Abstract
Protein synthesis is a complex and highly coordinated process requiring many different protein factors as well as various types of nucleic acids. All translation machinery components require multiple maturation events to be functional. These include post-transcriptional and post-translational modification steps and methylations are the most frequent among these events. In eukaryotes, Trm112, a small protein (COG2835) conserved in all three domains of life, interacts and activates four methyltransferases (Bud23, Trm9, Trm11 and Mtq2) that target different components of the translation machinery (rRNA, tRNAs, release factors). To clarify the function of Trm112 in archaea, we have characterized functionally and structurally its interaction network using Haloferax volcanii as model system. This led us to unravel that methyltransferases are also privileged Trm112 partners in archaea and that this Trm112 network is much more complex than anticipated from eukaryotic studies. Interestingly, among the identified enzymes, some are functionally orthologous to eukaryotic Trm112 partners, emphasizing again the similarity between eukaryotic and archaeal translation machineries. Other partners display some similarities with bacterial methyltransferases, suggesting that Trm112 is a general partner for methyltransferases in all living organisms.
INTRODUCTION
In all living cells, the ribosome nanomachine assisted by tRNAs and translation factors decodes the genetic information contained within messenger RNAs (mRNAs) to synthesize the corresponding proteins. mRNA translation is a highly complex process which in addition to the numerous factors involved, requires RNA and protein maturation events for faithful and timely protein production. Among those events, post-transcriptional modifications are found on all components and in the three domains of life. Indeed, transfer RNAs (tRNAs) are heavily modified with various different chemical structures added on nucleotides, thereby contributing to their stability and to translation accuracy (1). In eukaryotic and archaeal ribosomal RNAs (rRNAs), modifications such as pseudouridylation or 2′-O-methylation are introduced by RNA-guided multi-protein enzymes (2) while modifications on the bases such as methylation are introduced by protein-only enzymes (3). These rRNA modifications cluster mostly to functionally important regions of the ribosome (peptidyl transferase center, tRNA and mRNA binding sites, subunit-subunit interface) and hence contribute to its optimal activity (4,5). More recently, the N6-methyladenosine (m6A) mRNA modification has been the focus of many studies aimed at clarifying the role of this epitranscriptomics modification on mRNA splicing, transport, translation and decay (6–9). Finally, translation factors are also subject to many post-translational events such as methylation. This is indeed the case for uS13, uL5 and eL42 ribosomal proteins ((10); ribosomal proteins are numbered according to the naming proposed by Ban et al. (11)) as well as translation elongation factors eEF1A, eEF2 and eEF3 (this later is only present in yeasts; (12–17)). Bacterial (RF1 and RF2) and eukaryotic (eRF1) class I translation termination factors are also methylated on the glutamine side chain of the universally conserved GGQ motif that interacts with the peptidyl transferase center to trigger the release of the newly synthesized proteins (18–23). The methylation of the GGQ motif of the bacterial RF1 and RF2 proteins is catalyzed by PrmC. In eukaryotes, the same motif of eRF1 is modified by a complex formed by the Mtq2 methyltransferase (MTase) catalytic subunit and its activator Trm112 subunit (19,20,24–27). Interestingly, in Saccharomyces cerevisiae, the Trm112 protein also interacts with and activates three other MTases, namely Bud23, Trm9 and Trm11, which modify factors involved in translation (28). The Bud23–Trm112 complex and its human ortholog (BUD23-TRMT112 or WBSCR22-TRMT112) catalyze the N7-methylation of G nucleotide (m7G) at positions 1575 and 1639 on the yeast and human 18S rRNA, respectively, and these complexes are essential for 40S biogenesis (29–35). The Trm11–Trm112 complex catalyzes the addition of a methyl group on nucleotide G10 from several tRNAs to form N2-methylguanosine (m2G) thereby most likely contributing to their stability (36,37). The Trm9–Trm112 complex and its human ortholog ALKBH8-TRMT112 are involved in the 5-methoxycarbonylmethyluridine (mcm5U) modification of the wobble position from the anticodon loop of some tRNAs and hence enhance translational fidelity and efficiency (38–47). It is noteworthy that human TRMT112 partners have been associated with diseases and are potential targets for drug developments. Indeed, ALKBH8 protein is highly expressed in a variety of human cancer cells including bladder cancer. Furthermore, ALKBH8 silencing induces apoptosis of urothelial carcinoma cells thereby suppressing tumor growth, angiogenesis and metastasis and also renders the cells sensitive to methyl methane sulfonate (MMS) and to the anti-cancer drug bleomycin (41,48). Human BUD23 could be a tumor marker for invasive breast cancer, myeloma cells and hepatocarcinoma (49–51) and could contribute to lung pathologies (52).
Bioinformatics analyses have revealed the presence of Trm112 orthologues in bacteria and archaea suggesting that its role might extend outside eukaryotic organisms (25,28,36). While nothing is known on bacterial Trm112 orthologues, the detection of Mtq2, Trm9 and Trm11 orthologues in archaeal genomes together with the strong similarity between eukaryotic and archaeal translation machineries suggest that archaeal Trm112 might play a role similar to the eukaryotic Trm112 (53–55). Indeed, Trm11 orthologues (hereafter named aTrm11) from Pyrococcus abyssii and Thermococcus kodakarensis have been biochemically characterized as enzymes methylating guanine nucleotide at position 10 of some tRNAs (56,57). However, these two enzymes not only catalyze the formation of N2-methylguanosine but also of N2,2-dimethylguanosine and do not require Trm112 to be active. Regarding Trm9, several observations argue in favor of its presence in some archaea. First, an initial survey of Haloferax volcanii genome suggested that the HVO_0574 gene encodes for a potential Trm9 orthologue (58) but a more recent analysis proposed HVO_1032 gene as a better candidate (59). Second, genes encoding for proteins displaying some sequence similarity with the various enzymes (Elp3, Tuc1 and Trm9) involved in the formation of mcm5s2U modification at position 34 of tRNAs are present in H. volcanii and genes for Elp3 and Trm9 orthologues cluster in Sulfolobus solfataricus (58). Third, studies in the early 90’s revealed the presence of unknown modifications at position U34 of some tRNAs from H. volcanii (60). Regarding class I release factors, it is striking that despite radically different 3D-structures of the bacterial and eukaryotic factors, dedicated machineries have evolved to methylate the glutamine side chain of the universally conserved GGQ motif. Hence, one can imagine that such modification also exists on archaeal aRF1. Considering the structural similarity between aRF1 and eukaryotic eRF1 factor (61,62), the enzyme responsible for this modification is very likely to be orthologous to Mtq2. This is further supported by the presence of an Mtq2 ortholog in all archaeal phyla (25,28). Finally, so far, no m7G modification of the nucleotide corresponding to S. cerevisiae G1575 in archaeal 16S rRNA has been found, in agreement with the absence of proteins with significant sequence homology with Bud23 in archaeal genomes (28).
