Abstract
Given our previous demonstration that RBPJ binds a methylated repressor element and regulates smooth muscle cell (SMC)-specific gene expression, we used genome-wide approaches to identify RBPJ binding regions in human aortic SMC and to assess RBPJ’s effects on chromatin structure and gene expression. RBPJ bound to consensus cis elements, but also to TCmGGGA sequences within Alu repeats that were less transcriptionally active as assessed by DNAse hypersensitivity, H3K9 acetylation, and Notch3 and RNA Pol II binding. Interestingly, RBPJ binding was frequently detected at the borders of open chromatin, and a large fraction of genes induced or repressed by RBPJ depletion were associated with this cluster of RBPJ binding sites. RBPJ binding dramatically co-localized with serum response factor (SRF) and RNA seq experiments in RBPJ- and SRF-depleted SMC demonstrated that these factors interact functionally to regulate the contraction and inflammatory gene programs that help define SMC phenotype. Finally, we showed that RBPJ bound preferentially to phased nucleosomes independent of active chromatin marks and to cis elements positioned at the beginning and middle of the nucleosome dyad. These novel findings add important insight into RBPJ’s role in chromatin structure and gene expression in SMC.
INTRODUCTION
The evolutionarily conserved intercellular Notch signaling pathway plays a fundamental role in the regulation of cell-fate, proliferation, and patterning in many cell-types (1–7). Interaction between integral membrane Notch receptors (Notch1–4) and Delta-like or Jagged ligands results in proteolytic cleavage of the receptor, release of the transcriptionally active Notch intracellular domain (NICD), and translocation of the NICD to the nucleus where it interacts with the multifunctional DNA binding protein, RBPJ. In the absence of NICD, RBPJ binds a consensus site (GTGGGA) within the promoters of Notch target genes and inhibits gene expression by recruiting transcription repressors. NICD binding to RBPJ displaces the repressive factors but also leads to the recruitment of Mastermind-like (MAML) which, in turn, helps recruit other transcription activators such as the histone acetyltransferase, p300. The best characterized Notch targets are the inhibitory bHLH transcription factors of the Hes and Hey families that contain high affinity RBPJ binding sites within their promoters and that are up-regulated relatively quickly following Notch activation. Evidence suggests that many Notch effects are mediated by Hes/Hey-dependent repression of cell fate determinants and cell cycle regulators.
A variety of global and tissue-specific gain and/or loss of function models have been used to demonstrate that Notch/RBPJ-dependent transcription plays a particularly important role in cardiovascular development (7,8). Notch receptors and ligands are expressed throughout the developing heart where they regulate specification and differentiation of myocardial cells, as well as the complex morphogenic events that are required for chamber formation, valve development, and outflow track remodeling and alignment. Notch/RBPJ signaling is also required for smooth muscle cell (SMC) differentiation from several embryonic origins including cardiac neural crest cells (9), epicardial cells (10) and Tie-1 expressing stem cells (11). RBPJ binding sites have been described in the smooth muscle α-actin and smooth muscle myosin heavy chain (SM MHC) promoters (5,12), but the direct effects of Notch/RBPJ on the SMC differentiation marker gene expression are somewhat controversial and seem to be context dependent (13).
It is well known that SMCs are not terminally differentiated and under various stresses (including cell culturing) modulate their phenotype by repressing SMC differentiation marker gene expression. We have recently shown that down regulation of SM MHC expression in cultured SMCs was associated with progressive methylation of a GC rich repressor element (GCGGGA) within the SM MHC promoter (14). Importantly, using a non-biased affinity chromatography/mass spectroscopy-based approach, we identified RBPJ as a methylated GC-repressor binding protein and went on to show that structural similarities between methylated cytosine and thymine allow for RBPJ binding to this sequence. Importantly, RBPJ depletion in phenotypically modulated SMCs ‘derepressed’ the SMC markers by promoting the binding of serum response factor (SRF) to their promoters.
Several groups have now used ChIP-seq approaches in C2C12 skeletal myoblasts (15), immortal lymphoblast cell lines (16,17) and embryonic neural cortices (18) to better characterize Notch/RBPJ-dependent gene expression. In addition to identifying novel direct Notch/RBPJ targets in these cell-types, it was also shown that DNA from RBPJ ChIPs was significantly enriched for other transcription factor binding sites (e.g. ZNF143, Ets, Runx, among others) suggesting that RBPJ/Notch-dependent transcription is modulated by combinatorial interactions with additional factors (16). In addition, in lymphoblasts many RBPJ only binding sites were detected within transcriptionally inactive regions providing further support for RBPJ’s role as a repressor (19).
To further investigate the role of RBPJ in the regulation of SMC phenotype and to evaluate the potential role of methylation-dependent RBPJ binding, we performed RBPJ ChIP-seq experiments in cultured human aortic (HuAo) SMCs. Our data indicate that RBPJ interacted not only with regulatory regions containing the canonical RBPJ site, but also with methylated CpG-containing motifs located within the Alu family of repeat elements. When coupled with additional genome-wide determinations, our data indicate that RBPJ and SRF cooperate to regulate gene expression in HuAo SMCs and that nucleosome phasing is a critical determinant of RBPJ DNA binding.
MATERIALS AND METHODS
Cell culture and knockdowns
700 000 HuAo SMCs (Lonza) were plated in 10 cm dishes in SmGM-2 media. On each of the next two days, cells were transfected with siRNAs targeting RBPJ, SRF or GFP using standard Dharmacon protocols. After another 24 h cells were used for ChIP, western blot and RNA seq determinations. For experiments involving TGF-β treatment, cells were serum-starved for 16 h and then treated with TGF-β (1 ng/ml) for an additional 24h. SMARTpool siRNAs for RBPJ (M-007772-00) and SRF (M-009800-02) were obtained from Dharmacon. siRNA against GFP was as follows: GGUGCGCUCCUGGACGUAGCCGGCUACGUCCAGGAGCGCACC.
ChIP-seq and ChIP-PCR
Chromatin immunoprecipitation was performed according to Enzymatic Simple ChIP magnetic kit (Cell Signaling) except 0.7% formaldehyde was used for crosslinking. Antibodies were RBPJ (D10A4) from Cell Signaling, SRF(G-20) from Santa Cruz biotechnology, RNA Pol II (8WG16) from Millipore and H3K9ac from Abcam. 1% of input from control cells was also sequenced. DNA samples for high throughput sequencing were prepared by the UNC genomics core and 50bp or 100 bp paired ends sequencing was performed using the Illumina platform. For ChIP-PCR and for RNA Pol II and H3K9ac ChIP-seq, the MNAse digestion step was replaced with a sonication step in 0.12% SDS followed by 5× dilution with concentrated ChIP buffer. To quantify RBPJ binding to nucleosomes, Flag-RBPJ was transfected into 293 cells. Cells were fixed with 0.7% formaldehyde and DNA was digested by 10 ul MNAse (New England Biolabs) to an average length of 120 bp. Flag-RBPJ was precipitated using Flag-M2 agarose beads (Sigma) and washed immunoprecipates were subjected to western blotting using Abs to Flag or HistoneH3 (sc-8654) from Santa Cruz.
