Abstract
Bacterial transcription termination proceeds via two main mechanisms triggered either by simple, well-conserved (intrinsic) nucleic acid motifs or by the motor protein Rho. Although bacterial genomes can harbor hundreds of termination signals of either type, only intrinsic terminators are reliably predicted. Computational tools to detect the more complex and diversiform Rho-dependent terminators are lacking. To tackle this issue, we devised a prediction method based on Orthogonal Projections to Latent Structures Discriminant Analysis [OPLS-DA] of a large set of in vitro termination data. Using previously uncharacterized genomic sequences for biochemical evaluation and OPLS-DA, we identified new Rho-dependent signals and quantitative sequence descriptors with significant predictive value. Most relevant descriptors specify features of transcript C>G skewness, secondary structure, and richness in regularly-spaced 5′CC/UC dinucleotides that are consistent with known principles for Rho-RNA interaction. Descriptors collectively warrant OPLS-DA predictions of Rho-dependent termination with a ∼85% success rate. Scanning of the Escherichia coli genome with the OPLS-DA model identifies significantly more termination-competent regions than anticipated from transcriptomics and predicts that regions intrinsically refractory to Rho are primarily located in open reading frames. Altogether, this work delineates features important for Rho activity and describes the first method able to predict Rho-dependent terminators in bacterial genomes.
INTRODUCTION
In bacteria, the Rho factor mediates one of the two major pathways that lead to the termination of transcription (reviewed in ref. (1–3)). Rho is a ring-shaped homo-hexameric enzyme with RNA-dependent ATP hydrolase activity. This ATPase activity fuels the translocation of Rho along nascent transcripts and the ensuing displacement of RNA polymerases [RNAPs] halted along the DNA template at Rho-dependent termination sites (Figure 1A). Rho-induced dissociation of transcriptional complexes contributes to the orchestration of gene expression at the genome scale but also helps maintain genome integrity by preventing conflicts between the transcription and replication machineries (3–6). Rho activity is tightly controlled in vivo and usually requires uncoupling of translation from transcription (or lack of translation) which exposes nascent RNA Rut (Rho utilization) sites to binding by Rho (Figure 1A). RNA capture is thus the primary determinant of Rho action (7–10) even though subsequent steps along the termination pathway can also be subjected to tight regulation (11,12).
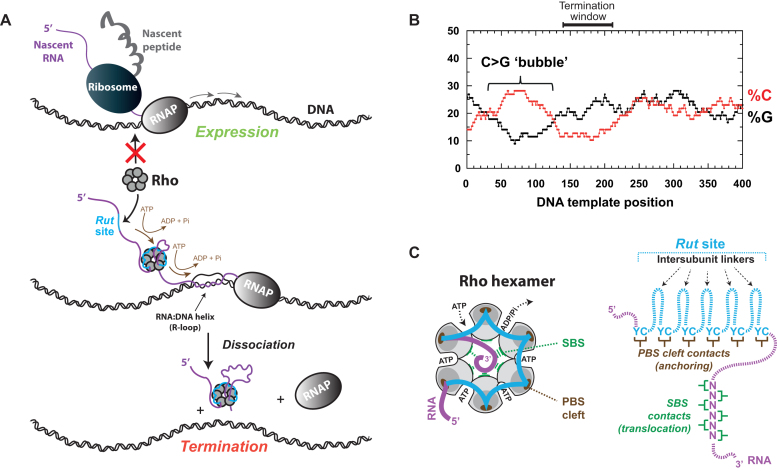
Figure 1.
Rho-dependent termination of transcription. (A) Schematic representation of the termination process. Putative contacts between Rho and RNAP (74) are not depicted. Transcriptional R-loops, sometimes formed behind RNAPs, are also dissociated by the Rho factor (68), as depicted. (B) Genomic regions encoding Rho-dependent terminators usually contain a C>G bubble upstream from the termination sites (10,13,29,37). The case of the pgaA terminator from E. coli is shown as a representative example (10). (C) Configuration of the RNA interaction network within the Rho hexamer based on crystal structures of E. coli’s Rho (16,75). The tethered tracking mechanism used by Rho implies that PBS-RNA contacts are preserved while other contacts change as the RNA chain is translocated within the SBS, leading to the progressive lengthening of the PBS→SBS linker (see (76) and references therein).
Despite the importance of the initial RNA binding step, there are no known rules or consensus features that allow precisely defining (and detecting) Rut sites. It has been proposed that Rho-dependent termination sites lie downstream from so-called C>G ‘bubbles’, i.e. strand regions rich and poor in C and G residues, respectively (13) (Figure 1B). This agrees well with biochemical data showing that single-stranded C-rich-and-G-poor RNA ligands can proficiently bind and activate Rho (reviewed in (14)), thereby minimally defining the composition of Rut sites within nascent transcripts. The compositional bias can be better understood in the light of crystal structures where the Rho hexamer binds short C-rich oligonucleotides using a specific 5′-YC (Y being a pyrimidine) binding cleft located in the N-terminus of each Rho subunit (15,16). These structures, and a wealth of additional experimental data, support the idea that 60–80 nucleotides (nt) of single-stranded RNA are required to span the entire N-terminal periphery of the Rho ring, which composes the Primary Binding Site (PBS), including 9–13 nucleotide (nt)-long ‘linkers’ between the 5′-YC binding PBS clefts of adjacent subunits (16–19) (Figure 1C). Little else is known about Rut sites except that their effectiveness might be increased by the presence of pyrimidine residues in the intersubunit linkers (20), which themselves might be advantageously replaced by hairpin-like RNA motifs (21,22).
One possible reason for the lack of more precise rules to define Rut sites is that previous biochemical/biophysical studies have focused on a very limited set of Rho-dependent terminator sequences or have used artificial RNA ligands that may not fully recapitulate Rut features (1,14). The lack of reliable predictive tool is regrettable given that bacterial genomes can harbor hundreds of Rho-dependent terminators, the abundance of which requires elaborate transcriptomics/genomics strategies for discovery (23–28). For instance, ∼1300 Rho-dependent termination loci have been uncovered by transcriptomics in the genome of the Escherichia coli MG1655 strain (25). Even in this case, however, the precise location of the terminators (or Rut sites) could not be determined with certainty due to the endogenous trimming of the transcripts by essential exoribonucleases (25). Moreover, this type of approach is not ideal to detect terminators regulating low-abundant transcripts and probably misses conditional terminators that are active (and thus detectable) only under specific growth conditions (many effector-controlled Rho-dependent termination mechanisms have now been identified (9,10,28–35)). The great genomic variations among bacterial strains (notably pathogenic ones) due to horizontal transfers, mutations, and recombination events (36) are also expected to impact the Rho-dependent transcriptome (23,24,37) in proportions that would be too burdensome to systematically investigate in vivo. Thus, a method able to quickly outline genome-wide Rho-dependent termination in a given strain (or species) as efficiently as the algorithms developed to detect intrinsic (Rho-independent) terminators (38–43) could prove particularly useful.
In this work, we show that it is possible to develop such a method. Based on the information described above, we hypothesized that the major fraction of Rho-dependent terminators present in a given genome can be detected by seeking C>G bubble regions containing adequately spaced 5′-YC dinucleotides; only the minor fraction of Rho-dependent terminators activated by the NusG cofactor (∼20% in E. coli MG1655), which lack a canonical Rut site (25,44), may escape detection. Likewise, the rules described here may not apply to species harboring Rho factors too divergent from that of E. coli. Notably, caution should be exerted with the ∼35% of Rho factors (in different species) that contain N-terminal domain (NTD) insertions (45) susceptible of altering PBS specificity (46–48). With this in mind, we focused our analysis on the genomes of two representative γ-Proteobacteria, E. coli MG1655 and Salmonella enterica LT2, whose Rho factors share 99.5% (417/419 residues) sequence identity (10). We systematically searched both genomes for the presence of C>G bubbles and [(YC)N9→13]1→6 sequence motifs (where N is any nucleotide). Based on this screen, we selected 104 genomic regions of diverse compositions (having a length of ∼500 base pairs [bp], on average) that we tested for the presence of Rho-dependent signals using in vitro transcription termination assays. The vast majority of the regions devoid of C>G bubbles (28 out of the 32 tested) showed no detectable in vitro termination activity, supporting the idea that a ‘naked’ RNA is not a sufficient condition to initiate Rho-dependent termination. Significantly, we also found that not all C>G bubbles promote Rho-dependent termination and, using multivariate statistical analysis and classification approaches, we identified explanatory variables related to the size, length, and YC-content of the C>G bubbles that have significant predictive value for Rho-dependent termination. Other major explanatory variables stemming from our analysis include the occurrence of small G-rich motifs within the non-template DNA strand as well as the template potential to encode stable RNA secondary structure. Using this information, we have built a multivariate prediction model for Rho-dependent termination based on OPLS-DA classification. The model provides reliable Rho-dependent termination prediction scores for major fractions of the MG1655 and LT2 genomes. Interestingly, sequences predicted to be refractory to Rho are overwhelmingly located within open reading frames where they may contribute to protect gene expression from unwanted transcription termination. Conversely, regions of ‘Strong’ termination probability are frequently located antisense of genes, consistent with Rho involvement in suppressing pervasive antisense transcription (25). Importantly, most (∼90%) of the Rho-dependent termination loci identified by transcriptomics (25) fall within regions of ‘Strong’ termination probability within the model-fitting portion of the MG1655 genome, thereby confirming the value of our approach. The model also predicts many termination-prone regions than were not anticipated from previous work, suggesting that additional layers of Rho-dependent regulation remain to be characterized. Taken together, the biochemical and computational data presented here identify new Rho-dependent termination signals in the E. coli and Salmonella genomes and provide the first predictive model for the automated detection of Rho-dependent terminators in bacterial genomes.
