Abstract
Although microscopic examination of stool samples remains the reference method for the diagnosis of intestinal protozoal infections, these techniques are time-consuming and require operators who are experienced and well trained. Molecular biology seems to offer performances at least equivalent in terms of sensitivity and specificity for certain parasites. This study aimed to compare three multiplex PCR assays on 93 prospectively collected positive stools (prospective cohort) and a panel of 12 more Cryptosporidium-positive samples (Cryptosporidium panel). On the prospective cohort, the sensitivity was 89%, 64% and 41% for Giardia sp. detection for BD MaxTM, G-DiaParaTM and RIDA®GENE, respectively and 75%, 100% and 100% for C. parvum/hominis detection. The sensitivity of the RIDA®GENE assay for all Cryptosporidium species was 100%, and for D. fragilis 71%. All the techniques obtained the same results for E. histolytica detection, with one positive sample. All species in the Cryptosporidium panel were identified by the RIDA®GENE PCR. The BD MaxTM and G-DiaParaTM assays detected only C. parvum/hominis with the exception of one positive sample for C. meleagridis. No assay showed satisfactory results for all parasites simultaneously, and the DNA extraction seems to be the critical step. More studies are needed to standardize this procedure.
Keywords: multiplex PCR, intestinal protozoa, Giardia intestinalis, Entamoeba histolytica, Cryptosporidium sp., Dientamoeba fragilis
Abstract
Bien que l’examen microscopique des selles reste la méthode de référence pour le diagnostic des protozooses intestinales, ces techniques sont chronophages et demandent une grande expérience et des opérateurs entrainés. La biologie moléculaire semble offrir des performances au moins équivalentes en termes de sensibilité comme de spécificité pour certains parasites. Cette étude visait à comparer trois techniques de PCR multiplex sur une cohorte de 93 selles positives collectées prospectivement et un panel de 12 échantillons positifs à Cryptosporidium. Respectivement pour BD MaxTM, G-DiaParaTM et RIDA®GENE la sensibilité était de 89 %, 64 % et 41 % pour la détection de Giardia sp. et 75 %, 100 % et 100 % pour la détection de C. parvum/hominis. La sensibilité de la technique RIDA®GENE pour l’ensemble des espèces de Cryptosporidium était de 100 % et de 71 % pour D. fragilis. Toutes les techniques ont obtenu les mêmes résultats pour la détection d’E. histolytica (1 échantillon positif). Toutes les espèces de Cryptosporidium ont été détectées par la PCR RIDA®GENE. Les techniques BD MaxTM et G-DiaParaTM ont détecté seulement C. parvum/hominis en dehors d’un échantillon positif à C. meleagridis. Aucun essai n’a montré de résultats satisfaisants pour l’ensemble des parasites simultanément et l’extraction d’ADN semble être l’étape critique. Plus d’études sont nécessaires afin de standardiser cette procédure.
Introduction
Because the world is in the era of globalization, laboratories are confronted with a growing number of parasitic diseases affecting people living in endemic areas, migrants, travellers and international workers [19]. Intestinal protozoan parasites are responsible for a broad spectrum of clinical manifestations, ranging from mild gastrointestinal symptoms to life-threatening watery or haemorrhagic diarrhoea, and can even lead to complications with extra-intestinal localizations. Among them, giardiasis and dientamoebiasis are a major cause of disease in terms of frequency [1, 22], and cryptosporidiosis and amoebiasis are, respectively, the third and fourth parasitic causes of death worldwide [8]. However, these infections are often neglected and underreported.
Diagnosis of protozoan intestinal infections mainly relies on microscopic examination after stool concentration, which remains the reference method, as it enables detection of all parasites. However, it is time-consuming and requires special skills and well-trained operators, since protozoan parasites are somewhat difficult to identify, particularly when they are present in low numbers. Therefore, new diagnostic methods are needed. While the development of molecular techniques marked a turning point in the diagnosis of bacterial and viral diseases, these techniques are more difficult to apply to the diagnosis of intestinal protozoosis, due to the thick wall of parasite (oo)cysts, making DNA extraction difficult, and due to the high density of PCR inhibitors in stool samples. Many procedures have been developed, often based on pre-treatment of stool samples by mechanical, chemical or thermic lysis, and followed by various extraction systems, like silica-column-based or magnetic extraction, with variable performances. Today, molecular biology techniques could help to save time, provided that they are able to detect in a same run most protozoan parasites infecting humans, and that they can be automated.
In this study, we evaluated three commercial multiplex PCR assays, BD MaxTM Enteric Parasite Panel (Becton Dickinson, Pont-de-Claix, France), G-DiaParaTM (Diagenode Diagnostics, Liege, Belgium), and RIDA®GENE Parasitic Stool Panel I (R-BioPharm, Darmstadt, Germany), and compared them to microscopy, using the same panel of stool samples found positive for protozoa by microscopic examination. The BD MaxTM assay is fully automated, and the two other methods were evaluated after automated extraction using MagNA Pure 96 (Roche Diagnostics, Meylan, France), with the aim of adapting amplification steps on a flow system (Roche).