High throughput sequencing data analysis
Raw sequencing reads were mapped to hg19 using bowtie2 http://bowtie-bio.sourceforge.net/bowtie2/index.shtml (20). Reads from replicate experiments were concatenated into a single file before peak calling. ChIP peaks were called with MACS software http://liulab.dfci.harvard.edu/MACS/ version macs14 1.4.1 (21) using the following parameters—macs14 -t ChIP.bed -c INP.bed -n ChIP -g hs -p 0.000001 -m 5,30 -s 20. Danpos 2 was used to calculate nucleosome positions and phasing scores proportional to the quantity of DNA protected from MNAse digestion for each calculated nucleosome position https://sites.google.com/site/danposdoc/ (22). Homemade awk scripts were used to calculate distances between nucleosome borders and Alu start sites and motif positions that were extracted from the genome hg19 using bowtie. Motif analysis was done using CisFinder https://lgsun.irp.nia.nih.gov/CisFinder/ (23). We used SeqMiner for clustering of ChIP-seq datasets https://sourceforge.net/projects/seqminer/ (24). Homer softeware was used for calculation of normalized RNA levels, DNA ends densities, and scores of RNA Pol II in 2000 bp regions surrounding summits of RBPJ peaks http://homer.ucsd.edu/homer/ (25). For facilitating comparisons between experiments, Homer-counted reads were normalized to total read counts in each experiment. RNA-seq datasets were processed using standard cufflinks workflow http://cole-trapnell-lab.github.io/cufflinks/ (26) or Homer (25) with similar results obtained by both programs. When plotting RNA Pol II binding data only those regions with average RNA Pol II Homer read scores over 20 were used. Similarly, we analyzed 26 762 genes isoforms with RNA levels scores more than 20. Metascape software was used to analyze gene annotations http://metascape.org/gp/index.html#/main/step1 (27). All data is presented in supplementary excel file with appropriate 0–1 filters.
EMSA
RBPJ clone BC051387 was purchased from OriGene and cloned into mammalian expression pcDNA3 vector (Invitrogen) with a Flag epitope on the C-terminus. Flag-RBPJ was expressed in Cos cells by transient transfection and nuclear extracts were prepared by Pierce NE-PER kit. 1.5 ul of nuclear extract was incubated with 0.12 pmol of P-32 labeled probe (see Figure 1 for sequences) and 2.5 pmol of random 20 bp oligos in binding buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 mM MgCl2, 8% glycerol, 0.05% Triton X-100, 1 mM DTT). Samples where run on a native gel, dried, and exposed to film.
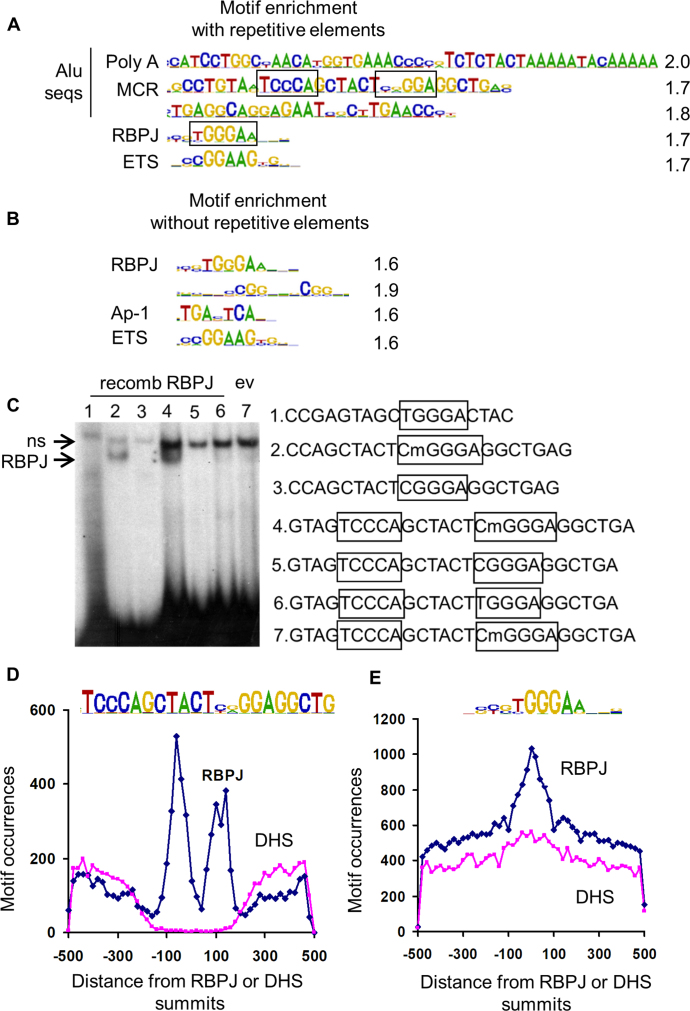
Figure 1.
ChIP-seq analysis of RBPJ binding sites in HuAo SMC identified consensus and methylated Alu motifs. Motif over-representation at RBPJ ChIP-seq peaks summits (±100 bp) when repetitive elements were included (A) or not included (B) in the analysis. Quantification of motif enrichment is listed at right, and putative RBPJ binding sites are boxed. (C) Gel shift assays were performed by incubating RBPJ-containing COS cell lysates with the methylated or unmethylated double stranded DNA probes listed at right. Note that RBPJ interacted only with probes containing the CmGGGA sequence (lower arrow). Note that the binding reaction in lane 7 contained lysates from COS cells transfected with empty expression vector, and the upper arrow marks a non-specific, non-RBPJ-containing binding complex. (D) Distribution of Methylated Cytosine RBPJ binding elements near RBPJ peak summits or 30 000 random DNase hypersensitivity (DHS) regions summits. (E) Distribution of consensus RBPJ binding elements near RBPJ peak summits or 30 000 random DHS regions summits.
RESULTS
RBPJ associated with methylated Alu repetitive elements in addition to consensus sequences
Our recent demonstration that RBPJ binds a methylated repressor element within the SM MHC promoter (14) suggested a novel mechanism for RBPJ-dependent gene regulation. To begin to examine this possibility, we identified RBPJ binding sites in HuAoSMCs using ChIP-seq approaches. By comparison of read densities in RBPJ ChIP DNA and DNA samples before immunoprecipitation, we identified 28 220 RBPJ binding sites with MACS using a stringent cut off at P = 10−6 (Supplemental excel table). We next used CisFinder to search for over-represented sequences in the 200 bp region centered at RBPJ peak summits (Figure 1A). Although consensus RBPJ and ETS factor binding sites were enriched in the RBPJ ChIP data set, the top 3 scoring motifs were sequences within the Alu family of repetitive elements that consist of two nearly identical repeat sequences of the ancestral 7SL RNA gene (28) (Supplementary Figure S1A). When repetitive elements were excluded from our enrichment analysis, we identified two additional over-represented sequences, one of which was a consensus AP-1 element (Figure 1B).