To clarify the roles of archaeal Trm112, we identified potential partners of Trm112 in the H. volcanii model organism by co-immunoprecipitation followed by mass spectrometry-based proteomic identification. We validated some of these partners, characterized the in vivo and in vitro biochemical functions of two of these and solved the crystal structure of another one. Altogether, our results show that H. volcanii Trm112 (hereafter named HvoTrm112) displays striking similarities with its eukaryotic orthologs but is also able to interact with a much larger number of MTases than the eukaryotic Trm112.
MATERIALS AND METHODS
Deletion of TRM112 and TRM9 genes in H. volcanii
The chromosomal copy of the HVO_1131 gene (hereafter named HVO_TRM112) encoding for HvoTrm112 protein was deleted by the pop-in/pop-out method to generate the Haloferax volcanii H98 trm112Δ strain (HvoMG5; Supplementary Table S1) using published protocols (63,64). First, two 400 bp fragments covering the upstream (US) and downstream (DS) regions of the HVO_TRM112 gene were amplified from H. volcanii genome by PCR using oligonucleotides oMG86/oMG87 and oMG88/oMG89, respectively (Supplementary Table S2). The PCR products were then digested with BamHI/XbaI and XhoI/BamHI restriction enzymes, respectively and then cloned into XbaI/XhoI digested plasmid pTA131 to obtain pTA131 containing US-DS of HVO_TRM112 (pMG613) used for the pop-in/pop-out procedure.
Colonies resulting from excision of the plasmid (pop-out) were screened by colony lift to check for insertion of the Flag-tag sequence. A DIG-labelled PCR product using oligonucleotides oMG320 and oMG321 on genomic DNA was used as a probe, and detected using the DIG Luminescent Detection kit (Roche) and a ChemiDoc MP (BioRad). 52% of the colonies proved to be deleted for HVO_TRM112 gene (Supplementary Figure S1A).
An additional screening was performed by PCR on some colonies using oMG163 and oMG164 oligonucleotides (Supplementary Table S2) to amplify the whole upstream and downstream parts of HVO_TRM112 gene (Supplementary Figure S1B). Parts of colonies were picked and lysed by mixing in 50 μl water, and then used as DNA templates for PCR reaction. The PCR was performed with Q5 High-Fidelity DNA Polymerase (Biolabs), according to manufacturer's instruction (PCR conditions: one 30 s cycle at 98°C followed by 30 cycles (10 s at 98°C; 20 s at 62°C and 30 s at 72°C) and a final step of 2 min at 72°C).
To study the enzymatic activity of HvoTrm9–Trm112 complex, an H. volcanii H26 strain deleted for the HVO_1032 gene (HvoMG2; Supplementary Table S1), which encodes for the putative Trm9 orthologue, was generated by the pop-in/pop-out method. First, two 400 bp fragments of upstream (US) and downstream (DS) regions surrounding HVO_1032 gene in H. volcanii genome were amplified by PCR using oligonucleotides oMG72/oMG73 and oMG74/oMG75, respectively (Supplementary Table S2). The PCR products were then digested with BamHI/XbaI and XhoI/BamHI restriction enzymes, respectively and then cloned into XbaI/XhoI digested plasmid pTA131 to obtain pTA131 harboring US-DS of HVO_1032 (pMG612). This plasmid was then used for pop-in/pop-out to delete HVO_1032 gene as described above for the generation of the H. volcanii H98 trm112Δ strain. PCR were performed on genomic DNA from different colonies using oMG73 and oMG162 (Supplementary Table S2) and revealed that 50% of the colonies were deleted for HVO_1032 gene (Supplementary Figure S2).
Co-immunoprecipitation of HvoTrm112-Flag protein
To identify HvoTrm112 protein partners, we expressed a Trm112-Flag protein from a plasmid and performed co-immunoprecipitation (co-IP) under cross-linking conditions as described below. First, the HVO_TRM112 gene was amplified from H. volcanii genomic DNA using oligonucleotides oMG320/oMG321 (this later includes a sequence encoding for a Flag-tag; Supplementary Table S2). The PCR product was digested by NdeI and NotI restriction enzymes and later inserted into pTA962 digested with the same enzymes to yield pMG772. The pMG772 or pMG807 (pTA927-derived plasmid expressing only the Flag-tag as a control; generous gift from Prof. Anita Marchfelder, Ulm University, Germany) plasmids purified from competent dam− E. coli cells were transformed into HvoMG5 strain according to the pop-in procedure described above. The transformants were plated on YPC and incubated at 45°C for at least 5 days. This resulted in the H. volcanii H98 trm112Δ HvoTrm112-Flag and trm112Δ Flag-only strains.
