Abstract
Background and Aim:
Methylated SEPT9 is a novel circulating tumor DNA marker for colorectal cancer, while the effects of various colorectal cancer clinicopathological factors on its detection performance have not been fully evaluated. This study aims to investigate the significance of the clinicopathological factors on methylated SEPT9 performance in a symptomatic endoscopy cohort, with a specific focus on colorectal cancer.
Methods:
A total of 1160 participants were recruited in this study, including 300 patients with colorectal cancer, 122 patients with adenoma, 103 patients with hyperplastic polyps, 568 normal participants (no evidence of disease), and 67 patients with other gastrointestinal diseases. Peripheral blood samples of these participants were collected from 3 Chinese hospitals, and the methylated SEPT9 level was measured using the Epi proColon 2.0 assay.
Results:
Cancer stage, size, and invasion depth were positively correlated with the detection sensitivity, while no difference in sensitivity was identified among cancers at various locations. Infiltrative colorectal cancer exhibited higher sensitivity than ulcerative and protrude colorectal cancer, while no difference in sensitivity was observed among assessed histological types. The colorectal cancer differentiation showed a clear correlation with the cancer stage, and moderate and poorly differentiated colorectal cancer exhibited higher sensitivity than well-differentiated colorectal cancer. Furthermore, colorectal cancer with distal metastasis (M1) showed higher sensitivity than those without any metastasis, while colorectal cancer with lymph node metastasis (N1 and N2) did not show statistical significance compared to those without it. Finally, local vessel or nerve invasion did not affect the sensitivity.
Conclusion:
Factors that reflect the colorectal cancer intrinsic properties, including cancer stage, size, invasion depth, classification, differentiation, and metastasis, exhibited significant effect on the mSEPT9 detection performance.
Keywords: SEPT9, septin 9, colorectal cancer, methylation, adenoma, polyps
Introduction
Colorectal cancer (CRC) is the third most common cancer in the world.1 Early detection is now the most effective way to reduce CRC mortality. However, 60% to 70% patients with CRC were found at middle or late clinical stage when they were diagnosed, leading to the high mortality.1 Statistics from US Preventive Services Task Force showed that about 60% deaths could be spared if a regular periodic screening was carried out, and the average 5-year survival rate could be increased from 46% to 73%.2 Therefore, effective early detection methods for CRC can prolong survival time and reduce mortality.
The circulating tumor DNA (ctDNA) has been proved to be a source of biomarkers for early of cancer detection.3,4 The SEPT9 gene methylation assay detects the aberrant hypermethylation at the promoter region of SEPT9. As the fragments of genomic DNA were released from necrotic and apoptotic CRC cells into the peripheral blood, it was suggested that the risk of CRC in screening settings can be determined by detecting the methylated SEPT9 gene (mSEPT9) in the peripheral blood.3,4
The SEPT9 gene methylation assay was approved by the US Food and Drug Administration (FDA) for CRC screening in average-risk population aged from 50 to 75 and by the European Union (EU) and the Chinese Food and Drug Administration (CFDA) for CRC detection (EU) and CRC early detection, respectively. Although more than 20 independent clinical studies have proved that mSEPT9 is a specific biomarker for CRC early detection and screening,5 limited details are available on the relationship between detection sensitivity and various CRC intrinsic properties, mostly focused on the sensitivity at different locations.6,7 Here, we recruited 300 patients with CRC from a total of 1160 participants from 3 Chinese hospitals in a symptomatic endoscopy cohort study and evaluated the correlation between mSEPT9 detection performance and clinicopathological parameters, including CRC stage, location, size, invasion depth, gross classification, pathological classification, differentiation, vessel or nerve invasion, and lymph node or distal metastasis. Our results showed that factors except cancer location and vessel or nerve invasion all influenced the detection performance of the mSEPT9 assay, indicating the crucial roles of these CRC clinicopathological features in mSEPT9-based CRC detection.
Materials and Methods
Sample Size Estimation, Patient Recruitment, and Ethics
Sample size estimation was based on the following equation for known positive detection rate (PDR): N = Z2*[p (1−p)]/E2. The parameters were defined as follows: Z is a statistical parameter (Z = 1.96 for 95% confidence interval); E represented the error (8% was chosen in this study); and p represented the putative PDR). The P value (.75) was obtained from a previous study looking at the sensitivity of Epi proColon assay in CRC screening. From this, an estimated 113 CRC cases were required. The P value of .25 was used for calculation of number of patients with adenoma, and an estimated 113 cases were required. Similarly, P value of .1 was used for other gastrointestinal disease (GID) and hyperplastic polyps (HP), leading to an estimation of 54 cases for both the groups. From the estimation that CRC accounts for 15% of all patients, at least 753 patients should be included; therefore, the study goal was to recruit 941 patients, anticipating a 20% loss of follow-up rate due to patient quit, incomplete diagnosis information, loss of contact or unqualified samples for assay, and so on.