Alu elements encompass ∼10% of the human genome, contain over 25% of all CpGs, and are heavily cytosine methylated in most cell types including SMC (29,30). One of these Alu sequences, herein referred to as the Methylated Cytosine RBPJ binding element (MCR), contained a potential methylation-dependent RBPJ binding sequence (CGGGA) positioned head to head with a second nearly consensus RBPJ binding sequence (TGGGA). Since this arrangement is similar to that of the RBPJ binding sites within the Hes1 promoter (31), we characterized RBPJ binding to this sequence using gel shift assays with methylated and non-methylated probes. As shown in Figure 1C, RBPJ did not bind to the more consensus RBPJ binding sequence TGGGA under any condition tested. In contrast, RBPJ did bind to the CGGGA sequence, but only when this cytosine was methylated. We and others have shown that thymine and methylated cytosine can function interchangeably within an RBPJ binding site (14,32) most likely because both of these pyrimidines are methylated at the 5′ position. However, this was not the case for RBPJ binding to the MCR (Figure 1C compare lanes 4 and 6).
To further address methylation-dependent RBPJ binding within Alu sequences, we characterized four RBPJ binding peaks whose summits were within 200 bp of an MCR (see below and Methods). As shown in Supplementary Figure S1B, targeted ChIP assays in control and RBPJ knockdown cells confirmed that all four regions were bound to RBPJ and bisulfite sequencing demonstrated that the CpG was fully methylated in three out of four of these MCR sequences (Supplementary Figure S1C). Interestingly, ChIP assays for the Notch3 NICD demonstrated little if any Notch3 binding to the Alu regions even though we detected significant levels of cleaved NICD3 in these cells (Supplementary Figure S1D) and precipitated ∼30% of total NICD3 protein (data not shown). As expected, analysis of the distribution of MCR and classical RBPJ binding motifs revealed enrichment of these sequences within 200 bp of RBPJ summits (Figure 1D and E). Even though Alu sequence varies at this position, the CGGGA sequence was much more prevalent in the Alu data set than the TGGGA sequence (Supplementary Figure S1E). Taken together, these results strongly support our previous demonstration that RBPJ interacts with methylated DNA and suggest that methylated Alu elements are a major sub-set of RBPJ interaction sites.
Differential chromatin structure at RBPJ-binding subsets
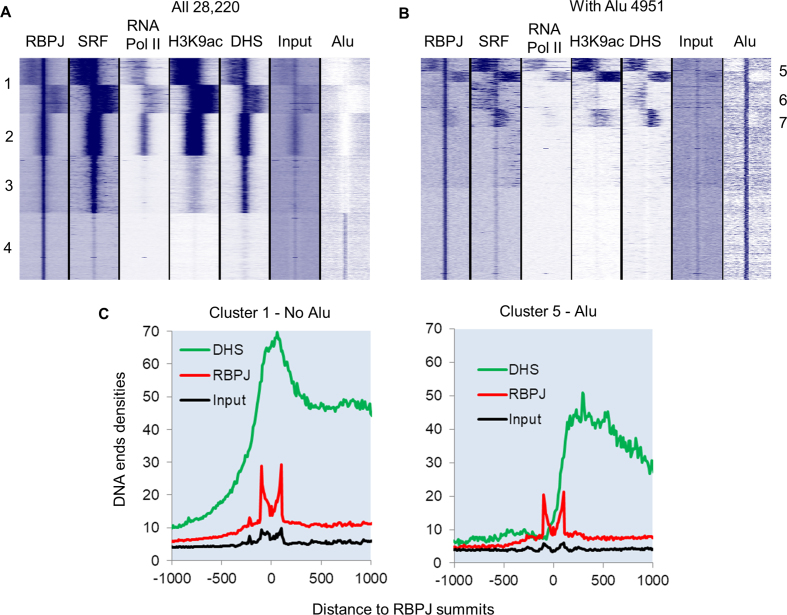
To better understand the role of RBPJ binding on gene expression in HuAo SMC, we compared our RBPJ ChIP-seq data set with other genome-wide determinations including DNase hypersensitivity and ChIP-seq data for RNA polymerase II, H3K9ac, and serum response factor (SRF), a transcription factor required for SMC marker gene expression (14). As shown in Figure 1D, RBPJ-binding MCR sequences were not found within 30,000 randomly selected DNase hypersensitive (DHS) sites, consistent with the observation that Alu repeats are excluded from active promoter regions (33). In contrast, DHS sites were not depleted from RBPJ binding regions that contain the classical RBPJ binding sites (Figure 1E). Indeed, cluster analysis of our genome-wide data sets revealed that RBPJ binding peak summits co-localized either with positive regulatory marks or with Alus, but not both (Figure 2A, compare clusters 1–3 with cluster 4). Interestingly, RBPJ binding to the non-Alu subset was enriched at promoter regions (Supplementary Figure S2A) and the majority of these sites overlapped (74%) or was directly adjacent to DNAse hypersensitivity and SRF-binding regions (63%). The fact that 59% of the non-Alu subset also exhibited high H3K9 acetylation and RNA Pol II binding (Figure 2A, clusters 1 and 2) strongly suggested that RBPJ and SRF factors cooperate to regulate gene expression in SMC. We also found that RBPJ binding sites tended to cluster in promoter regions (Supplementary Figure S2B) and that SRF binding was not detected within Alu elements.
Figure 2.
Differential chromatin structure and transcription factor binding between consensus and Alu RBPJ binding sites. (A) Heat map of clustered reads densities for the indicated genome-wide determination or DNA sequence feature centered around RBPJ peak summits for all 28 220 RBPJ binding sites. (B) The same analysis as in A for the 4921 RBPJ peak summits that intersected with an Alu element within 200 bp. (C) Quantification of DNA ends densities for MNAse digested input, RBPJ immunoprecipitated, and DNase hypersensitive DNA for cluster 1 (without Alu) and cluster 5 (with Alu) regions.
Specific clustering of the Alu-containing subset of RBPJ binding sites revealed that some were associated with open chromatin (Figure 2B, clusters 5–7). However, these open chromatin regions were always offset from RBPJ peak summits, and quantification of DNA end densities with Homer software revealed that they were less open than those associated with consensus RBPJ binding sites (Figure 2C). The Alu-containing RBPJ binding sites were not significantly enriched at promoters and only 11% were associated with RNA Pol II binding (cluster 5 in Figure 2A). These data suggest that RBPJ binding to methylated Alu elements may contribute to the repressive function of Alu elements. Moreover, the frequent adjacent positioning of RBPJ binding to other positive chromatin regions in both the Alu and non-Alu subsets (clusters 1, 5, 6 and 7 in Figure 2A and B) suggests that RBPJ may serve as a boundary transcription factor under certain conditions.