Next, one colony of H. volcanii strain H98 trm112Δ HvoTrm112-Flag was used to over-express HvoTrm112-Flag. First, the colonies were inoculated to 10 mL YPC (supplemented with thymidine 60 μg/mL), incubated O/N at 45°C and 150 rpm. In the next day, 2.5 ml O/N culture was applied to 1 L YPC (+thymidine 60 μg/ml) and grown O/N at 45°C and 150 rpm. On the third day, 18% SW-dissolved tryptophan was added to the O/N culture to the final concentration of 5 mM, followed by 6 h incubation at 45°C and 150 rpm for protein over-expression. The cells were harvested by centrifugation at 4000 rpm for 30 min. For the Flag-only control, the same protocol was used for Hvo H98 trm112Δ Flag-only strain.
Co-IP experiments of HvoTrm112-Flag and of Flag-only were carried out as described in Fischer et al. (65), with some slight modifications (see supplementary materials for details). Proteomic analysis details are given in the supplementary materials.
In vitro studies of HvoTrm112-MTase complexes
The procedures for cloning, expression and purification of proteins and protein complexes used for in vitro studies (enzymology, SEC-MALLS and X-ray crystallography) are described in supplementary materials. The tRNAs from S. cerevisiae or H. volcanii strains were purified as previously described (47).
The MTase assays were performed in a total volume of 10 μl (for HvoMtq2-Trm112) or 20 μl (for HvoTrm9–Trm112) containing 400 mM potassium phosphate buffer pH 7.5, 3 M KCl, 2.5 mM EDTA, 5 mM MgCl2, 5 mM NH4Cl, 0.25 mg/ml Bovine Serum Albumin, 50 μM S-adenosyl-l-methionine (SAM; containing 0.87 Ci/mmol of [3H]-SAM; Perkin Elmer) and 5 pmol of HvoMtq2–Trm112 complex or 2 pmol of HvoTrm9–Trm112 complex. The reaction was initiated by adding 100 pmol of substrate (HvoaRF1, HvoaRF3 or total tRNAs) to the mixture. After incubation for 2 h at 45°C, the reaction was stopped by precipitation with cold trichloroacetic acid (5%), followed by filtration on Whatman GF/C filters. The [3H] incorporation was measured using a Beckman Coulter LS6500 scintillation counter.
Crystallization and structure determination of the Hvo_0019–Trm112 complex
Crystals of the Hvo_0019–Trm112 complex were obtained by mixing 1 μL of protein complex (15 mg/ml in buffer B: 1 M NaCl, 20 mM Tris–HCl pH 7.5, 5 mM β-mercaptoethanol, 10 μM ZnCl2) with an equal volume of crystallization solution (0.1–0.3 M NaCl; 0.1 M Bis–Tris pH 5.5; 20–25% w/v PEG3350) at both 4°C and 24°C. Crystals appeared within 4–5 h and reached their maximum size (400 μm length) within 3 days. Crystals were cryo-protected by transfer into their crystallization condition supplemented with progressively higher glycerol concentration up to 30% and then flash-frozen in liquid nitrogen.
All datasets were collected on beam-line Proxima-2A (Synchrotron SOLEIL, Saint-Aubin, France) at 100 K. The structure was solved by Sulfur-SAD (Single Anomalous Dispersion) using a highly redundant dataset (seven datasets of 1400° each collected on different regions of a single crystal) collected at 2.0664 Å to get strong anomalous signal from sulfur atoms (see Supplementary Table S3 for statistics). A 1.35 Å resolution dataset was obtained by merging two datasets collected at different crystal-detector distances on another crystal (Supplementary Table S3). Data were processed with XDS (66) and scaled using XSCALE. All crystals belonged to space group P212121 with two copies of Hvo_0019–Trm112 complex per asymmetric unit. Twenty-four sulfur sites (eleven per Hvo_0019–Trm112 complex and two from S-adenosyl-l-homocysteine (SAH) molecules bound to Hvo_0019 methyltransferase) were successfully located using SHELXD (67). Experimental phasing followed by density modification were performed with the PHASER_EP and RESOLVE programs implemented in the PHENIX suite (68–71). The final model was obtained by iterative cycles of building and refinement performed using COOT (72) and BUSTER (73) programs, respectively (Supplementary Table S3). The final model for Hvo_0019–Trm112 complex contains Hvo_0019 residues 2–227 (including the first histidine from the His6-tag) and 2–12, 15–231 (including five histidine residues from the His6-tag) for protomers A and B, respectively as well as HvoTrm112 residues 1–59 and 1–25, 29–58 for protomers C and D, respectively. In addition, 387 water molecules, two SAH molecules and two glycerol molecules from the cryoprotectant solution have also been modeled. The coordinates and structure factors files are available from the Protein Data Bank (PDB) under accession code 6F5Z.
RESULTS
Identification of several partners of HvoTrm112 protein by co-immunoprecipitation and proteomics
HvoTrm112 protein partners were isolated by co-immunoprecipitation (co-IP) of Flag-tagged HvoTrm112 (Figure 1A). For this purpose, we deleted the TRM112 gene in the H. volcanii H98 background by the pop-in/pop-out method as described in the methods section. Among all colonies, 52% lacked this gene indicating that TRM112 is not an essential gene (Supplementary Figure S1A and B). The deletion of this gene did not affect generation time but resulted in much smaller colonies (Supplementary Figure S1C and D). The H. volcanii trm112Δ strain was then transformed with a plasmid encoding a C-terminally Flag-tagged version of HvoTrm112 under the control of tryptophan inducible promoter. The trm112Δ strain was also transformed with a plasmid expressing only the Flag-tag (kind gift from Prof. Anita Marchfelder, Ulm University, Germany) as negative control. Before cell lysis, an in vivo cross-linking step was performed in the co-IP protocol by adding 1% formaldehyde as described by Fischer and colleagues (65). This step was added for the following reasons. First, an initial co-IP experiment with no cross-linking resulted in a low number of specific peptide spectra in the mass spectrometry analysis (data not shown). Second, HvoTrm112 could interact with some of its partners in a transient manner. Third, protein–protein interactions in this organism might be maintained by high salt concentration while the low salt concentration used during the co-IP protocol could induce complex dissociation and/or result in protein aggregation (Figure 1A). Proteins co-immunoprecipitated with HvoTrm112-Flag or with the Flag-tag control were identified by mass spectrometry as described in the Supplemental methods. This revealed that the cross-linking step resulted in an increased number of proteins identified by mass spectrometry coupled to a higher number of specific peptide MS/MS spectra.
