Abstract
With the daily release of data from whole genome sequencing projects, tools to facilitate comparative studies are hard-pressed to keep pace. Graphical software solutions can readily recognize synteny by measuring similarities between sequences. Nevertheless, regions of dissimilarity can prove to be equally informative; these regions may harbor genes acquired via lateral gene transfer (LGT), signify gene loss or gain, or include coding regions under strong selection. Previously, we developed the software S-plot. This tool employed an alignment-free approach for comparing bacterial genomes and generated a heatmap representing the genomes’ similarities and dissimilarities in nucleotide usage. In prior studies, this tool proved valuable in identifying genome rearrangements as well as exogenous sequences acquired via LGT in several bacterial species. Herein, we present the next generation of this tool, S-plot2. Similar to its predecessor, S-plot2 creates an interactive, 2-dimensional heatmap capturing the similarities and dissimilarities in nucleotide usage between genomic sequences (partial or complete). This new version, however, includes additional metrics for analysis, new reporting options, and integrated BLAST query functionality for the user to interrogate regions of interest. Furthermore, S-plot2 can evaluate larger sequences, including whole eukaryotic chromosomes. To illustrate some of the applications of the tool, 2 case studies are presented. The first examines strain-specific variation across the Pseudomonas aeruginosa genome and strain-specific LGT events. In the second case study, corresponding human, chimpanzee, and rhesus macaque autosomes were studied and lineage specific contributions to divergence were estimated. S-plot2 provides a means to both visually and quantitatively compare nucleotide sequences, from microbial genomes to eukaryotic chromosomes. The case studies presented illustrate just 2 potential applications of the tool, highlighting its capability to identify and investigate the variation in molecular divergence rates across sequences. S-plot2 is freely available through https://bitbucket.org/lkalesinskas/splot and is supported on the Linux and MS Windows operating systems.
Keywords: comparative genomics, alignment-free, gene transfer, gene loss
Background
Modern sequencing technologies can quickly and affordably produce genomic sequences for species across the tree of life. Consequently, many new lineages and poorly resolved areas of the tree have been identified.1–3 With tens of thousands of bacterial genomes now publicly available, comparative genomics has produced numerous insights into microbial life.4 Several tools are currently used to detect genome similarity through sequence alignment.5–8 In addition, tools employing a graphical “dot plot” approach, such as Gepard,9 Serolis,10 and SeqTools’ Dotter,11 can highlight genomic similarities and rearrangements as well as gene duplications. These tools, however, have their limitations: Serolis10 is limited in the size of sequence it can analyze (4 kbp), and Dotter11 is significantly slower than Gepard9 for larger sequences. Nevertheless, alignment-free approaches, including the aforementioned “dot plot” tools, have a significant advantage over alignment-based methods: they are less computationally expensive (regarding both time and resources) and impervious to synteny-related problems (see reviews of Vinga and Almeida12 and Bonham-Carter et al13).
In addition to sequence similarities, the dissimilarities between genomic sequences can be equally informative.14 These dissimilarities can indicate strain-specific genes horizontally/laterally acquired rather than vertically inherited. Lateral gene transfer (LGT) is an important force in the evolution of prokaryotes,15 including the exchange of defense mechanisms and virulence factors.4,15,16 Although certainly less prevalent (and fiercely debated), LGT between eukaryotes and prokaryotes can also occur.17–20 Disparities between genomic sequences can also be the result of gene loss, another pervasive and often significant driver of evolution in prokaryotic21,22 and eukaryotic species23 (see review Albalat and Cañestro24). Moreover, recognition of substantial sequence divergence between orthologous gene sequences can signify genes under strong selection (see review Long et al25). Such genes can provide insight into phenotypic differences between species.26,27
Previously we developed S-plot, a tool for the rapid analysis and visualization of bacterial genomic sequences.28 This tool was applied to the examination of Escherichia,28 Bacillus,29 and Neisseria30 genomes, identifying regions of unusual nucleotide composition corresponding to LGT events. Herein, we present the next generation of this tool: S-plot2. Similar to its predecessor, S-plot2 creates an interactive, 2-dimensional heatmap. Similar to the aforementioned dot-plot tools, S-plot2 captures the similarities in nucleotide usage between genomic sequences, but unique to this tool is the fact that it also captures the dissimilarities in nucleotide usage between genomic sequences. Through the examination of nucleotide usage, phylogenetic signals can be uncovered.31 In S-plot2, whole eukaryotic chromosomes and smaller prokaryotic genomes can be efficiently compared. Furthermore, the new version includes functionality to extract, analyze, and automate BLAST queries of regions of interest within the heatmap. This facilitates the investigation of quickly evolving coding regions, novel coding regions, and laterally transferred elements.