RBPJ depletion enhanced the differentiation program of HuAoSMCs
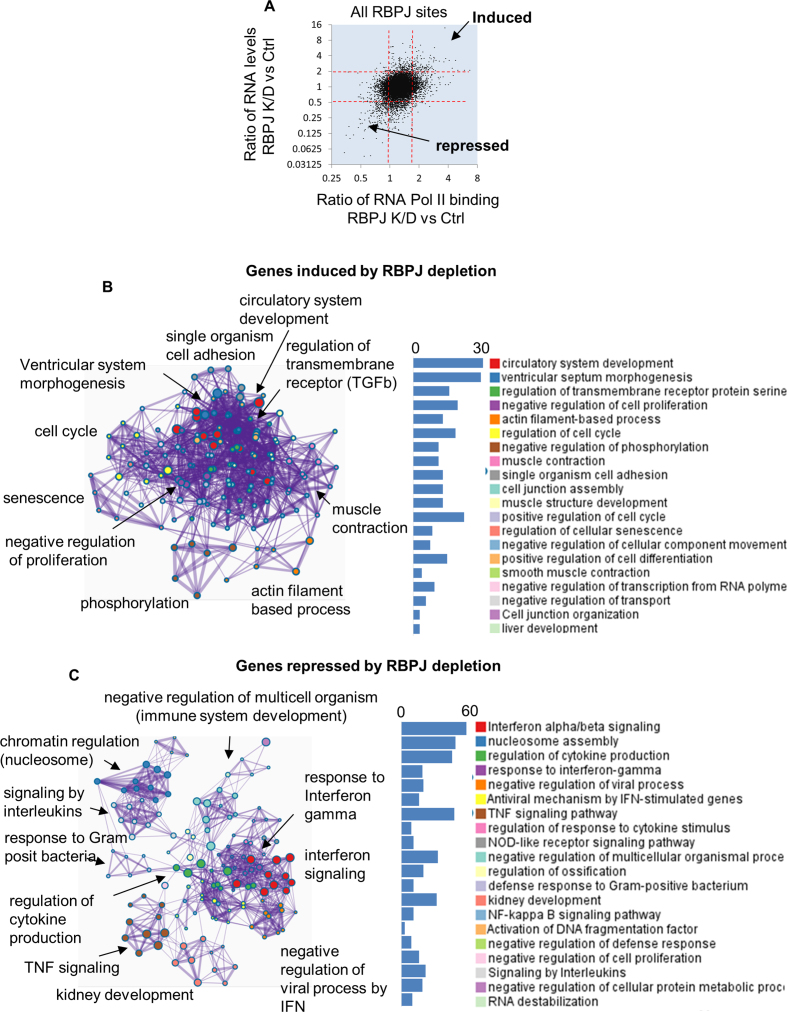
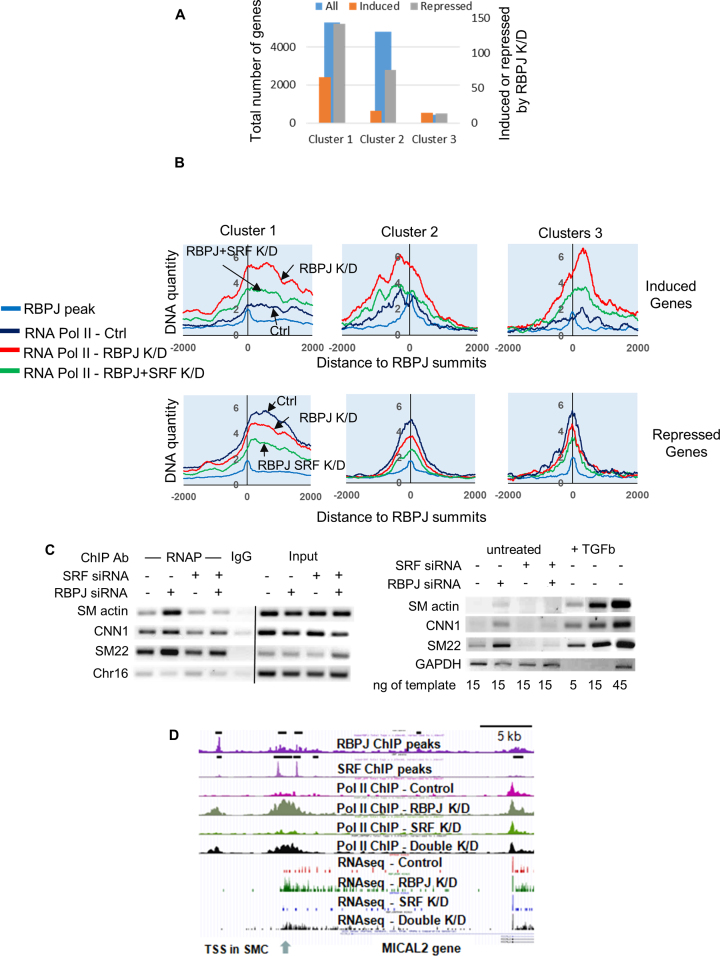
To help identify direct RBPJ targets in HuAo SMCs we identified genes that were up- or down-regulated by RBPJ knockdown by at least 2-fold (as measured by RNA seq) that also exhibited a similarly directed change in RNA Pol II ChIP signal at the RBPJ binding region nearest to that gene's transcription start site. Based upon the above criteria, we identified 82 genes that were up-regulated and 187 genes that were down-regulated by RBPJ knockdown (Figure 3A). Gene ontology (GO) analysis of the 82 induced genes revealed programs related to vascular development, TGF-β signaling, ventricle septum morphogenesis, muscle/smooth muscle contraction, actin assembly, and regulation of cell cycle (Figure 3B). The 187 genes that were repressed by RBPJ knockdown included antiviral response, interferon, cytokine, TNF and NF-κB signaling components (Figure 3C). These data are consistent with our previous demonstration that RBPJ repressed SMC differentiation in phenotypically modulated cultured HuAo SMCs (14). In further support of this idea, the changes in gene expression and H3K9 acetylation observed in RBPJ depleted cells were similar to those induced by treatment of cells with TGF-β, an agonist known to promote the differentiated SMC phenotype (Supplementary Figures S3A and S5B). Moreover, knockdown of RBPJ resulted in increased H3K9 at a large number of TGF-β signaling and matrix-associated genes (Supplementary Figure S3B). Of interest, as quantified in Figure 4A, a majority of the genes that were induced or repressed by RBPJ knockdown were associated with the cluster of RBPJ binding sites that localized to the edges of transcriptionally active regions (cluster 1 from Figure 2A,) providing additional support for RBPJ’s role as a potential boundary factor.
Figure 3.
RBPJ depletion in HuAo SMC promoted a differentiation gene program. (A) RNA Pol II binding and RNA expression in control and RBPJ depleted cells genes were assessed by ChIP-seq and RNA-seq respectively. The RBPJ-depletion-induced change in RNA Pol II binding within the 2000 bp around RBPJ summits was plotted against the RBPJ-depletion-induced change in RNA levels for the genes closest to that specific RBPJ binding site. Genes with RNA level measurements of <20 normalized read counts or RNA Pol II binding of <20 normalized read counts were excluded from the analysis. (B) GO annotation clustering for 82 genes up-regulated by RBPJ knockdown as defined as those that exhibited a >2-fold increase in expression and a >1.7-fold increase in RNA Pol II binding. (C) GO annotation clustering for 187 genes inhibited by RBPJ knockdown as defined by a >50% reduction in expression coupled with a decrease in RNA Pol II binding.
Figure 4.