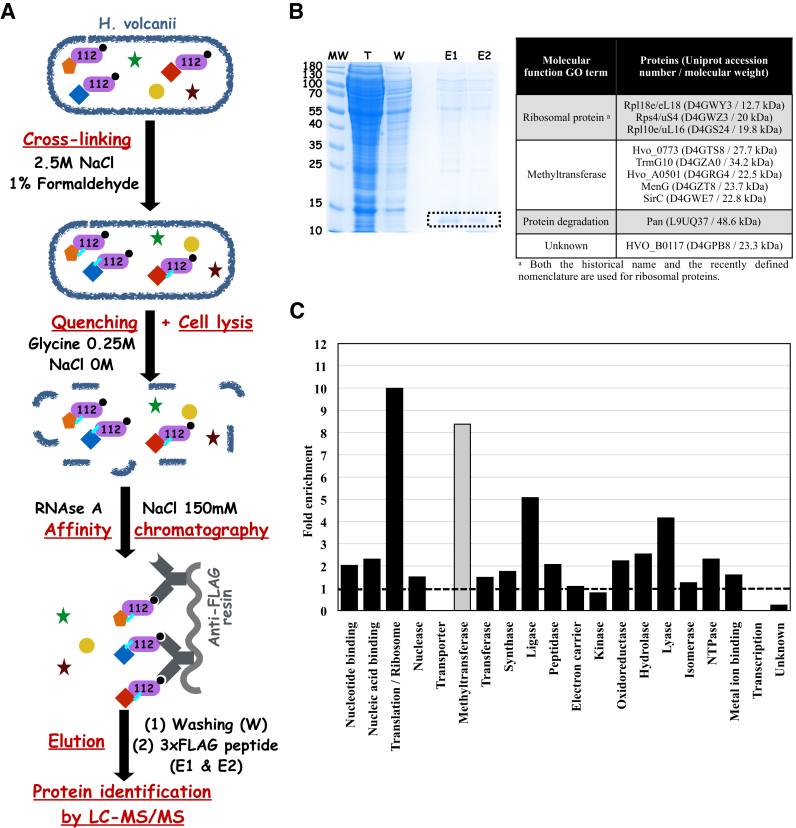
Figure 1.
Trm112 interaction network in H. volcanii. (A) Schematic representation of the protocol used for co-immunoprecipitation of HvoTrm112-Flag (purple shape) under cross-linking conditions. H. volcanii proteins are depicted as colored and filled geometric shapes. Cross-links are depicted by cyan lines. (B) SDS-PAGE analysis of the different purification steps. MW corresponds to molecular weight marker, T to Total extract,W to Washing fraction, E1 and E2 to Elution fractions. The bands corresponding to HvoTrm112-Flag in the E1 and E2 fraction are highlighted by a black dashed box. The table analyses the molecular function gene ontologies of the 10 proteins exhibiting the higher NSAF values (excluding HvoTrm112). (C) Enrichment of major ‘molecular function’ GO terms in the 100 proteins exhibiting the higher NSAF values in the HvoTrm112-Flag co-IP experiments. Fold enrichment was calculated as the ratio between the percentage of proteins from a given ‘molecular function’ GO terms in the list of 100 proteins exhibiting the higher NSAF values and the percentage of proteins from the same ‘molecular function’ GO terms in the entire H. volcanii proteome. The dashed line shows an enrichment of one fold. Methyltransferase ‘molecular function’ GO term is shown in grey and was not included in the transferase ‘molecular function’ GO term in this analysis.
As suggested by the SDS-PAGE analysis of the co-IP results (Figure 1B and Supplementary Figure S3), >1000 proteins were identified by the LC–MS/MS analyses of four independent co-immunoprecipitation experiments of HvoTrm112-Flag whereas only a few hundreds were identified in the negative control experiments with the Flag-only construct (Supplementary Table S4). To reduce this extensive list, a series of stringent MS-based criteria (see supplemental information) were applied to select the most confident protein candidates. In addition, proteins already identified by Fischer et al. (65) in their co-immunoprecipitation of HvoLsm-Flag (performed in identical conditions to those used in our study) were considered as non-specific interacting partners (most probably, highly abundant proteins) and were removed. Those strict filtering criteria led to a final list of 499 proteins (Supplementary Table S5).
We then ranked these proteins based on their Normalized Spectral Abundance Factor (NSAF; (74)) and observed that HvoTrm112 exhibited the higher NSAF value in agreement with its role as bait protein (Table 1). Next, the analysis of the molecular functions (as defined by Gene Ontology terms) of the 100 proteins with the highest NSAF values in the HvoTrm112-Flag co-IP experiments highlighted two molecular functions, i.e. translation/ribosome and methyltransferase (MTase), as strongly enriched among the potential Trm112 partners (Figure 1B–C). Proteins endowed with ‘ligase’ molecular function are also enriched, although to a lesser extent, and more than half of these ligases (16 out of 29) are involved in protein synthesis process (i.e. amino-acyl tRNA synthetases or enzymes involved in amino acid metabolism). This fits with the function of the eukaryotic ortholog Trm112 as an activator of MTases modifying factors involved in translation (rRNAs, tRNAs, release factors). Further analysis revealed that 23 MTases out of the 62 predicted in the H. volcanii proteome are present in the list of 499 proteins (Table 1). Seventeen of those are members of the class I MTase family and include homologs to known eukaryotic Trm112 MTase partners : Mtq2 (PrmC or Hvo_2744, hereafter termed HvoMtq2), Trm9 (Hvo_1032, named HvoTrm9; (59)) but also Trm11 (TrmG10/HvoTrm11). Five are of the class III family (75) and one is not a SAM-dependent enzyme (MetE1). Hence, the HvoTrm112 interaction network shares common features with the eukaryotic Trm112 interactome but might be more complex.
