Abstract
The ATR kinase is crucial for genome maintenance, but the mechanisms by which ATR controls the DNA repair machinery are not fully understood. Here, we find that long-term chronic inhibition of ATR signaling severely impairs the ability of cells to utilize homologous recombination (HR)-mediated DNA repair. Proteomic analysis shows that chronic ATR inhibition depletes the abundance of key HR factors, suggesting that spontaneous ATR signaling enhances the capacity of cells to use HR-mediated repair by controlling the abundance of the HR machinery. Notably, ATR controls the abundance of HR factors largely via CHK1-dependent transcription, and can also promote stabilization of specific HR proteins. Cancer cells exhibit a strong dependency on ATR signaling for maintaining elevated levels of HR factors, and we propose that increased constitutive ATR signaling caused by augmented replication stress in cancer cells drives the enhanced HR capacity observed in certain tumor types. Overall, these findings define a major pro-HR function for ATR and have important implications for therapy by providing rationale for sensitizing HR-proficient cancer cells to PARP inhibitors.
INTRODUCTION
ATR (Ataxia telangiectasia and Rad3-related) is a member of the phosphatidylinositol-3-kinase-like kinase (PIKKs) family involved in genome maintenance. In response to DNA replication stress or DNA damage, ATR is activated and phosphorylates an extensive network of substrates, evoking a coordinated DNA damage response (1–3). While the related kinases ATM and DNA-PKcs are activated upon double strand breaks (DSBs), the ATR kinase specifically responds to exposure of single stranded DNA (ssDNA) resulting from a broad spectrum of DNA lesions (4). Upon replication stress or detection of replication-associated lesions, ATR is recruited to RPA-coated ssDNA and becomes activated through the action of the ATR activators TOPBP1 and ETAA1 (5–10). In response to replication stress, ATR has been shown to mediate a global cellular response that promotes cell cycle arrest, inhibition of late origin firing, stabilization of replication forks, transcriptional regulation and DNA repair (11,12). ATR kinase exerts its function in genome maintenance by targeting and phosphorylating the key effector kinase CHK1, which mediates cell cycle arrest through the phosphorylation and degradation of the CDC25 phosphatase (13–15). In addition, ATR-CHK1 signaling plays a prominent role in controlling E2F-dependent transcription (16–18), which includes a large set of genes with important roles in DNA replication, DNA repair and cell cycle control (19). Mechanistically, CHK1 has been shown to phosphorylate and inhibit the E2F6 repressor (20). Additional mechanisms may also couple ATR and CHK1 to the control of E2F-dependent transcription (16,21).
ATR also plays crucial roles in the control of DNA repair. It has been shown that ATR signaling regulates the repair of DNA interstrand cross-links and nucleotide excision repair by directly phosphorylating Fanconi Anemia (FA) or Xeroderma Pigmentosum (XP) proteins (22–24). In addition, others and we have recently proposed roles for ATR in homologous recombination (HR)-mediated repair (25–27), a crucial pathway to repair DSBs. Of note, HR-mediated repair occurs preferably during S/G2 phase of the cell cycle since sister chromatids can be used as a template for error-free DNA repair (28–30). As an alternative to HR, cells may repair DSBs using non-homologous end joining (NHEJ), which is relatively less favored in S/G2 as compared to in the G1 phase of the cell cycle (30,31). Since the improper use of NHEJ in S phase leads to chromosomal aberrations and decreased survival (32,33), balanced engagement of HR and NHEJ repair pathways is essential for maintaining genomic integrity. Recently, ATR was shown to promote HR by phosphorylating PALB2 and enhancing its localization to DNA lesions via an interaction with BRCA1 (26). Additionally, we proposed that ATR mediates BRCA1 phosphorylation and its interaction with TOPBP1 to promote HR by stabilizing BRCA1 at lesions during S-phase (25). Therefore, ATR seems to play a key role in promoting HR-mediated repair and suppressing improper NHEJ during replication stress.
The physiological importance of ATR is highlighted by the fact that mice lacking functional ATR are embryonic lethal (34,35). Also, homozygous mutations in human ATR that cause defective mRNA splicing and severely reduced ATR expression are associated with Seckel syndrome, a genetic disorder characterized by growth defect (dwarfism), microcephaly and mental retardation (36). Notably, Seckel syndrome cells show high genomic instability and increased micronuclei formation (37,38), supporting the role of ATR in genome integrity.
In the context of cancer, ATR is believed to be crucial for the ability of many cancer types to withstand the increased levels of replication stress generated by oncogene-induced de-regulation of DNA replication (18,39–42). While the inhibition of ATR activity leads to moderate cytotoxicity in normal cells due to increased fork stalling and collapse, this cytotoxicity is further exacerbated in cancer cells with higher replication stress, providing rationale for using ATR inhibitors (ATRi) in cancer treatment (43,44). Cancer cells frequently bear mutations in components of DNA damage response pathways, leading to increased dependency on ATR signaling (45). Consistent with this notion, it has been shown that inhibition of ATR kinase activity is synthetic lethal in tumor cells that have mutations in ATM, p53, ERCC1 and XRCC1 (46–52). Therefore, specific inhibition of ATR signaling is expected to selectively kill cancer cells with genetic defects in DNA damage response pathways and/or elevated oncogene-induced replication stress. Accordingly, in the last eight years, highly selective and potent ATR inhibitors have been developed and are currently under phase I/II clinical trials in cancer treatment (39,45).
Despite extensive work establishing ATR inhibitors as potential anti-cancer agents, it is not yet well understood precisely how ATR inhibition impacts DNA repair in the context of normal and cancer cells. As described above, ATR has been shown to control HR-mediated repair, however, the effects of short-term treatment with ATR inhibitors on HR are rather modest. Here we find that chronic ATR inhibition severely impairs the ability of cancer cells to utilize HR-mediated repair, leading to an effect that is significantly stronger when compared to acute ATR inhibition. Proteomic analysis reveals that chronic, but not acute, ATR inhibition depletes the abundance of key components of the HR machinery, including TOPBP1, BRCA1 and RAD51. We show that ATR-mediated control of HR factor abundance involves transcription and post-translational control mechanisms. Interestingly, cancer cells seem to exhibit a stronger dependency on ATR signaling for maintaining the levels of HR factors, providing rationale for the use of ATR inhibitors in combination with drugs known to preferentially target HR-deficient cells for anti-cancer therapy.
MATERIALS and METHODS
Mammalian cell culture
Human U2OS, HCT116, hTERT RPE-1, HEK293T, HeLa, U87, T98G, A2780 and human skin fibroblast cells (GM08398 and GM18366) were grown in DMEM media supplemented with 10% bovine calf serum, penicillin/streptomycin and non-essential amino acids. SV40 large T antigen transformed hTERT RPE-1 cells were generated by infecting hTERT RPE-1 cells with a SV40 large T antigen (SV40 LT)-expressing retrovirus and were selected using 500 μg/ml G418. The stable SV40 large T antigen transformed hTERT RPE-1 cell line was then maintained in culture media supplemented 500 μg/ml G418. For chronic treatment with ATR inhibitor (ATRi, VE-821) or ATM inhibitor (ATMi, KU-55933), cells were maintained in media with 5 μM ATR or ATM inhibitor respectively for 8 days before cells were subjected to western blot analysis, proteomic analysis, DR-GFP/EJ5-GFP assay or immunofluorescent staining. For CHK1 inhibitor (UCN-01), cells were treated for 5 days with 0.2 μM of inhibitor before the analysis. Primary human fibroblast cells from control subjects (GM08398) and patients with Seckel syndrome (GM18366, ATR-defective) were obtained from Coriell Institute for Medical Research, Camden, NJ, USA.
DR-GFP and EJ5-GFP assays
For the DR-GFP assay, cells (U2OS, HCT116, hTERT RPE-1, HEK293T, HeLa, U87, T98G, A2780, 293-T-REX-E2F6 cells and SV40 large T antigen transformed hTERT RPE-1) were transfected with the mCherry plasmid or DR-GFP reporter plasmid (pDR-GFP; addgene plasmid 26475) together with a plasmid coding for I-SceI (pCBASceI; addgene plasmid 26477; gifts from M. Jasin) or DR-GFP reporter plasmid together with an empty plasmid pCAGGS. Two days after transfection, cells were trypsinized, resuspended in PBS and then analyzed by flow cytometry using FACSAria Fusion or Accuri C6 cytometer (BD). In each experiment, the percentage of GFP-positive cells from the sample transfected with empty plasmid pCAGGS was subtracted from the percentage of GFP-positive cells in the sample transfected with I-SceI. The GFP percentage was then normalized by the mCherry transfection efficiency. Each graph was plotted based on at least three independent experiments showing mean ± SEM (N ≥ 3). For the EJ5-GFP assay, EJ5-GFP reporter plasmid (pimEJ5GFP, addgene plasmid 44026; gift from J. Stark) was used instead of DR-GFP reporter plasmid and the assay was performed as described for DR-GFP assay.