Implementation
Developed in Java, S-plot2 performs pairwise comparisons of genomic sequences (partial or complete) via a sliding window approach. Windows can be of a user-defined length (the “genome approach”) or confined to annotated coding regions (the “gene-by-gene approach”). The “genome approach” permits windows to be either adjacent or overlapping. Regardless of the approach selected, each window’s k-mer (subsequence of length k) frequencies are enumerated. The similarity/dissimilarity between 2 windows is calculated based on these k-mer frequencies, using either the Pearson (r) or Spearman rank (ρ) correlation coefficient. The resulting values for each pairwise window comparison are then graphed as a 2-dimensional heatmap using Glimpse32 (eg, Figure 1A). Windows with a similar k-mer usage are represented in the heatmap using colors at one end of the color spectrum, whereas windows with dissimilar k-mer usage are represented by colors at the other end of the spectrum. Draft genome sequences that include several scaffold sequences can be examined using the “genome approach” in S-plot2. The scaffolds can be concatenated, separated by, eg, Ns, into a single FASTA sequence. S-plot2 does not calculate frequencies for windows in which greater than half of the sequence is not A, T, C, or G; thus, windows containing more than one scaffold will be ignored. The “gene-by-gene approach” is a new feature released in S-plot2, as is the Spearman rank correlation coefficient metric for sequence comparison.
Figure 1.
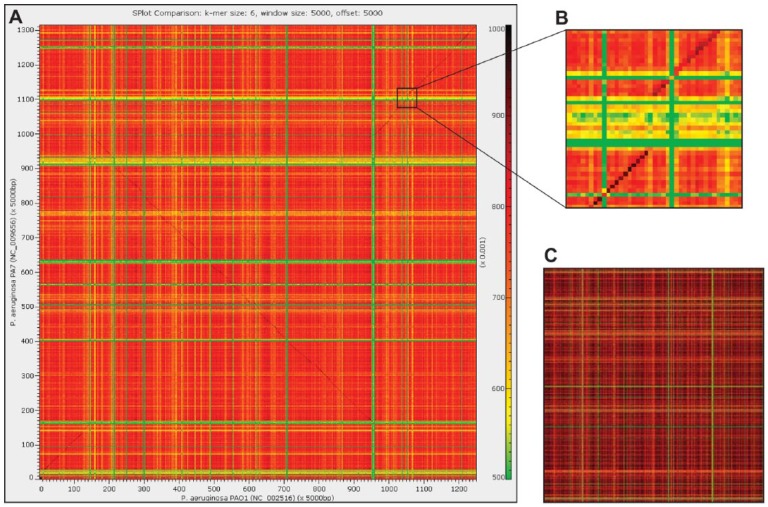
Comparison of Pseudomonas aeruginosa PAO1 (x-axis) and PA7 (y-axis) genomes. (A) “Genome approach” comparison with a window and offset of 5000 bp. (B) Genomic island present with the PA7 strain. (C) “Gene-by-gene approach” comparison of protein-coding gene sequences annotated for the 2 genomes in panel A (*.faa files). Here, the window size is equivalent to a single coding region and k = 3 is evaluated (the same color bar as shown in panel A). The comparisons conducted here for both approaches were done using the Pearson correlation coefficient. Sequence similarity is measured by the frequency of shared k-mers, with green signifying low similarity and red signifying high similarity.