Materials and methods
Stool samples
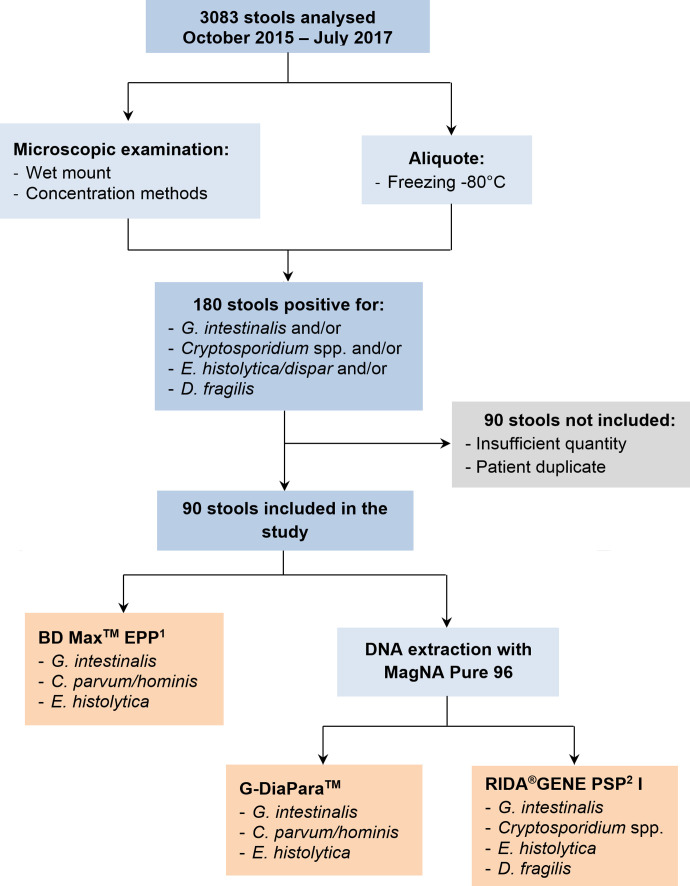
This study included 90 stool samples analysed prospectively at the Consultation Unit of the Laboratory of Parasitology of the University Hospital of Rennes (France) over an 18-month period (October 2015–July 2017) and tested positive for Giardia intestinalis, Cryptosporidium sp., Entamoeba histolytica/dispar and/or Dientamoeba fragilis by microscopic examination (Fig. 1), so-called “prospective cohort”. Three additional samples positive for commensal protozoa and/or helminths were also included in order to assess the specificity of the assays. These 93 samples were analysed by experienced operators, as quickly as permitted by the opening hours of the laboratory and the timeframe for transport of the stool. The procedure consisted in a microscopic examination of a fresh stool wet mount and in-house concentration methods (Bailenger’s, Thebault’s and merthiolate-iodine-formalin biphasic methods [5]). Cryptosporidium detection relied on Henriksen’s modified Ziehl-Neelsen staining on concentration pellets. An aliquot of stool sample was also frozen at −80 °C until DNA extraction. When Cryptosporidium sp. oocysts were detected, a stool sample was sent to the French National Reference Centre for Cryptosporidiosis (NRCC) (University Hospital of Rouen, France) for species identification by sequencing analysis. For all samples, parasites were semi-quantified as “rare”, “few”, “quite numerous”, “numerous” and “many”. Twelve samples provided by the NRCC were also included to extend our cohort with stool samples positive for various species of Cryptosporidium (“Cryptosporidium panel”). These specimens were stored frozen with potassium dichromate 2.5%. The final stool panel consisted of 44 G. intestinalis, 23 Cryptosporidium spp. (13 C. parvum, 5 C. felis, 3 C. hominis, 1 C. canis, and 1 C. meleagridis), 13 E. histolytica/dispar, and 28 D. fragilis. Six samples were poly-parasitized: 2 with D. fragilis and G. intestinalis, 2 with E. histolytica/dispar and G. intestinalis, and 2 with Cryptosporidium sp. and G. intestinalis.
Figure 1.
Study flow chart. 1EPP: Enteric Parasite Panel; 2PSP: Parasitic Stool Panel.