As a result, a total of 1160 participants from 3 Chinese hospitals at different regions (Xijing Hospital of Digestive Diseases, The Army General Hospital, and Fudan University Shanghai Cancer Center) were enrolled in a symptomatic endoscopy cohort, including 300 patients with CRC, 122 patients with adenoma, 103 patients with polyps, 568 controls (no evidence of disease, NED), and 67 participants with other gastrointestinal diseases (non-CRC GID; Table 1). The Non-CRC GIDs in this study are limited to colon and rectum, including 20 patients with inflammatory bowel disease, 29 patients with colonitis or rectitis, 16 patients with colonic or rectal diverticula, and 2 patients with colonic ulcer. Blood samples were obtained from all participants before colonoscopy was performed to confirm the diagnosis of colorectal diseases. The general clinical features of these participants were summarized in Table 1. In all cases, informed consent was obtained for the use of resected tumor specimens and blood samples. Written consent was signed by each participant recruited in this study. This study was approved by the moral and ethical committee of all 3 hospitals. The members of ethical committees included Rongya Yang, Tiansheng Sun, Shirong Li, Weiwei Zhang, and Mei Zhang from the Army General Hospital; Huaying Wang, Jiong Wu, Xiaoqiu Li, Haiyi Guo, Yanfei Liu from Fudan University Shanghai Cancer Center; and Zhaojiang Guo, Aidong Wen, Liqiang Song, Jian Yang, Xu Feng, Guichun Yuan from Xijing Hospital of Digestive Diseases. All members of ethical committees approved the study.
Table 1.
The Number of Enrolled Participants by Diagnosis Group.
| Diagnosis Group | Total | Sex | Age | ||||
|---|---|---|---|---|---|---|---|
| Male | Female | <50 | 50-59 | 60-69 | ≥70 | ||
| CRC | 300 | 152 | 148 | 51 | 83 | 88 | 78 |
| Stage 0 | 23 | 13 | 10 | 2 | 9 | 6 | 6 |
| Stage I | 42 | 19 | 23 | 7 | 10 | 14 | 11 |
| Stage II | 99 | 53 | 46 | 16 | 22 | 29 | 32 |
| Stage III | 124 | 61 | 63 | 25 | 38 | 35 | 26 |
| Stage IV | 12 | 6 | 6 | 1 | 4 | 4 | 3 |
| Adenoma | 122 | 80 | 42 | 26 | 38 | 36 | 22 |
| Other GID | 67 | 41 | 26 | 39 | 20 | 4 | 4 |
| HP | 103 | 72 | 31 | 39 | 42 | 13 | 9 |
| NED | 568 | 308 | 260 | 296 | 156 | 86 | 30 |
Abbreviations: CRC, colorectal cancer; GID, gastrointestinal diseases; HP, hyperplastic polyps; NED, no evidence of disease.
Colonoscopy and Pathological Examinations
All endoscopists were blinded to the recruitment of the participants in this study. To ensure the quality of colonoscopy, we asked all study sites to perform split-dose or same-day bowel preparation and spend >6 minutes during scope withdrawal. Achievement of cecal intubation was verified by the coprincipal investigators in the individual sites based on the photodocumentation of cecum. The diagnosis by colonoscopy was made by 2 independent gastrointestinal doctors based on videos of colonoscopy. All detailed findings, including neoplastic and nonneoplastic lesions, were recorded in a standard case report form. The size and location of the detected lesions were also recorded.
Pathological examination was performed, and diagnosis was made based on biopsy samples from colonoscopy if patients were recommended for colonoscopy examination or therapy without subsequent surgery. Biopsy samples from surgery were used for pathological diagnosis of patients who underwent surgery. All pathological samples were examined by 2 independent pathologists, and diagnosis was made independently. Inconsistent diagnosis was reassessed by a senior pathologist, and final diagnosis was made. All pathologists involved in this study followed the same guideline in pathological diagnosis.
Sample Collection, Processing, and Storage
Samples were collected from inpatients, and the sample information was recorded in sample collection forms. A peripheral blood sample of 10 mL was collected using 10-mL Vacutainer K2EDTA anticoagulant tubes (BD Biosciences, San Jose, California) for the mSEPT9 assay to ensure the accuracy of the assay. Blood samples were centrifuged for 12 minutes at 1350 rcf, and the plasma was collected in a fresh 15-mL collection tube. The plasma was centrifuged again for 12 minutes at 1350 rcf, and the supernatant was collected in a 5-mL collection tube. Blood samples were collected and processed on the same day within 8 hours. Plasma samples were stored at −15 to −25°C before subsequent ctDNA extraction. The mSEPT9 assay was performed within 2 weeks since the samples were collected.