Table 1.
Trm112 and methyltransferases detected in the HvoTrm112-Flag co-immunoprecipitation experiments
| Ranka | Symbol | UniProt accession number | NSAF | Putative function | Validated interaction with HvoTrm112? | Predicted SAM-dependent MTase class |
|---|---|---|---|---|---|---|
| 1 | Trm112 | D4GW82 | 0.0598 | Methyltransferase activator | - | - |
| 2 | HVO_0773 | D4GTS8 | 0.0193 | Unknown | YES | Class I |
| 6 | TrmG10 | D4GZA0 | 0.0088 | tRNA (Guanine(10).N(2))-dimethyltransferase | NOb | Class I |
| 7 | HVO_A0501 | D4GRG4 | 0.0084 | Unknown | Not tested | Class I |
| 8 | MenG | D4GZT8 | 0.0084 | Demethylmenaquinone methyltransferase | Not tested | Class I |
| 9 | SirC | D4GWE7 | 0.0083 | Uroporphyrin-III C-methyltransferase | Not tested | Class III |
| 17 | HVO_0475 | D4GS15 | 0.0064 | Unknown | YES | Class I |
| 18 | HVO_1475 | D4GYB0 | 0.0064 | DNA methylase | Not tested | Class I |
| 33 | CbiT | D4GP64 | 0.0051 | cobalt-precorrin-6B C(15)-methyltransferase | Not tested | Class I |
| 38 | CbiH2 | D4GP59 | 0.0045 | Precorrin-3B C17-methyltransferase | Not tested | Class III |
| 41 | HVO_1032 | D4GVK8 | 0.0044 | tRNA (Uracil(34)) methyltransferase | YES | Class I |
| 42 | HVO_0574 | D4GSG9 | 0.0043 | Unknown | YES | Class I |
| 59 | HVO_2875 | D4GXJ1 | 0.0037 | Unknown | YES | Class I |
| 64 | HVO_0019 | D4GYL4 | 0.0035 | 24-sterol C-methyltransferase | YES | Class I |
| 132 | AglP | D4GYG5 | 0.0023 | Hexuronic acid methyltransferase | Not tested | Class I |
| 140 | PrmC | D4GW96 | 0.0021 | Class I translation termination factor methyltransferase | YES | Class I |
| 149 | CbiH1 | D4GP60 | 0.0019 | Precorrin-3B C17-methyltransferase | Not tested | Class III |
| 211 | CbiF | D4GP62 | 0.0015 | Cobalamin biosynthesis precorrin-3 methylase | Not tested | Class III |
| 240 | HVO_1534 | D4GYH8 | 0.0014 | Unknown | Not tested | Class I |
| 260 | HVO_1715 | D4H060 | 0.0013 | Unknown | YES | Class I |
| 273 | MetE1 | D4GW90 | 0.0012 | Methionine synthase | Not tested | SAM-independent MTase |
| 289 | HVO_1093 | D4GW13 | 0.0011 | Protein-L-isoaspartate O-methyltransferase | Not tested | Class I |
| 310 | Dph5 | D4GUZ5 | 0.0010 | Diphtine synthase | Not tested | Class III |
| 341 | HVO_2664 | D4GV35 | 0.0009 | Unknown | Not tested | Class I |
aProteins were ranked from the highest to the lowest NSAF values.
bThe interaction between these two proteins was not confirmed as HvoTrm11 could not be expressed as a soluble protein in E. coli in the absence or the presence of HvoTrm112.
Validation of several HvoTrm112-MTase complexes
To validate the co-IP and proteomic results, a subset of the identified SAM-dependent class I MTases was selected to test whether they are indeed bona fide binding partners of HvoTrm112. In addition to the H. volcanii homologs of the known eukaryotic Trm112 partners, namely HvoMtq2, HvoTrm9 and HvoTrm11, six other MTases were chosen: Hvo_0773 (exhibiting the highest NSAF value among the detected MTases), Hvo_0475, Hvo_0574 (initially proposed to be the Trm9 archaeal orthologue (58)), Hvo_2875, Hvo_0019 and Hvo_1715 (Table 1). N-terminal His6-tagged versions of these MTases were expressed in E. coli either alone or with untagged HvoTrm112 and purified on Ni-NTA resin as described in Supplementary Materials and Methods section. Interestingly, similarly to S. cerevisiae Mtq2, Trm9 and Bud23 (25,30,42), most MTases tested (HvoMtq2, HvoTrm9, Hvo_0019, Hvo_1715, Hvo_0773 and Hvo_2875) can only be over-expressed as soluble proteins in the presence of HvoTrm112 (Supplementary Figure S4A). Only two (Hvo_0475 and Hvo_0574) do not require HvoTrm112 for efficient soluble expression (Supplementary Figure S4B). Next, we performed a 3-steps (Ni-NTA, ion-exchange and size-exclusion chromatographies) purification protocol from E. coli cultures co-expressing each His-tagged MTase with HvoTrm112. In all cases but HvoTrm11 (see below), at the end of this stringent purification protocol, we could observe a second major band, migrating at the expected size for HvoTrm112 in addition to the band corresponding to the tagged MTase. Mass spectrometry analyses confirmed that for each MTase tested, these bands correspond to both the MTase of interest and to HvoTrm112 (data not shown). This demonstrates that HvoTrm112 interacts individually with almost all the tested MTases and that the resulting complexes are stable. SEC-MALLS analyses further revealed that these HvoTrm112-MTase complexes adopt different oligomeric states (Table 2; Supplementary Figure S5), i.e. heterodimers (HvoTrm9–Trm112, HvoMtq2–Trm112, Hvo_0773–Trm112, Hvo_0574–Trm112 and Hvo_1715-Trm112), heterotetramers (Hvo_0019–Trm112) or heterohexamers (Hvo_0475-Trm112). Despite extensive efforts (optimized gene, fusion with GST, different E. coli expression strains or temperature), it was not possible to express HvoTrm11 as a soluble protein, neither alone nor in the presence of HvoTrm112 (Supplementary Figure S4C). Hence, we cannot conclude about the direct interaction between HvoTrm11 and HvoTrm112.