DNA extraction using MagNA Pure 96
Stool DNA was extracted with BD MaxTM (Becton Dickinson, Franklin Lakes, NJ, USA) and MagNA Pure 96 (Roche Diagnostics, Meylan, France). For each sample, a small amount (approximately pea-sized) of thawed stool was suspended in 500 μL of PBS and vortexed, except for liquid stools, which were used plain. In case of insufficient quantity of stool, the volume of PBS was reduced proportionally. Suspensions were centrifuged for 5 s at 500 g, and 90 μL of supernatant were added to 90 μL of MagNA Pure 96 Bacteria Lysis Buffer (Roche Diagnostics) and 20 μL of proteinase K (Roche Diagnostics). After homogenization, the mix was incubated for 10 min at 65 °C, followed by 10 min at 95 °C. Then, two cycles of freezing and thawing were performed (at least 10 min at −80 °C followed by 10 min at 95 °C). Each incubation step was followed by vortexing to homogenize the suspension. After a short centrifugation (few seconds at max speed), 20 μL and 10 μL of the RIDA®GENE Parasitic Stool Panel I (PSP I) (R-BioPharm) and DiaControlDNATM Cy5 (Diagenode Diagnostics) internal controls, respectively, were added. Mixes were then extracted with a MagNA Pure 96 system (Roche Diagnostics Ltd) using MagNA Pure 96 DNA and Viral NA Small Volume (Roche Diagnostics Ltd), and eluted in a 100 μL volume. Other pre-treatment procedures were tested on 17 samples: one with a bead-beating step before thermal and chemical lysis, and another with only chemical lysis. Bead-beating was done with MagNA Lyser Green Beads (Roche Diagnostics) using the MagNA Lyser device (Roche Diagnostics Ltd), for 35 sec at maximum speed and repeated twice, followed by centrifugation for 1 min at 8000 g, before lysing the supernatant as described above. DNA extracts were stored at −80 °C until amplification with the RIDA®GENE PSP I and the G-DiaParaTM assays.
Amplification
DNA extracts were amplified by the RIDA®GENE PSP I (R-BioPharm) and G-DiaParaTM (Diagenode Diagnostics) assays using a LightCycler 480 II device (Roche Diagnostics), following the manufacturer’s recommendations. As D. fragilis can be detected only by RIDA®GENE PSP I, a total of 65 and 93 samples were analysed with the G-DiaParaTM and the RIDA®GENE PSP I assays, respectively. The pathogens possibly detected by each assay are shown in Figure 1. Briefly, for the RIDA®GENE PSP I assay, 5 μL of DNA were added to 19.9 μL of mix and 0.1 μL of Taq polymerase solution, and amplified for 45 cycles with a 15 s extension step at 60 °C. For the G-DiaParaTM assay, 5 μL of DNA were added to 20 μL of mix composed of 5 μL of Master Mix 5X (Diagenode Diagnostics), 2.5 μL of probes and primers for parasite detection, 2.5 μL of probes and primers for internal control detection, and 10 μL of water suitable for molecular biology. Amplification was carried out for 45 cycles with an extension step at 60 °C for 60 s. Positive and negative controls were included in each run on the LC 480 II (provided by PCR kits). False-negative results with the RIDA®GENE PSPI and G-DiaParaTM assays were re-analysed using DNA extracts diluted at 1:10. For two stool specimens (1 C. parvum and 1 C. meleagridis), the remaining quantity was insufficient for extraction on MagNA Pure 96, thus PCR assays were performed using BD MaxTM extracts stored at −80 °C.
Extraction and amplification with the BD MaxTM system
A small quantity of thawed stool was collected with a 10 μL-calibrated loop and suspended in a tube containing 1.5 mL of lysis buffer, supplied in the BD MaxTM EPP kit. Tubes were heated for 52 min in a pre-warmed heater device, before being transferred into a BD MaxTM system rack containing unitized dried reagents. Processing by the BD MaxTM system includes a DNA extraction step with magnetic beads and multiplex amplification using Taqman® technology. Each rack allows the analysis of up to 24 samples in a same run. Positive controls (positive clinical samples) were included in the first run only. As with the G-DiaParaTM assay, 65 samples were analysed with this technique (exclusion of stools positive for D. fragilis only). After analysis, all remaining extracts were stored at −80 °C.
Statistics
Microscopy was considered as the reference method to calculate the sensitivity and specificity of each PCR assay.
Results
Impact of the pre-treatment method on PCR performance
As shown in Table 1, the bead-beating pre-treatment inhibited the detection of Cryptosporidium spp. in the selected samples. The detection of G. intestinalis was moderately affected, while E. histolytica and D. fragilis detection was apparently not affected, with the limitation of the small number of samples. Freezing and thawing cycles were maintained because of better amplification of internal controls (CT from 25–29 versus 30–33 without, data not shown), even if the number of positive stools was not impacted.
Table 1.
Results for tested pre-treatments.
| Results by RIDA®GENE assay (n/N) |
||
|---|---|---|
| Without bead-beating | With bead-beating | |
| G. intestinalis | 5/8 | 3/8 |
| Cryptosporidium sp. | 4/5 | 0/5 |
| E. histolytica | 1/1 | 1/1 |
| D. fragilis | 2/3 | 2/3 |
Evaluation of the 3 PCR assays’ performances
The detection of Giardia by the BD MaxTM, G-DiaParaTM and RIDA®GENE assays yielded respectively a sensitivity of 89%, 64% and 41%, respectively (Table 2). False-negative results were obtained with samples showing “rare” or “few” parasites by microscopy, except for 4 which were negative by G-DiaParaTM and/or RIDA®GENE PSP I assays while quantified as “quite numerous” or “numerous” by microscopy. Five false-negative samples were obtained with the BD MaxTM; in none of them the results of the internal control suggested the presence of PCR inhibitors. All three assays detected a positive stool which was negative by microscopy, leading to a specificity of 95%–98%. As illustrated in Figure 2, samples tested positive with the RIDA®GENE PSP I assay were also positive with the G-DiaParaTM and BD MaxTM assays, and almost all samples positive with the G-DiaParaTM assay were also positive with the BD MaxTM assay.