EpiproColon 2.0 CE Assay
Epi proColon 2.0 CE is a qualitative assay for the real time (RT) polymerase chain reaction (PCR) detection of methylated Septin9 DNA in bisulfite converted DNA from human plasma samples. The assay carried out in this study followed instructions and procedures in the instructions for use of Epi proColon 2.0 CE. The assay is comprised of the Epi proColon Plasma Quick Kit (M5-02-001), the Epi proColon Sensitive PCR Kit (M5-02-002), and the Epi proColon Control Kit (M5-02-003) (Epigenomics AG, Berlin, Germany). The 2/3 algorithm was used for data interpretation in this study. All kits were provided by BioChain (Beijing) Science and Technology, Inc. Beijing, China. The assays were performed using the ABI 7500 Fast Dx Real Time PCR device (Life Technologies, Applied Biosystems 7500/7500 Fast Real-Time PCR System, New York, USA). Assays were performed in 3 Chinese hospitals involved in this study by trained technicians.
Statistical Analysis
Numeric data are expressed as means and standard deviations, and categorical data are shown as number and proportion. Numeric variables were compared by independent sample t tests, and categorical variables were compared by χ2 test or Fisher exact test. The reported P values were for 2-sided statistical tests, and any P value <.05 was considered statistically significant. Statistics and data analysis were performed with PRISM 5.0 (GraphPad Software, Inc, La Jolla, California); figures were made using the same software.
Results
The PDR of the mSEPT9 Assay is Affected by Tumor Type and Stage But Not Tumor Location
The PDR or sensitivity of the mSEPT9 assay in various GIDs was examined first. The PDR for CRC, adenoma, other GID, HP, and NED groups was 73.7%, 27.0%, 16.4%, 8.7%, and 3.0%, respectively. It can be observed from Figure 1 and Table 2 that all examined diseases, including HP (χ2 = 7.73, P < .01), other GID (χ2 = 25.63, P < .001), adenoma (χ2 = 86.47, P < .001), and all stages of CRC (χ2 = 492.71, P < .001) exhibited significantly higher PDR than that of the NED group. Furthermore, adenoma showed significantly higher PDR than HP (χ2 = 12.33, P < .001), and CRC showed significantly higher PDR than adenoma (χ2 = 78.66, P < .001). The comparison of PDR between any 2 groups was shown in Table 2. The PDR for stage 0, I, II, III, and IV CRC was 60.9%, 54.8%, 80.8%, 75.0%, and 91.7%, respectively (Figure 1). Stage II, III, and IV showed significantly higher sensitivity than stage 0 and I (χ2 = 11.99, P < .001). The sensitivity and specificity of the assay is dependent on the definition of positive and negative groups. If CRC was defined as the positive group, and NED was defined as the negative group, the sensitivity was 73.7% (221/300), and the specificity was 97.0% (551/568). If CRC was defined as positive, while non-CRC was defined as negative, the sensitivity was 73.7% (221/300) while the specificity was 91.9% (790/860). If tumors (CRC and adenoma) were defined as positive and nontumors were defined as negative, the sensitivity was 60.2% (254/422) and the specificity was 95.0% (701/738).
Figure 1.
The positive detection rate (sensitivity) of the mSEPT9 assay for NED (normal control), HP, adenoma, and CRC stage 0-IV using the 2/3 algorithm. The number of patients for each group was 300, 122, 67, 103, and 568 for CRC, Adenoma, Other GID, HP, and NED, respectively, and was 23, 42, 99, 124, and 12 for Stage 0, I, II, III, and IV CRC, respectively. CRC indicates colorectal cancer; GID, gastrointestinal disease; HP, hyperplastic polyps; NED, no evidence of disease.
Table 2.
Results of χ2 Test Between Disease Groups in This Study.
| χ2 Value | P Value | Significance | |
|---|---|---|---|
| CRC vs Ade | 78.66 | <.001 | |
| CRC vs other GID | 77.19 | <.001 | *** |
| CRC vs HP | 131.94 | <.001 | *** |
| CRC vs NED | 492.71 | <.001 | *** |
| Ade vs other GID | 2.74 | .098 | NS |
| Ade vs HP | 12.33 | .0004 | *** |
| Ade vs NED | 86.47 | <.001 | *** |
| Other GID vs HP | 2.31 | .129 | NS |
| Other GID vs NED | 25.63 | <.001 | *** |
| HP vs NED | 7.73 | .005 | ** |
Abbreviations: Ade, adenoma; CRC, colorectal cancer; GID, gastrointestinal diseases; HP, hyperplastic polyps; NED, no evidence of diseases; NS, not significant. **P < 0.01, ***P < 0.001.