Immunofluorescence
Primary human fibroblast cells were grown on glass coverslips and irradiated at 10 Gy. Cells were then fixed with 3.7% formaldehyde/PBS for 15 min at room temperature 8 h post-irradiation. Fixed cells were then washed with PBS, permeabilized with 0.2% Triton X-100 for 5 min at room temperature and blocked with 5% BSA at 37°C for 30 min. Blocked samples were incubated with primary antibodies for 1 h at room temperature, followed by three PBS washes and secondary antibody incubation using Alexa Fluor488 donkey anti-rabbit (A-21206; Thermo Fisher Scientific) for 1 h. After secondary antibody incubation, cells were washed in PBS three times and mounted onto glass microscope slides using Vectashield mounting media (H1200; Vector Laboratory).
Microscopy analysis
Fixed cells were imaged at room temperature using a CSU-X spinning disc confocal microscope (Intelligent Imaging Innovations) on an inverted microscope (DMI600B; Leica), with 63×, 1.4 NA objective lens and a charge-coupled device camera (cool-SNAP HQ2, Photometrics). Z stack images were obtained and saved in Slidebook software (Intelligent, Imaging, Innovations), where maximum intensity projections were created. For RAD51 and BRCA1 foci analysis in control skin fibroblast and Seckel cells, >150 cells for each sample were imaged and analyzed per replicate. The fraction of cells with more than 10 distinct RAD51 foci or more than 5 BRCA1 foci were determined for each replicate. The graph was plotted using the arithmetic mean and SEM derived from four independent biological replicates. A two-tailed Student's t test with 95% confidence interval was used for the statistical analysis.
Immunoblotting analysis
Cells were harvested and lysed in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% tergitol, 0.25% sodium deoxycholate, 5 mM EDTA) supplemented with Complete EDTA-free protease inhibitor cocktail (Roche), 1 mM PMSF, 5 mM sodium fluoride, 10 mM β-glycerol-phosphate and 0.4 mM sodium orthovanadate. Whole cell lysates were cleared with 5 min centrifugation at 13 000 rpm at 4°C and mixed with 3× SDS sample buffer (bromophenol blue, stacking gel buffer, 50% glycerol, 3% SDS and 60 mM DTT). After resolved on SDS-PAGE gels, proteins were then transferred onto polyvinylidene difluoride (PVDF) membranes and detected with designated antibodies.
Proteomic analysis
For quantitative proteomic analysis, U2OS cells chronically treated with DMSO or ATR inhibitor were cultured respectively in ‘light’ or ‘heavy’ SILAC DMEM media (ThermoFisher Scientific 88425) supplemented with 10% dialyzed FBS and penicillin/streptomycin. ‘Light’ SILAC media were supplemented with ‘light’ (normal) arginine 12C6, 14N2 and lysine 12C6, 14N4, while ‘heavy’ SILAC media contained ‘heavy’ lysine 13C6, 15N2 and ‘heavy’ arginine 13C6, 15N4. From SILAC cells chronically treated with DMSO or ATR inhibitor, nuclei were isolated using hypotonic buffer and further lysed in modified RIPA buffer. The nuclear lysates were cleared by centrifugation for 5 min at 4 °C. The nuclear proteins were reduced, alkylated, precipitated and trypsin-digested. The peptides were then desalted, dried and resuspended in 80% acetonitrile and 1% formic acid and then fractionated using Hydrophilic Interaction Chromatography (HILIC). HILIC fractions were dried and reconstituted in 0.1% trifluoroacetic acid and analyzed using a Q-Exactive Orbitrap and Lumos mass spectrometer. Database search and quantitation of heavy/light peptide isotope ratios were performed using Sorcerer as previously described (53,54). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Proteomics Identifications) (55) partner repository with the data set identifier PXD010223.
Chemicals and antibodies
The following chemicals were used in the study: ATR inhibitor (VE-821) and ATM inhibitor (KU-55933) from Selleckchem, CHK1 inhibitor (UCN-01) from Sigma, Geneticin (G418 sulfate) from Thermo Fisher Scientific, and Cycloheximide (CHX) from MP Bio. The following antibodies were used in western blot analysis and immunofluorescent staining: anti-53BP1 and anti-FANCJ from Novus Biologicals, anti-phospho-CHK1 (Ser345) from Cell Signaling, anti-RAD51 from Millipore, anti-CHK1, anti-E2F6 and anti-E2F1 from Santa Cruz Biotechnology, anti-Actin and anti-Tubulin from Sigma, ECL HRP-linked secondary antibody from GE healthcare. Rabbit polyclonal anti-TOPBP1 and anti-BRCA1 antibodies were home-raised as previously described (56,57). Anti-RRM2 serum was raised in rabbit against an immunogen containing the first 250 amino acids of human RRM2. Dr. Kai Ge provided the antibody against PTIP.
Cell cycle analysis
Cells were harvested by standard trypsinization and fixed with 70% ethanol. After fixation, cells from each sample were resuspended in PBS, permeabilized using 0.1% Triton X-100 and treated with RNase A (0.8 mg/ml). Then cells were stained with propidium iodide and analyzed by flow cytometry using FACSAria Fusion (BD) or Accuri C6 cytometer (BD) to determine the fraction of cells in different stages of the cell cycle.
Real time PCR
Cells cultured from 10 cm dish were collected and lysed in 1 ml of TRIzol (Invitrogen). Chloroform was added to the lysed sample to separate RNA. Separated RNA was precipitated by isopropyl alcohol, followed by two washes with 75% ethanol. 1 μg of extracted RNA from each sample was then reverse transcribed with Bio-Rad iScript cDNA synthesis kit. The samples (∼100 ng) were analyzed in a Roche 480 light cycler in a total of 15 μl containing SYBR green mix and primers indicated in the figure. The values were calculated using ‘absolute quantification/2nd derivative max’ in lightcycler 480 program and then normalized to the level of GAPDH. Primers used in the study are as follows. BRCA1- F:ACCTTGGAACTGTGAGAACTCT, R:TCTTGATCTCCCACACTGCAATA, RAD51- F:CAACCCATTTCACGGTTAGAGC, R:TTCTTTGGCGCATAGGCAACA, 53BP1- F:ATGGACCCTACTGGAAGTCAG, R:TTTCTTTGTGCGTCTGGAGATT, GAPDH- F:TCATTGACCTCAACTACATGGTTT, R:GGAAGATGGTGATGGGATTTC.
Crystal violet staining survival assay
First, hTERT RPE-1, U2OS, HCT116, HeLa, A2780 or U87 cells were treated for 5 days with either DMSO or 5 μM ATR inhibitor. Next, 1 × 105 cells of each cell type were plated onto a 10 cm dish and further treated with DMSO, ATR inhibitor or/and PARP inhibitor for additional 3 days. Cells were then washed with PBS and either fixed in 100% methanol for 15 min at 4°C or fixed after additional 2 or 8 days of culture in drug-free media. Fixed plates were stained with 0.1% crystal violet solution overnight followed by a distilled water wash. Pictures were taken after the plates were dried. For quantitation of cell viability, total viable cells were counted using a hematocytometer after indicated treatment.