Table 2.
Performances of the multiplex PCR assays compared to microscopy on 93 positive samples analysed routinely na: not applicable; nd: not determined.
| Sensitivity n/N (%) |
Specificity n/N (%) |
|||||
|---|---|---|---|---|---|---|
| BD MaxTM | G-DiaParaTM | RIDA®GENE | BD MaxTM | G-DiaParaTM | RIDA®GENE | |
| G. intestinalis | 39/44 (89%) | 28/44 (64%) | 18/44 (41%) | 20/21 (95%) | 20/21 (95%) | 48/49 (98%) |
| Cryptosporidium sp. | 6/11 (55%) | 8/11 (73%) | 11/11 (100%) | 54/54 (100%) | 54/54 (100%) | 82/82 (100%) |
| C. parvum/hominis | 6/8 (75%) | 8/8 (100%) | 8/8 (100%) | na | na | na |
| D. fragilis | na | na | 20/28 (71%) | na | na | 63/65 (97%) |
| E. histolytica/dispar | nd1 | nd1 | nd1 | nd1 | nd1 | nd1 |
As microscopy does not enable species identification, sensitivity and specificity could not be determined for E. histolytica detection. However, concordant positive results were obtained with the 3 PCR assays for 1 sample.
Figure 2.
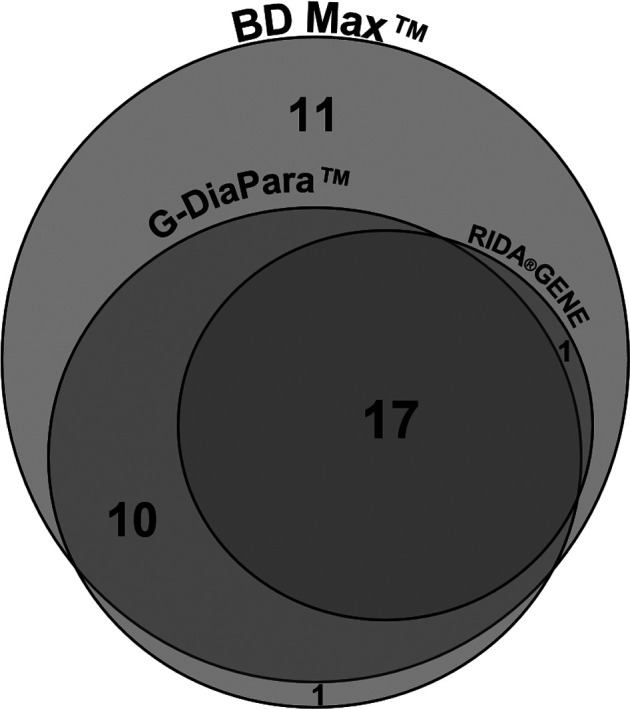
Venn diagram of results for Giardia detection. All Giardia-positive samples detected by the RIDA®GENE assay were detected by the G-DiaParaTM and the BD MaxTM assays, and almost all those detected by the G-DiaParaTM assay were detected by the BD MaxTM assay (1 exception).
The sensitivity for Cryptosporidium sp. detection by BD MaxTM, G-DiaParaTM and RIDA®GENE PSP I assays was for the prospective cohort 55%, 73% and 100%, respectively. When considering only samples positive for C. parvum and C. hominis, as targeted by their respective primers, the first two assays reached sensitivities of 75% and 100%, respectively. In this prospective cohort, the RIDA®GENE assay detected all positive stools, the G-DiaParaTM assay detected all C. parvum/hominis, and the BD MaxTM assay did not detect two stools positive for C. parvum/hominis because of PCR inhibitors (Table 2). Specificity was excellent for all techniques (100%). When considering the 12 samples from the NRCC Cryptosporidium panel (Table 3), the RIDA®GENE assay missed two Cryptosporidium sp. (one C. hominis sample containing PCR inhibitors and one C. felis), the BD MaxTM assay missed two C. hominis positive stools, and G-DiaParaTM detected all C. parvum/hominis. None of the G-DiaParaTM and BD MaxTM assays detected the other species except C. meleagridis. For three of the four false-negative results by the BD MaxTM assay, the internal control highlighted PCR inhibitors. The RIDA®GENE assay detected at least one sample for each Cryptosporidium species tested, confirming the wide detection of Cryptosporidium species.