MATERIALS AND METHODS
Materials
Unless specified otherwise, chemicals and enzymes were purchased from Sigma-Aldrich and New England Biolabs, respectively. Nucleoside triphosphates and radionucleotides were purchased from GE-Healthcare and PerkinElmer, respectively. Synthetic oligonucleotides were obtained from Eurogentec. Rho protein was prepared and purified as described previously (49). Rho concentration is expressed in hexamers throughout the manuscript.
Preparation of the DNA templates
DNA templates used in in vitro transcription reactions were prepared by standard PCR procedures, as described previously (50). Briefly, each DNA template containing a specific genomic region downstream from the strong pT7A1 promoter was prepared in two successive PCR rounds by amplification of genomic DNA from E. coli MG1655 or S. typhimurium LT2 using synthetic DNA primers (see Supplementary Table S1 for details). Templates were purified with the GeneJET PCR purification kit (Thermo Fisher Scientific) followed by G50 size exclusion chromatography and their sizes were verified by 1.5% agarose gel electrophoresis.
Transcription termination experiments
Standard transcription termination experiments were performed as described previously (12) with minor modifications. Briefly, DNA template (0.1 pmol), E. coli RNAP (0.45 pmol), Rho (1.4 pmol), and Superase-In (0.5 U/μl; Ambion) were mixed in 18 μl of transcription buffer (40 mM Tris–HCl, pH 8.0, 50 mM KCl, 5 mM MgCl2, 1,5 mM DTT) and incubated for 10 min at 37°C (in control reactions, Rho was omitted). Then, 2 μl of initiation mix (2 mM ATP, GTP and CTP, 0.2 mM UTP, 2.5 μCi/μl of 32P-αUTP and 250 μg/ml of rifampicin in transcription buffer) were added to the reaction mixtures before further incubation for 20 min at 37°C. Transcription reactions were stopped by adding 4 μl of EDTA (0.5 M), 6 μl of tRNA (0.25 mg/ml) and 80μl of sodium acetate (0.42 M) before precipitation at –20°C with 330 μl of ethanol. Reaction pellets were dissolved in denaturing loading buffer (95% formamide, 5 mM EDTA), and analyzed by denaturing 7% polyacrylamide gel electrophoresis (PAGE) and Typhoon-9500 imaging (GE-Healthcare). Note that, due to the different internal labeling of the terminated and runoff transcripts, the efficiency of Rho-dependent termination cannot be precisely measured with this assay. Termination signals were thus categorized as ‘None’, ‘Weak’ or ‘Strong’ based on the visual changes in the transcription profiles induced by Rho (see legend to Figure 3 for details).
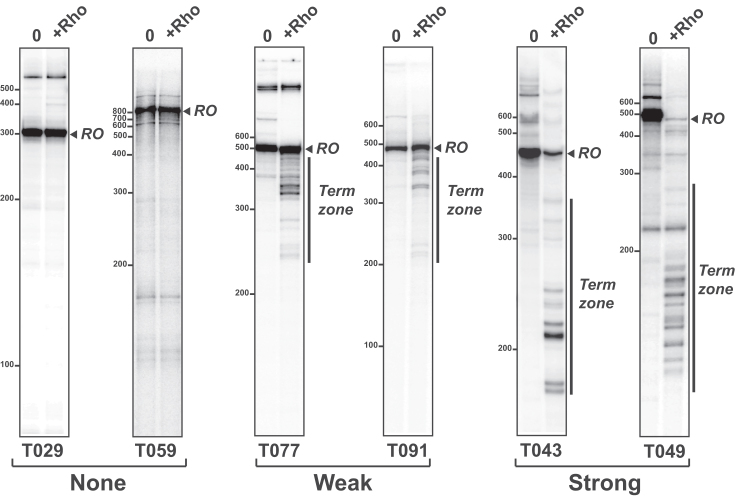
Figure 3.
Rho-dependent termination signals encoded by DNA templates containing C>G bubbles. Representative denaturing PAGE gels illustrate the different classes of transcription termination signals. Rho did not change the transcription profiles for 5.6% of the tested ‘C>G-plus’ DNA templates (‘None’ category). Rho-dependent signals were considered ‘Weak’ when bands migrating faster than runoff transcripts appeared in the presence of Rho while the intensity of the ‘runoff’ band was hardly affected (47.2% of the ‘C>G-plus’ templates). They were considered ‘Strong’, when Rho elicited the appearance of fast-migrating bands and a sharp decrease of the intensity of the runoff band (47.2% of the ‘C>G-plus’ templates).
Ideally, termination efficiencies are determined from single-run transcription experiments wherein uniformly-labeled, stalled TECs are ‘chased’ with a mixture of unlabeled NTPs (51). However, the method is tedious and unpractical for the analysis of a large set of DNA templates and often requires template-specific modifications of the upstream region of the DNA template (to halt TECs) which may themselves alter the termination signals. On-beads transcription experiments require the same type of modifications and were thus only performed with selected DNA template controls (See results). To this effect, the sequences of the T049, T077 and T091 templates were modified to include the sequence 5′-AGATTGTATATGGTAAT between the T7A1 promoter and genomic sequences. This modification allowed preparation of TECs halted at position +26 (or +27 for the modified T091) of the templates. A 5′-biotinylated forward primer was also used during PCR amplification of the DNA templates to allow immobilization of the TECs on streptavidin-coated beads. Preparation of the bead-immobilized TECs and transcription chase reactions, with or without Rho, were then performed as described previously (10,48). We note that, to date, single-run transcriptions with bead-affixed TECs always supported the idea that the Rho-dependent signals observed with our standard transcription termination assay stem from termination events (this work and ref. (10,48)), which attests to the robustness of this simple assay.
All standard and bead-immobilized transcription termination experiments were at least performed twice with each DNA template. For DNA templates yielding no (or very weak) termination signals in the presence of Rho, experiments were also repeated with completely distinct batches of reactants and buffers to rule out the presence of inhibitory contaminants.
Bioinformatic analysis and model building
Reference genomes used for bioinformatics analyses are U00096.3 for E. coli MG1655 and NC_003197.1 for Salmonella LT2.
Dedicated scripts for the detection of C>G bubbles and [(YC)N9→13]n motifs in genomic sequences and for the production of sequence descriptors for a given DNA template (or genomic region) were written in language Python 2.7 (the definitions of the descriptors and Python scripts are provided in Supplementary information). The C>G bubbles were identified using a 78-nt sliding window, as described previously (13). DNA regions were defined as C>G bubbles if they comply with %C ≥ %G (percentages calculated over the 78-nt window) at all positions and with %C > %G at one position at least. Regions were considered devoid of C>G bubbles if they comply with %C ≤ %G at all positions.
Minimum RNA folding free energies were determined with the Mfold software (52) using a locally installed 3.2 version and the RNA Quickfold option (standard RNA setting rules: 37°C; 1 M Na+, 0 M Mg2+; structures: 5% suboptimal, default window size, 100 folding max; no limit on maximum distance between paired bases).
The 104 DNA templates tested in vitro were divided into three classes (‘Strong’, ‘Weak’ or ‘None’) according to their capacity to elicit Rho-dependent termination (see legend to Figure 3 for details). The capacity of each sequence descriptor to discriminate between classes was evaluated with ANOVA (Analysis of Variance) and post-hoc Student–Newman–Keuls (P ≤ 0.05) significance tests using Kaleidagraph 4.0 software (Synergy). Multivariate analyses, including Principal Component Analysis (PCA), Projection to Latent Structures Discriminant Analysis (PLS-DA), and OPLS-DA were performed with SIMCA 14.1 software (Umetrics) using a training set composed of the termination responses (‘Strong’, ‘Weak’ or ‘None’) measured with the 104 DNA templates used in this work (Supplementary Table S1). Explanatory variables (descriptors) were centered and auto-scaled to unit variance (UV scaling) using the default settings of SIMCA. The significance of PCA, PLS-DA and OPLS-DA components and the performance of resulting models were tested by jackknife cross validation (CV) with the training set as implemented in SIMCA (1/7th of the data held out and used as test set per CV round). Quality assessment for fit and prediction was based, respectively, on values of R2, the fraction of the Sum of Squares (SS) explained by the component (or R2cum for the several components of a model), and Q2, the fraction of the total variation of ‘descriptors’ or ‘response’ (i.e. dummy Ypred variables in (O)PLS-DA) that can be predicted by a component (Q2cum for several components). The reliability and degree of overfitting of the (O)PLS-DA models were assessed, respectively, with CV-ANOVA of the cross-validated predictive residuals (53) and permutation plots (100 permutations) as implemented in SIMCA. Receiver Operating Characteristic (ROC) curves were calculated in SIMCA with the full training set of 104 observations and distinct moving threshold parameters for PCA-class (proprietary PModXPS probability) and (O)PLS-DA (dummy Ypred response variable) models. For validated models with Q2cum < 0.5, we verified that the quality parameters remain stable after permutation of the rows in the dataset, as recommended (54).
Following guidelines from SIMCA’s manufacturer, sequences with a ‘probability of model membership’ (proprietary PmodXPS+ parameter in SIMCA) lower than 0.05 (95% confidence level) were considered to be OPLS-DA model outliers (‘out-of-model’ sequences). Methods used to define the predicted ‘None’, ‘Weak’, ‘Strong’ and ‘Out-of-model’ regions for genomic predictions as well as template sequence descriptor values used for PCA and (O)PLS-DA model training are provided in Supplementary Information.