Table 2.
Oligomeric states of HvoTrm112-MTase complexes
| Trm112-MTase complex | Theoretical MW of heterodimer (kDa) | Experimental MW determined by SEC-MALLS (kDa) | Oligomeric states |
|---|---|---|---|
| HvoTrm9–Trm112 | 31 | 29.5 | Heterodimer |
| HvoMtq2-Trm112 | 28.9 | 27.3 | Heterodimer |
| Hvo_0019–Trm112 | 33.2 | 59.8 | Heterotetramer |
| Hvo_0574-Trm112 | 36.8 | 34.6 | Heterodimer |
| Hvo_0475-Trm112 | 40.1 | 117 | Heterohexamer |
| Hvo_0773-Trm112 | 35.4 | 33.9 | Heterodimer |
| Hvo_1715-Trm112 | 34.6 | 32.3 | Heterodimer |
| Hvo_2875-Trm112 a | 28.6 | ND | ND |
aDue to low yields, SEC-MALLS analysis could not be performed on this complex.
ND: Not determined.
We thus conclude that in H. volcanii, Trm112 interacts directly with at least eight different MTases (HvoMtq2, HvoTrm9, Hvo_0019, Hvo_0773, Hvo_0574, Hvo_0475, Hvo_2875 and Hvo_1715). Its interaction network is then much more complex than for its eukaryotic orthologs studied so far (28).
Characterization of HvoMtq2–Trm112 and HvoTrm9–Trm112 enzymatic activities
Among all these HvoTrm112 partners, there are clear orthologues for two eukaryotic Trm112 partners, namely Mtq2 and Trm9, paving the way towards their functional characterization.
In eukaryotes, the Mtq2-Trm112 complex is enzymatically active on class I translation termination factor eRF1 but only when this later exists as a complex with the GTP-bound form of class II translation termination factor eRF3 (25,26). To characterize the enzymatic activity of the recombinant HvoMtq2-Trm112 complex in vitro, we over-expressed in E. coli and purified both HvoaRF1 and HvoaRF3 (also known as HvoaEF1A). First, the HvoaRF1 methylation activity of the HvoMtq2-Trm112 complex, in the presence of HvoaRF3 and GTP, was measured both in the absence and presence of KCl (3M), the latter corresponding to physiological conditions (76). In agreement with several reports on H. volcanii enzymes, a strong methylation activity was detected only in the presence of KCl (Figure 2A). In these conditions, HvoMtq2-Trm112 complex catalyzes the methylation of nearly 26 pmol of HvoaRF1 out of 100 pmol in 2 h. To rule out the possibility that the detected enzymatic activity resulted from an E. coli contaminant, we substituted Tyr111 from the NPPY signature in HvoMtq2 by Ala with the aim of inactivating the HvoMtq2-Trm112 complex as the corresponding S. cerevisiae Mtq2 mutant is completely inactive (27). No enzymatic activity could indeed be detected for this Y111A HvoMtq2–Trm112 mutant (Figure 2A). In addition, substitution of Gln187 in the HvoaRF1 GGQ motif to Ala also resulted in undetectable activity. Finally, both HvoaRF3 and GTP were necessary for methylation of HvoaRF1 by the HvoMtq2–Trm112 complex.
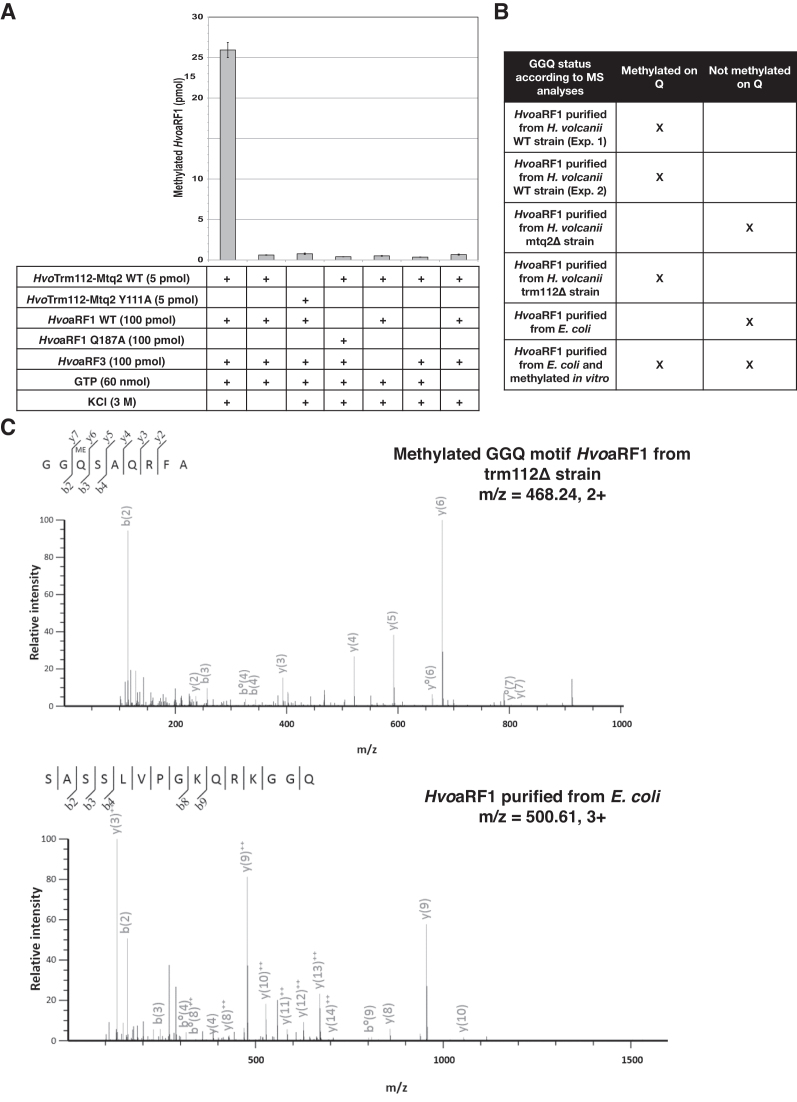
Figure 2.