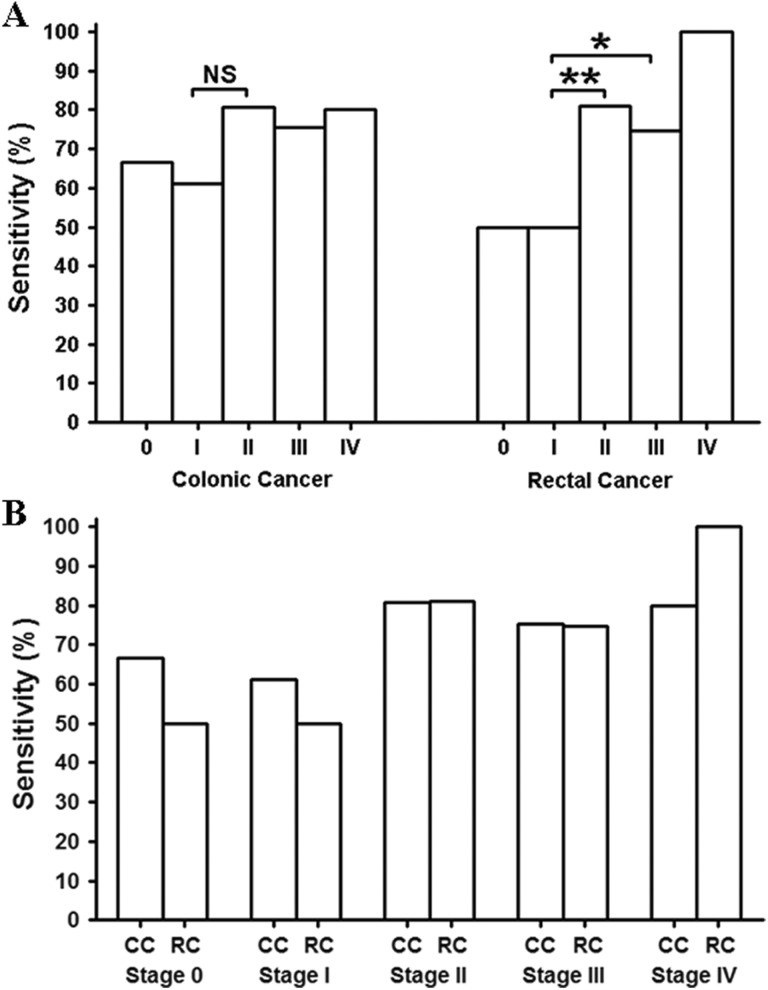
We then studied the sensitivity for rectal cancer (RC) and colonic cancer (CC) at each stage separately. Figure 2A shows the stage-dependent sensitivity for RC and CC. No statistical difference in sensitivity was found among the stages in CC, although there was a trend that the sensitivity for stage 0 and I might be lower than that of later stages (χ2 test, P = .09). In contrast, the sensitivity for stage II, III, and IV in RC was found to be significantly higher than that of stage 0 and I (χ2 test, P = .0015). Overall, the sensitivity of early-stage CRC (RC and CC, Stage 0 and I) was significantly lower than that of the later stage CRC (RC and CC, stage II, III, and IV; χ2 test, P < .001). We further compared the sensitivity between RC and CC at each stage. Figure 2B shows the comparison, and no statistical difference in sensitivity was found between RC and CC at each stage.
Figure 2.
The sensitivity for CC and RC at each stage. The stage-dependent sensitivity was compared in panel A for CC and RC, respectively, and the sensitivity of each stage of CC or RC was compared in panel B. The number of patients for Stage 0, I, II, III, and IV was 15, 18, 57, 61, and 5, respectively, for CC and was 8, 24, 42, 63 and 7, respectively, for RC. CC indicates colonic cancer; NS, not significant; RC, rectal cancer; *significant difference; **highly significant difference.
Colorectal cancer may happen anywhere along the colorectal tract. We therefore studied the sensitivity for CRC at each main segment of colorectal tract. It can be observed from the analysis in Figure 3 that no difference in sensitivity was found among CRC at ascending colon, transverse colon, descending colon, sigmoid colon, and rectum.
Figure 3.
The detection sensitivity for colorectal cancer (CRC) at ascending, transverse, descending, sigmoid colon and rectum. The number of participants was 60, 20, 17, 49, and 154 for ascending, transverse, descending, sigmoid colon and rectum, respectively.
Colorectal Cancer With Bigger Size and Deeper Invasion Exhibited Higher Sensitivity
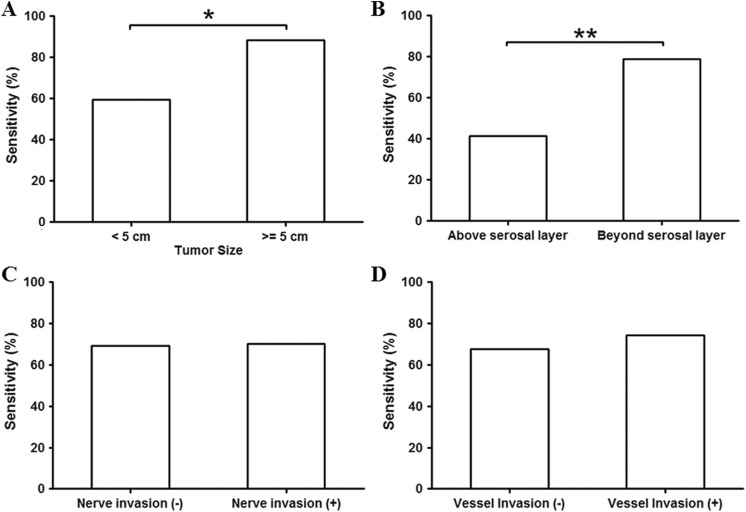
Colorectal cancer with bigger size and deeper invasion commonly represent higher stage of lesion. Here, we studied the relationship between tumor size or depth of invasion and the mSEPT9 assay detection sensitivity. The sensitivity for participants with tumor size <5 cm was shown to be significantly lower than those with tumor size >5 cm (Figure 4A). Similarly, tumors not reaching serosal layer (representing those with <T4a primary tumor) exhibited less sensitivity than tumors reaching or growing beyond serosal layer (representing those with T4a or T4b primary tumor; Figure 4B). We further studied the sensitivity of the mSEPT9 assay on tumors with or without nerve or vessel invasion. Invasions were determined by pathological examinations on CRC removed from surgery. These invasions may happen at early stages of CRC and represent a risk of metastasis or poor prognosis. However, no significant difference was observed between the detection sensitivity of tumors with or without nerve or vessel invasion (Figure 4 C and D).