RESULTS
Genomic regions devoid of C>G bubbles do not contain significant Rho-dependent signals
Rho-dependent terminators often trigger transcript release at multiple, heterogeneous sites (1). It is usually unclear whether such a wide termination window reflects a diversity of potential Rho ‘entry’ points on the transcript (for instance, due to loose Rut site rules), a distribution of transcriptional pause sites where Rho is able to dissociate RNAP, or a combination of both. Notwithstanding, analysis of a handful of Rho-dependent terminators suggested that termination windows are invariably located downstream from so-called C>G bubble regions where the non-template DNA strand is richer in C than in G residues (13) (Figure 1B). These C>G bubble regions were identified upon scanning of the non-template DNA strand with a 78 nt-long sliding sequence window, assumed to be an ideal Rut size (13). We have used this ‘bubble’ methodology (see Supplementary Figure S1 and methods) and other sequence descriptors to probe the importance of C>G skewness in Rho-dependent termination.
If the presence of a C>G bubble is a prerequisite for Rho-dependent termination (13), then DNA sequences devoid of C>G bubbles should not contain Rho-dependent signals. Surprisingly, this prediction has never been tested thoroughly despite the fact that many genomic regions are free of C>G bubbles (hereafter named ‘C>G-free’ regions). For instance, the E. coli MG1655 genome contains 3381 ‘C>G-free’ regions having a length of at least 200 bp (Supplementary Figure S1) while ‘C>G-free’ regions ≥ 100 bp represent 25.2% of the genome (proportions are similar for the Salmonella LT2 genome).
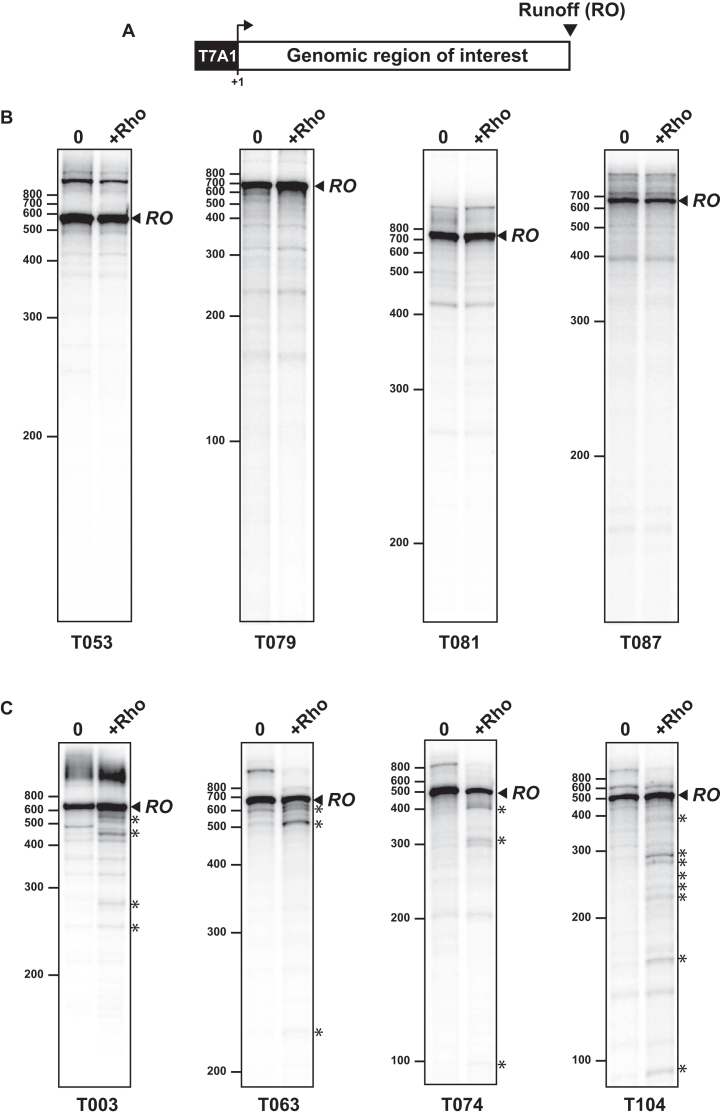
To test the assumption that ‘C>G-free’ sequences cannot elicit Rho-dependent termination (at least in the absence of NusG), we prepared thirty-two DNA templates containing a genomic ‘C>G-free’ region fused to the T7A1 promoter (Figure 2A). The sizes of these genomic segments ranged from 572 to 1130 bp, representing a total of 22 083 bp (Supplementary Table S1). The templates were used in standard in vitro transcription experiments in the presence or absence of Rho (see methods). For twenty-eight of the DNA templates, transcription patterns were not affected by the presence of the Rho protein (representative examples are shown in Figure 2B), indicating that the corresponding sequences do not encode strong Rut sites or Rho-dependent terminators. For the last four DNA templates (T003, T063, T074 and T104 templates), we detected shorter-than-runoff transcripts formed in low amounts in the presence of Rho (Figure 2C, black stars), implying that these templates contain weak Rho-dependent signals. Similar profiles were observed upon using 32P-αCTP (instead of 32P-αUTP) to label the transcripts, ruling out effects dependent on which NTP is at a subsaturating concentration in the transcription assay (see Materials and Methods). Using RSAT oligo analysis (55), we found that the T003, T063, T074, and T104 templates are significantly enriched in AT-rich motifs in the non-template strand (e.g. TTTAAA, TTTTA, TATT, TTAT; P < 10−5) as compared to the other ‘C>G-free’ templates. These motifs may facilitate productive interactions between Rho and the nascent transcript (for instance, by limiting the formation of stable RNA secondary structures) but are unlikely to represent primary determinants of Rho activation given the weakness of the associated Rho-dependent signals (see Figure 2C and below). Hence, our results support the view that genomic regions devoid of C>G bubbles do not encode canonical Rut sites or strong Rho-dependent terminators.
Figure 2.
C>G-less genomic regions are devoid of significant Rho-dependent signals. (A) Schematic depiction of the DNA templates used in our standard in vitro transcription termination experiments. (B) Representative denaturing PAGE gels illustrate the absence of formation of Rho-specific truncated transcripts during transcription of C>G-less templates. The RO bands correspond to runoff transcripts. RNAs migrating more slowly than the RO bands result from template switching events (once RNAP has reached a template end; see (12) and references therein). (C) Representative transcription experiments with the four unusual C>G-less templates yielding low amounts of truncated transcripts (identified by stars next to the gels) in the presence of Rho.
Distribution of C>G bubbles and 5′-YC dimers in the E. coli and Salmonella genomes
Next, we looked at the distribution of C>G bubbles within the genomes of the E. coli MG1655 and Salmonella LT2 strains. We found that both genomes contain a high density of C>G bubbles evenly distributed (in numbers and sizes) between both genomic strands (Supplementary Table S2). For instance, the E. coli genome contains 20 882 C>G bubbles having a length of at least 80 bp (∼2.2 per kilobase). This is an order of magnitude higher than the number of Rho-dependent termination loci that were identified by transcriptomics (25). This difference is all the more notable considering that more than half of the aforementioned C>G bubbles (61%) are located antisense or outside of open reading frames (ORFs) where they should not be hindered by translating ribosomes.
We also scanned the MG1655 and LT2 genomes for the presence of groups of YC dimers adequately spaced for interaction with Rho's PBS. We limited the search to [(YC)N9→13]n sequences encoding RNA motifs containing ‘intersubunit linkers’ (Figure 1C) of 9 to 13 nt. This criterion is a tradeoff between the proposal (based on Rho crystal structures) that a length of 12–13 nt of single-stranded RNA is required to span adjacent subunit PBS clefts (56) and biophysical observations suggesting that even shorter linkers (9–10 nt) are compatible for binding (18,19). For instance, a footprint of (57 ± 2) nt of single-stranded RNA for the full PBS has been estimated from single-molecule force extension experiments (19). Assuming that the six PBS clefts were occupied by YC dimers, this leaves (45 ± 2) nt for the five intersubunit linkers (Figure 1C), i.e. (9 ± 0.4) nt per linker on average. Of note, our [(YC)N9→13]n motif search cannot detect special cases where hairpin-forming sequences can de facto bring the distance between YC dimers inside the accepted range (21,22).
As expected, the number of genomic [(YC)-N9→13]n motifs sharply decreases as a function of the number of repeats, n (Supplementary Table S3). There are 32,351 [(YC) N9→13]6 sequences in the MG1655 genome (∼3.5 per kilobase on average) encoding RNA segments that could theoretically span the six Rho subunits (and thus the entire PBS). This is slightly less than for the LT2 genome (∼3.9 [(YC) N9→13]6 motifs per kilobase) but still much more than the ∼1300 Rho-dependent termination loci (∼0.14 per kilobase) identified by transcriptomics (25). We note, however, that three of our ‘C>G-free’ templates unable to elicit Rho-dependent termination (T025, T056 and T088; see above) encode [(YC) N9→13]6 motifs, suggesting that the presence of such presumably ideal motifs in transcripts is not a sufficient determinant for termination.
Next, we attempted to estimate the number of C>G bubbles containing [(YC)N9→13]n motifs that are sufficiently long to encode candidate Rut sites. We found that known Rho-dependent terminators contain C>G bubbles of at least ∼80 bp located upstream from (or comprising) the termination sites and that all of these bubbles contain at least one [(YC)N9→13]4 motif (Supplementary Table S4). The later observation suggests that the RNA chain needs to contact at least four distinct Rho subunits to make a productive interaction with the hexameric enzyme. The MG1655 and LT2 genomes harbor >16 000 C>G bubbles with these characteristics (∼80 bp or longer and containing a [(YC)N9→13]4 motif) (Supplementary Table S5), a number which is still much higher than that of experimentally validated terminators. This suggests that the number of Rho-dependent terminators is largely underestimated or that C>G bubbles with the aforementioned characteristics are not sufficient (or adequate) predictors for the occurrence of termination.