RBPJ and SRF cooperated to regulate gene expression in SMC. (A) Coordinately induced and repressed genes were categorized based upon the cluster analysis shown in Figure 2A. (B) Quantification of RNA Pol II binding centered at RBPJ summits in control, RBPJ and RBPJ/SRF double knockdown cell lines for the indicated cluster from Figure 2A. Genes induced (top) or repressed (bottom) by RBPJ knockdown were analyzed separately. (C) RNA Pol II binding to and mRNA expression of the indicated SMC-specific genes was measured by targeted ChIP and semi-quantitative RT assays, respectively, in RBPJ knockdown and RBPJ/SRF double knockdown cells. (D) UCSC browser screenshot highlighting changes in RNA Pol II binding and mRNA induction at the MICAL2 gene in RBPJ and RBPJ/SRF double knockdown Hu Ao SMC. A previously un-annotated transcription start was identified and is marked by an arrow.
Due to the longer distances between RBPJ-binding Alu elements and transcription start sites, the lower number of Alu-associated genes, and the relatively smaller changes in gene expression and RNA Pol II binding observed following RBPJ knockdown, it was more difficult to conclude that RBPJ binding at these regions directly regulated nearby genes. Nevertheless, similar groups of genes were up-regulated (transcriptional activity of SMADs, muscular development) and down-regulated (inflammatory genes) by RBPJ depletion (Supplementary Figure S4).
RBPJ and SRF co-operated to regulate gene expression and RNA Pol II recruitment
We have previously shown that RBPJ depletion promoted SMC differentiation marker gene expression in phenotypically modulated HuAo SMCs by enhancing SRF binding (14). To further examine the functional relationship between RBPJ and SRF in HuAoSMC we compared RNA seq data between RBPJ knockdown, SRF knockdown, and RBPJ/SRF double knockdown cells. In general, RBPJ knockdown enhanced the expression of a variety of developmental genes, consistent with its involvement in the control of cell fate decisions. This increase was SRF-dependent for a subset of these genes, and in agreement with our previous studies (14), this subset included many genes related to vascular development and SMC contraction (Supplementary Figure S5; black dots and bars). These SRF-dependent genes were also preferentially up-regulated by TGF-β (Supplementary Figure S5B and S5C). RBPJ knockdown inhibited inflammatory and immune response gene expression and this effect was increased by co-depletion of SRF (Supplementary Figures S5 and S6; red and pink dots, respectively) suggesting that these transcription factors cooperate to promote the activity of these programs. Finally, SRF depletion on its own inhibited many genes associated with cell proliferation (Supplemental Figure S6), consistent with its involvement in this process.
Importantly, we observed a similar relationship between RBPJ and SRF in regard to RNA Pol II binding. For those genes where RNA Pol II binding was induced by RBPJ depletion, this signal was attenuated by co-depletion of SRF (Figure 4B, upper panels). In contrast, when RNA Pol II binding was repressed by RBPJ depletion, we observed further decreases in RNA Pol II binding in double knockdown cells (Figure 4B, lower panels). This functional interaction between RBPJ and SRF was confirmed by RNA Pol II ChIP assays specifically targeted to the SMC marker gene promoters and RT-PCR measurements of SMC marker gene expression (Figure 4C). An additional excellent example of this regulatory mechanism is the RBPJ/SRF-dependent regulation of protein-methionine sulfoxide oxidase (MICAL2), an atypical actin-regulatory protein that promotes the activity of the Myocardin-related SRF co-factors (Figure 4D).
RBPJ bound to phased nucleosomes
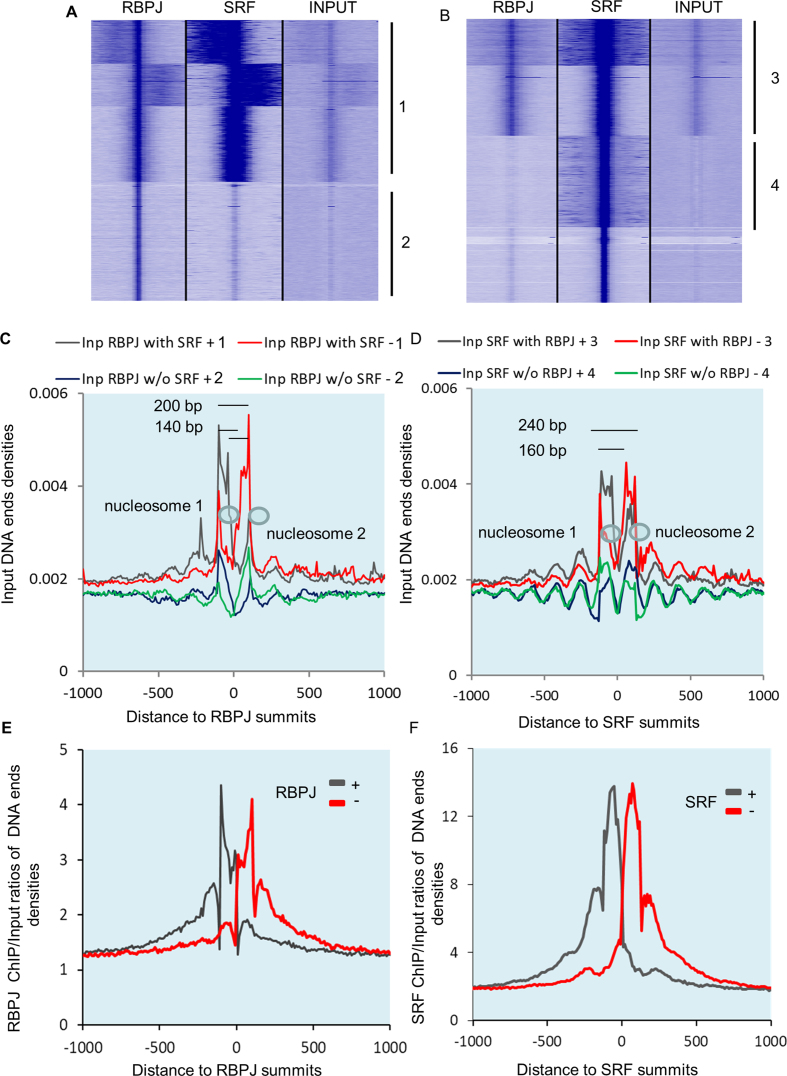
During the analysis of our RBPJ ChiP seq data, we noticed that RBPJ binding peaks were always associated with increases in the quantity of input DNA (see Figure 2A and B) and input DNA ends density (Figure 2C), suggesting nucleosome-mediated protection from MNase cleavage in these regions. Given that nucleosome structure is thought to influence the positioning of transcription factor binding in general (34) and the positioning of RBPJ binding specifically (35), we examined this relationship more closely by clustering RBPJ, SRF and Input DNA around RBPJ and SRF summits (Figure 5A and B). Several observations are worth noting. First, when input DNA ends densities were centered at RBPJ binding peak summits, we observed peak maxima on opposite strands that were separated by ∼140 bp consistent with nucleosome protection from MNAse digestion (Figure 5C). The fact that peak maxima on the same strand were separated by ∼200 bp suggests the existence of a 60 bp linker between nucleosomes. Assuming, that the maxima of DNA ends densities on separate strands correspond to the maxima of nucleosome protection, this would localize the RBPJ binding site to ∼40–100 bp within nucleosomes. Second, when centered at SRF summits, we observed a 160 bp distance between input DNA ends density maxima on opposite strands with 40 bp intersection with SRF summits suggesting that SRF binds at 20–120 bp within the nucleosome core placing it closer to the edges of or in between nucleosomes (Figure 5D). Interestingly, we also observed a wider array of fixed nucleosomes positioning around SRF summits. Third, we observed higher DNA ends densities of input material at sites that bound both RBPJ and SRF than at sites that bound either transcription factor alone. Finally, the maxima of DNA ends enrichment for RBPJ and SRF ChIP DNA (–100 bp and −80 bp, respectively) corresponded to DNA ends of phased nucleosomes (Figure 5E and F). To verify RBPJ binding at nucleosomes biochemically, we over-expressed Flag-RBPJ in 293 cells, and after fixation and MNAse over-digestion, immunoprecipitated Flag according to our ChIP protocols. As shown in Supplementary Figure S7, ∼1% of histones co-immunoprecipitated with Flag RBPJ.