RESULTS
Chronic ATR inhibition severely impairs HR-mediated repair
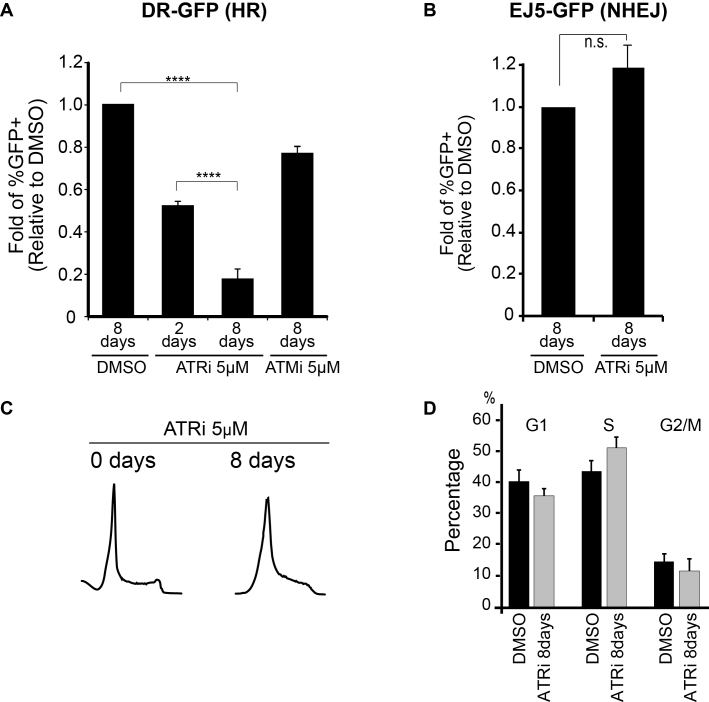
Despite extensive work investigating the roles of ATR in genome maintenance and establishing inhibitors as potential anti-cancer agents, the extent to which ATR inhibition impacts DNA repair is not well understood. ATR has been shown to control HR-mediated repair, in part, by promoting interactions between HR factors, such as BRCA1 interactions with PALB2 and TOPBP1 (25,26). Here, using the DR-GFP reporter system (58), we found that long-term ATR inhibition (8 days) in U2OS cells leads to a strikingly more severe impairment in HR-mediated repair as compared to short-term ATR inhibition (Figure 1A and Supplementary Figure S1), and hypothesized that ATR also controls HR through mechanisms other than the regulation of protein-protein interactions. The concentration of ATR inhibitor (ATRi) used in these experiments (5 μM) resulted in almost complete ATR inhibition, as monitored by CHK1 phosphorylation in HU-treated cells (Supplementary Figure S2). Of note, chronic ATM inhibition did not result in major changes in HR-mediated repair efficiency (Figure 1A), consistent with previous reports (59,60). Analysis using the EJ5-GFP (NHEJ) assay (61) revealed that chronic ATR inhibition does not result in a significant change in NHEJ efficiency (Figure 1B and Supplementary Figure S3), supporting that after 8 days treatment with 5 μM of ATRi, cells are still able to utilize NHEJ-mediated repair. Furthermore, the severe reduction in HR-mediated repair upon chronic ATR inhibition was not due to major changes in the cell cycle, as it results in a minor increase in the number of S-phase cells upon chronic ATR inhibition (Figure 1C and D). Of note, DNA damage was not detectable in cells undergoing chronic ATR inhibition alone. Taken together, these findings are consistent with a pro-HR-mediated repair function for ATR. Since ATR-mediated protein-protein interactions are expected to be impaired by short-term treatment, our results suggest that the severe effect of long-term ATR inhibition on HR-mediated repair occurs through alternative mechanisms.
Figure 1.
Chronic inhibition of ATR impairs HR-mediated repair in U2OS cells. (A) HR efficiency measured in U2OS cells using the DR-GFP reporter system. Relative fold change of GFP positive cells in ATRi or ATMi treated cells over DMSO (error bars = SEM, N ≥ 5, ****P-value < 0.0001; one sample-t-test was used for the statistical analysis). (B) NHEJ efficiency measured in U2OS cells using the EJ5-GFP reporter system. Relative fold change of GFP positive cells in ATRi or ATMi treated cells over DMSO (error bars = SEM, N = 5, one sample-t-test was used for the statistical analysis). (C) Representative FACS data and (D) quantitation of G1, S and G2/M population for cell cycle analysis of ATRi treated cells (error bars = SD, N = 3).
Chronic ATR inhibition severely reduces the abundance of HR proteins
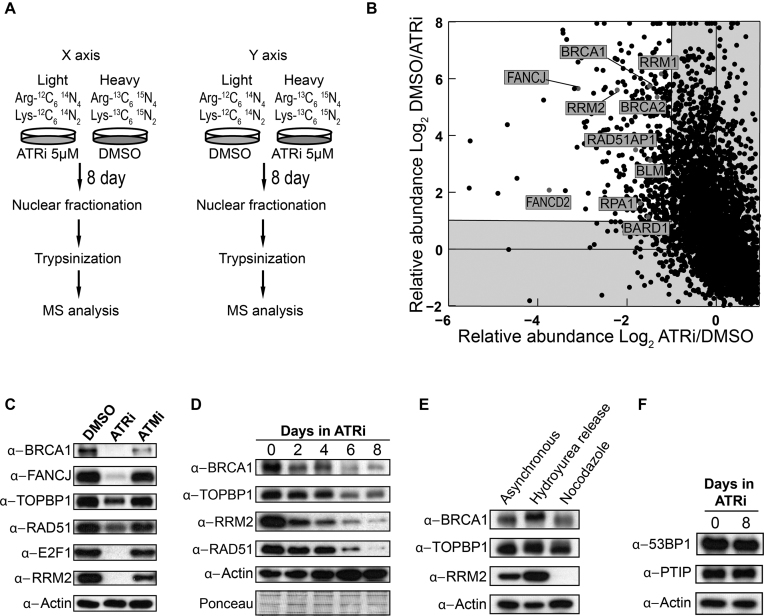
We hypothesized that chronic ATR inhibition might strongly impair HR-mediated repair by altering the abundance of proteins required for HR. While the ATR-CHK1 pathway has been shown to control gene expression (16,18,20,62,63), little is understood about the impact of ATR inhibition in shaping DNA repair mechanisms via control of protein abundance. To test the above hypothesis, we performed proteomic analysis of nuclear proteins extracted from U2OS cells, comparing the nuclear proteome of untreated cells with the nuclear proteome of cells treated for 8 days with 5 μM of ATRi. We used stable isotope labeling with amino acids in cell culture (SILAC) and performed two experiments inverting the light and heavy isotopes (Figure 2A). Consistent with our hypothesis, we found that the abundance of proteins required for HR-mediated repair, including BRCA1 and FANCJ, was reduced in cells treated chronically with ATRi (Figure 2B and Supplementary Table S1). Notably, BRCA1 and FANCJ expression is regulated by E2F (64,65), which in turn can be controlled by the ATR-CHK1 pathway (16,20). Gene Ontology-analysis of the proteomic dataset revealed that the majority of the proteins whose abundance is reduced upon chronic ATR inhibition belong to categories associated with DNA replication and repair (Supplementary Table S2 and Supplementary Figure S4). Down-regulated proteins include many known E2F targets, such as RRM2, RFCs, MCMs and POLE. Western blot (WB) analysis validated these findings (Figure 2C). Moreover, we found that protein levels of RAD51 and TOPBP1, two other E2F targets required for HR-mediated repair (25,66), were also reduced upon chronic ATR inhibition (Figure 2C). Time-course analysis revealed that minor changes in protein abundance could be observed after 2 and 4 days of continuous treatment with ATRi, and that the changes become more striking after 6 and 8 days of chronic drug treatment (Figure 2D). Taken together, these findings suggest a model that ATR signaling occurring physiologically during every cell cycle is important to maintain the abundance of key proteins required for HR-mediated repair.
Figure 2.
Chronic inhibition of ATR depletes the abundance of key HR factors. (A) Diagram of the SILAC/MS experiment. (B) Quadrant diagram of SILAC/MS experiment. Each dot represents a different identified protein. Proteins with abundance reduced two-fold or more after ATRi treatment are shown in the upper-left corner (white quadrant). (C) WB analysis of U2OS cells after 8 days of chronic treatment with 5 μM of ATRi or ATMi. (D) Time course WB analysis of U2OS cells during chronic treatment with 5 μM ATRi. (E) WB analysis of U2OS cells at distinct cell cycle stages. Cells were treated with 1 mM hydroxyurea (HU) or 50 ng/ml nocodazole (Noc) for 24 h. HU treated cells were released for 2.5 h in HU-free media. (F) WB analysis of U2OS cells untreated or treated for 8 days with 5 μM ATRi.
The observed changes in protein abundance are not due to cell cycle changes since chronic treatment with 5 μM of ATRi leads only to minor changes in cell cycle distribution, as seen by the slight increase in S phase cells (Figure 1C). Importantly, the reduced expression of HR factors upon chronic ATRi treatment cannot be attributed to such slight increase in S phase cells since cells in S phase actually express more HR factors and E2F targets (see Figure 2E and Supplementary Figure S5). Moreover, while the abundance of several proteins involved in HR-mediated repair is severely reduced upon chronic ATR inhibition, changes in the abundance of the NHEJ factors 53BP1 and PTIP were minimal (Figure 2F). This finding is consistent with our result using the EJ5-GFP assay showing that chronic ATR inhibition does not impair NHEJ efficiency (Figure 1B). Collectively, the findings above show that chronic ATR inhibition has a major impact on the abundance of key proteins required for HR-mediated repair, which is correlated with the severe reduction in HR-mediated repair efficiency observed in U2OS cells.
To investigate whether the role of ATR in maintaining the abundance of HR-mediated repair factors is consistently observed in a physiological setting, we analyzed a Seckel syndrome patient cell line containing the A210G mutation in ATR that generates a splicing defect (36). Analysis of a primary fibroblast cell line GM18366, which has attenuated ATR signaling (3,36), revealed reduced abundance of HR-mediated repair factors compared to control fibroblasts (Supplementary Figure S6A). Although we were not able to detect BRCA1 in WB for technical reasons, we found that the abundance of FANCJ, TOPBP1 and RAD51 is decreased. This Seckel cell line did not show any major differences in cell cycle distribution, consistent with a previous report (67), indicating that the abundance changes are not due to effects on the cell cycle (Supplementary Figure S6B). To examine the ability of Seckel cells to utilize HR-mediated repair, we imaged BRCA1 and RAD51 foci after irradiation (IR) treatment and observed a significant reduction in RAD51 and BRCA1 foci formation post IR (Supplementary Figure S6C and D). The results above reveal similarities between these Seckel cells with the U2OS cells chronically treated with ATRi. In both cases, the abundance of important HR-mediated repair factors is reduced and markers of HR-mediated repair are impaired (37,38). Overall, these findings support a key role for ATR in maintaining the abundance of HR-mediated repair factors and suggest that the severe impairment in HR-mediated repair upon chronic ATR inhibition is caused by depletion of key components of the HR-mediated repair machinery.