Testing the Rho-dependent termination potential of genomic C>G bubble regions
To better identify features defining a productive Rut site, we compared the Rho-dependent transcriptional responses of a large set of DNA templates probed under the same standard in vitro transcription conditions. We selected 72 genomic sequences (average length ∼490 bp, representing a total of 35 171 bp) from the E. coli and Salmonella genomes (Supplementary Table S1), each bearing at least one C>G bubble starting no closer than 130 bp from the downstream edge of the sequence (hereafter named ‘C>G-plus’ regions). These ‘C>G-plus’ regions were empirically selected, yet were relatively diverse in terms of C>G bubble length (ranging from 20 to 355 bp) and YC content (ranging from 0 to 18 [(YC)N9→13]4 motifs per region). All ‘C>G-plus’ regions were fused to the T7A1 promoter (Figure 2A) and transcribed in the presence or absence of Rho as described above for the C>G-less templates. Rho had no detectable effect on the transcription of four of the ‘C>G-plus’ DNA templates (Figure 3, ‘None’ class). For the vast majority (∼94%) of the templates, however, the presence of Rho induced the formation of shorter-than-runoff transcripts (representative examples are shown in Figure 3), consistent with the presence of Rho-dependent signals within the corresponding sequences. We empirically classified the signals as ‘weak’ or ‘strong’ (34 ‘C>G-plus’ templates in each class) based on their apparent strengths estimated from transcription termination gel band profiles (Figure 3 and Supplementary Figure S2). Details on categorization of termination signals are provided in the legend to Figure 3.
We selected a few ‘C>G-plus’ DNA templates from the weak and strong classes to verify that the Rho-dependent signals arise from true termination events (rather than from Rho-induced transcriptional arrest). We modified the upstream sequences of the selected DNA templates in order to prepare halted transcription elongation complexes (TECs) immobilized on streptavidin-coated beads, which were then used in single-run ‘chase’ transcriptions (see methods). In all tested instances, we observed that the presence of Rho in the reaction mixture triggers the release of shorter-than-runoff transcripts in the supernatant (Supplementary Figure S3). This observation is consistent with the dissociation of the TECs upon transcription termination (whereas transcriptional arrest would not have disrupted the TECs).
Using RSAT oligo analysis (55), we found that the set of DNA templates unable to elicit Rho-dependent termination (T005, T029, T059, and T075) is significantly enriched in G-rich motifs in the non-template strand (e.g. CAGGG, GGGCA; P < 10−4) as compared to the other ‘C>G-plus’ templates. However, many of these motifs are located in the T059 template alone and a significant enrichment is no longer detected (P > 0.05) if the T059 sequence is removed from the analysis. We note that the T059 template also contains the smallest C>G bubble of all ‘C>G-plus’ region templates (20 bp; Supplementary Figure S4) which could contribute to its ‘unresponsiveness’ to Rho. The other three ‘unresponsive’ templates, however, contain C>G bubbles that are no smaller than those found in some of the ‘C>G-plus’ region templates eliciting Rho-dependent termination. Notwithstanding, there appears to be some relationship between the length/area of the C>G bubble and the strength of the Rho-dependent signal to the point that all tested DNA templates containing C>G bubbles longer than 78 bp (or with an area over 700% × bp) elicit Rho-dependent termination (Supplementary Figure S4).
Seeking relevant sequence descriptors of Rho-dependent termination
To characterize the ‘None’, ‘Weak’ and ‘Strong ‘signals further, we systematically compared the 104 DNA templates (regardless of their C>G bubble content and discounting the T7A1 promoter region) using 111 distinct sequence explanatory variables, herein named ‘descriptors’ (see Supplementary methods and Supplementary Table S6). The percentages of individual monomers, dimers and trimers of nucleotides found in the non-template DNA strands (or runoff transcripts) make a first group of descriptors (larger motifs were not considered because none were evident from pairwise RSAT (55) comparisons of the termination classes of templates). A second group of descriptors provides scores for selected features of the C>G bubbles (length or area of the longest C>G bubble in template, cumulated length or area of all C>G bubbles in template, etc.). A third group of descriptors counts [(YC)N9→13]n motifs found in the C>G bubbles alone (none for ‘C>G-free’ templates) or in the full non-template DNA strands. The last descriptor represents the minimum free energy (per kilobase) for RNA secondary structure formation as determined for runoff transcripts with Mfold software (52).
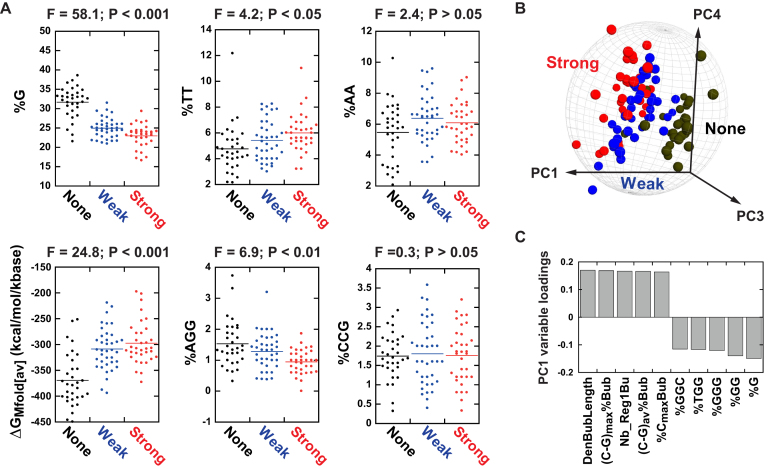
Next, we examined how each individual descriptor is able to distinguish the three classes of templates (i.e. templates triggering ‘None’, ‘Weak’, or ‘Strong’ termination signals). Representative dotplots taken from this analysis are shown in Figure 4A. For 28 of the descriptors, we did not detect statistically significant differences among template classes (ANOVA, F ≤ 3 and P > 0.05; Figure 4A and Supplementary Table S6). Among the 83 remaining descriptors, 27 descriptors were able to differentiate the three template classes (Student-Newman-Keuls post hoc test P ≤ 0.05 for all pairwise comparisons), 49 descriptors were meaningful for only two pairwise comparisons, and 7 descriptors for only one pairwise comparison (Figure 4A and Supplementary Table S6). Among the 27 ‘most-differentiating’ descriptors, 13 are C>G bubble descriptors (including numbers of [(YC)N9→13]1 –aka YC dimers- and [(YC)N9→13]2 motifs in the longest C>G bubble) while 14 are nucleotide (%C, %G), dinucleotide (%CA, %CT, %YC, %GA, %GT, %GG), and trinucleotide (%AAC, %ACA, %CTT, %CAC, %GTG, %GGT) sequence descriptors (Supplementary Table S6).
Figure 4.
Statistical analysis of the Rho-dependent signals detected with the training set of DNA templates (Supplementary Table 1). (A) Dot plots for representative sequence descriptors. ANOVA F- and P-values are shown above each plot. (B) Unsupervised PCA 3D score plot obtained for the 104 DNA templates of the training set using the complete set of 111 descriptors. Q2cum = 0.464 and R2cum = 0.561 for the four PCA components. The gray sphere represents the Hotelling's T2 = 0.05 limit. (C) The bar graph shows the best positive and negative variable loadings for the first principal component (PC1). DenBubLength: cumulated length of all C>G bubbles in non-template DNA strand relative to full strand length (density); (C–G)max%Bub: maximal difference between %C and %G in longest C>G bubble of non-template DNA strand; Nb_Reg1Bu: Number of YC dimers in longest C>G bubble; (C–G)av%Bub: average difference between %C and %G in longest C>G bubble; %CmaxBub: highest %C in longest C>G bubble. %GGC, %TGG, %GGG, %GG and %G are percentages of respective motifs in the non-template DNA strand.
We observed that the minimal RNA folding energies calculated for the full-length transcripts are significantly higher for the ‘Strong’ and ‘Weak’ classes than for the ‘None’ class (Figure 4A and Supplementary Table S6). This suggests that the ‘Strong’ and ‘Weak’ transcripts are usually less structured than the ‘None’ transcripts, in agreement with models proposing that Rut sites are generally poor in RNA secondary structures (1–3). Differences between the ‘Weak’ and ‘Strong’ RNA folding energies are, however, not significant (Student–Newman–Keuls post hoc test P > 0.05). This holds true when RNA folding energies are calculated for sequences restricted to the longest C>G bubble regions (data not shown), suggesting that RNA structure content is not a discriminant between the ‘Weak’ and ‘Strong’ classes.
Overall, we found a number of sequence descriptors that vary significantly with the strength of the Rho-dependent signal (Supplementary Table S6), suggesting that Rho-dependent termination can be predicted upon scanning and detection of specific sequence features/motifs.
A predictive multivariate model for the presence of Rho-dependent termination signals
We cannot rule out that relevant descriptors have escaped detection due to insufficient statistical power or multivariate interactions. Increasing statistical power is difficult because it would require increasing sample sizes significantly, a task beyond the scope of the present work wherein an already large number (>100) of DNA templates have been characterized biochemically. By contrast, the multivariate structure of the data can be explored with statistical approaches such as Principal Component Analysis (PCA) (57). In PCA and related approaches, complex sets of variables, which are often correlated, are reduced into much smaller sets of uncorrelated variables called principal components (note that all original variables are used to build each principal component but differ in their respective ‘loadings’, i.e. component weights) (57,58). In this way, patterns of similarities among variables and/or observations are more easily identified and can be used to generate predictive models.