Functionality has been developed in S-plot2 to aid in the interpretation of the heatmap. Users can specify regions of interest based on window coordinates or select windows meeting specific criteria (eg, regions exhibiting aberrant k-mer usage) and then output or BLAST33 these regions. For instance, a cluster of genes which appears in one genome and not the other (indicative of a gene loss/gain), such as that shown in Figure 1B, can be queried; in the case in which a gene was acquired via LGT, the putative source can be identified. Queries to National Center for Biotechnology Information’s (NCBI) eUtils API were automated using JEutils.34 All BLAST queries in S-plot2 use the blastn algorithm and remotely query the NCBI nucleotide collection (nr/nt) database. Users can also output statistics computed for the heatmap as well as generate multi-FASTA format files for windows with an r or ρ value within a user-defined range. The heatmap image itself can be saved to file as a TIF file, implemented using the iCafe package.35
An executable jar file, sample sequence data, and a tutorial are freely available through https://bitbucket.org/lkalesinskas/splot. S-plot2 was tested thoroughly on the Windows and Ubuntu operating systems. Due to the lack of support for compatibility profiles on MacOS, rendering and maneuvering within the S-plot2 heatmap are suboptimal (due to incompatibilities with the Glimpse visualization version used) on MacOS. Exploration of the S-plot2 heatmap (scrolling through a sequence, zooming in/out, etc) was optimized for use with the mouse on Windows and Ubuntu.
As the similarity between windows is calculated based on the correlation (either the Pearson or Spearman rank correlation coefficient) of the frequency of shared k-mers, the condition 4k<<w where w is the window size must be followed. If w ⩾ 4k, then most k-mer frequencies will be 0 or 1 and thus unsuitable for the correlation analysis. Run time and memory usage are dependent on the number of windows. For a genome of size M, the number of windows, n, is M/w. Thus, for smaller window sizes, a larger heatmap will be generated. For each window, k-mer frequencies are enumerated for the original and reverse-complement sequences. Calculation of k-mer usage is linear and values are stored in a sorted array. The run time and memory usage estimate is Ο(n2). For instance, the heatmap generated in Figure 1A was generated in 42 seconds. It is important to note that for large sequences, the required RAM may exceed the RAM allocated or available for the Java Virtual Machine (particularly if the user has a 32-bit version installed) in which case the application will not execute. Nevertheless, a complete human chromosome can be compared using less than 8 GB of RAM in a matter of minutes; S-plot2’s performance is significantly faster than other graphical alignment-free available graphical tools.9,11
Results and Discussion
To illustrate the functionality and utility of S-plot2, we conducted 2 case studies. In addition to providing a visualization of the genomic sequences under investigation, the new functionality developed in S-plot2 can lead to a deeper understanding of the variation in molecular divergence rates across sequences.
Case study 1: exploring the evolution of bacterial genomes
The genomes of the opportunistic bacterial pathogen Pseudomonas aeruginosa are highly mosaic and include regions of genomic plasticity.36 The P aeruginosa accessory genome exceeds that of its core genome.37 Figure 1 shows the pairwise comparison of the P aeruginosa strains PAO1 (NC_002516) and the known “taxonomic outlier” for the species, PA7 (NC_009656).38 Two comparisons were conducted: the “genome approach” using a fixed window size (Figure 1A) and the “gene-by-gene approach” in which each window is an individual gene (Figure 1C). As even closely related P aeruginosa strains can be distinguished by single-nucleotide polymorphisms, indels, and inversions,39,40 it is thus not surprising to observe genomic variation between the PAO1 and PA7 genomes (Figure 1A and C). The nucleotide sequence of the PA7 region shown in Figure 1B was investigated using S-plot2’s automated BLAST functionality. This region includes numerous transposases and integrases as well as plasmid- and phage-associated genes. It corresponds to the previously identified genomic island RGP42 within the P aeruginosa PA7 genome.38 The region shown in Figure 1B is but one of the many genomic islands within these 2 strains. Users can recognize windows of unusual composition visually via the “genome approach” or individual genes of interest via the “gene-by-gene approach” and BLAST the sequences. Furthermore, S-plot2 can automatically identify such regions and BLAST their sequences.