Figure 4.
The sensitivity for colorectal cancer (CRC) categorized by size, invasion depth, nerve or vessel invasion. Panel A shows the sensitivity of CRC < 5 cm or ≥5 cm. Panel B shows the sensitivity for CRC not reaching the serosal layer (above serosal layer) or reaching or growing beyond serosal layer (beyond serosal layer). The sensitivity for CRC with or without nerve or vessel invasion was shown in panel C and D, respectively. The number of patients was 193 for tumor size <5 cm, and was 107 for tumor size >5 cm. The number of patients was 70 for tumors above serosal layer, and was 230 for tumors beyond serosal layer. The number of patients was 53 for those without nerve invasion, and was 247 for those with nerve invasion. The number of patients was 189 for those with no vessel invasion, and was 111 for those with vessel invasion. *significant difference; **highly significant difference.
Colorectal Cancer With Infiltrative Growth and Low Differentiation Exhibited Higher Sensitivity
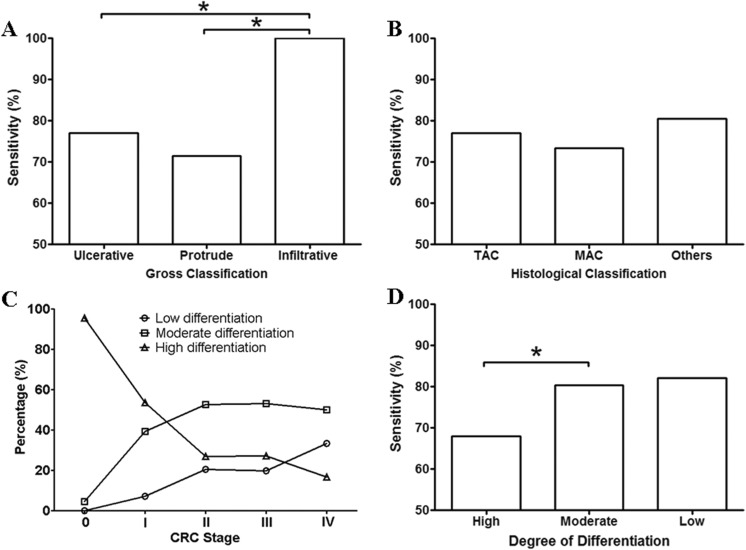
The CRC growth pattern, pathological typing, and differentiation are crucial factors in prognosis prediction and choice of therapeutic strategy. We categorized the participants collected in this study and examined the detection sensitivity of the mSEPT9 assay in each group. As shown in Figure 5A, CRC with infiltrative growth pattern exhibited significantly higher sensitivity than those with ulcerative or protrude growth pattern, suggesting potential higher degree of malignancy and poorer prognosis. We also examined the sensitivity of the most common types of CRC, including the tubular adenocarcinoma (TAC), the mucinous adenocarcinoma (MAC), and other types of CRC (papillary adenocarcinoma, signet ring cell carcinoma, undifferentiated carcinoma, and neuroendocrine tumors, and so on). No significant difference was observed between TAC and MAC and between either of them and other types of CRC (Figure 5B).
Figure 5.
The sensitivity for CRC categorized by gross classification, histological classification, and degree of differentiation. Panel A shows the sensitivity of CRC categorized by gross classification, including ulcerative, protrude, and infiltrative CRC. Panel B shows the sensitivity of CRC categorized by histological classification, including the TAC, the MAC, and other types of CRC (others). Panel C shows the relationship between the ratio of low, moderate or high differentiation and the clinical stage of CRC. Panel D shows the detection sensitivity of CRC with high, moderate, and low differentiation. The number of patients was 231, 53, and 16 for ulcerative, protrude, and infiltrate CRC, respectively. The number of patients was 198, 85, and 17 for TAC, MAC, and other types, respectively. The number of patients for Stage 0-IV CRC is shown in Table 1. The number of patients with high, moderate, and low differentiation was 110, 140, and 50, respectively. CRC indicates colorectal cancer; MAC, mucinous adenocarcinoma; TAC, tubular adenocarcinoma, *significant difference (P < .05).