The HvoMtq2-Trm112 complex modifies the GGQ motif of HvoaRF1 protein. (A) In vitro enzymatic activity of HvoMtq2–Trm112 complex. The conditions (proteins, salt and ligand) used for each experiment are indicated in the table below each graph. The amount of methylated substrate (in pmol) after a 2 h reaction is indicated for every condition. Error bars have been calculated from the results of three independent experiments. (B) Mass spectrometry analysis of the methylation status of the GGQ motif of HvoaRF1 proteins purified from different H. volcanii strains or methylated in vitro. (C) MS/MS spectrum of the 185GGQSAQRFA193 methylated peptide from HvoaRF1 protein purified from trm112Δ strain. The methylated glutamine is highlighted in bold. (D) MS/MS spectrum of the 173SASSLVPGKQRKGGQ187 peptide from HvoaRF1 protein purified from E. coli and not incubated with HvoMtq2-Trm112 complex. The glutamine of interest is highlighted in bold.
Next, we investigated the in vivo methylation state of the HvoaRF1 GGQ motif in different H. volcanii strains. First, we deleted HvoMTQ2 gene in H. volcanii and observed that this deletion severely affects the generation time (251 minutes compared to 128 minutes for wild-type; Supplementary Figure S1C) and results in small size colonies (similarly to trm112Δ strains). Hence, as previously described in S. cerevisiae (20,42), the MTQ2 gene is also important for optimal growth in H. volcanii. Second, we introduced a Flag-tag at the C-terminal extremity of the HvoaRF1 protein in the three following strains: WT, mtq2Δ and trm112Δ and purified HvoaRF1-Flag from these three strains. Using mass spectrometry, we analyzed for the presence of a methyl group on the GGQ motif of these endogenous HvoaRF1 proteins. We could detect the presence of a methyl group on the glutamine residue of the GGQ motif on the HvoaRF1 protein purified from WT and trm112Δ strains but not from the mtq2Δ strain (Figure 2B, C and Supplementary Figure S6). As control, we expressed HvoaRF1 in E. coli, purified it and incubated it with the HvoMtq2-Trm112 holoenzyme under optimal conditions for in vitro enzymatic activity. Mass spectrometry analyses revealed the presence of methylated glutamine only on HvoaRF1 protein incubated with the HvoMtq2–Trm112 holoenzyme (Figure 2B and Supplementary Figure S6).
Altogether, this demonstrates that HvoMtq2–Trm112 holoenzyme catalyzes the methylation of the glutamine side chain from the HvoaRF1 GGQ motif in an HvoaRF3- and GTP-dependent manner similarly to its yeast and human orthologs (25,26). This also reveals that in H. volcanii, Mtq2 is mandatory for HvoaRF1 methylation in vivo while Trm112 is not, explaining the strong differences in generation times between the trm112Δ and mtq2Δ strains (251 ± 13 min versus 123 ± 6 min, respectively; Supplementary Figure S1C).
In parallel, we investigated whether the HvoTrm9–Trm112 acts as a tRNA modification enzyme similarly to its eukaryotic ortholog. To obtain putative tRNA substrates, we generated an H. volcanii H26 strain deleted for the HVO_1032 gene (hereafter termed H. volcanii trm9Δ), which encodes for HvoTrm9, by the pop-in/pop-out method as described above for the H. volcanii trm112Δ strain construction. 50% of the colonies obtained after plasmid excision (pop-out) were deleted for HvoTRM9 gene (3 out of 6 tested, Supplementary Figure S2). Our ability to obtain this deletion mutant indicates that the gene encoding for HvoTrm9 is not essential. Contrary to the deletion of H. volcanii TRM112 gene, the deletion of HvoTRM9 gene did not affect the size of the colonies but resulted in an affected generation time (178 min compared to 128 min for wild-type H. volcanii strain; Supplementary Figure S1C, D). Total tRNAs purified from this strain as well as those purified from S. cerevisiae trm9Δ or elp1Δ strains (47) were used for in vitro enzymatic assays. Surprisingly, the HvoTrm9–Trm112 complex was active on total tRNAs from S. cerevisiae trm9Δ strain as substrates but not from H. volcanii trm9Δ strain (Figure 3A). HvoTrm9–Trm112 complex (2 pmol) modifies up to 5.5 pmol of tRNAs from S. cerevisiae trm9Δ strain out of 100 pmol while S. cerevisiae Trm112-Trm9 complex (1.5 pmol) was modifying up to 7 pmol (out of 75 pmol) of the same tRNAs (47). As observed for HvoMtq2–Trm112 complex, HvoTrm9–Trm112 complex is significantly more active in the presence of 3 M KCl than in the absence of KCl, where only ∼1.5 pmol of tRNAs are modified. Its activity depends on the presence of the cm5U (5-carboxymethyluridine) modification at position 34 of tRNA anticodon loop as tRNAs purified from S. cerevisiae elp1Δ strain are not substrates of this complex. Finally, as Hvo_0574 gene product, which also interacts with HvoTrm112, was initially predicted to be orthologous to eukaryotic Trm9 (58), we have included this purified complex in our enzymatic assays. As shown in Figure 3A, this complex does not exhibit enzymatic activity on total tRNAs extracted from either S. cerevisiae trm9Δ or H. volcanii trm9Δ strains.