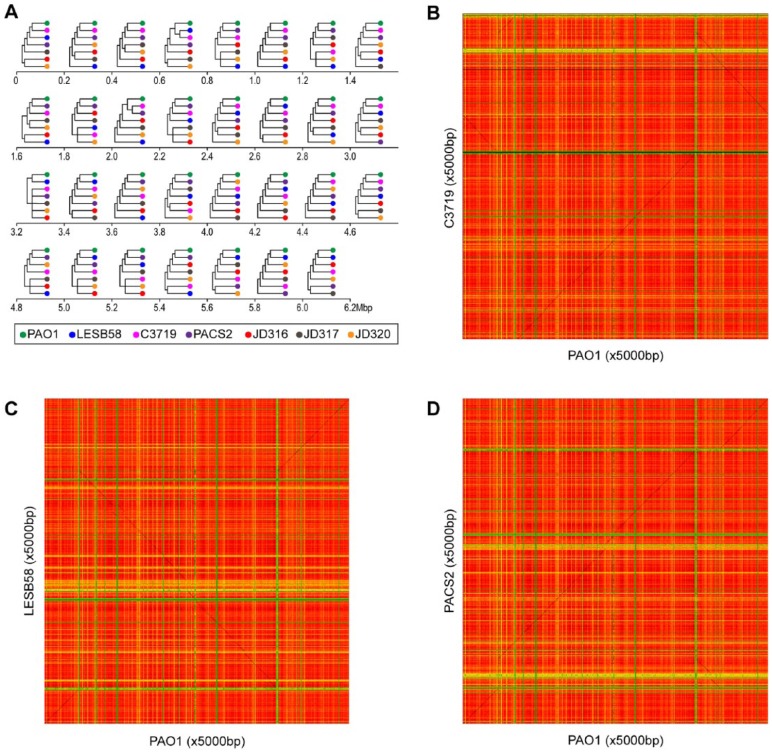
Recombination within P aeruginosa species is frequent and previous research has found variation in the evolutionary histories of regions of the P aeruginosa genome.41 To exemplify how S-plot2 can be used to investigate recombination, 7 genomes included in the comparative genomic study of Dettman et al41 were selected (Table 1) and pairwise comparisons were performed. Sequence similarity was assessed for each window size of 5000 bp (base pairs) for k = 6 using the Pearson correlation coefficient. Figure 2 (panels B, C, and D) shows the pairwise comparisons for PAO1 and C3719, LESB58, and PACS2, respectively. These heatmaps illustrate the presence/absence of unique regions within the genomes and, most notably, rearrangements. The matrices generated by S-plot2 were saved and contiguous 0.2 Mbp regions along the PAO1 genome were evaluated. Thus, an alignment-free approach was used to identify and quantify similarity/dissimilarity between homologous regions of the PAO1 genome and other P aeruginosa strains. As shown in Figure 2A, different regions of the PAO1 genome are represented by different topologies. Consistent with prior alignment-based analyses,41 we find that the evolution of the P aeruginosa genome is not uniform across the entire genome sequence. In this fashion, S-plot2 can provide evidence of evolution across a genome sequence both visually and quantitatively.
Table 1.
Seven Pseudomonas aeruginosa genomes examined.
| Strain | Genome size, Mbp | No. of scaffolds | No. of coding regions | Assembly |
|---|---|---|---|---|
| PAO1 | 6.26 | 1 | 5572 | GCA_000006765 |
| LESB58 | 6.60 | 1 | 6041 | GCA_000026645 |
| C3719 | 6.22 | 1 | 5648 | GCA_000152525 |
| PACS2 | 6.49 | 1 | 5913 | GCA_000168335 |
| JD316 | 6.19 | 1882 | 6590 | GCA_000506125 |
| JD317 | 6.49 | 2043 | 6979 | GCA_000506145 |
| JD320 | 6.41 | 2038 | 6876 | GCA_000506165 |
Sequences were retrieved for genomes (*_genomic.fna.gz) and coding sequences (*_cds_from_genomic.fna.gz).42
Figure 2.
Evolution of the Pseudomonas aeruginosa chromosome. (A) Comparison of cluster topologies based on sequence similarity based on 6-mer usage for window size = offset size = 5000 bp over 0.2 Mbp regions of the PAO1 genome. Heatmaps for (B) PAO1 vs C3719, (C) LESB58, and (D) PACS2. The same color scale as Figure 1 is used here: sequence similarity is measured by the frequency of shared k-mers, with green signifying low similarity and red signifying high similarity.