We first performed PCA with the whole set of 104 templates (i.e. observations) and 111 descriptors (i.e. variables) using the dedicated SIMCA software (see methods). The resulting PCA model contains four significant principal components which are able to separate ‘None’, ‘Weak’ and ‘Strong’ observations in fairly distinct populations even though some population overlap remains (Figure 4B and Supplementary Figure S5). As expected, PCA separation is most effective along principal component 1 for ‘None’ versus ‘Weak’ or ‘Strong’ populations (Figure 4B and Supplementary Figure S5), which is in large part due to C>G bubble and G-rich motif descriptors (Figure 4C).
The significant separation obtained with PCA (Figure 4B), where specific classes are not defined a priori (unsupervised approach), encouraged us to test several supervised classification approaches suited for prediction (58). Using the SIMCA software, we generated prediction models based on (i) PCA-class analysis where disjoint (one per class) PCA models are used, (ii) Projections to Latent Structures Discriminant Analysis (PLS-DA) and (iii) Orthogonal PLS-DA (OPLS-DA). The PLS-DA and OPLS-DA methods rely on similar principles to build (O)PLS components representing the best possible compromise between the description of the explanatory variables (here ‘descriptors’) and the prediction of the response (here, ‘None’, ‘Weak’ or ‘Strong’). In OPLS-DA, orthogonal components are also produced to isolate the variation in the explanatory variables that is uncorrelated (orthogonal) to the response (58).
We found the PLS-DA method to be suboptimal, especially to predict the ‘Weak’ class, whereas PCA-class (Supplementary Figure S6) and OPLS-DA (Figure 5A) methods yield comparably high prediction efficiencies for the three Rho-dependent termination classes with overall success rates in the order of 85% upon jackknife cross-validation (Table 1). Since the OPLS-DA model yields slightly better ROC diagnostics than the PCA-class model (Supplementary Figure S7) and does not seem to be subjected to data overfitting (Supplementary Figure S8) (54), we selected this model (OPLS-DA-111 model in Table 1 and Figure 5A) for subsequent analyses. We note that the descriptors of C>G bubbles and G-rich motifs and, to a lesser extent, RNA folding stability (ΔGMfold[av]) and richness in [(YC)N9→13]1→3 motifs provide the largest descriptor contributions to the OPLS-DA-111 model (Supplementary Table S7). We also note that OPLS-DA models built with reduced sets of descriptors (including one with only C>G bubble descriptors) were outperformed by the OPLS-DA-111 model (higher misclassification rates upon jackknife cross-validation and poorer ROC diagnostics; see Table 1 and Supplementary Figure S7).
Figure 5.
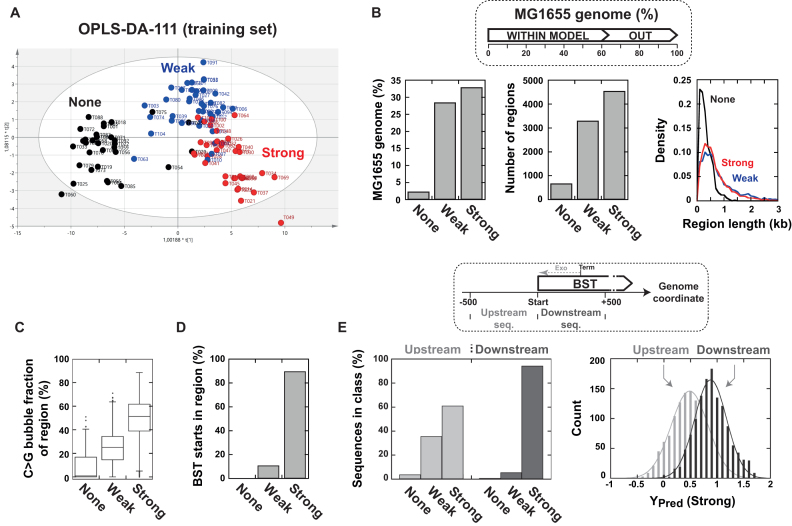
OPLS-DA modelling of Rho-dependent termination of transcription. (A) Standard 2D score plot obtained for the OPLS-DA-111 model with the training set of DNA templates. (B) Analysis of the E. coli MG1655 genome with the OPLS-DA-111 model predicts that regions refractory to Rho-dependent termination (Predicted ‘None’ regions) are fewer and smaller than regions eliciting termination (see Supplementary methods for definition of predicted regions). The proportions of genomic positions for which model predictions are reliable (in-model), or not (out-of-model), according to the PmodXPS+ = 0.05 threshold are shown inset. (C) The C>G bubble content strongly correlates with the predicted termination strength of the regions (ANOVA P < 0.001; F = 2713). Regions as in panel B. (D) The Bicyclomycin Sensitive Transcript (BST) start points (25) fall overwhelmingly within predicted ‘Strong’ regions. (E) Comparison of the termination classes predicted with the OPLS-DA-111 model for 500 nt-long sequences located either upstream or downstream from BST start points (see diagram inset). The distributions of the predicted response scores (Ypred) from SIMCA for the ‘Strong’ class are also shown.
Table 1.
Main features of the multivariate predictive models of Rho-dependent termination
| CV-ANOVAb | Classification of observations into known classesc | ROC area under curve (AUC)e | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | PCsa | R 2 Xcum | R 2 Ycum | Q 2 cum | F | P | Weak | Strong | None | Total | P d | Weak | Strong | None | Remark | |
| PCA-class-111 | N | 3 | 0.546 | n.a. | 0.277 | n.a. | n.a. | 89.5% | 79.4% | 90.6% | 86.5% | 1.2 x 10−6 | 0.861 | 0.928 | 0.956 | One PCA per class with all 111 descriptors |
| W | 3 | 0.480 | n.a. | 0.302 | ||||||||||||
| S | 5 | 0.580 | n.a. | 0.233 | ||||||||||||
| PLS-DA-111 | 1 | 0.271 | 0.359 | 0.345 | 11.5 | 2.1 x 10−8 | 21.0% | 91.2% | 90.6% | 65.4% | 3.0 x 10−7 | 0.578 | 0.876 | 0.973 | PLS-DA with all 111 descriptors | |
| OPLS-DA-111 | 2 (2)f | 0.513 | 0.639 | 0.410 | 4.4 | 2.1 x 10−7 | 86.8% | 82.3% | 87.5% | 85.6% | 1.1 x 10−6 | 0.927 | 0.968 | 0.989 | OPLS-DA with all 111 descriptors | |
| OPLS-DA-40 | 2 (1) | 0.709 | 0.536 | 0.454 | 7.9 | 4.3 x 10−12 | 71% | 76.5% | 84.4% | 76.9% | 9.6 x 10−7 | 0.861 | 0.942 | 0.976 | VIPs < 1 in OPLS-DA-111 removedg | |
| OPLS-DA-83 | 2 (2) | 0.572 | 0.627 | 0.453 | 5.9 | 2.4 x 10−10 | 81.6% | 82.4% | 90.6% | 84.6% | 1.1 x 10−6 | 0.918 | 0.965 | 0.991 | OPLS-DA with only the 83 descriptors that pass ANOVAg | |
| OPLS-DA-6 | 2 (0) | 0.962 | 0,398 | 0,372 | 8.6 | 4.4 x 10−10 | 44.7% | 67.7% | 96.9% | 68.3% | 8.7 x 10−7 | 0.734 | 0.904 | 0.961 | OPLS-DA with C>G Bubble descriptors onlyh | |
aOrthogonal Principal Components (PCs) are in parentheses when relevant. R2Xcum values do not include contributions from orthogonal PCs.
bANOVA of the cross-validated predictive residuals as implemented in SIMCA software.
cClassification based on jackknife cross-validation (see methods).
dFisher's probability of the classification occurring by chance.
eThe AUC of the ROC curve varies from 0.5 (random prediction) to 1.0 (perfect prediction).
fSources of orthogonal variance in OPLS-DA-111 uncorrelated to termination are discussed in Supplementary information.
gAlthough excluding low ranking descriptors based on ANOVA or VIP (variable Importance in the Projection) scores sometimes improves OPLS-DA predictions, this strategy was detrimental in our case (see also Supplementary Figure S5).
hDescriptors used were SumBubLength, SumBubSurf, DenBubLength, DenBubSurf, BublongLength and BublongSurf (see Supplementary information for details).
To further evaluate OPLS-DA modelling of Rho-dependent termination, we looked at OPLS-DA-111 predictions for a test set of known Rho-dependent terminators that were not used for model training (Supplementary Table S4). The OPLS-DA-111 model unambiguously assigned most of the test terminators (83.3%) to the ‘Strong’ category and none to the ‘None’ category (Supplementary Table S4), thereby illustrating the model predictive value. Taken together, these data support the idea that supervised multivariate (e.g. OPLS-DA) models can be built to predict the presence/absence of Rho-dependent termination signals within DNA sequences of interest.
Genome-wide OPLS-DA model prediction of Rho-dependent termination
We compiled OPLS-DA-111 probability scores as a function of genomic position for the complete E. coli MG1655 genome. To facilitate interpretation of the data, we divided both genome strands into successions of distinct regions wherein probability scores for either ‘Out-of-model’, ‘None’, ‘Weak’, or ‘Strong’ termination prevailed at all positions (see methods). Using this strategy, we delineated 8464 regions of interest (i.e. predicted ‘Strong’, ‘Weak’, and ‘None’ regions) representing ∼65% of the genomic positions (Supplementary Table S8). The predicted ‘None’ regions (i.e. regions where probability for ‘no termination’ is highest throughout) were markedly fewer and smaller than the predicted ‘Weak’ or ‘Strong’ termination regions (Figure 5B). This suggests that the portion of the MG1655 transcriptome that is refractory to Rho-dependent termination is limited. This ‘refractory’ portion likely corresponds to genomic regions devoid of extensive C>G bubbles. In line with this proposal, we observed that the predicted ‘None’ regions are characterized by a much smaller C>G bubble content than the other predicted regions (Figure 5C). Similar observations were made upon analysis of the Salmonella LT2 genome with the OPLS-DA-111 model (Supplementary Figure S9 and Supplementary Table S8).