ATR controls HR factor abundance through CHK1-mediated transcription
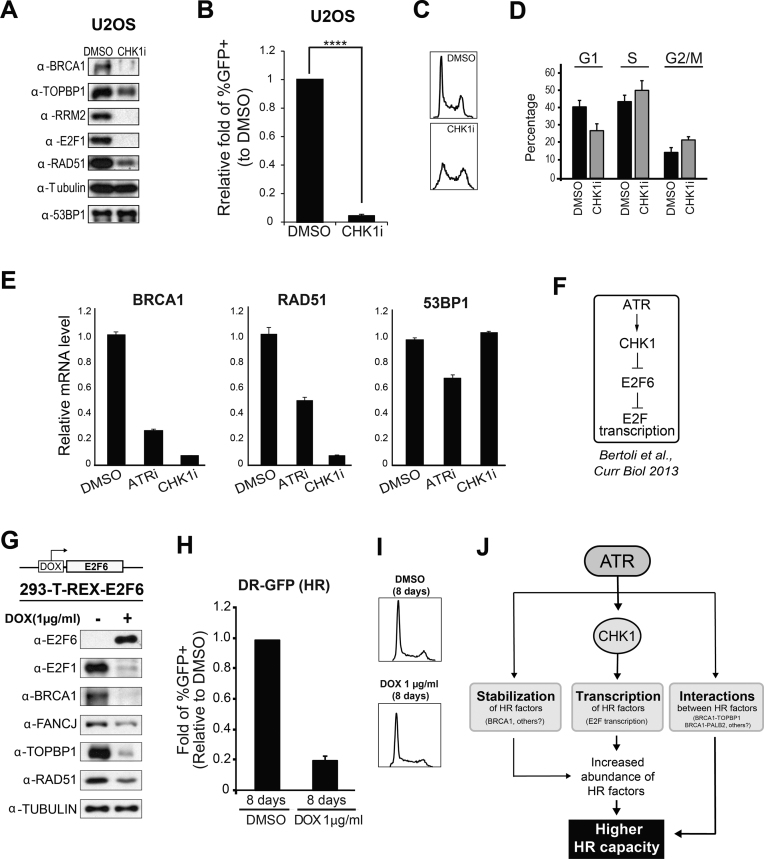
ATR has been shown to control the transcription of E2F targets via activation of the downstream checkpoint kinase CHK1 (20,26,68). We therefore examined if chronic CHK1 inhibition results in a similar depletion of HR factors as we observed after chronic ATR inhibition. As shown in Figure 3A, chronic CHK1 inhibition severely reduced the abundance of the HR factors BRCA1 and RAD51, as well as the abundance of the canonical E2F target RRM2, and E2F1 itself. The depletion of HR factors after CHK1 inhibition was correlated with a sharp decrease in HR efficiency (Figure 3B and Supplementary Figure S7), and both effects were not a consequence of a reduction in the number of S phase cells since CHK1 inhibition actually results in an increase in the relative number of S phase cells (Figure 3C and D). Of importance, CHK1 as well as ATR inhibition results in a reduction in the mRNA levels of BRCA1 and RAD51 (Figure 3E). Taken together, these results are consistent with the model that ATR controls the transcription of key HR factors through the ATR-CHK1 pathway. Notably, we did not observe major changes in the mRNA levels of 53BP1 (Figure 3E), suggesting that ATR inhibition preferentially depletes factors involved in HR, while not changing the protein machinery required for NHEJ, and therefore not impairing NHEJ-mediated repair (Figure 1B).
Figure 3.
ATR controls HR factor abundance via CHK1-mediated transcription. (A) WB analysis of U2OS cells after chronic treatment with CHK1 inhibitor (0.2 μM for 5 days). (B) Analysis of HR-mediated repair using the DR-GFP system. Relative fold change of GFP positive U2OS cells treated with CHK1i compared to DMSO treatment control (error bars = SEM, N = 4, ****P-value < 0.0001; one sample-t-test was used for the statistical analysis). (C) Representative FACS data and (D) quantitation of G1, S and G2/M population for cell cycle analysis of CHK1i treated cells (error bars = SD, N = 3). (E) Real time PCR analysis of total RNA extracted from U2OS cells treated with DMSO, ATRi (5 μM for 8 days) or CHK1i (0.2 μM for 5 days). Values were normalized to GAPDH mRNA levels (error bars = SD, N = 3). (F) Proposed model of how E2F transcription is regulated through ATR-CHK1 signaling. (G) WB analysis of 293-T-REX-E2F6 cells after 8 days of treatment with DMSO or doxycycline. (H) DR-GFP analysis of 293-T-REX-E2F6 cells after 8 days of treatment with DMSO or doxycycline (error bars = SEM, N = 4). (I) Cell cycle analysis of 293-T-REX-E2F6 cells after 8 days of treatment with DMSO or doxycycline. (J) A model depicting multiple roles of ATR in the control of HR capacity.
Mechanistically, ATR-CHK1 signaling has been shown to control E2F transcription by inhibiting the E2F6 repressor (Figure 3F) (20). We therefore tested whether chronic overexpression of E2F6 leads to the same effect as ATR inhibition on the HR machinery. As shown in Figure 3G and H, we used a 293-T-REX-E2F6 system for doxycycline-induced expression of E2F6 and found that, as expected, overexpression of E2F6 does result in severe reduction in HR factor abundance and HR capacity. Congruent with these effects not being consequences of differences in cell cycle distribution, we did not observe any substantial change in the cell cycle upon overexpression of E2F6 (Figure 3I). Of note, we also examined if the stability of HR proteins can be affected independently of translational control. We treated U2OS cells with the translation inhibitor cycloheximide (CHX) and asked if ATR inhibition results in changes in the abundance of some HR factors. As shown in Supplementary Figure S8, we noticed that the protein levels of BRCA1 are decreased upon CHX treatment and ATR inhibition compared to CHX treatment alone, suggesting that ATR signaling promotes BRCA1 protein stabilization. In U2OS cells, such transcription-independent mode of protein stabilization was only observed for BRCA1 as the abundance of the other HR proteins monitored did not change upon ATR inhibition in CHX-treated cells.
Overall, our findings indicate that ATR controls the abundance of HR proteins through the control of transcription and, in the case of BRCA1, also through the control of protein stability (Figure 3J). Since BRCA1 has been proposed to be a target of ATR (25,26,69), we speculate that ATR-mediated phosphorylation sites in BRCA1 promote BRCA1 stabilization. Overall, we propose that ATR plays multiple pro-HR roles by controlling the abundance of a range of HR proteins and interactions between them (Figure 3J).
Correlation of HR factor abundance and HR capacity
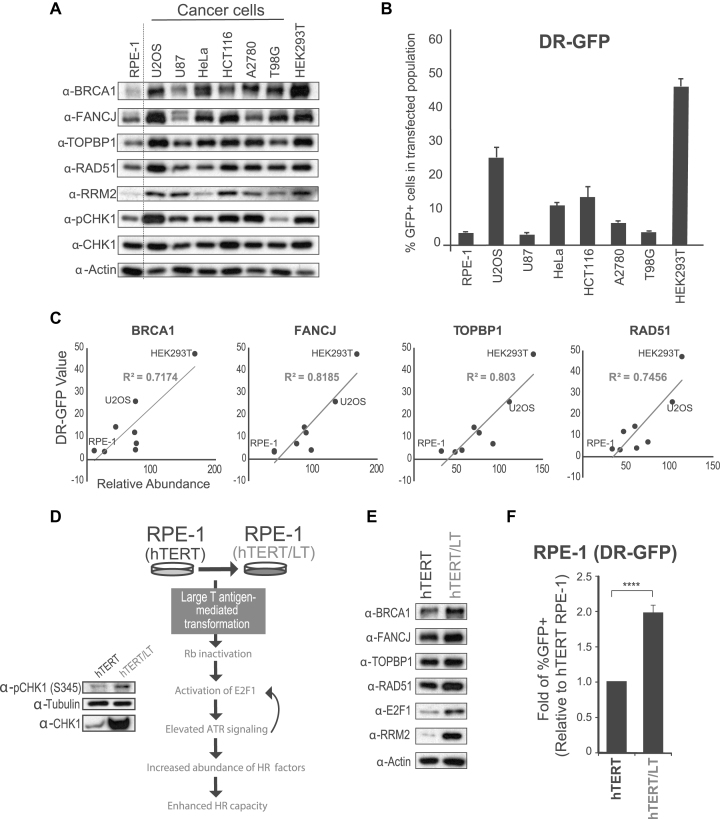
To explore the hypothesis that HR factor abundance is a major determinant of HR capacity, we monitored the abundance of selected HR factors (BRCA1, FANCJ, RAD51 and TOPBP1) and HR capacity in a panel of cell lines. As shown in Figure 4A, most cancer cells tested had higher levels of the HR factors compared to untransformed hTERT RPE-1 cells, consistent with the fact that increased CHK1 signaling, RB mutation or E2F1 overexpression are often correlated with cancer (70–73). Analysis of HR using DR-GFP assay revealed major differences in HR capacity in the cells monitored (Figure 4B). While hTERT RPE-1 cells had the lowest level of HR factors and displayed the lowest HR capacity, HEK293T cells had the highest level of HR factors and displayed the highest HR capacity (Figure 4A and B). Overall, there was a reasonable correlation between HR capacity and the abundance of HR factors (Figure 4C). These findings are consistent with a previous report showing that untransformed hESC have lower HR efficiency than cancer cells (74).