Table 3.
Results for the Cryptosporidium sp. panel from the French NRCC.
| Sensitivity: n/N
|
|||
|---|---|---|---|
| BD MaxTM | G-DiaParaTM | RIDA®GENE | |
| Cryptosporidium sp. (N = 12) | 7/12 | 9/12 | 10/12 |
| C. parvum (N = 5) | 5/5 | 5/5 | 5/5 |
| C. hominis (N = 3) | 1/3 | 3/3 | 2/3 |
| C. felis (N = 2) | 0/2 | 0/2 | 1/2 |
| C. canis (N = 1) | 0/1 | 0/1 | 1/1 |
| C. meleagridis (N = 1) | 1/1 | 1/1 | 1/1 |
Because microscopy cannot be used to identify E. histolytica/dispar species, neither sensitivity nor specificity could be calculated. However, results were identical for the three assays, which detected one sample positive for E. histolytica among the stools positive for E. histolytica/dispar. Another sample was collected from the same patient 4 days before this one (not included): microscopic examination showed only commensal protozoa (Entamoeba coli and Blastocystis hominis) but all PCR assays were positive for E. histolytica.
Among the 28 stools positive for D. fragilis, 20 tested positive with the RIDA®GENE assay, yielding a sensitivity of 71%. Among the eight false-negative samples observed, all were quantified as “rare” or “few”, except one quantified “quite numerous” by microscopy. No PCR inhibitor was detected by internal controls. Two samples negative by microscopy were positive by the RIDA®GENE assay (specificity 97%), one contained Endolimax nanus, Chilomastix mesnili and Pentatrichomonas intestinalis, and the other contained Blastocystis hominis, G. intestinalis and C. mesnili.
PCR inhibitors were detected in 26% of stools by the BD MaxTM assay (17/65), impacting only detection of Cryptosporidium sp., and 0% (0/65) and 1% (1/93) for the G-DiaParaTM and RIDA®GENE assays, respectively. The extract with inhibitors detected by the RIDA®GENE assay was associated with a false-negative result for Giardia sp. The amplification of diluted extracts did not enable recovery of false-negative results.
Discussion
The aim of this study was to evaluate and compare three PCR assays for the detection of intestinal protozoa. Performances obtained on our prospective cohort of 93 consecutive frozen stool samples from 93 patients, positive by microscopic examination, were heterogeneous among assays and targets. Overall, we observed high specificity, but variable sensitivities for Giardia intestinalis and Cryptosporidium sp. detection. BD MaxTM was the most sensitive for G. intestinalis detection (89%), followed by G-DiaparaTM (64%) and RIDA®GENE (41%). These two latter sensitivities are poorer than previously observed in other studies. Indeed, Laude et al. observed 92% sensitivity with the G-DiaParaTM assay on a smaller series of 38 Giardia-positive stool samples after extraction with the QiaSymphony device (Qiagen, Courtaboeuf, France) [12]. To our knowledge, there are no evaluations of the performances of the RIDA®GENE Parasitic Stool Panel I in the literature, but similar panels from the same manufacturer showed sensitivities of 95%–100% in some reports [13, 15, 21]. Nevertheless, in these reports, only few data are available about the microscopic examination performed: it is therefore difficult to interpret the observed performances. It is well known that the extraction process influences the results of amplification. These disappointing results cannot therefore be solely attributed to the PCR assay. As discussed further, even though we tried to optimize DNA extraction from cysts by using various pre-treatment protocols, many factors involved in this step could interfere in our study. However, it can be stated that the G-DiaParaTM assay performed better than the RIDA®GENE assay for Giardia detection, using our extraction technique. The BD MaxTM system yielded results in agreement with the 93.5%–100% sensitivity observed in the literature [14, 17, 18]. Interestingly, one sample was negative by microscopy and positive with the three assays. This patient was previously diagnosed with G. intestinalis, so this result could either reflect the presence of specific DNA without living parasite, or parasite recrudescence, or resistance to treatment. This highlights that, while molecular techniques are adapted for the diagnosis of intestinal protozoosis, more studies are needed to define their role in post-treatment follow-up.
Regarding Cryptosporidium detection, the G-DiaParaTM and RIDA®GENE assays showed very good sensitivity and specificity using routine specimens. The BD MaxTM assay had a slightly lower sensitivity mainly impacted by PCR inhibitors, leading to 25% (2/8 C. parvum/hominis) false-negative results. Despite this, the BD MaxTM assay was the only to detect C. parvum DNA in a stool negative by microscopy (sample not included in the panel, but tested retrospectively). The patient had been previously diagnosed with cryptosporidiosis and this negative stool was sampled before the onset of antiparasitic treatment, thus this could rather be a “true positive” undetected by microscopy. A possible explanation for this apparently variable performance might be that the BD MaxTM system performs better than the MagNA Pure 96 system for nucleic extraction, although it is more susceptible to PCR inhibitors. Interestingly, the BD MaxTM and G-DiaParaTM assays detected C. meleagridis DNA, whereas they are designed to detect only C. parvum/hominis, which could be explained by the fact that these species are genetically close. Such cross-detection is not systematic, as in the study of Laude et al. a stool positive for C. meleagridis was undetected by the G-DiaParaTM assay [12]. Finally, even though C. parvum/C. hominis are the species mainly responsible for cryptosporidiosis worldwide, the use of PCR assays restricted to these species should be followed by microscopic examination of stools, if the PCR result is negative. In fact, in France, species other than C. parvum/C. hominis account for about 10% of cases [4], which would remain undiagnosed by these assays.