Figure 5.
RBPJ, SRF, and phased nucleosomes colocalized. (A) Heat map of RBPJ ChIP, SRF ChIP and input DNA densities clustered around RBPJ summits. (B) Heat map of RBPJ ChIP, SRF ChIP and input DNA densities clustered around SRF summits. Note the increase of input DNA density under RBPJ summits and the two peaks of input DNA density at regions where SRF binds without RBPJ (cluster 4). (C) DNA ends densities for input material for DNA ends clusters centered around RBPJ with or without SRF binding. (D) DNA ends densities for input material for DNA ends clusters centered around SRF summits with or without RBPJ binding. (E and F) Plot of RBPJ and SRF ChIP signals relative to input DNA centered at corresponding RBPJ and SRF summits.
Modeling RBPJ binding to nucleosomes
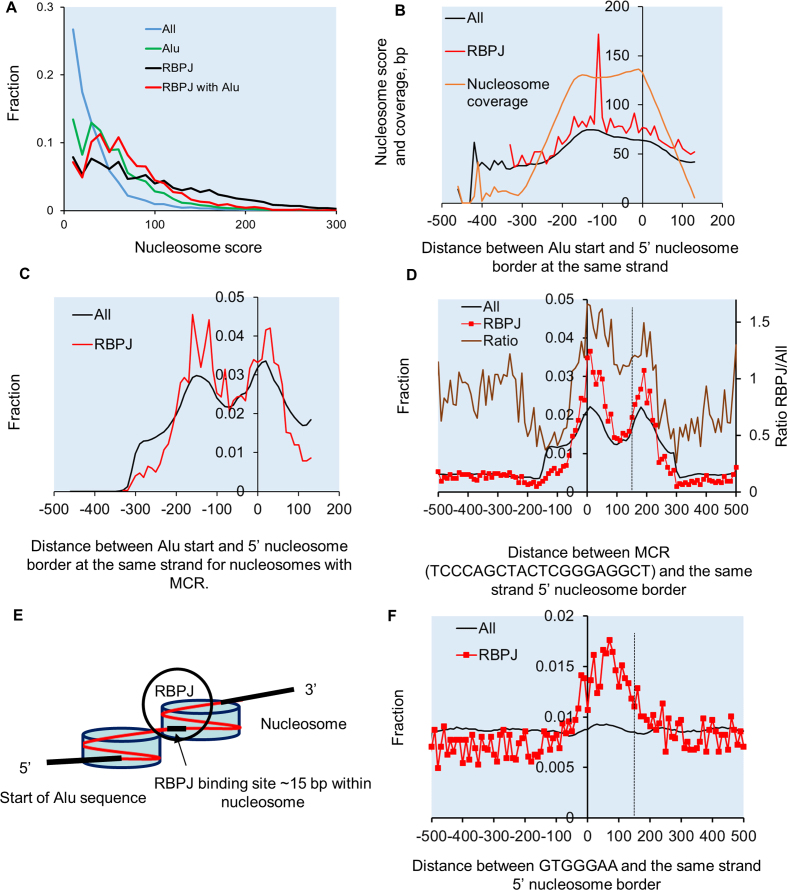
We hypothesized that nucleosome positioning could influence RBPJ binding by locating the RBPJ binding element at specific positions around the nucleosome core. To address this, we first used DANPOS2 software to determined nucleosome scores for specific data subsets (i.e. Alus, RBPJ-binding regions, etc.). RBPJ-binding regions had relatively high nucleosomes scores that were on par with the 2.4 million Alu regions analyzed (Figure 6A). We next plotted the nucleosome scores for Alu elements in relation to their position within nucleosome structure. The highest scores were for Alu's whose start sites ranged from −150 bp to the nucleosome border (Figure 6B, black line). The subset of Alus that were bound to RBPJ exhibited a similar pattern, although the nucleosome scores for these regions were slightly higher and a bit more variable (Figure 6B, red line). Our data also suggested that RBPJ preferentially bound Alus that started around −150 relative to nucleosomes borders (Figure 6C, red line). Based upon the known positioning of the MCR within Alus, we predicted that the MCR should preferentially localize to either 12 or 62 bp within nucleosomes. Indeed, actual measurements of MCR positioning relative to nucleosome borders for all nucleosome and the RBPJ bound subset confirmed this prediction (Figure 6D, brown enrichment line) Taken together these data support the RBPJ binding model show Figure 6E. Importantly, analyses of the classical RBPJ binding site GTGGGA revealed a similar pattern of nucleosome positioning for these elements (Figure 6F).
Figure 6.
RBPJ preferentially bound at 15 and 65 bp from nucleosome borders. (A) Nucleosome scores and positions were calculated by Danpos using default parameters and were plotted for the indicated groups as a fraction of their distribution. (B) Average nucleosome scores and Alu nucleosome coverage in base pairs (orange line) plotted verses Alu start position relative to the 5′ end of bound nucleosome reveals highest nucleosomes scores and coverage for Alus that start at around −150 or closer relative to the 5′ of nucleosomes for All nucleosomes with MCR (black line) and RBPJ bound subset (red line). (C) Distribution of Alu start sites relative to 5′end of bound nucleosome for all Alu with MCR and RBPJ bound subset reveals that Alus preferentially start at either −150 bp or +10 bp relative to nucleosome border and these are overrepresented in RBPJ bound group. (D) Distribution of MCR motifs relative to nucleosome borders for all Alu bound nucleosomes (black) and the RBPJ bound subset (red line). (E) Model depicting probable Alu positioning and RBPJ binding locations within nucleosome structure. (F) Distribution of the consensus GTGGGA sequence relative to nucleosome borders for all nucleosomes (black) and RBPJ bound subset (red line).
DISCUSSION
Our genome-wide analysis of RBPJ in HuAo SMC has led to several novel and potentially important insights into RBPJ’s role in the regulation of gene expression and chromatin structure. Importantly, our ChIP seq data indicated that RBPJ bound not only to consensus elements within transcriptionally active chromatin regions, but also to methylated sequences within Alu repeats. These results add to a growing body of evidence supporting the idea that cytosine methylation can promote the binding of various transcription factors such as RBPJ, C/EBPb, and KLF4 (14,32,36–39) and significantly complicate the role of DNA methylation in the regulation of gene expression.