Cancer cell differentiation is a crucial factor related to cancer malignancy. Poorly differentiated cancer commonly represents higher malignancy and worse prognosis, while well-differentiated cancer normally exhibits lower malignancy and better prognosis. Here, we first examined the relationship between CRC stage and differentiation. Figure 5C shows the percentage of cases with low, moderate, or high differentiation at each CRC stage. It can be seen that most cases at stage 0 belonged to high differentiation group with little moderate differentiation and no low differentiation. As the stage moved from 0 to IV, the ratio of high differentiation dropped sharply, while the ratio of moderate and low differentiation substantially increased. The ratio of high differentiation reached its lowest, while the ratio of low differentiation reached its highest at stage IV. These observations clearly suggest a correlation between the CRC stage and the degree of differentiation. We further studied the sensitivity for CRC with high, moderate, and low differentiation. It clearly shows in Figure 5D that CRC with moderate and low differentiation exhibited significantly higher sensitivity than those with high differentiation, indicating a higher detection rate with more malignant CRC subtypes.
Colorectal Cancer With Distal Metastasis Exhibited Higher mSEPT9 Assay Detection Sensitivity
Distal metastasis and lymph node metastasis in CRC are factors leading to poor prognosis. Patients with distal or lymph node metastasis normally represent higher clinical stage than those without metastasis, even if the primary tumor grading (T grading) is identical. Distal metastasis (M1) leads to clinical staging of IV, no matter what the T or N grading is, therefore, it can be expected that participants with distal metastasis (M1) exhibited higher detection sensitivity than those without it, as shown in Figure 6A. In contrast, participants with 1 to 3 lymph node metastasis (N1) or more than 4 lymph node metastasis (N2) did not exhibit statistically significantly higher sensitivity than those without it (N0; Figure 6B), although there is a trend that the sensitivity with N2 grading might be higher than that of the N0 and N1.
Figure 6.
Sensitivity of CRC categorized by the grade distal or LN metastasis and vessel or nerve invasion. Panel A shows the sensitivity of CRC with (M1) or without (M0) distal metastasis. Panel B shows the sensitivity of CRC without (N0), or with 1-3 (N1), or with ≥4 (N2; LN) metastasis. The number of patients without distal metastasis (M0) was 288, and 12 had distal metastasis (M1). The number of patients with no LN metastasis (N0), 1 to 3 LN metastasis (N1) and ≥4 LN metastasis was 166, 96, and 38, respectively. CRC indicates colorectal cancer; LN, lymph node; *significant difference (P < .05).
Discussion
The Relationship Between Detection Sensitivity and Colorectal Tumor Progression
The mSEPT9 assay (Epi proColon, Epigenomics AG, Berlin, Germany) was approved by the US FDA in 2016 as a CRC screening assay for average-risk population aged from 50 to 75 years old. Many validation studies were performed before and after the approval.5 The CFDA approved the use of the assay as an in vitro diagnostic products (IVD) product in early detection of CRC, and clinical validation and clinical trial were performed for approval purposes.8 Although the application of the assay in screening is widely accepted, studies for the potential uses of mSEPT9 as a biomarker for disease progress monitoring, therapeutic effect assessment, and prognosis prediction never stopped. Our study investigated the possibility of the assay for the above applications by looking at the relationship between mSEPT9 detection and clinicopathological features of CRC and provided some evidence for future application expansion.
In this study, we found that mSEPT9 detection sensitivity was positively correlated with CRC stage, tumor size, invasion depth, and metastasis. Since tumor size, invasion depth, and metastasis are all positively correlated with CRC stage, our observation suggests that SEPT9 methylation could be a marker for CRC progression. Generally speaking, higher stage of CRC would present bigger tumor size and deeper invasion, and these features are closely correlated with each other. It can be suggested that more mSEPT9 molecules may be released into peripheral circulation when CRC goes into higher stages, and the chance to detect the mSEPT9 would be higher in later than in earlier stage CRC, which leads to higher detection sensitivity. However, since this study is a qualitative study and no quantitative statistical analysis has been done to investigate the correlation between mSEPT9 level or detection rate and clinicopathological features, we believe that the mSEPT9 positivity is related to CRC progression but only as a consequence of the relationship with other pathological progression features that lead to higher ctDNA release. Therefore, further quantitative study, such as logistic regression, is needed to elucidate the relationship between mSEPT9 level or detection rate with the stratified CRC progression factors.
