: 276-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côté RA, Rothwell DJ, Palotay JL, Beckett RS, Brochu L. The Systematized Nomenclature of Human and Veterinary Medicine: SNOMED International. Northfield, Ill: College of American Pathologists, 1993.
- 9.Rector AL, Glowinski AJ, Nowlan WA, Rossi-Mori A. Medical concept models and medical records: an approach based on GALEN and PEN&PAD. J Am Med Inform Assoc. 1995;2: 19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spackman KA, Campbell KE, Côté RA. Snomed RT: a reference terminology for health care. Proc AMIA Fall Symp. 1997: 640-4. [PMC free article] [PubMed]
- 11.Stuart-Buttle CDG, Brown PJB, Price C, O'Neil MJ, Read JD. The Read Thesaurus: creation and beyond. In: Pappas C, et al. (eds). Proceedings of the 14th Medical Informatics Europe '97 Conference. Oxford, UK: IOS Press, 1997: 416-20. [PubMed]
- 12.Rodgers JE, Rector AL. Terminology systems: bridging the generation gap. Proc AMIA Fall Symp. 1997: 610-4. [PMC free article] [PubMed]
- 13.Sönksen P, Williams C. Information technology in diabetes care “Diabeta”: 23 years of development and use of a computer-based record for diabetes care. Int J Biomed Comput. 1996;42: 67-77. [DOI] [PubMed] [Google Scholar]
- 14.National Health Service Centre for Coding and Classification. Read Codes File Structure Version 3.0: Overview and Technical Description. Technical Report. Loughborough, UK: NHS: CCC, 1994.
- 15.World Health Organization. International Classification of Disease and Related Health Problems (vol 1), 10th rev. Geneva, Switzerland: WHO, 1992.
- 16.Severs MP. The Clinical Terms Project. Bull R Coll Physicians. 1993;27(2): 9-10. [Google Scholar]
- 17.Buckland R. The language of health. BMJ. 1993;306: 287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Office of Population Censuses and Surveys. Tabular list of the classification of surgical operations and procedures, 4th rev. London, UK: Her Majesty's Stationery Office, 1990.
- 19.National Health Service Centre for Coding and Classification. Read Codes File Structure Version 3.1: The Qualifier Extensions. Technical Report. Loughborough, UK: NHS CCC, 1994.
- 20.Brown PJB, O'Neil M, Price C. Semantic definition of disorders in version 3 of the Read Codes. Methods Inf Med. 1998;37: 415-9. [PubMed] [Google Scholar]
- 21.Brown PJB. An examination of the conceptual components of disorders and their use in analysis of clinical records [MD thesis]. London, UK: University of London, 1999.
- 22.Brown PJB, Price C. Semantic-based concept differential retrieval and equivalence detection in Clinical Terms Version 3 (Read Codes). Proc AMIA Annu Symp. 1999: 27-31. [PMC free article] [PubMed]
- 23.Brown PJB, Price C, Sönksen P. Evaluating the terminology requirements to support multi-disciplinary diabetic care. Proc AMIA Fall Symp. 1997: 645-9. [PMC free article] [PubMed]
- 24.Steib SA, Reichley RM, McMullin ST, et al. Supporting adhoc queries in an integrated clinical database. Proc 19th Annu Symp Comput Appl Med Care. 1995: 62-6. [PMC free article] [PubMed]
- 25.National Health Service Centre for Coding and Classification. The Read Codes: Demonstrators, Oct 1997 [CD-ROM]. Loughborough, UK: NHSCCC, 1997.
- 26.Friedman CP, Wyatt JC. Evaluation Methods in Medical Informatics. New York, Springer, 1997: 185-203.
- 27.Brown PJB, Sönksen P, Price C, Young P. A standard for evaluating the retrieval performance of clinical terminologies. Proc AMIA Fall Symp. 1999: 1031.
- 28.Cohen J. Weighted kappa: nominal scale agreement with provision for scaled disagreement or partial credit. Psychol Bull. 1968;70: 213-20. [DOI] [PubMed] [Google Scholar]
- 29.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33: 159-74. [PubMed] [Google Scholar]
- 30.National Health Service Executive. Information for Health: An Information Strategy for the Modern NHS. London, UK: Her Majesty's Stationery Office, 1998.
- 31.National Health Service Information Authority and College of American Pathologists. Snomed Clinical Terms: A Global Leader in Healthcare Terminology. 1999. Available from: http://www.coding.nhsia.nhs.uk. Accessed Jan 16, 2000.
- 32.Lussier YA, Bourque M. Comparing SNOMED and ICPC retrieval accuracies using relational database models. Proc AMIA Fall Symp. 1997: 514-8. [PMC free article] [PubMed]
- 33.Schulz EB, Price C, Brown PJB. Symbolic anatomical knowledge representation in the Read Codes version 3: structure and application. J Am Med Inform Assoc. 1997;4: 38-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mant J, Mant F, Winner S. How good is routine information? Validation of coding for acute stroke in Oxford hospitals. Health Trends. 1998;(4)29: 96-9. [Google Scholar]
- 35.Friedman C. Toward a comprehensive medical language processing system: methods and issues. Proc AMIA Fall Symp. 1997: 595-9. [PMC free article] [PubMed]
- 36.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996;35: 285-301. [PubMed] [Google Scholar]
- 37.Haug PJ, Christensen L, Gundersen M, Clemons B, Koehler S, Bauer K. A natural language parsing system for admitting diagnoses. Proc AMIA Fall Symp. 1997: 814-8. [PMC free article] [PubMed]
- 38.Lin R, Lanert L, Middleton B, Shiffman S. A free-text processing system to capture physical findings: canonical phrase identification system (CAPIS). Proc 15th Annu Symp Comput Appl Med Care. 1992: 843-7. [PMC free article] [PubMed]