To assess the value of the OPLS-DA-111 predictions at the genome scale, we compared the map of predicted regions for the MG1655 genome (Supplementary Table S8) with that of Bicyclomycin Sensitive Transcripts (BSTs) identified by transcriptomics (25). BSTs arise nearby Rho-dependent termination loci owing to the inhibition of Rho activity by Bicyclomycin (25). About 60% of these BSTs can be used for comparison purposes as they begin within model-compatible regions (PmodXPS+ > 0.05) of the E. coli genome (Supplementary Table S8). Remarkably, most of these BSTs (90%) begin within a ‘Strong’ termination region and none within a ‘None’ region (Figure 5D). This distribution differs significantly from a random distribution of BST start points among the predicted regions (P <10−5; Fisher's exact test).
Since BST start points are expected to be shifted upstream from the bona fide Rho-dependent termination sites due to posttranscriptional [3′→5′]-exonucleolytic trimming of the transcripts (25), we also compared OPLS-DA-111 prediction scores calculated for the 500 nt-long sequences located either upstream or downstream from the BST starts (see Figure 5E, inset). We found that the downstream sequences are more frequently and more strongly associated with the ‘Strong’ termination class than the upstream sequences (Figure 5D), which is consistent with 3′-exonucleolytic trimming of the Rho-dependent transcripts. Taken together, these data strongly suggest that a ‘Strong’ region status (as determined with the OPLS-DA-111 model) is a good prognostic for in vivo Rho-dependent termination. This status likely reflects a sequence composition that is intrinsically favorable to Rho activity but does not provide information on the potential presence of other regulatory signals, such as intrinsic terminators, within the same region. For instance, 2.5% of the ‘Strong’ regions predicted for the MG1655 genome also contain intrinsic terminators listed in regulonDB (59), accounting for ∼40% of the terminators in this database (Supplementary Table S8). Moreover, given their relatively large sizes (Figure 5B), some predicted ‘Strong’ regions are likely to bear several Rho-dependent signals. We thus cannot exclude that some Rho-dependent signals are silent unless transcription read through their upstream (intrinsic or Rho-dependent) terminator partner(s) occurs.
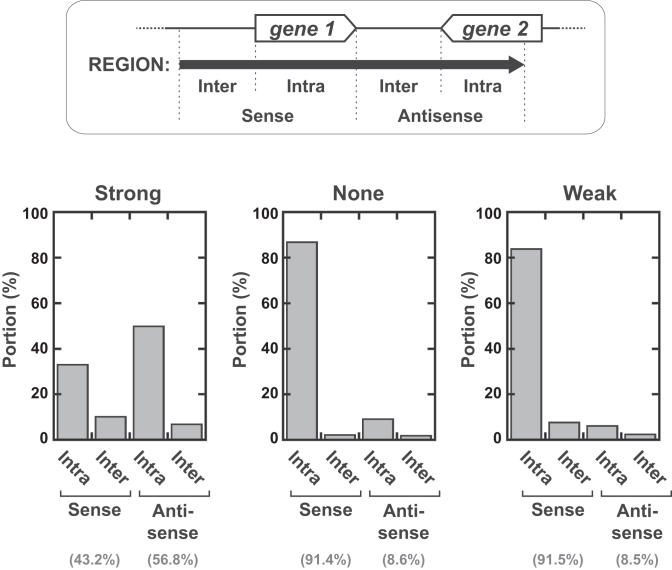
Interestingly, predicted ‘Strong’ regions are frequently found antisense to genes (Figure 6), which is consistent with a prevalent role of Rho-dependent termination in silencing antisense transcription (25). By contrast, ‘None’ and ‘Weak’ regions are primarily associated to intragenic sense sequences (Figure 6), most of which (∼95%) correspond to open reading frames (not shown). Thus, the absence of strong Rho-dependent signals may contribute to protect the expression of some protein-coding genes, possibly to compensate for poor mRNA translation (which would limit mRNA shielding by ribosomes). In agreement with this proposal, we note that both Rho-dependent termination (Figure 4A) and mRNA translation (60) efficiencies are inversely correlated with mRNA folding stability.
Figure 6.
Distribution of the predicted ‘Strong’, ‘None’, and ‘Weak’ regions as a function of gene location in the MG1655 genome. Four distinct categories (Sense-Intragenic, Sense-Intergenic, Antisense-Intragenic and Antisense-Intergenic) were defined with respect to the arrangement of regions and genes (key is inset). Categorization of the intergenic regions as sense/antisense was done with respect to the next downstream gene (‘sense’ if the region and gene are in same strand orientation, ‘antisense’ otherwise). Contributions to these categories were calculated for each predicted region and then summed up for all regions of the same class, as shown in the diagrams.
DISCUSSION
Elucidation of sequence features governing Rho-dependent termination is notoriously difficult (1–3). Early attempts based on the meticulous dissection of known terminators uncovered very few consensus features (reviewed in ref. (1)). These features are: (i) a minimal transcript length of ∼100 nt, (ii) the presence of a Rut site, which is at least ∼80 nt-long and usually rich in C residues and poor in G residues, upstream from the termination endpoint(s), (iii) the paucity of RNA secondary structure in the Rut site region, and (iv) the requirement for signals (of highly variable compositions) triggering RNA pausing at the termination sites. Recent genomics and transcriptomics analyses of Rho-dependent termination in E. coli (23–25,44) essentially confirmed these observations. They also showed that the subset of NusG-dependent terminators (∼20% in E. coli MG1655) are, on average, less C>G-skewed (25) and less dependent on Rut-PBS interactions (44) than the other Rho-dependent terminators but, disappointingly, did not unveil additional termination features of significant predictive value. Although this lack of strong consensus features likely reflects the flexibility necessary to accommodate Rho-dependent terminators within a variety of coding (and regulatory signal) regions (61), it complicates the development of computational models to predict Rho-dependent termination.
To get around this difficulty, we surmised that a quantitative rather than qualitative description of Rho-dependent termination could prove beneficial. We thus defined quantitative descriptors of DNA/RNA composition and examined univariate and multivariate relationships between these descriptors and the in vitro transcription termination responses of a training set of DNA templates. With this approach, we identified descriptors of significant predictive value (Figure 4 and Supplementary Table S7) and were able to build a multivariate prediction model of Rho-dependent termination using OPLS-DA (Figure 5A and Table 1, OPLS-DA-111 model), a supervised classification approach widely used in analytical chemistry and ‘omics’ (54,58). The most relevant descriptors deduced from our analyses gauge traits (e.g. length, area) of the C>G bubbles present in the DNA templates (Supplementary Table S7 and Supplementary Figure S10), thereby lending quantitative support and validation to the C>G bubble concept (13) (Supplementary Figure S1). This concept is important because it stipulates that Rho-dependent termination requires non-template DNA strand (or transcript) regions of uninterrupted (rather than diffuse) C>G skewness. This condition likely reflects the requirement for Rho binding to a largely unstructured and YC-rich Rut site (2,3,14). We note, however, that not only descriptors of the longest C>G bubble but also descriptors of the sum of all C>G bubbles dispersed in the non-template DNA strand stand are among the most relevant descriptors (Supplementary Table S7). This suggests that Rho-dependent termination is a probabilistic event that not only depends on the quality but also on the density of potential Rut sites within the transcribed DNA region. The nature of other significant descriptors supports this view. For instance, richness in [(YC)N9→13]1→3 motifs (which encode 5′-YC dimers suitably spaced for collective interactions with Rho's PBS (16)), not only in the longest C>G bubble but also in the whole DNA template sequence, yields high ANOVA and OPLS-DA ranking descriptors (Supplementary Table S7). The RNA secondary structure potential and density of G-rich motifs (e.g. %G, %GG, %GGG, %GGC, %CGG, %GCG, %GGA, %TGG, %GGT) encoded by the full-length DNA templates also make significant descriptors (Supplementary Table S7), which are inversely correlated with termination (Supplementary Figure S10). This agrees well with transcription experiments showing that aberrant Rho-dependent termination sites can be activated artificially by reducing the secondary structure of transcripts upon incorporation of inosine instead of guanosine (62,63).
We note that a high frequency of pyrimidines (or YC dimers) may not only favor Rut recognition by Rho but may also facilitate subsequent, sequence-sensitive steps such as RNA interaction with the SBS and allosteric closure of the Rho ring required for catalytic activation ((64) and references therein). Moreover, sequences promoting RNAP pausing at the points of Rho-mediated TEC release are usually considered necessary (1–3). The negative correlation observed between the density of G-rich motifs (%G, %GG, %CG or %TG) and termination (Figure 4B and Supplementary Figure S10) argues against a favorable role of elemental pause site motifs (GN8YG or GGN8YG consensus) (65,66). In fact, the frequency of GN8YG (or GGN8YG) motif occurrence is also negatively correlated with termination (Supplementary Figure S11). Thus, RNAP pause sites favoring Rho-dependent termination (1–3) likely follow other sequence rules that are not readily apparent from our analysis.