The stage-dependent sensitivity is a common feature in RT-qPCR-based blood methylation assay. This is because the release of tumor DNA into blood is dependent on the severity of the primary tumor, and bigger tumors with higher stages generally release more DNA into blood than smaller tumors at lower stages. If the detection capability of the RT-qPCR assay remains stable, the detection rate is positively correlated with the level of ctDNA. The mSEPT9 assay was reported to detect early-stage CRC (stage 0 and I) with sensitivity higher than 50%,6,7,9-11 which is so far the best among the commercial blood-based cancer detection assay. This is based on its capability of detecting as low as 1 to 2 genomics copies of abnormally mSEPT9 DNA.12 Many previous reports have confirmed the claim that the mSEPT9 positivity is associated with higher CRC stage, and generally speaking, higher stage is correlated with larger CRC tumor size, deeper invasion, and metastasis. A couple of meta-analysis of mSEPT9 screening, cohort, and case–control studies showed clear trend of correlation between detection sensitivity and CRC stage.5,13 The pooled mSEPT9 detection sensitivity for stage I, II, III, and IV CRC was 44.6%, 75.2%, 80.1%, and 83.7%, respectively.13 It is obvious that the detection sensitivity for stage I was much lower than that of the later stages (II, III, and IV), reflecting the intrinsic tumor biology of CRC, in which early stage cancer is smaller, less invasive, and therefore, more difficult to be detected. Furthermore, our group recently confirmed that the CRC tumor size (maximal diameter) was correlated with the blood mSEPT9 level, and the blood mSEPT9 level decreased to undetectable level after the cancer was removed by radical surgery (data not published). Furthermore, patients with stage IV CRC treated with chemo- and/or radiotherapy also showed decrease in blood mSEPT9 level before tumor shrinkage can be observed with computer tomography (data not published). The findings in this study are in line with all these reports.
It was previously found that in colon biopsy tissues, laser-microdissected epithelial cells and stroma, SEPT9 mRNA level decreased in the progression of colon neoplastic disease from adenoma to dysplasia to carcinoma, and SEPT9 protein was significantly underexpressed in patients with CRC compared to healthy controls.14 Significant correlation between the SEPT9 hypermethylation and the loss of mRNA expression in CRC strongly suggests that downregulation of SEPT9 mRNA and decrease in SEPT9 expression may account for the pathological progression from benign to malignant lesions in colon tissues.14 The reduction in SEPT9 protein expression may be positively correlated with the progression of the disease from benign adenoma to malignant CRC.
Our observations suggest that higher degree of colorectal tumors exhibited higher detection sensitivity. The benign hyperplastic polyps exhibited higher sensitivity than normal people, and adenoma exhibited higher sensitivity than the benign polyps, and CRC exhibited much higher overall sensitivity than adenoma. The characteristic increase in sensitivity correlated with the progression of colorectal diseases very well. This correlation was based on the level of mSEPT9 DNA detected in blood, indicating that lower degree of tumors released less DNA into the blood.15 The amount of DNA released into blood may be related to the rate of cell turnover, apoptosis or necrosis in a certain disease, and benign lesions with lower growth speed and noninvasive growth pattern normally exhibited lower rate of cell turnover, apoptosis or necrosis, and vice versa.
Correlation Between Sensitivity and Pathological Features
Progressive CRC can be categorized by its growth features into 4 types: the ulcerative, protrude, infiltrate, and colloid type. We examined 3 most common types in this study and found that the infiltrative type exhibited significantly higher sensitivity than the ulcerative and protrude types. As the infiltrative type causes the most server tissue damage compared to the other 2 types, its prognosis is the worst among all types. As mentioned earlier, the mSEPT9 detection sensitivity may be related to the rate of cell turnover, apoptosis, or necrosis; it can be expected that the infiltrative type, due to its intrinsic nature of higher cell turnover rate, may release more SEPT9 DNA into the blood and therefore exhibit higher sensitivity. It can also be suggested that higher detection of SEPT9 methylation in a subgroup could be a prognostic marker for a certain type of CRC, but this assumption still needs to be verified before the mSEPT9 can be used as a prognostic marker for individual prognosis.16 Quantified measurement may be needed to achieve this goal. In this study, we also studied the sensitivity of the most common histological type of CRC, the TAC, and the MAC. Other types were also studied, and they were pooled together to show the sensitivity due to limited number of cases. No difference was observed between any 2 of the 3 groups of CRC, suggesting that pathological type may not be a factor affecting the detection sensitivity of the mSEPT9 assay. However, as only 2 specific types were examined in this study, whether other rare types may show different sensitivity still needs further investigation.
One of the most interesting observations in this study was the correlation between the ratio of various differentiation and the CRC stage. The correlation clearly indicated that early-stage CRC was composed of higher ratio of well-differentiated CRC, while later stage CRC was composed of higher ratio of poorly differentiated CRC. As CRC progressed from stage 0 to stage IV, the overall differentiation became worse and worse. It was not clear whether the change in differentiation ratio was the cause or the result of stage progression. However, both of them were definite prognostic factors. Clinically, cancers with high differentiation are believed to be less malignant than those with moderate or low differentiation. However, apart from pathological criteria, it is lack of biomarkers to evaluate the degree of differentiation that makes it hard to predict the outcome and prognosis Since correlation was observed between the mSEPT9 detection and differentiation, stratification of differentiation could be achieved if the quantitative mSEPT9 level can be related to the degree of differentiation. The mSEPT9, combined with pathological stratification of differentiation and stage, could be used in assessing the prognosis and establishing the therapeutic strategies. The higher sensitivity found in moderate and low differentiation again suggested that SEPT9 methylation could be a prognostic factor in accordance with the progression of CRC. Although some evidence suggested that the plasma mSEPT9 level reflects the prognosis for CRC patient as it is an indirect measure for cancer stage,16 more evidence is needed to confirm the roles of mSEPT9 in progression assessment of prognosis prediction. The abovementioned suggestion on the roles of mSEPT9 in prognosis prediction is speculative, and a study should be conducted where CRC cases of the same stage with the same CRC features are compared, and clear association between mSEPT9 positivity and the time to event should be investigated.