Detection of E. histolytica was entirely concordant among the three assays. Results were coherent in that the positive sample came from a patient who was positive for E. histolytica/dispar by microscopy. This patient had another stool positive by PCR for E. histolytica, but negative by microscopic examination, suggesting that microscopy missed one positive sample. Today, molecular biology is critical in the diagnosis of E. histolytica and allows us to distinguish real cases of amoebiasis from carriage of E. dispar, E. moshkovskii or E. bangladeshi, which are poorly detected using antigenic assays [3, 20, 23, 24]. This discrimination is crucial to rationalize drug consumption and to avoid misdiagnosis.
The detection of D. fragilis by the RIDA®GENE assay showed a sensitivity of 71%, and was mainly affected by low parasitic loads. Diagnosis of dientamoebiasis by microscopy is highly difficult, principally because trophozoites are pleomorphic and degrade rapidly outside the intestinal lumen. Therefore, PCR is considered to be the reference method for D. fragilis detection [22]. However, available molecular assays are at risk of false positivity because of cross-reactions [6]. In our cohort, two samples were positive whereas microscopic examination of the stool sample was negative and these patients had no previous history of dientamoebiasis. These samples contained Pentatrichomonas hominis and Chilomastix mesnili, two flagellated protozoa, which are likely to induce cross-reactions. Although the stool containing P. hominis was unique in this cohort, other stools contained C. mesnili, sometimes in high quantity, and did not yield false-positive results by the RIDA®GENE assay. This suggests that P. hominis could be responsible for cross-reactions, which is concordant with the fact that, unlike C. mesnili, it belongs to the Trichomonada, together with D. fragilis. Nevertheless, the only way to confirm this hypothesis would be to sequence PCR products obtained with the RIDA®GENE assay primers targeting D. fragilis. As the pathogenicity of D. fragilis remains debated [22, 25], the need to include this protozoa in multiplex PCR panels may be questionable. However, even though the relationship between D. fragilis carriage and diarrhoea is unclear, reports of gastrointestinal symptoms related to heavy parasitic loads are frequent [22]. Furthermore, many studies on D. fragilis pathogenicity relied on microscopic examination of stools, which is not an ideal method as discussed above, and could impact the strength of the observed association between parasite carriage and symptoms. For these reasons, molecular biology could add value for further epidemiological studies on the pathogenicity of this protozoon.
The most striking observation in this study is the heterogeneous sensitivity, especially for G. intestinalis detection. The employed DNA extraction process could be insufficient in this respect, explaining the lower performances of the G-DiaParaTM and RIDA®GENE assays. For example, Adamska et al. compared various methods for extraction of DNA from G. intestinalis cysts and concluded that each stage before amplification was crucial [2]. They also reported their best results with an overnight proteinase K treatment instead of short incubations. While some studies reported better DNA extraction of G. intestinalis after a bead-beating step of stools, many others observed similar results without mechanical lysis [7, 10, 12, 24]. Interestingly, some authors also reported poorer DNA extraction after this step, depending of the extraction technique used and on the type of beads. For example, in the study of Kaisar et al. [11], mechanical lysis diminished the number of positive samples for G. intestinalis and D. fragilis. In the same way, a multicentric study showed highly variable effects of the bead-beating step on the extraction of Cryptosporidium DNA, especially with the MagNA Lyser® Green Beads which impacted it negatively [16]. The other major factor is the extraction device itself. In the literature, few studies employed the MagNA Pure 96 for protozoa detection. The most interesting one is that of Gotfred-Rasmussen et al., describing sensitivity of 91% for Giardia detection by an in-house PCR, compared to direct fluorescent assay. [9] No pre-treatment was applied, except incubation in a transport medium. The extraction reagent was the DNA and Viral NA Large Volume, which enables treatment of higher sample input than the Small Volume reagent we used.
Finally, sensitivity could also be affected by PCR inhibitors. Internal control DNA is supposed to detect them, but using the recommended volumes, the Ct of amplification was always low, thus unlikely to be able to detect moderate inhibition. However, a dilution to 1:10 of DNA extracts from false-negative samples did not allow us to recover the positivity of any sample, which seems to rule out the hypothesis that PCR inhibitors would be the source of false-negative results. Another hypothesis could be that long storage at −80 °C (up to one year before extraction) could have altered DNA and impact performances. However, the BD MaxTM assay detected most Giardia sp. positive stools, showing that DNA quality was unlikely an issue.