The mechanistic basis of other significant descriptors is more enigmatic. For instance, positively correlated descriptors %CA, %CAC, %AC, %ACA, %AAC, and %CAA (Supplementary Table S7 and Supplementary Figure S10) may reflect the presence of CA-rich auxiliary motifs in some terminators (1). However, these motifs, which are 6 to 7 nt-long (1), were not detected by RSAT oligo analysis (data not shown). Such motifs may not be frequent or conserved enough to allow detection with our training set of DNA templates which, despite its reasonable size (104 distinct templates having an average length of ∼500 bp), represents less than 0.4% of the MG1655 or LT2 sequence. We anticipate that additional relevant descriptors, of possibly higher sequence complexity, will be found with a larger training set. Increasing the number of observations would also lend higher statistical power to OPLS-DA modelling of Rho-dependent termination. In this respect, sampling genomic sequences that are not reliably modeled by the current OPLS-DA-111 model (Figure 5B, inset) could prove particularly effective. Using new observations to build a ‘naïve’ set of data sufficiently large and representative of each termination class to better evaluate the false positive rate of the model could also prove useful. However, a major hindrance to the significant expansion of the training set or assembly of a naïve dataset (or to the deployment of our method in species harboring potentially divergent Rho-dependent machineries (6,45,48)) rests with the relatively high cost and low-throughput of current transcription termination assays. It will thus be necessary to develop cheaper and higher throughput quantitative termination assays, directly in vivo if possible, to better assess the contribution of accessory factors and to achieve more comprehensive predictions of Rho-dependent termination.
Despite the abovementioned shortcomings, our OPLS-DA-111 model constitutes the first and only computational tool to probe Rho-dependent termination at the genome scale. Most notably, the model predicts that the portion of the MG1655 (or LT2) genome that is intrinsically refractory to Rho action is limited (Figure 5B and Supplementary Figure S9, ‘None’ regions). This suggests that Rho-dependent termination is more constrained by indirect factors such as transcript shielding by ribosomes or formation of antitermination complexes (3,67) than by a lack of termination signals. A wealth of termination signals throughout the genome would best suit Rho acting as a global curator of unwanted events such as transcription-translation uncoupling (24,35) or formation of transcriptional R-loops (68). Notwithstanding, we cannot formally exclude that termination-resistant sequences are underrepresented in OPLS-DA-111 predictions because such sequences could be predominantly located in out-of-model regions (Figure 5B, inset) and thus remain undetected. We also cannot exclude that a fraction of the ‘Weak’ termination signals predicted by the OPLS-DA-111 model are actually too weak to trigger significant effects in vivo. In line with this proposal is the observation that a predicted ‘Strong’ region status is the best prognostic for the Rho-dependent termination loci (Figure 5D and E) identified in E. coli by BST transcriptomics (25). We note, however, that there are 3.6-fold more ‘Strong’ regions than BSTs in the MG1655 genome and that many ‘Strong’ regions are long enough (Figure 5B) to contain several Rho-dependent terminators. Moreover, a number of the Rho-dependent terminators characterized in this work (Supplementary Table S1) or in other studies (28,35,69) were not detected by BST transcriptomics (25). Taken together, these observations strongly suggest that the MG1655 genome contains significantly more Rho-dependent terminators than envisioned from previous work. Such terminators may be latent in the growth conditions used for BST transcriptomics (25) or reside in poorly transcribed regions complicating experimental detection. Rho-dependent signals arranged in tandem with upstream (intrinsic or Rho-dependent) terminators are also likely to be quiescent or undetectable, yet may constitute failsafe signals against upstream terminator bypass. Considering the pervasiveness of transcription in E. coli and Salmonella (25,70,71), we thus propose that additional, unexplored layers of Rho-dependent regulation likely take effect under specific environmental/genetic backgrounds. In support of this proposal is the recent discovery of additional Rho-dependent loci in transcriptomic analyses relying on new identification (28) or genetic perturbation (72) strategies.
Among the tested DNA sequences that yield ‘Strong’ Rho-dependent termination signals in vitro (Supplementary Table S1), several are located in (or comprise) the 5′-untranslated regions (5′UTRs) of genes where they may be involved in specific attenuation mechanisms. This conjecture has already been proven true for a couple of specimens (T031 and T043 templates in Supplementary Table S1) participating in the complex regulation of the rpoS and corA genes, as revealed by studies published since we started the present work (28,31). Specimens that may also be worth investigating include sequences upstream from other highly regulated genes such as moaA (T064), for which there is also in vivo evidence of Rho-dependent termination (28), as well as hcp (T008), gapA (T023), hmp (T028), or acs (T049) (Supplementary Figure S12). More intriguing is the case of the riboswitch-regulated btuB gene, which bears an in vitro termination signal in the beginning of its coding region (T047 template; Supplementary Figure S12 and data not shown). Rho-dependent terminators at similar locations contribute to the in vivo regulation of other riboswitch-dependent genes but could not be detected in the case of btuB (69). We suspect that detection of the btuB terminator was hindered by an experimental limitation such as adenosyl-cobalamin contamination of the growth medium (69) or use of a terminator-selective Rho mutant (44). Indeed, Rho-dependent regulation in the well-conserved 5′UTR of mgtA (9,73) was also not detected with this system (69). Thus, it may be worth looking deeper into the potential involvement of Rho in btuB regulation.
In summary, we have built the largest collection of DNA templates for in vitro characterization of Rho-dependent termination and identified new terminators that could play an important role in the regulation of E. coli, Salmonella, and related species. The resulting dataset was used to develop sequence descriptors and a multivariate OPLS-DA model that fits experimental data (including transcriptomics) reasonably well. This work thus establishes the proof of principle and provides a solid groundwork for computational modelling of Rho-dependent termination (the strategy might even be tested with the NusG-dependent fraction of terminators once a relevant dataset is available for model training). The interest of alternative supervised classification approaches, such as decision tree-based methods (random forest, gradient boosted trees, etc.), may also be assessed in the future. At present, our OPLS-DA-111 model may be used directly to predict Rho-dependent termination in other E. coli or Salmonella strains as well as in species having similar Rho and transcription machineries (computer scripts for descriptors and training set descriptor values necessary for OPLS-DA-111 parametrization are provided in Supplementary Information). Moreover, our method should be easily tuned to in vivo data training provided that a sufficiently large and accurate (i.e. not plagued by posttranscriptional processing) set of transcript endpoints can be constituted. Future efforts should also aim at minimizing the fraction of out-of-model predictions, possibly by developing additional descriptors, and at extending predictions to other, potentially divergent bacterial species.
Supplementary Material
ACKNOWLEDGEMENTS
We warmly thank Lionello Bossi for helpful discussions and critical reading of the manuscript, Yannick Berteaux for help with computer software/hardware installation and maintenance, Norbert Garnier for access to the CBM computer cluster for computationally intensive calculations, and Marylène Bertrand for help with statistical analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Agence Nationale de la Recherche [ANR-15-CE11-0024-02 to M.B., ANR-3-BSV3-0005 to N.F.B.] (in part); PhD fellowship from Région Centre-Val de Loire (to C.N.). Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ciampi M.S. Rho-dependent terminators and transcription termination. Microbiology. 2006; 152:2515–2528. [DOI] [PubMed] [Google Scholar]
- 2. Boudvillain M., Figueroa-Bossi N., Bossi L.. Terminator still moving forward: expanding roles for Rho factor. Curr. Opin. Microbiol. 2013; 16:118–124. [DOI] [PubMed] [Google Scholar]
- 3. Ray-Soni A., Bellecourt M.J., Landick R.. Mechanisms of bacterial transcription Termination: All good things must end. Annu. Rev. Biochem. 2016; 85:319–347. [DOI] [PubMed] [Google Scholar]
- 4. Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012; 149:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Washburn R.S., Gottesman M.E.. Regulation of transcription elongation and termination. Biomolecules. 2015; 5:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grylak-Mielnicka A., Bidnenko V., Bardowski J., Bidnenko E.. Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology. 2016; 162:433–447. [DOI] [PubMed] [Google Scholar]
- 7. Richardson L.V., Richardson J.P.. Rho-dependent termination of transcription is governed primarily by the upstream Rho utilization (rut) sequences of a terminator. J. Biol. Chem. 1996; 271:21597–21603. [DOI] [PubMed] [Google Scholar]
- 8. Guerin M., Robichon N., Geiselmann J., Rahmouni A.R.. A simple polypyrimidine repeat acts as an artificial Rho-dependent terminator in vivo and in vitro. Nucleic Acids Res. 1998; 26:4895–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hollands K., Proshkin S., Sklyarova S., Epshtein V., Mironov A., Nudler E., Groisman E.A.. Riboswitch control of Rho-dependent transcription termination. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Figueroa-Bossi N., Schwartz A., Guillemardet B., D’Heygere F., Bossi L., Boudvillain M.. RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 2014; 28:1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kriner M.A., Sevostyanova A., Groisman E.A.. Learning from the Leaders: Gene regulation by the transcription termination factor rho. Trends Biochem. Sci. 2016; 41:690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rabhi M., Espeli O., Schwartz A., Cayrol B., Rahmouni A.R., Arluison V., Boudvillain M.. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011; 30:2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alifano P., Rivellini F., Limauro D., Bruni C.B., Carlomagno M.S.. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991; 64:553–563. [DOI] [PubMed] [Google Scholar]
- 14. Rabhi M., Rahmouni A.R., Boudvillain M.. Jankowsky E. RNA Helicases. 2010; 19:Cambridge: RSC Publishing; 243–271. [Google Scholar]
- 15. Bogden C.E., Fass D., Bergman N., Nichols M.D., Berger J.M.. The structural basis for terminator recognition by the Rho transcription termination factor. Mol Cell. 1999; 3:487–493. [DOI] [PubMed] [Google Scholar]
- 16. Skordalakes E., Berger J.M.. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003; 114:135–146. [DOI] [PubMed] [Google Scholar]