Case study 2: exploring the evolution of primate chromosomes
S-plot2 is also capable of evaluating whole eukaryotic chromosomes. As such, it can be used to estimate chromosome-specific molecular divergence rates, estimate lineage specific contributions to divergence, and identify regions that are significant contributors to observed divergence. As a case study of S-plot2, we performed pairwise comparisons for all homologous human, chimpanzee, and rhesus autosomes (window size = offset size = 100 Kbp for k = 6 using the Pearson correlation coefficient). Each chromosome was also compared with itself using the same window size, offset size, and k. This self-sequence comparison provides a baseline for the variation within a chromosome relative to that observed between species (see Supplemental File 1). Prior whole genome comparison studies between human and chimpanzee found ≈1.4% sequence divergence43 and 23 inversions,44 as well as other differences (for a review, see the work by Kehrer-Sawatzki and Cooper45). Sequence analysis of human-chimpanzee chromosome pairs suggests that recombination, proximity to telomeres, bias in repair mechanisms, and GC content are all exerting influence on genetic variation.46–50
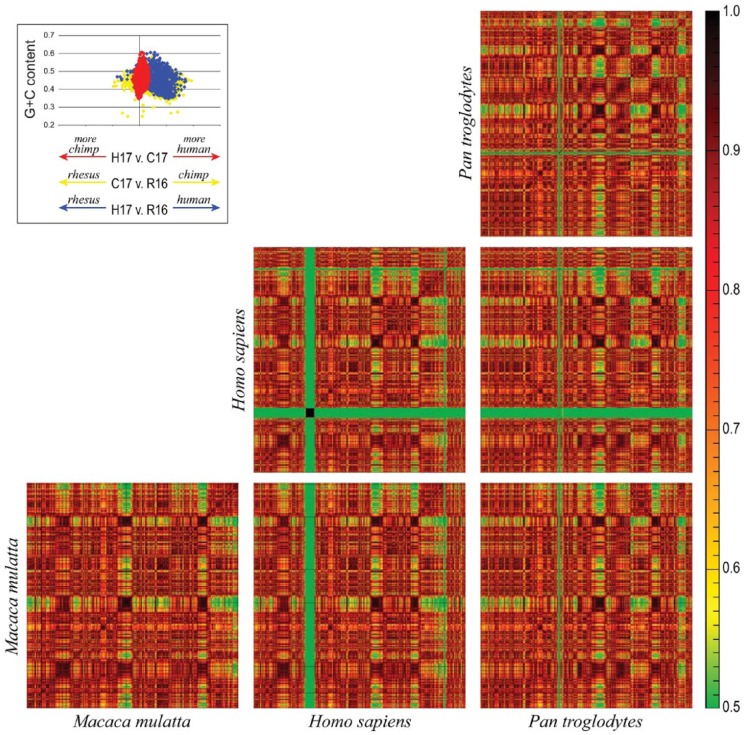
Here, we present a comparison between human chromosome 17, chimpanzee chromosome 17, and rhesus chromosome 16. As the heatmaps in Figure 3 show, the pericentric inversion previously found between these sequences44 can be identified through the pairwise comparisons of the human, chimpanzee, and rhesus autosomes. The heatmaps for these 3 pairwise comparisons, however, do not readily present how these chromosomes are evolving. For instance, the differences observed between the homologous human and chimpanzee chromosomes may be the result of changes within the chimpanzee chromosome or changes within the human chromosome. Comparisons of both chromosomes to the rhesus chromosome let us distinguish between these 2 scenarios. If we oversimplify the process of species divergence to a single point in time (thus ignoring subsequent gene flow), one could assume that the chromosomal sequences are essentially identical. Thus, for a window in the human chromosome, its homologous window in the chimpanzee genome would have the same sequence (and thus nucleotide composition). As such, the heatmap for an individual chromosome compared with itself would be indiscernible from the comparison of the chromosome to its homolog. Post-speciation, the 2 genomes would begin to diverge and this divergence can be quantified by the cross-species comparison value (eg, human vs chimpanzee) relative to the intraspecies comparison (eg, human vs human). The matrices of r values were retrieved for each of the plots shown in Figure 3 and used to calculate the divergence between species (see Supplemental File 1 for details regarding this calculation). The inlay in Figure 3 shows the results of this calculation for human vs chimpanzee (red), chimpanzee vs rhesus (yellow), and human vs rhesus (blue). In this figure, the x-axis is representative of the divergence calculated for a window relative to its GC content. As shown in the inlay in Figure 3, regions in the human genome with a GC content ≈45% are the most divergent windows from chimpanzee; these regions are evolving within the human lineage.