Basically, no assay showed a sufficiently outstanding performance for the detection of all parasites in routine use to discard microscopic examination of stools. Although multiplex PCR techniques would be of great interest in diagnostic labs with automated platforms, more studies are needed to standardize procedures for DNA extraction, which is the critical step. Furthermore, the pathogens targeted by these assays are the most common and most pathogenic protozoa and fit with 90% of screened patients in routine diagnosis, but the sole use of these assays could lead to the non-diagnosis of other protozoa such as Cystoisospora belli, Cyclospora cayetanensis, and Blastocystis hominis. Moreover, no marketed multiplex PCR assay targets helminths, which rules out the possibility of abandoning microscopy. Nevertheless, a reliable molecular assay for protozoa detection would lighten microscopic examination of stools, as helminths are easily detected. In this context, particularly for laboratories with a strong migrant clientele, single tests with enlarged parasitic panels would be welcome.
Acknowledgments
The authors gratefully thank Professor Loïc Favennec and the Laboratory of Parasitology of Rouen University Hospital for the Cryptosporidium panel, and Roche Diagnostics and Becton Dickinson for their technical support. They declare that they have no conflicts of interest; however, Roche Diagnostics, RBioPharm, Diagenode Diagnostics and Becton Dickinson provided some of the reagents.
Cite this article as: Autier B, Belaz S, Razakandrainibe R, Gangneux JP & Robert-Gangneux F. 2018. Comparison of three commercial multiplex PCR assays for the diagnosis of intestinal protozoa. Parasite 25, 48.
References
- 1. Adams DA, Thomas KR, Jajosky RA, Foster L, Baroi G, Sharp P, Onweh DH, Schley AW, Anderson WJ, Nationally Notifiable Infectious Conditions Group. 2017. Summary of notifiable infectious diseases and conditions – United States, 2015. MMWR, Morbidity and Mortality Weekly Report, 64, 1–143. [DOI] [PubMed] [Google Scholar]
- 2. Adamska M, Leońska-Duniec A, Maciejewska A, Sawczuk M, Skotarczak B. 2010. Comparison of efficiency of various DNA extraction methods from cysts of Giardia intestinalis measured by PCR and TaqMan real time PCR. Parasite, 17, 299–305. [DOI] [PubMed] [Google Scholar]
- 3. Al-Areeqi MA, Sady H, Al-Mekhlafi HM, Anuar TS, Al-Adhroey AH, Atroosh WM, Dawaki S, Elyana FN, Nasr NA, Ithoi I, Lau Y-L, Surin J. 2017. First molecular epidemiology of Entamoeba histolytica, E. dispar and E. moshkovskii infections in Yemen: different species-specific associated risk factors. Tropical Medicine & International Health: TM & IH, 22, 493–504. [DOI] [PubMed] [Google Scholar]
- 4. ANOFEL Cryptosporidium National Network. 2010. Laboratory-based surveillance for Cryptosporidium in France, 2006-2009. Euro Surveillance: Bulletin Européen sur les Maladies Transmissibles = European Communicable Disease Bulletin, 15, 19642. [PubMed] [Google Scholar]
- 5. Bailenger J. 1966. Les méthodes diphasiques de concentration parasitaire en coprologie: Explication de leurs divergences par l’énoncé de leur principe. Annales de Parasitologie Humaine et Comparée, 41, 607–622. [PubMed] [Google Scholar]
- 6. Chan D, Barratt J, Roberts T, Phillips O, Šlapeta J, Ryan U, Marriott D, Harkness J, Ellis J, Stark D. 2016. Detection of Dientamoeba fragilis in animal faeces using species specific real time PCR assay. Veterinary Parasitology, 227, 42–47. [DOI] [PubMed] [Google Scholar]
- 7. Elwin K, Fairclough HV, Hadfield SJ, Chalmers RM. 2014. Giardia duodenalis typing from stools: a comparison of three approaches to extracting DNA, and validation of a probe-based real-time PCR typing assay. Journal of Medical Microbiology, 63, 38–44. [DOI] [PubMed] [Google Scholar]
- 8. GBD 2013 Mortality and Causes of Death Collaborators. 2015. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet, 385, 117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gotfred-Rasmussen H, Lund M, Enemark HL, Erlandsen M, Petersen E. 2016. Comparison of sensitivity and specificity of 4 methods for detection of Giardia duodenalis in feces: immunofluorescence and PCR are superior to microscopy of concentrated iodine-stained samples. Diagnostic Microbiology and Infectious Disease, 84, 187–190. [DOI] [PubMed] [Google Scholar]