Repetitive elements are thought to affect gene expression by a variety of mechanisms including transcription factor recruitment, induction of DNA methylation, nucleosome positioning, and chromatin structure (40–43). Our data indicate that RBPJ binds preferentially to nucleosomal DNA and support previous findings that Alu sequences and Alu CpG methylation promote nucleosome phasing (44,45). Importantly, our analysis of RBPJ ChIP seq data in T-cells generated by Wang et al. (16) revealed a similar increase in input material centered under RBPJ binding maxima suggesting that RBPJ binds to phased nucleosomes in other cell-types as well (Supplementary Figure S8). When coupled with our observation that RBPJ binding was frequently observed at the edges of transcriptionally active chromatin regions, these findings suggest that RBPJ may have effects on chromatin state and/or be a reader of chromatin structure. Our demonstration that RBPJ binds to phased nucleosomes (See model in Figure 6E) is similar to observations made for FOXA2 (46,47), GATA3 (48) and the progesterone receptor (49). Like these ‘pioneering’ transcription factors, RBPJ may be involved in recruiting positive remodelers to condensed chromatin. In support of this role, Lake et al. have shown that RBPJ remains associated with chromatin during mitosis by a mechanism that involves DNA binding and that RBPJ binds to nucleosomes in vitro (35). Based upon the similar RNA Pol II and H3K9 acetylation ChIP footprints that we observed in control and RBPJ knockdown cells (Supplementary Figure S9), we do not think that RBPJ has significant effects on nucleosome positioning. It is well known that CpGs are significantly underrepresented in mammalian genomes due to the conversion of methylated cytosine into thymine by spontaneous deamination. Given the data presented above, it is possible that RBPJ binding to phased nucleosomes within Alus could protect against deamination or functionally contribute to conservation of these CpG rich regions. Our bioinformatics analyses of RBPJ binding within Alu sequences and nucleosome structure revealed preferential localization of the RBPJ binding site at the beginning and middle of the nucleosome dyad. This would tend to place the RBPJ cis element facing the outside of nucleosome structure, a positioning that has been shown to promote binding of other transcription factors to nucleosomes (50–52). Interestingly, we observed widespread correlation between RBPJ and SRF binding and our demonstration that SRF binds further away from the nucleosome core or between nucleosomes supports co-occupancy by these factors (34). RBPJ and SRF colocalization also correlated with increased RNAP recruitment and nucleosome phasing consistent with the detection of a phased nucleosome positioned just past transcription start sites (53,54).
By comparing genome-wide measurements in control and RBPJ siRNA-treated SMC, we demonstrated that RBPJ inhibits the SMC differentiation gene program but is required for the expression of the inflammatory gene program. These changes closely parallel those observed in SMC in vivo during the development of atherosclerosis or during post-angioplasty restenosis suggesting that regulation of RBPJ function is important under these conditions. Interestingly, ∼75% of the genes induced by RBPJ knockdown had RBPJ binding sites positioned on the edges of active chromatin (Figure 2 A, cluster 1) perhaps supporting an inhibitory boundary function for RBPJ under these conditions. The observation that these changes in gene expression and Pol II binding were dependent upon SRF further supports cooperation between these transcription factors and that the presence of RBPJ likely controls the recruitment of SRF as we have shown previously (14). For those genes with binding sites that overlapped with other positive chromatin marks (Figure 2A, cluster 2) the predominant change in expression in RBPJ knockdown cells was repression. The mechanisms by which these differences in RBPJ-associated chromatin structure result in differences in Pol II recruiting and gene expression are unknown. Previous studies have shown that differential recruitment of the NICD was important for context-dependent activation of Notch/RBPJ-dependent gene expression (55,15) and this mechanism is the focus of continuing studies.
Although our data suggest that RBPJ and SRF interact in different ways to affect distinct gene programs in HuAo SMC, it will be interesting to determine whether these interactions occur in other cell types, or whether they are promoted by transcription signals or chromatin states that are unique to SMC. To our knowledge, there are no other cell-types in which both RBPJ and SRF binding have been examined by ChIP seq. However, SRF and RBPJ binding have been studied in separate primary B-cell lines that were immortalized similarly; GM12878 cells by EBV infection and IB4 cells by expression of the EBV viral component EBNA2. Thus, we compared SRF and RNA Pol II peaks in GM12878 cells (available on the UCSC ENCODE database) with RBPJ peaks identified in IB4 cells (17). As seen in Supplementary Figure S10, the percentage of RBPJ sites that also bound SRF in B-cells was much smaller than that observed in HuAo SMC suggesting that cooperation between these two transcription factors is at least partially cell-type-specific. In addition, the extent to which RBPJ, SRF and RBPJ/SRF binding sites overlapped with RNA Pol II binding was much less in B-cells suggesting that RNA Pol II recruitment requires additional mechanisms. It is important to note that our analyses demonstrated that most of binding sites for each factor were unique to that cell-type (Supplementary Figure S10B) which could reflect cell-type specific differences in transcription factor levels or chromatin accessibility.
In summary, our data suggest that RBPJ plays a critical and multifunctional role in the regulation of gene expression and chromatin structure in HuAo SMC. The fact that RBPJ interacted with consensus and methylated Alu elements significantly expands the mechanisms by which RBPJ could affect gene expression. Given the importance of RBPJ in the regulation of many developmental and disease processes, especially in the cardiovascular system, it will be critical to further examine the mechanisms that control RBPJ binding, its interactions with SRF, and the consequences of RBPJ binding to very different chromatin landscapes.
DATA AVAILABILITY
All raw sequencing data was deposited into SRA: SRP047467. DNAse hypersensitivity data for HuAo SMCs is available in the UCSC browser: UCSC genome browser tracks.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [HL109607 to C.P.M., HL130367 to J.M.T., C.P.M.]. Funding for open access charge: National Heart, Lung, and Blood Institute [R01HL130367].
Conflict of interest statement. None declared.