Figure 4.
Correlation between HR factor abundance and HR-mediated repair. (A) WB analysis of HR factor abundance in a panel of cell lines. (B) Analysis of HR-mediated repair using the DR-GFP system. Relative fold change of GFP positive population from different cell lines (error bars = SEM, N ≥ 3). (C) R2 correlation between protein abundance and DR-GFP level. Abundance of each protein was measured by densitometry and normalized to corresponding abundance of actin. (D) Diagram of the experimental setup and rationale for using LT transformation to induce increased replication stress, HR factor abundance and HR capacity. (E) WB analysis of hTERT RPE-1 and LT transformed hTERT RPE-1 cells. (F) Analysis of HR-mediated repair using the DR-GFP system. Relative fold change of GFP positive hTERT RPE-1 cells over LT transformed hTERT RPE-1 cells (error bars = SEM, N ≥ 3, ****P-value < 0.0001; one sample-t-test was used for the statistical analysis).
Consistent with the hypothesis that E2F transcription impacts the level of HR-mediated repair, hTERT RPE-1 cells transformed with Large T (LT) antigen (Figure 4D), which was previously shown to result in high expression of E2F targets via impairment of RB function (75,76), resulted in increased HR factor abundance and HR capacity (Figure 4E and F). It has also been reported that LT antigen transformation and E2F1 overexpression can activate the ATR/ATM pathway (77–79), and as expected, phospho-CHK1 levels increased upon LT transformation, suggesting that ATR is more active in LT transformed cells (Figure 4D). LT transformation did not result in major cell cycle changes, confirming that the protein abundance changes are not a cell cycle effect (Supplementary Figure S9). Taken together, these findings are consistent with the model that HR factor abundance is a key determinant of HR capacity and further supports the importance of the role of ATR signaling in controlling HR capacity via modulation of HR factor abundance.
Cancer cell lines heavily rely on ATR to sustain the abundance of HR factors
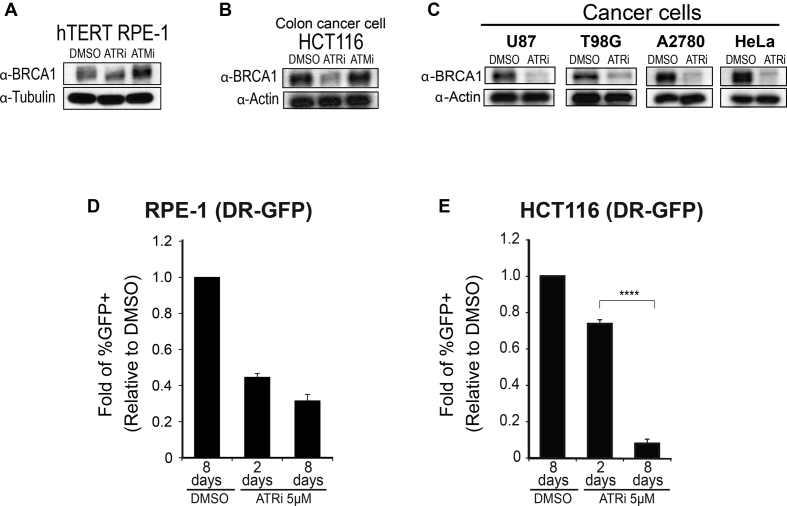
Many cancer cells are known to have de-regulated DNA replication and, as a consequence, higher levels of replication stress (80–82). Since ATR signaling is responsive to intrinsic levels of replication stress, we reasoned that higher levels of basal ATR signaling in cancer cells result in increased abundance of HR factors and higher HR capacity. Consistent with this model, it was recently proposed that cells become addicted to E2F transcription to cope with high levels of replication stress (18). As shown in Figure 4A, phospho-CHK1 and the abundance of key HR proteins were indeed higher in a panel of cancer cell lines compared to an hTERT RPE-1 cell typically used as a non-cancer cell line reference. We predicted that these cancer cell lines would be highly dependent on ATR signaling to maintain the abundance of HR proteins. Indeed, while chronic ATRi treatment did not result in major changes in the abundance of BRCA1 in hTERT RPE-1 cells (Figure 5A), severe reduction in the abundance of BRCA1 was found in cancer cells (Figure 5B and C). Consistent with this finding, chronic ATR inhibition in hTERT RPE-1 cells did not result in a striking drop in HR-mediated repair efficiency compared to short-term (2 day) treatment (Figure 5D and Supplementary Figure S10), whereas in the HCT116 colon cancer cell line, the effect of chronic ATR inhibition was significantly more severe than that of short-term treatment (Figure 5E and Supplementary Figure S11). Taken together, the results reveal cell type-specific responses to long-term ATR inhibition, with cancer cells often displaying stronger dependency on ATR signaling for sustaining the abundance of HR factors. The sharp correlation between observed changes in HR protein abundance and measured changes in HR-mediated repair efficiency strongly suggests that reduction in HR factor abundance is the major cause of the reduction in HR-mediated repair efficiency.
Figure 5.
Cancer cell lines display high dependency on ATR signaling for sustaining the abundance of HR factors and HR-mediated repair. (A and B) WB analysis of hTERT RPE-1 or HCT116 (colon cancer) cells after 8 day chronic treatment with 5 μM ATRi or ATMi. (C) WB analysis of indicated cancer cell lines upon 8 day chronic treatment with 5 μM ATRi. (D and E) Analysis of HR-mediated repair using the DR-GFP system. Relative fold change of GFP positive hTERT RPE-1 or HCT116 cells over DMSO after ATRi treatment (error bars = SEM, N≥3, ****P-value < 0.0001; one sample-t-test was used for the statistical analysis comparing 2 days versus 8 days of treatment).