Figure 3.
Comparison of human (Homo sapiens) chromosome 17 (H17), chimpanzee (Pan troglodytes) chromosome 17 (C17), and rhesus (Macaca mulatta) chromosome 16 (R16). Sequence similarity is measured by the frequency of shared k-mers, with green signifying low similarity and red signifying high similarity. The inlay shows the divergence between H17 and C17 (red), C17 and R16 (yellow), and H17 and R16 (blue), relative to the window’s GC content. The x-axis is representative of the divergence calculated for a window relative to its GC content.
Conclusions
S-plot2 provides a means to visually and quantitatively compare genomic sequences ranging from microbial genomes to eukaryotic chromosomes. These comparisons can be generated in a matter of seconds to minutes (depending on the size of the sequence under consideration). S-plot2 includes functionally to aid in the analyses of genomic sequences, allowing users to quickly investigate their data and test hypotheses based on either observed patterns or statistics capturing both the similarities and dissimilarities of sequences. The case studies presented highlight just some of the applications of S-plot2. Furthermore, the analyses performed for the Pseudomonas genomes and human-chimpanzee-rhesus autosomes illustrate the variation in molecular divergence rates across sequences.
Supplemental Material
Supplemental material, SupplementalFile1 for S-plot2: Rapid Visual and Statistical Analysis of Genomic Sequences by Laurynas Kalesinskas, Evan Cudone, Yuriy Fofanov and Catherine Putonti in Evolutionary Bioinformatics
Acknowledgments
The authors would like to thank Mr Arya Mehrtash and Matt Lewczuk for their assistance in early software development and also thank Dr Heather Wheeler and Jon Brenner for their feedback during development. The authors also greatly appreciate the many conversations with Dr Michael Travisano on the analyses of the primate chromosomes.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Science Foundation (grant number 1149387 to C.P.). E.C. and L.K. were supported through a Mulcahy Research Fellowship (Loyola University Chicago). The funding bodies had no role in the design of the study nor the collection, analysis, and interpretation of data.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: LK and EC implemented the application. YF and CP designed the project. LK, EC, and CP performed the analyses of the case studies. CP was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
References
- 1. Hinchliff CE, Smith SA, Allman JF, et al. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci U S A. 2015;112:12764–12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saw JH, Spang A, Zaremba-Niedzwiedzka K, et al. Exploring microbial dark matter to resolve the deep archaeal ancestry of eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hug LA, Baker BJ, Anantharaman K, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. [DOI] [PubMed] [Google Scholar]
- 4. Land M, Hauser L, Jun S-R, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choudhuri JV, Schleiermacher C, Kurtz S, Giegerich R. GenAlyzer: interactive visualization of sequence similarities between entire genomes. Bioinformatics. 2004;20:1964–1965. [DOI] [PubMed] [Google Scholar]
- 6. Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. [DOI] [PubMed] [Google Scholar]
- 9. Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on Genome Scale. Bioinformatics. 2007;23:1026–1028. [DOI] [PubMed] [Google Scholar]
- 10. Serolis. http://www.code10.info/index.php?option=com_content&view=article&id=63:serolis-software-package-for-dot-plot-creation&catid=50:cat_coding_software_serolis&Itemid=75.