It is also intriguing to find that the mSEPT9 can detect adenoma, other GID, and HP. The sensitivity for these non-CRC diseases was much lower than that of the CRC; however, this did not hinder its use in the detection of these diseases. Under the screening setting of average-risk population, the chance to detect CRC is much lower than the chance to detect these non-CRC diseases. This is because the incidence of these diseases is much higher than that of the CRC in population, and some of them can still be detected even if the detection sensitivity is low. In practice, we did identify many adenomas, HP, and some early-stage CRC participants with the mSEPT9 assay. Early intervention can be performed to prevent the diseases from progressing, and the risk of CRC can be minimized. However, it should be noted that the detection of nontumor colorectal diseases, such as inflammatory bowel disease, colonitis/rectitis, diverticula and ulcer, may lead to high false-positive rate and cause unnecessary examination or intervention. Therefore, risk stratification for patients may be needed. The mSEPT9 level quantification, combined with careful history collection and other routine physical and blood examination, could be one way to reach the purpose.
Conclusions
The PDR of the SEPT9 gene methylation assay was positively correlated with indicators of tumor malignancy, including cancer stage, tumor size, invasion depth, degree of differentiation, tumor classification, and severity of metastasis. SEPT9 methylation may be used as an indicator to assess the clinicopathological status of CRC in future.
Acknowledgments
We thank Kaichun Wu, Yongzhan Nie, Jianqiu Sheng, and Sanjun Cai for support, advice, and organizing the clinical trials in this study. This work was supported by BioChain (Beijing) Science and Technology, Inc and the National Natural Science Foundation Innovation Research Group Science Foundation project “key molecular events in the carcinogenesis and development of gastrointestinal cancer and the clinical significance: 2015.01-2020.12” (grant number: 81421003).
Abbreviations
- CC
colonic cancer
- CFDA
Chinese Food and Drug Administration
- CRC
Colorectal cancer
- ct DNA
circulating tumor DNA
- EU
European Union
- FDA
Food and Drug Administration
- GID
gastrointestinal disease
- HP
hyperplastic polyps
- mSEPT9
methylated SEPT9
- NED
no evidence of disease
- PDR
positive detection rate
- RC
rectal cancer
- TAC
tubular adenocarcinoma.
Footnotes
Authors’ Contribution: Na He and Lele Song contributed equally to this study.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Lele Song, Guangpeng Zhou, Jianming Wang, and Xiaoliang Han are current employees of BioChain (Beijing) Science and Technology, Inc. BioChain is a collaborator of Epigenomics AG, a Germany-based company that launched the first commercial SEPT9 assay. Other authors claim no conflict of interests.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by BioChain (Beijing) Science and Technology, Inc and the National Natural Science Foundation Innovation Research Group Science Foundation project “key molecular events in the carcinogenesis and development of gastrointestinal cancer and the clinical significance: 2015.01-2020.12” (grant number: 81421003).
References
- 1. Schreuders EH, Ruco A, Rabeneck L, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637–1649. [DOI] [PubMed] [Google Scholar]
- 2. National Cancer Institute, PDQ, Treatment, Health Professionals. Survival Rate for Colorectal Cancer by Stage. Bethesda, MD: National Cancer Institute; 1999. [Google Scholar]
- 3. Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54(2):414–423. [DOI] [PubMed] [Google Scholar]
- 4. Gruètzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3(11):e3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song L, Li Y. SEPT9: a specific circulating biomarker for colorectal cancer. Adv Clin Chem. 2015;72:171–204. [DOI] [PubMed] [Google Scholar]
- 6. Tóth K, Sipos F, Kalmár A, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. Plos One. 2012;7(9):e46000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu D, Zhou G, Jin P, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18(4):535–545. [DOI] [PubMed] [Google Scholar]
- 9. Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol. 2015;30(5):830–833. [DOI] [PubMed] [Google Scholar]
- 10. Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. Plos One. 2014;9(6):e98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song L, Jia J, Yu H, et al. The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J Cancer Res Clin Oncol. 2017;143(6):1093–1101. doi:10.1007/s00432-017-2363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60(9):1183–1191. [DOI] [PubMed] [Google Scholar]
- 13. Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep. 2017;7(1):3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toth K, Galamb O, Spisak S, et al. The Influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res. 2011;17(3):503–509. [DOI] [PubMed] [Google Scholar]
- 15. Tóth K, Wasserkort R, Sipos F, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. Plos One. 2014;9(12):e115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tham C, Chew M, Soong R, et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer. 2014;120(20):3131–3141. [DOI] [PubMed] [Google Scholar]