Rationale for synergistic sensitivity of cancer cells to combined ATR and PARP inhibition
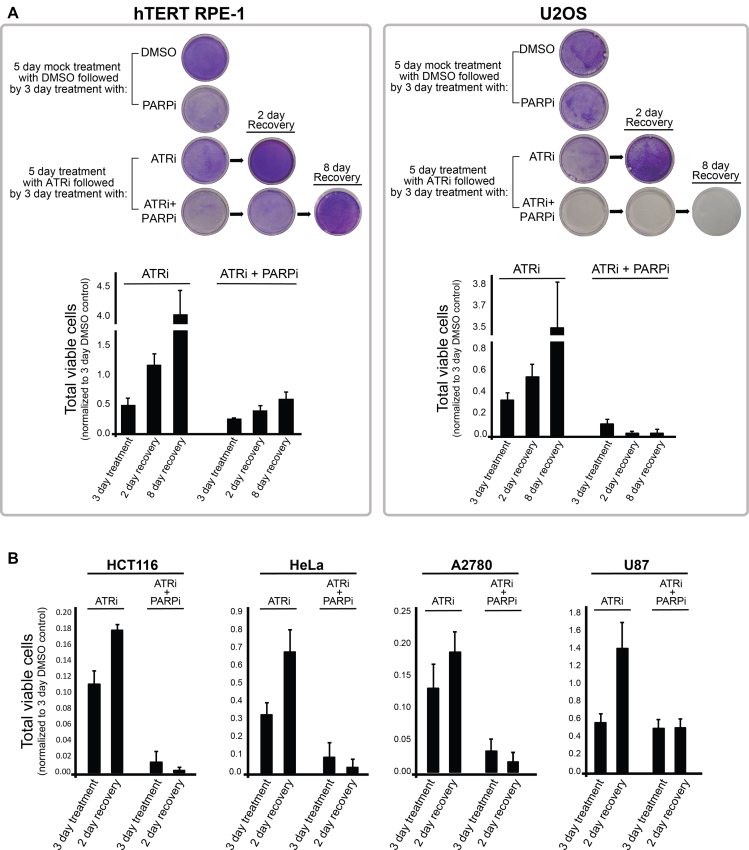
Our results reveal that chronic ATR inhibition mimics ‘BRCAness’, a term used to define a state of cancer cell lines with dysfunctional BRCA1 or BRCA2, and therefore, dysfunctional HR-mediated repair (83). Of note, BRCAness cells with BRCA1 or BRCA2 mutations are highly sensitive to PARP inhibitors (84–87), which are clinically effective drugs under FDA approval for ovarian and breast cancer treatment. We therefore reasoned that long-term treatment with ATRi could be used to hypersensitize HR-proficient cancer cells to PARP inhibitors. Based on our rationale, cells should first undergo a long-term treatment with a low dose of ATRi to deplete key HR components (such low dose would have a minor effect on non-cancer cells), and PARP inhibitor should only be added after this first treatment phase. In this manner, depletion of the HR machinery in HR-proficient cancer cells will make them particularly sensitive to a second treatment phase in which PARP inhibitors are also added. To test the efficacy of this strategy, we subjected hTERT RPE-1 and HR-proficient U2OS cancer cells to 5 days of chronic ATRi treatment. We then treated these cells with ATR and/or PARP inhibitors for an additional 3 days (Figure 6A). Crystal violet cell staining assay showed that hTERT RPE-1 cells did not show major changes after inhibitor treatment, whereas U2OS cancer cells showed dramatic loss of viability upon combined treatment with ATR and PARP inhibitors. Strikingly, U2OS cells treated only with ATRi for 8 days recovered after switching to drug-free media, whereas U2OS cells undergoing the full treatment regime including the combination of ATRi and PARPi, could not recover. Interestingly, other cancer cells that have relatively higher HR capacity compared to hTERT RPE-1 cells (see Figure 4B), including HeLa, HCT116 and A2780, also did not recover well after the full ATRi/PARPi combination treatment (Figure 6B and Supplementary Figure S12). We note that U87 cancer cells did not recover as well as hTERT-RPE-1 cells, but for reasons that remain unknown, seem not as impacted as the other cancer cells by the combined ATRi/PARPi treatment. Overall, the described findings are consistent with previous reports that showed synthetic lethality of ATR and PARP inhibition in cancer cells and tumors (46,88). Collectively, our work provides the mechanistic explanation for why the ATRi/PARPi combination can be so effective at sensitizing HR-proficient cancer cells.
Figure 6.
Chronic treatment with ATR inhibitor hypersensitizes several HR-proficient cancer cells, but not hTERT RPE-1 cells, to PARP inhibition. (A) Cell viability analysis for testing the synergistic effect of ATRi and PARPi following chronic treatment with ATRi. First, cells were treated for 5 days with either DMSO or 5 μM ATR inhibitor. Next, 1 × 105 cells of each cell type were plated onto a 10 cm dish and further treated with DMSO, ATR inhibitor and/or PARP inhibitor for additional 3 days. Cells were either fixed or allowed to recover after additional 2 or 8 days in drug-free media. Crystal violet stainings of representative results are shown at the top and quantitation of viable cells from multiple experiments is shown at the bottom (error bars = SD, N = 3). (B) Indicated cancer cell lines were subjected to the same experimental protocol described in A. These cells did not display noticeable sensitivity to treatment with PARP inhibitor only (Supplementary Figure S12A).
DISCUSSION
The work presented here reveals a major role for ATR signaling in modulating HR capacity by controlling the abundance of the recombination machinery. Our results have important implications to understand how cancer cells acquire enhanced HR capacity. Furthermore, the reported findings provide rationale for the therapeutic use of ATR inhibitors in sensitizing HR-proficient cancer cells to drugs known to target HR-deficient cancer cells.
A model for the control of HR capacity via ATR signaling
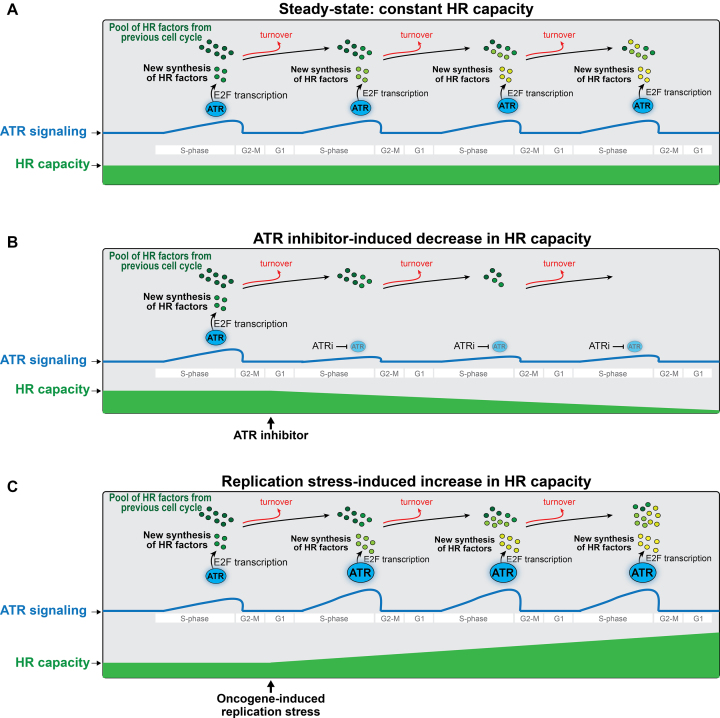
ATR signaling mediates key interactions between HR proteins that are important for HR-mediated repair (25,26,89). In this context, short-term inhibition of ATR is expected to impair such pro-HR function of ATR. Indeed, we attribute the modest reduction in HR observed upon short-term ATR inhibition to loss of specific key pro-HR interactions (for example: BRCA1-TOPBP1 and PALB2-BRCA1), although we note that it is possible that ATM may partially compensate for the loss of ATR signaling in mediating these interactions. While it is possible that the impairment of protein interactions contributes partially to the severe inhibition of HR observed upon long-term (over 4 days) ATR inhibition, we favor the model that the severe reduction in HR capacity is mostly caused by a pre-conditioned state of HR factor depletion. In this model, the role of ATR in controlling transcription of E2F targets during every S-phase is crucial to maintain the proper abundance of HR proteins and, therefore, sustain the capacity of cells to utilize HR (Figure 7A). Consistent with this model, E2F targets, which include many HR factors, are induced every S phase as part of a G1/S wave of cell cycle transcription (16,19). Furthermore, the ATR-CHK1 pathway is supposedly active every S-phase (53,90,91) and is known to prolong E2F transcription (16,18,20,40,62,63). Therefore, our work presented here suggests that chronic ATR inhibition during multiple cell cycles gradually decreases the abundance of HR factors (Figure 7B). A logical explanation is that under chronic ATR inhibition the amount of protein loss due to constitutive protein turnover surpasses the amount of HR factors produced by de novo protein synthesis, generating a deficit in HR factor abundance that is exacerbated after multiple cell cycles. As a consequence, depletion of the HR machinery severely impairs HR capacity. While it is currently unclear whether it is the level of a specific HR factor or global levels that are behind the synergistic sensitivity to ATRi/PARPi, we favor the model that the strong reduction in HR-mediated repair is due to the combined reduction in the abundance of several repair factors.
Figure 7.
Model depicting how modulation of ATR signaling alters HR capacity in cancer cell growth and ATRi-mediated cancer therapy. See text in the discussion for details.
Increased ATR signaling enhances the HR capacity in cancer cells
Many cancer cells experience high levels of replication stress (41,82,92–94), which results in higher levels of spontaneous ATR signaling (40,95,96). Based on our model presented in Figure 7C, we propose that such constitutively higher ATR signaling in cancer cells during every S-phase will create a surplus in HR factor abundance and lead to higher HR capacity. In fact, many cancers have increased HR capacity (74). It is worth mentioning that a correlation between ATR signaling and HR capacity could be lost due to mutations or alterations that directly deregulate the RB-E2F pathway. For example, we noticed that HEK293T cells exhibit one of the highest levels of HR factors and the highest HR capacity of all cells examined, but not the highest levels of CHK1 phosphorylation (Figure 4A and B). This is consistent with the fact that LT-transformation is known to directly inhibit RB function, and therefore maintain high E2F transcription (75,76), which would bypass the need for CHK1 signaling for enhanced HR factor abundance. Interestingly, we also noted the potential for an ATR-dependent and CHK1-independent mechanism for HR factor control in HCT116 cells. As shown in Supplementary Figure S13, differently than ATR inhibition, CHK1 inhibition did not lower the level of E2F1 nor the abundance of other HR proteins in HCT116. This result suggests that there is an alternative pathway, other than CHK1, that regulates E2F transcription in these cells. Finally, we note that ATR may also control BRCA1 protein stability, revealing transcription-independent mechanisms for ATR-dependent regulation of HR factor abundance. Overall, despite possibilities for alternative rewiring of the mechanisms for control of E2F transcription, the model for enhanced ATR signaling promoting increased HR factor abundance seems applicable in many cases and provides a mechanistic explanation for the high HR capacity observed in many cancers.