- 10. Jeddi F, Piarroux R, Mary C. 2013. Application of the NucliSENS easyMAG system for nucleic acid extraction: optimization of DNA extraction for molecular diagnosis of parasitic and fungal diseases. Parasite, 20, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaisar MMM, Brienen EAT, Djuardi Y, Sartono E, Yazdanbakhsh M, Verweij JJ, Supali T, Van Lieshout L. 2017. Improved diagnosis of Trichuris trichiura by using a bead-beating procedure on ethanol preserved stool samples prior to DNA isolation and the performance of multiplex real-time PCR for intestinal parasites. Parasitology, 144, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laude A, Valot S, Desoubeaux G, Argy N, Nourrisson C, Pomares C, Machouart M, Le Govic Y, Dalle F, Botterel F, Bourgeois N, Cateau E, Leterrier M, Le Pape P, Morio F. 2016. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clinical Microbiology and Infection, 22, 190.e1–190.e8. [DOI] [PubMed] [Google Scholar]
- 13. Lloyd T, Carson J, Chan W, Pillai D. 2014. C01: RIDAGENE parasitic stool panel and xTAG Gastrointestinal pathogen panel for the detection of common intestinal parasites. Canadian Journal of Infectious Diseases & Medical Microbiology, 25, e20–e70. [Google Scholar]
- 14. Madison-Antenucci S, Relich RF, Doyle L, Espina N, Fuller D, Karchmer T, Lainesse A, Mortensen JE, Pancholi P, Veros W, Harrington SM. 2016. Multicenter evaluation of BD Max Enteric Parasite Real-Time PCR assay for detection of Giardia duodenalis, Cryptosporidium hominis, Cryptosporidium parvum, and Entamoeba histolytica. Journal of Clinical Microbiology, 54, 2681–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martens D, von Thien H, Tannich E. 2013. eP721. Evaluation of a novel RIDA GENE Parasitic Stool Panel assay for the detection of Giardia lamblia, Cryptosporidium parvum, Entamoeba histolytica and Dientamoeba fragilis in stool specimens. 23rd ECCMID.
- 16. Mary C, Chapey E, Dutoit E, Guyot K, Hasseine L, Jeddi F, Menotti J, Paraud C, Pomares C, Rabodonirina M, Rieux A, Derouin F, ANOFEL Cryptosporidium National Network. 2013. Multicentric evaluation of a new real-time PCR assay for quantification of Cryptosporidium spp. and identification of Cryptosporidium parvum and Cryptosporidium hominis. Journal of Clinical Microbiology, 51, 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parčina M, Reiter-Owona I, Mockenhaupt FP, Vojvoda V, Gahutu JB, Hoerauf A, Ignatius R. 2018. Highly sensitive and specific detection of Giardia duodenalis, Entamoeba histolytica, and Cryptosporidium spp. in human stool samples by the BD MaxTM Enteric Parasite Panel. Parasitology Research, 117, 447–451. [DOI] [PubMed] [Google Scholar]
- 18. Perry MD, Corden SA, Lewis White P. 2017. Evaluation of the BD MAX Enteric Parasite Panel for the detection of Cryptosporidium parvum/hominis, Giardia duodenalis and Entamoeba histolytica. Journal of Medical Microbiology, 66, 1118–1123. [DOI] [PubMed] [Google Scholar]
- 19. Ponce C, Kaczorowski F, Perpoint T, Miailhes P, Sigal A, Javouhey E, Gillet Y, Jacquin L, Douplat M, Tazarourte K, Potinet V, Simon B, Lavoignat A, Bonnot G, Sow F, Bienvenu A-L, Picot S. 2017. Diagnostic accuracy of loop-mediated isothermal amplification (LAMP) for screening patients with imported malaria in a non-endemic setting. Parasite, 24, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Royer TL, Gilchrist C, Kabir M, Arju T, Ralston KS, Haque R, Clark CG, Petri WA. 2012. Entamoeba bangladeshi nov. sp., Bangladesh. Emerging Infectious Diseases, 18, 1543–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shakir MJ. 2015. Evaluation of multiplex real-time PCR for detection of three diarrhea causing intestinal protozoa. Indian Journal of Applied Research, 5, 783–786. [Google Scholar]
- 22. Stark D, Barratt J, Chan D, Ellis JT. 2016. Dientamoeba fragilis, the neglected trichomonad of the human bowel. Clinical Microbiology Reviews, 29, 553–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stark D, van Hal S, Fotedar R, Butcher A, Marriott D, Ellis J, Harkness J. 2008. Comparison of stool antigen detection kits to PCR for diagnosis of amebiasis. Journal of Clinical Microbiology, 46, 1678–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van den Bossche D, Cnops L, Verschueren J, Van Esbroeck M. 2015. Comparison of four rapid diagnostic tests, ELISA, microscopy and PCR for the detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in feces. Journal of Microbiological Methods, 110, 78–84. [DOI] [PubMed] [Google Scholar]
- 25. Wong ZW, Faulder K, Robinson JL. 2018. Does Dientamoeba fragilis cause diarrhea? A systematic review. Parasitology Research, 117, 971–980. [DOI] [PubMed] [Google Scholar]