- 11. Barson G, Griffiths E. SeqTools: visual tools for manual analysis of sequence alignments. BMC Res Notes. 2016;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vinga S, Almeida J. Alignment-free sequence comparison—a review. Bioinformatics. 2003;19:513–523. [DOI] [PubMed] [Google Scholar]
- 13. Bonham-Carter O, Steele J, Bastola D. Alignment-free genetic sequence comparisons: a review of recent approaches by word analysis. Brief Bioinformatics. 2014;15:890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. [DOI] [PubMed] [Google Scholar]
- 15. Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–482. [DOI] [PubMed] [Google Scholar]
- 16. Darmon E, Leach DRF. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunning Hotopp JC, Clark ME, Oliveira DC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. [DOI] [PubMed] [Google Scholar]
- 18. Boothby TC, Tenlen JR, Smith FW, et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A. 2015;112:15976–15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luan J-B, Chen W, Hasegawa DK, et al. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol. 2015;7:2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wybouw N, Pauchet Y, Heckel DG, Van Leeuwen T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol Evol. 2016;8:1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole ST, Eiglmeier K, Parkhill J, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. [DOI] [PubMed] [Google Scholar]
- 22. Li N, Wang K, Williams HN, et al. Analysis of gene gain and loss in the evolution of predatory bacteria. Gene. 2017;598:63–70. [DOI] [PubMed] [Google Scholar]
- 23. Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. [DOI] [PubMed] [Google Scholar]
- 25. Long M, VanKuren NW, Chen S, Vibranovski MD. New gene evolution: little did we know. Annu Rev Genet. 2013;47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Long Q, Rabanal FA, Meng D, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45:884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cagan A, Blass T. Identification of genomic variants putatively targeted by selection during dog domestication. BMC Evol Biol. 2016;16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Putonti C, Luo Y, Katili C, et al. A computational tool for the genomic identification of regions of unusual compositional properties and its utilization in the detection of horizontally transferred sequences. Mol Biol Evol. 2006;23:1863–1868. [DOI] [PubMed] [Google Scholar]
- 29. Alcaraz LD, Olmedo G, Bonilla G, et al. The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment. Proc Natl Acad Sci U S A. 2008;105:5803–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Putonti C, Nowicki B, Shaffer M, Fofanov Y, Nowicki S. Where does Neisseria acquire foreign DNA from: an examination of the source of genomic and pathogenic islands and the evolution of the Neisseria genus. BMC Evol Biol. 2013;13:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan CX, Bernard G, Poirion O, Hogan JM, Ragan MA. Inferring phylogenies of evolving sequences without multiple sequence alignment. Sci Rep. 2014;4:6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glimpse. http://metsci.github.io/glimpse/.
- 33. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 34. JEutils. http://www.algosome.com/resources/JeUtils/JeUtils.html.
- 35. iCafe Package. https://github.com/dragon66/icafe.
- 36. Valot B, Guyeux C, Rolland JY, Mazouzi K, Bertrand X, Hocquet D. What it takes to be a Pseudomonas aeruginosa? the core genome of the opportunistic pathogen updated. PLoS ONE. 2015;10:e0126468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mosquera-Rendón J, Rada-Bravo AM, Cárdenas-Brito S, Corredor M, Restrepo-Pineda E, Benítez-Páez A. Pangenome-wide and molecular evolution analyses of the Pseudomonas aeruginosa species. BMC Genomics. 2016;17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy PH, Tetu SG, Larouche A, et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE. 2010;5:e8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klockgether J, Munder A, Neugebauer J, et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol. 2010;192:1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winstanley C, O’Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dettman JR, Rodrigue N, Kassen R. Genome-wide patterns of recombination in the opportunistic human pathogen Pseudomonas aeruginosa. Genome Biol Evol. 2015;7:18–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. NCBI Genome Database. ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/.
- 43. Scally A, Dutheil JY, Hillier LW, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feuk L, MacDonald JR, Tang T, et al. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kehrer-Sawatzki H, Cooper DN. Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons. Hum Mutat. 2007;28:99–130. [DOI] [PubMed] [Google Scholar]
- 46. Chimpanzee Sequencing Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. [DOI] [PubMed] [Google Scholar]
- 47. Nusbaum C, Mikkelsen TS, Zody MC, et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. [DOI] [PubMed] [Google Scholar]
- 48. Spencer CCA, Deloukas P, Hunt S, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, SupplementalFile1 for S-plot2: Rapid Visual and Statistical Analysis of Genomic Sequences by Laurynas Kalesinskas, Evan Cudone, Yuriy Fofanov and Catherine Putonti in Evolutionary Bioinformatics