A chemo-therapeutic strategy to sensitize HR proficient cancer cells to PARP inhibitors
Our finding that chronic treatment of cancer cells with sub-lethal doses of ATRi leads to severe impairment of HR-mediated repair has important implications for cancer therapy. As described above, many cancer cells seem to rely on constitutive ATR hyper-signaling to overexpress components of the HR machinery and acquire enhanced HR capacity, which would improve their ability to deal with increased levels of replication stress. In this context, we propose that partial inhibition of ATR over long-term treatment protocol is particularly deleterious for cancer cells, since the compromised HR machinery is expected to be unable to efficiently deal with enhanced replication stress. Of importance, since the treatment uses relatively low doses of ATRi, it is possible that cells such as hTERT RPE-1 (and potentially other non-cancer cells), that do not explore ATR hyper-signaling, are not subjected to the deleterious effects of a high dose of ATR or CHK1 inhibitor, which include the well-established effects on origin firing and fork integrity.
Since chronic ATR inhibition is very effective at depleting the HR machinery and reducing HR capacity, we further predicted that this rationale could be explored in combination therapy to sensitize HR-proficient cancer cells to the treatment of PARP inhibitors, which are typically used to treat HR-deficient cancers such as BRCA1 or BRCA2 mutated cancers (84–87). Indeed, our data on a panel of cancer lines revealed that our prediction is correct for most of the cancer cell lines tested. Further analyses involving a larger panel of cancer cell lines and in organismal contexts will be required to further evaluate the effectiveness of this approach. We note that a recent study showed that ATR inhibition and knockdown of HR factors, such as RAD51, leads to synergistic lethality in cancer cells (73). While this is an interesting and potentially effective strategy, it is likely that this synergism arises from the short-term effects of ATR inhibitors in causing fork collapse, which would then require the HR machinery for fork restart. In this context, ATR inhibitors are being used as a DNA damage-generating drug. Such rationale is fundamentally distinct from our proposed rationale, in which treatment with sub-lethal doses of ATR inhibitors is used to reduce HR capacity and compromise the ability of cancer cells to respond to DNA damage.
Overall, the findings reported here provide a novel strategy for how the HR capacity can be modulated in cancer cells via controlled ATR inhibition. In addition to helping improve therapy, we expect that the generated knowledge could have far reaching implications to better understand how cancer cells develop an optimized machinery for robust genome replication and maintenance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Beatriz S. Almeida and Haiyuan Yu for technical support. We also thank Robert S. Weiss, Robertus A. de Bruin and members of the Smolka lab for fruitful discussions. We thank Robertus A. de Bruin for generously providing the 293-T-REX-E2F6 cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
M.B.S. grant from the National Institute of Health (NIH) [R01GM097272]; R.F. grants from the Spanish Ministry of Economy and Competitiveness [SAF2016-80626-R, BFU2016-81796-REDC]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 2. Smolka M.B., Albuquerque C.P., Chen S.H., Zhou H.. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:10364–10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stokes M.P., Rush J., Macneill J., Ren J.M., Sprott K., Nardone J., Yang V., Beausoleil S.A., Gygi S.P., Livingstone M. et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19855–19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 5. Bass T.E., Luzwick J.W., Kavanaugh G., Carroll C., Dungrawala H., Glick G.G., Feldkamp M.D., Putney R., Chazin W.J., Cortez D.. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haahr P., Hoffmann S., Tollenaere M.A., Ho T., Toledo L.I., Mann M., Bekker-Jensen S., Raschle M., Mailand N.. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- 7. Kumagai A., Lee J., Yoo H.Y., Dunphy W.G.. TopBP1 activates the ATR-ATRIP complex. Cell. 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- 8. Lee Y.C., Zhou Q., Chen J., Yuan J.. RPA-Binding protein ETAA1 Is an ATR activator involved in DNA replication stress response. Curr. Biol. 2016; 26:3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mordes D.A., Nam E.A., Cortez D.. Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18730–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin S.J., Wardlaw C.P., Morishita T., Miyabe I., Chahwan C., Caspari T., Schmidt U., Carr A.M., Garcia V.. The Rad4(TopBP1) ATR-activation domain functions in G1/S phase in a chromatin-dependent manner. PLoS Genet. 2012; 8:e1002801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nam E.A., Cortez D.. ATR signalling: more than meeting at the fork. Biochem. J. 2011; 436:527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao Z., Chen Z.H., Gunasekera A.H., Sowin T.J., Rosenberg S.H., Fesik S., Zhang H.Y.. Chk1 mediates S and G(2) arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 2003; 278:21767–21773. [DOI] [PubMed] [Google Scholar]
- 14. Jin J.P., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S.J., Harper J.W.. SCF beta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Gene Dev. 2003; 17:3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sorensen C.S., Syljuasen R.G., Falck J., Schroeder T., Ronnstrand L., Khanna K.K., Zhou B.B., Bartek J., Lukas J.. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003; 3:247–258. [DOI] [PubMed] [Google Scholar]
- 16. Bertoli C., Skotheim J.M., de Bruin R.A.. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013; 14:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buisson R., Boisvert J.L., Benes C.H., Zou L.. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell. 2015; 59:1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertoli C., Herlihy A.E., Pennycook B.R., Kriston-Vizi J., de Bruin R.A.M.. Sustained E2F-Dependent transcription is a key mechanism to prevent replication-stress-induced DNA damage. Cell Rep. 2016; 15:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bracken A.P., Ciro M., Cocito A., Helin K.. E2F target genes: unraveling the biology. Trends Biochem. Sci. 2004; 29:409–417. [DOI] [PubMed] [Google Scholar]
- 20. Bertoli C., Klier S., McGowan C., Wittenberg C., de Bruin R.A.. Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell-cycle transcription. Curr. Biol. 2013; 23:1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin W.C., Lin F.T., Nevins J.R.. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001; 15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 22. Collis S.J., Ciccia A., Deans A.J., Horejsi Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J.. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the fanconi anemia core complex. Mol. Cell. 2008; 32:313–324. [DOI] [PubMed] [Google Scholar]
- 23. Collins N.B., Wilson J.B., Bush T., Thomashevski A., Roberts K.J., Jones N.J., Kupfer G.M.. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009; 113:2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L.C., Gautier J.. The Fanconi anemia pathway and ICL repair: implications for cancer therapy. Crit. Rev. Biochem. Mol. Biol. 2010; 45:424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y., Cussiol J.R., Dibitetto D., Sims J.R., Twayana S., Weiss R.S., Freire R., Marini F., Pellicioli A., Smolka M.B.. TOPBP1(Dpb11) plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1(Rad9). J. Cell Biol. 2017; 216:623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buisson R., Niraj J., Rodrigue A., Ho C.K., Kreuzer J., Foo T.K., Hardy E.J., Dellaire G., Haas W., Xia B. et al. Coupling of homologous recombination and the checkpoint by ATR. Mol. Cell. 2017; 65:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H., Wang H., Powell S.N., Iliakis G., Wang Y.. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004; 64:7139–7143. [DOI] [PubMed] [Google Scholar]
- 28. Aylon Y., Liefshitz B., Kupiec M.. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004; 23:4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004; 431:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Branzei D., Foiani M.. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008; 9:297–308. [DOI] [PubMed] [Google Scholar]
- 31. Heidenreich E., Novotny R., Kneidinger B., Holzmann V., Wintersberger U.. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 2003; 22:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 34. Brown E.J., Baltimore D.. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000; 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A. et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000; 14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 36. O’Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., Goodship J.A.. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003; 33:497–501. [DOI] [PubMed] [Google Scholar]
- 37. Casper A.M., Durkin S.G., Arlt M.F., Glover T.W.. Chromosomal instability at common fragile sites in Seckel syndrome. Am. J. Hum. Genet. 2004; 75:654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alderton G.K., Joenje H., Varon R., Borglum A.D., Jeggo P.A., O’Driscoll M.. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 2004; 13:3127–3138. [DOI] [PubMed] [Google Scholar]
- 39. Karnitz L.M., Zou L.. Molecular pathways: targeting ATR in cancer therapy. Clin. Cancer Res. 2015; 21:4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herlihy A.E., de Bruin R.A.. The role of the transcriptional response to DNA replication stress. Genes (Basel). 2017; 8:E92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hills S.A., Diffley J.F.X.. DNA replication and Oncogene-Induced replicative stress. Curr. Biol. 2014; 24:R435–R444. [DOI] [PubMed] [Google Scholar]