- 17. McSwiggen J.A., Bear D.G., von Hippel P.H.. Interactions of Escherichia coli transcription termination factor rho with RNA. I. Binding stoichiometries and free energies. J Mol Biol. 1988; 199:609–622. [DOI] [PubMed] [Google Scholar]
- 18. Geiselmann J., Yager T.D., von Hippel P.H.. Functional interactions of ligand cofactors with Escherichia coli transcription termination factor rho. II. Binding of RNA. Protein Sci. 1992; 1:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koslover D.J., Fazal F.M., Mooney R.A., Landick R., Block S.M.. Binding and translocation of termination factor rho studied at the single-molecule level. J Mol Biol. 2012; 423:664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hitchens T.K., Zhan Y., Richardson L.V., Richardson J.P., Rule G.S.. Sequence-specific interactions in the RNA-binding domain of escherichia coli transcription termination factor rho. J. Biol. Chem. 2006; 281:33697–33703. [DOI] [PubMed] [Google Scholar]
- 21. Vieu E., Rahmouni A.R.. Dual role of boxB RNA motif in the mechanisms of termination/antitermination at the lambda tR1 terminator revealed in vivo. J. Mol. Biol. 2004; 339:1077–1087. [DOI] [PubMed] [Google Scholar]
- 22. Schwartz A., Walmacq C., Rahmouni A.R., Boudvillain M.. Noncanonical interactions in the management of RNA structural blocks by the transcription termination rho helicase. Biochemistry. 2007; 46:9366–9379. [DOI] [PubMed] [Google Scholar]
- 23. Cardinale C.J., Washburn R.S., Tadigotla V.R., Brown L.M., Gottesman M.E., Nudler E.. Termination factor rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008; 320:935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peters J.M., Mooney R.A., Kuan P.F., Rowland J.L., Keles S., Landick R.. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peters J.M., Mooney R.A., Grass J.A., Jessen E.D., Tran F., Landick R.. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012; 26:2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nicolas P., Mader U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S. et al. . Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012; 335:1103–1106. [DOI] [PubMed] [Google Scholar]
- 27. Mader U., Nicolas P., Depke M., Pane-Farre J., Debarbouille M., van der Kooi-Pol M.M., Guerin C., Derozier S., Hiron A., Jarmer H. et al. . Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 2016; 12:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sedlyarova N., Shamovsky I., Bharati B.K., Epshtein V., Chen J., Gottesman S., Schroeder R., Nudler E.. sRNA-Mediated control of transcription termination in E. coli. Cell. 2016; 167:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bossi L., Schwartz A., Guillemardet B., Boudvillain M., Figueroa-Bossi N.. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 2012; 26:1864–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brandis G., Bergman J.M., Hughes D.. Autoregulation of the tufB operon in Salmonella. Mol. Microbiol. 2016; 100:1004–1016. [DOI] [PubMed] [Google Scholar]
- 31. Kriner M.A., Groisman E.A.. The bacterial transcription termination factor Rho coordinates Mg homeostasis with translational signals. J. Mol. Biol. 2015; 427:3834–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sevostyanova A., Groisman E.A.. An RNA motif advances transcription by preventing Rho-dependent termination. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6835–E6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takemoto N., Tanaka Y., Inui M.. Rho and RNase play a central role in FMN riboswitch regulation in Corynebacterium glutamicum. Nucleic Acids Res. 2015; 43:520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Ji S.C., Jeon H.J., Lee Y., Lim H.M.. Two-level inhibition of galK expression by Spot 42: Degradation of mRNA mK2 and enhanced transcription termination before the galK gene. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7581–7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sedlyarova N., Rescheneder P., Magan A., Popitsch N., Rziha N., Bilusic I., Epshtein V., Zimmermann B., Lybecker M., Sedlyarov V. et al. . Natural RNA polymerase aptamers regulate transcription in E. coli. Mol. Cell. 2017; 67:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., Bingen E., Bonacorsi S., Bouchier C., Bouvet O. et al. . Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009; 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menouni R., Champ S., Espinosa L., Boudvillain M., Ansaldi M.. Transcription termination controls prophage maintenance in Escherichia coli genomes. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14414–14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lesnik E.A., Sampath R., Levene H.B., Henderson T.J., McNeil J.A., Ecker D.J.. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001; 29:3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Hoon M.J., Makita Y., Nakai K., Miyano S.. Prediction of transcriptional terminators in bacillus subtilis and related species. PLoS Comput. Biol. 2005; 1:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kingsford C.L., Ayanbule K., Salzberg S.L.. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007; 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gardner P.P., Barquist L., Bateman A., Nawrocki E.P., Weinberg Z.. RNIE: genome-wide prediction of bacterial intrinsic terminators. Nucleic Acids Res. 2011; 39:5845–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D.. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011; 8:11–13. [DOI] [PubMed] [Google Scholar]
- 43. Unniraman S., Prakash R., Nagaraja V.. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 2002; 30:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shashni R., Qayyum M.Z., Vishalini V., Dey D., Sen R.. Redundancy of primary RNA-binding functions of the bacterial transcription terminator Rho. Nucleic Acids Res. 2014; 42:9677–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. D’Heygere F., Rabhi M., Boudvillain M.. Phyletic distribution and conservation of the bacterial transcription termination factor Rho. Microbiology. 2013; 159:1423–1436. [DOI] [PubMed] [Google Scholar]
- 46. Nowatzke W.L., Burns C.M., Richardson J.P.. Function of the novel subdomain in the RNA binding domain of transcription termination factor Rho from Micrococcus luteus. J. Biol. Chem. 1997; 272:2207–2211. [DOI] [PubMed] [Google Scholar]
- 47. Mitra A., Misquitta R., Nagaraja V.. Mycobacterium tuberculosis Rho Is an NTPase with distinct kinetic properties and a novel RNA-Binding subdomain. PLoS One. 2014; 9:e107474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. D’Heygere F., Schwartz A., Coste F., Castaing B., Boudvillain M.. ATP-dependent motor activity of the transcription termination factor Rho from Mycobacterium tuberculosis. Nucleic Acids Res. 2015; 43:6099–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boudvillain M., Walmacq C., Schwartz A., Jacquinot F.. Simple enzymatic assays for the in vitro motor activity of transcription termination factor Rho from Escherichia coli. Methods Mol. Biol. 2010; 587:137–154. [DOI] [PubMed] [Google Scholar]
- 50. Rabhi M., Gocheva V., Jacquinot F., Lee A., Margeat E., Boudvillain M.. Mutagenesis-based evidence for an asymmetric configuration of the ring-shaped transcription termination factor Rho. J. Mol. Biol. 2011; 405:497–518. [DOI] [PubMed] [Google Scholar]
- 51. Artsimovitch I., Henkin T.M.. In vitro approaches to analysis of transcription termination. Methods. 2009; 47:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lennart E., Johan T., Svante W.. CV‐ANOVA for significance testing of PLS and OPLS® models. J. Chemometrics. 2008; 22:594–600. [Google Scholar]
- 54. Triba M.N., Le Moyec L., Amathieu R., Goossens C., Bouchemal N., Nahon P., Rutledge D.N., Savarin P.. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. bioSyst. 2015; 11:13–19. [DOI] [PubMed] [Google Scholar]
- 55. Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C. et al. . RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015; 43:W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Skordalakes E., Berger J.M.. Structural Insights into RNA-Dependent ring closure and ATPase activation by the rho termination factor. Cell. 2006; 127:553–564. [DOI] [PubMed] [Google Scholar]
- 57. Abdi H., Williams L.J.. Principal component analysis. Wiley Interdiscipl. Rev.: Comput. Stat. 2010; 2:433–459. [Google Scholar]
- 58. Bylesjö M., Rantalainen M., Cloarec O., Nicholson J.K., Holmes E., Trygg J.. OPLS discriminant analysis: combining the strengths of PLS‐DA and SIMCA classification. J. Chemometrics. 2006; 20:341–351. [Google Scholar]
- 59. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muniz-Rascado L., Garcia-Sotelo J.S., Alquicira-Hernandez K., Martinez-Flores I., Pannier L., Castro-Mondragon J.A. et al. . RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Richardson J.P. Rho-dependent transcription termination. Biochim. Biophys. Acta. 1990; 1048:127–138. [DOI] [PubMed] [Google Scholar]
- 62. Morgan W.D., Bear D.G., von Hippel P.H.. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J. Biol. Chem. 1983; 258:9553–9564. [PubMed] [Google Scholar]
- 63. Zhu A.Q., von Hippel P.H.. Rho-dependent termination within the trp t' terminator. I. Effects of rho loading and template sequence. Biochemistry. 1998; 37:11202–11214. [DOI] [PubMed] [Google Scholar]
- 64. Lawson M.R., Dyer K., Berger J.M.. Ligand-induced and small-molecule control of substrate loading in a hexameric helicase. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:13714–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vvedenskaya I.O., Vahedian-Movahed H., Bird J.G., Knoblauch J.G., Goldman S.R., Zhang Y., Ebright R.H., Nickels B.E.. Interactions between RNA polymerase and the "core recognition element" counteract pausing. Science. 2014; 344:1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. et al. . A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014; 344:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Santangelo T.J., Artsimovitch I.. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011; 9:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leela J.K., Syeda A.H., Anupama K., Gowrishankar J.. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bastet L., Chauvier A., Singh N., Lussier A., Lamontagne A.M., Prevost K., Masse E., Wade J.T., Lafontaine D.A.. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 2017; 45:7474–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raghavan R., Sloan D.B., Ochman H.. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio. 2012; 3:e00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G.. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015; 197:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Raghunathan N., Kapshikar R.M., Leela J.K., Mallikarjun J., Bouloc P., Gowrishankar J.. Genome-wide relationship between R-loop formation and antisense transcription in Escherichia coli. Nucleic Acids Res. 2018; 46:3400–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hollands K., Sevostiyanova A., Groisman E.A.. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E1999–E2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Epshtein V., Dutta D., Wade J., Nudler E.. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010; 463:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thomsen N.D., Berger J.M.. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009; 139:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gocheva V., Le Gall A., Boudvillain M., Margeat E., Nollmann M.. Direct observation of the translocation mechanism of transcription termination factor Rho. Nucleic Acids Res. 2015; 43:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.