REFERENCES
- 1. Buono K.D., Robinson G.W., Martin C., Shi S., Stanley P., Tanigaki K., Honjo T., Hennighausen L.. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev. Biol. 2006; 293:565–580. [DOI] [PubMed] [Google Scholar]
- 2. Iso T., Hamamori Y., Kedes L.. Notch signaling in vascular development. Arterioscler. Thromb. Vasc. Biol. 2003; 23:543–553. [DOI] [PubMed] [Google Scholar]
- 3. Lin H.Y., Kao C.H., Lin K.M., Kaartinen V., Yang L.T.. Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis. PLoS One. 2011; 6:e15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu H., Kennard S., Lilly B.. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ. Res. 2009; 104:466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang Y., Urs S., Boucher J., Bernaiche T., Venkatesh D., Spicer D.B., Vary C.P., Liaw L.. Notch and transforming growth factor-beta (TGFbeta) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J. Biol. Chem. 2010; 285:17556–17563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu X., Xu K., Zhang L., Deng Y., Lee P., Shapiro E., Monaco M., Makarenkova H.P., Li J., Lepor H. et al. Differentiation of the ductal epithelium and smooth muscle in the prostate gland are regulated by the Notch/PTEN-dependent mechanism. Dev. Biol. 2011; 356:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de la Pompa J.L., Epstein J.A.. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell. 2012; 22:244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gridley T. Notch signaling in the vasculature. Curr. Top. Dev. Biol. 2010; 92:277–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. High F.A., Zhang M., Proweller A., Tu L., Parmacek M.S., Pear W.S., Epstein J.A.. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 2007; 117:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grieskamp T., Rudat C., Lüdtke T.H., Norden J., Kispert A.. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ. Res. 2011; 108:813–823. [DOI] [PubMed] [Google Scholar]
- 11. Kurpinski K., Lam H., Chu J., Wang A., Kim A., Tsay E., Agrawal S., Schaffer D.V., Li S.. Transforming growth factor-beta and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells. 2010; 28:734–742. [DOI] [PubMed] [Google Scholar]
- 12. Doi H., Iso T., Sato H., Yamazaki M., Matsui H., Tanaka T., Manabe I., Arai M., Nagai R., Kurabayashi M.. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J. Biol. Chem. 2006; 281:28555–28564. [DOI] [PubMed] [Google Scholar]
- 13. Mack C.P. Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler. Thromb. Vasc. Biol. 2011; 31:1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rozenberg J.M., Tesfu D.B., Musunuri S., Taylor J.M., Mack C.P.. DNA methylation of a GC repressor element in the smooth muscle myosin heavy chain promoter facilitates binding of the Notch-associated transcription factor, RBPJ/CSL1. Arterioscler. Thromb. Vasc. Biol. 2014; 34:2624–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castel D., Mourikis P., Bartels S.J., Brinkman A.B., Tajbakhsh S., Stunnenberg H.G.. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013; 27:1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W.S., Schug J., Blacklow S.C., Arnett K.L. et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao B., Zou J., Wang H., Johannsen E., Peng C.W., Quackenbush J., Mar J.C., Morton C.C., Freedman M.L., Blacklow S.C. et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14902–14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Hibbs M.A., Gard A.L., Shylo N.A., Yun K.. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells. 2012; 30:741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koelzer S., Klein T.. A Notch-independent function of Suppressor of Hairless during the development of the bristle sensory organ precursor cell of Drosophila. Development. 2003; 130:1973–1988. [DOI] [PubMed] [Google Scholar]
- 20. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014; 1150:81–95. [DOI] [PubMed] [Google Scholar]
- 22. Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J., Dent S., He X., Li W.. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013; 23:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharov A.A., Ko M.S.. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res. 2009; 16:261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye T., Krebs A.R., Choukrallah M.A., Keime C., Plewniak F., Davidson I., Tora L.. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011; 39:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A., Moulton H.M., DeJesus P., Che J., Mulder L.C. et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015; 18:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vassetzky N.S., Kramerov D.A.. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 2013; 41:D83–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yideng J., Jianzhong Z., Ying H., Juan S., Jinge Z., Shenglan W., Xiaoqun H., Shuren W.. Homocysteine-mediated expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol. 2007; 26:603–611. [DOI] [PubMed] [Google Scholar]
- 30. Kochanek S., Renz D., Doerfler W.. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993; 12:1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nam Y., Sliz P., Song L., Aster J.C., Blacklow S.C.. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006; 124:973–983. [DOI] [PubMed] [Google Scholar]
- 32. Bartels S.J., Spruijt C.G., Brinkman A.B., Jansen P.W., Vermeulen M., Stunnenberg H.G.. A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One. 2011; 6:e25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Estécio M.R., Gallegos J., Vallot C., Castoro R.J., Chung W., Maegawa S., Oki Y., Kondo Y., Jelinek J., Shen L. et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010; 20:1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vierstra J., Wang H., John S., Sandstrom R., Stamatoyannopoulos J.A.. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat. Methods. 2014; 11:66–72. [DOI] [PubMed] [Google Scholar]
- 35. Lake R.J., Tsai P.F., Choi I., Won K.J., Fan H.Y.. RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLos Genet. 2014; 10:e1004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rozenberg J.M., Bhattacharya P., Chatterjee R., Glass K., Vinson C.. Combinatorial recruitment of CREB, C/EBPβ and c-Jun determines activation of promoters upon keratinocyte differentiation. PLoS One. 2013; 8:e78179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., Fitzgerald P., Vinson C.. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu B., Gharaee-Kermani M., Wu Z., Phan S.H.. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am. J. Pathol. 2010; 177:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017; 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Estécio M.R., Gallegos J., Dekmezian M., Lu Y., Liang S., Issa J.P.. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol. Cancer Res. 2012; 10:1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Häsler J., Strub K.. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006; 34:5491–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaur S., Pociot F.. Alu elements as novel regulators of gene expression in type 1 diabetes susceptibility genes. Genes (Basel). 2015; 6:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanaka Y., Yamashita R., Suzuki Y., Nakai K.. Effects of Alu elements on global nucleosome positioning in the human genome. BMC Genomics. 2010; 11:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salih F., Salih B., Kogan S., Trifonov E.N.. Epigenetic nucleosomes: Alu sequences and CG as nucleosome positioning element. J. Biomol. Struct. Dyn. 2008; 26:9–16. [DOI] [PubMed] [Google Scholar]
- 46. Ye Z., Chen Z., Sunkel B., Frietze S., Huang T.H., Wang Q., Jin V.X.. Genome-wide analysis reveals positional-nucleosome-oriented binding pattern of pioneer factor FOXA1. Nucleic Acids Res. 2016; 44:7540–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J.A., Mahony S., Pugh B.F., Lee D., Kaestner K.H., Zaret K.S.. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell. 2016; 62:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takaku M., Grimm S.A., Shimbo T., Perera L., Menafra R., Stunnenberg H.G., Archer T.K., Machida S., Kurumizaka H., Wade P.A.. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016; 17:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ballaré C., Castellano G., Gaveglia L., Althammer S., González-Vallinas J., Eyras E., Le Dily F., Zaurin R., Soronellas D., Vicent G.P. et al. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell. 2013; 49:67–79. [DOI] [PubMed] [Google Scholar]
- 50. Li Q., Wrange O.. Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol. Cell. Biol. 1995; 15:4375–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sahu G., Wang D., Chen C.B., Zhurkin V.B., Harrington R.E., Appella E., Hager G.L., Nagaich A.K.. p53 binding to nucleosomal DNA depends on the rotational positioning of DNA response element. J. Biol. Chem. 2010; 285:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hapala J., Trifonov E.N.. Nucleosomal TATA-switch: competing orientations of TATA on the nucleosome. Gene. 2013; 527:339–343. [DOI] [PubMed] [Google Scholar]
- 53. Voong L.N., Xi L., Sebeson A.C., Xiong B., Wang J.P., Wang X.. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016; 167:1555–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang H., Zang C., Taing L., Arnett K.L., Wong Y.J., Pear W.S., Blacklow S.C., Liu X.S., Aster J.C.. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data was deposited into SRA: SRP047467. DNAse hypersensitivity data for HuAo SMCs is available in the UCSC browser: UCSC genome browser tracks.