- 42. Liu E., Lee A.Y.L., Chiba T., Olson E., Sun P.Q., Wu X.H.. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J. Cell Biol. 2007; 179:643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gilad O., Nabet B.Y., Ragland R.L., Schoppy D.W., Smith K.D., Durham A.C., Brown E.J.. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010; 70:9693–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Syljuasen R.G., Hasvold G., Hauge S., Heland A.. Targeting lung cancer through inhibition of checkpoint kinases. Front. Genet. 2015; 6:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weber A.M., Ryan A.J.. ATM and ATR as therapeutic targets in cancer. Pharmacol. Therapeut. 2015; 149:124–138. [DOI] [PubMed] [Google Scholar]
- 46. Mohni K.N., Thompson P.S., Luzwick J.W., Glick G.G., Pendleton C.S., Lehmann B.D., Pietenpol J.A., Cortez D.. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PLoS One. 2015; 10:e0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pires I.M., Olcina M.M., Anbalagan S., Pollard J.R., Reaper P.M., Charlton P.A., McKenna W.G., Hammond E.M.. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br. J. Cancer. 2012; 107:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reaper P.M., Griffiths M.R., Long J.M., Charrier J.D., MacCormick S., Charlton P.A., Golec J.M.C., Pollard J.R.. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011; 7:428–430. [DOI] [PubMed] [Google Scholar]
- 49. Josse R., Martin S.E., Guha R., Ormanoglu P., Pfister T.D., Reaper P.M., Barnes C.S., Jones J., Charlton P., Pollard J.R. et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014; 74:6968–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huntoon C.J., Flatten K.S., Wahner Hendrickson A.E., Huehls A.M., Sutor S.L., Kaufmann S.H., Karnitz L.M.. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013; 73:3683–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mohni K.N., Kavanaugh G.M., Cortez D.. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014; 74:2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sultana R., Abdel-Fatah T., Perry C., Moseley P., Albarakti N., Mohan V., Seedhouse C., Chan S., Madhusudan S.. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PLoS ONE. 2013; 8:e57098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bastos de Oliveira F.M., Kim D., Cussiol J.R., Das J., Jeong M.C., Doerfler L., Schmidt K.H., Yu H., Smolka M.B.. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol. Cell. 2015; 57:1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bastos de Oliveira F.M., Kim D., Lanz M., Smolka M.B.. Quantitative analysis of DNA damage signaling responses to chemical and genetic perturbations. Methods Mol. Biol. 2018; 1672:645–660. [DOI] [PubMed] [Google Scholar]
- 55. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Danielsen J.M.R., Larsen D.H., Schou K.B., Freire R., Falck J., Bartek J., Lukas J.. HCLK2 is required for activity of the DNA damage response kinase ATR. J. Biol. Chem. 2009; 284:4140–4147. [DOI] [PubMed] [Google Scholar]
- 57. Kakarougkas A., Ismail A., Katsuki Y., Freire R., Shibata A., Jeggo P.A.. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 2013; 41:10298–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kass E.M., Helgadottir H.R., Chen C.C., Barbera M., Wang R., Westermark U.K., Ludwig T., Moynahan M.E., Jasin M.. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5564–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen C.C., Kass E.M., Yen W.F., Ludwig T., Moynahan M.E., Chaudhuri J., Jasin M.. ATM loss leads to synthetic lethality in BRCA1 BRCT mutant mice associated with exacerbated defects in homology-directed repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:7665–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bastos de Oliveira F.M., Harris M.R., Brazauskas P., de Bruin R.A., Smolka M.B.. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 2012; 31:1798–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Travesa A., Kuo D., de Bruin R.A., Kalashnikova T.I., Guaderrama M., Thai K., Aslanian A., Smolka M.B., Yates J.R. 3rd, Ideker T. et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012; 31:1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang A., Schneider-Broussard R., Kumar A.P., MacLeod M.C., Johnson D.G.. Regulation of BRCA1 expression by the Rb-E2F pathway. J. Biol. Chem. 2000; 275:4532–4536. [DOI] [PubMed] [Google Scholar]
- 65. Eelen G., Vanden Bempt I., Verlinden L., Drijkoningen M., Smeets A., Neven P., Christiaens M.R., Marchal K., Bouillon R., Verstuyf A.. Expression of the BRCA1-interacting protein Brip1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008; 27:4233–4241. [DOI] [PubMed] [Google Scholar]
- 66. Baumann P., West S.C.. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 1998; 23:247–251. [DOI] [PubMed] [Google Scholar]
- 67. Wilsker D., Bunz F.. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol. Cancer Ther. 2007; 6:1406–1413. [DOI] [PubMed] [Google Scholar]
- 68. D’Angiolella V., Donato V., Forrester F.M., Jeong Y.T., Pellacani C., Kudo Y., Saraf A., Florens L., Washburn M.P., Pagano M.. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012; 149:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tibbetts R.S., Cortez D., Brumbaugh K.M., Scully R., Livingston D., Elledge S.J., Abraham R.T.. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000; 14:2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen H.Z., Tsai S.Y., Leone G.. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009; 9:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sherr C.J., McCormick F.. The RB and p53 pathways in cancer. Cancer Cell. 2002; 2:103–112. [DOI] [PubMed] [Google Scholar]
- 72. Ma X., Gao Y., Fan Y., Ni D., Zhang Y., Chen W., Zhang P., Song E., Huang Q., Ai Q. et al. Overexpression of E2F1 promotes tumor malignancy and correlates with TNM stages in clear cell renal cell carcinoma. PLoS One. 2013; 8:e73436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Krajewska M., Fehrmann R.S., Schoonen P.M., Labib S., de Vries E.G., Franke L., van Vugt M.A.. ATR inhibition preferentially targets homologous recombination-deficient tumor cells. Oncogene. 2015; 34:3474–3481. [DOI] [PubMed] [Google Scholar]
- 74. Fung H., Weinstock D.M.. Repair at single targeted DNA double-strand breaks in pluripotent and differentiated human cells. PLoS One. 2011; 6:e20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ahuja D., Saenz-Robles M.T., Pipas J.M.. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005; 24:7729–7745. [DOI] [PubMed] [Google Scholar]
- 76. Cantalupo P.G., Saenz-Robles M.T., Rathi A.V., Beerman R.W., Patterson W.H., Whitehead R.H., Pipas J.M.. Cell-type specific regulation of gene expression by simian virus 40 T antigens. Virology. 2009; 386:183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rohaly G., Korf K., Dehde S., Dornreiter I.. Simian virus 40 activates ATR-Delta p53 signaling to override cell cycle and DNA replication control. J. Virol. 2010; 84:10727–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Forero A., Giacobbi N.S., McCormick K.D., Gjoerup O.V., Bakkenist C.J., Pipas J.M., Sarkar S.N.. Simian virus 40 large T antigen induces IFN-stimulated genes through ATR kinase. J. Immunol. 2014; 192:5933–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Powers J.T., Hong S., Mayhew C.N., Rogers P.M., Knudsen E.S., Johnson D.G.. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol. Cancer Res. 2004; 2:203–214. [PubMed] [Google Scholar]
- 80. Hook S.S., Lin J.J., Dutta A.. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007; 19:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Blow J.J., Gillespie P.J.. Replication licensing and cancer—a fatal entanglement. Nat. Rev. Cancer. 2008; 8:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Halazonetis T.D., Gorgoulis V.G., Bartek J.. An oncogene-induced DNA damage model for cancer development. Science. 2008; 319:1352–1355. [DOI] [PubMed] [Google Scholar]
- 83. Turner N., Tutt A., Ashworth A.. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004; 4:814–819. [DOI] [PubMed] [Google Scholar]
- 84. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 85. Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 86. Rottenberg S., Jaspers J.E., Kersbergen A., van der Burg E., Nygren A.O., Zander S.A., Derksen P.W., de Bruin M., Zevenhoven J., Lau A. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17079–17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009; 361:123–134. [DOI] [PubMed] [Google Scholar]
- 88. Kim H., George E., Ragland R.L., Rafail S., Zhang R.G., Krepler C., Morgan M.A., Herlyn M., Brown E.J., Simpkins F.. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-Mutant ovarian cancer models. Clin. Cancer Res. 2017; 23:3097–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dubois J.C., Yates M., Gaudreau-Lapierre A., Clement G., Cappadocia L., Gaudreau L., Zou L., Marechal A.. A phosphorylation-and-ubiquitylation circuitry driving ATR activation and homologous recombination. Nucleic Acids Res. 2017; 45:8859–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sorensen C.S., Syljuasen R.G.. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012; 40:477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Enders G.H. Expanded roles for Chk1 in genome maintenance. J. Biol. Chem. 2008; 283:17749–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bartek J., Mistrik M., Bartkova J.. Thresholds of replication stress signaling in cancer development and treatment. Nat. Struct. Mol. Biol. 2012; 19:5–7. [DOI] [PubMed] [Google Scholar]
- 93. Macheret M., Halazonetis T.D.. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol.-Mech. 2015; 10:425–448. [DOI] [PubMed] [Google Scholar]
- 94. Murga M., Campaner S., Lopez-Contreras A.J., Toledo L.I., Soria R., Montana M.F., Artista L., Schleker T., Guerra C., Garcia E. et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011; 18:1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Flynn R.L., Zou L.. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 2011; 36:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Marechal A., Zou L.. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.