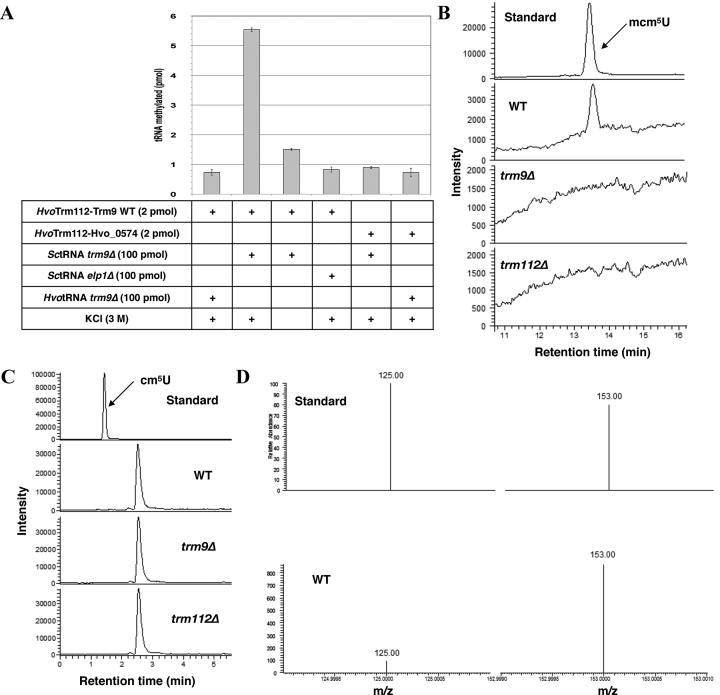
Figure 3.
The HvoTrm9–Trm112 complex catalyzes the formation of mcm5U on tRNAs. (A) In vitro enzymatic activity of HvoTrm9–Trm112 complex. The conditions (proteins, salt and tRNAs) used for each experiment are indicated in the table below each graph. The amount of methylated substrate (in pmol) after a 2 h reaction is indicated for every condition. Error bars have been calculated from the results of three independent experiments. (B) Reversed phase LC–MS/MS elution profiles measured from 50 pmol of synthesized mcm5U standard or a 2 μg injection of a total tRNA digest from WT, trm9Δ or trm112Δ strains. (C) Reversed phase LC–MS/MS elution profiles measured from 50 pmol of synthesized cm5U standard or a 2 μg injection of a total tRNA digest from WT, trm9Δ or trm112Δ strains. (D) SRM product ions (Supplementary Table S9) measured for the cm5U standard (top) and for the unknown molecule eluted 1 minute later than cm5U in the digested WT tRNAs sample (bottom).
Next, we have tried to clarify our surprising observation that tRNAs extracted from the H. volcanii trm9Δ strain are not substrates of the HvoTrm9–Trm112 complex and to document the chemical nature of the yet uncharacterized modifications found at U34 on several H. volcanii tRNAs (tRNAArg(UCG), tRNAGlu(UUC), tRNAGly(UCC), tRNALys(UUU), tRNALeu(UAG); (60,77)). We then performed nucleoside analyses following total digestion of tRNAs extracted from H. volcanii WT, trm9Δ or trm112Δ strains by reversed phase LC–MS/MS. This revealed that the mcm5U (5-methoxycarbonylmethyluridine) nucleoside exists only in tRNAs from WT but neither from trm9Δ nor trm112Δ strains (Figure 3B). In parallel, the assay showed no presence of cm5U in the tRNAs extracted from all these strains at the retention time of the standard (Figure 3C). Interestingly, a second peak was detected in the assay having the same transition as cm5U yet eluting a minute later. We found the later eluting peak to have a different ion ratio to that of the cm5U standard indicating this was not cm5U (Figure 3C–D). To further verify the absence of cm5U in the original sample, a 500 ng spike of cm5U standard was added to the WT strain and subjected to analysis (Supplementary Figure S7). This showed the presence of the cm5U spike as well as the later eluting peak with their respective ion ratio. While the later eluting peak has not been identified in this work, it is clear that all samples lacked the cm5U modification. To our knowledge, these results demonstrate for the first time that the mcm5U modification exists in H. volcanii and most probably in other archaea with a Trm9 ortholog. They also show that both HvoTrm9 and HvoTrm112 proteins are necessary for the formation of mcm5U in vivo. Finally, these analyses rationalize the lack of enzymatic activity of the HvoTrm9–Trm112 complex on tRNAs purified from the trm9Δ H. volcanii strain due to the absence of the cm5U Trm9 substrate in those tRNAs. This strongly indicates that in the absence of HvoTrm9 or HvoTrm112, cm5U is rapidly converted into other chemical structures (such as for instance, ncm5U as previously observed in Δtrm9 S. cerevisiae strain (44)) that remain to be identified. Altogether, these results demonstrate that the HvoTrm9–Trm112 complex is indeed a tRNA methyltransferase, which, similarly to its eukaryotic ortholog, modifies cm5U into mcm5U (5-methoxycarbonylmethyluridine) at position 34 of the tRNAs as the cm5U modification, catalyzed by the Elp1-6 complex in eukaryotes (78,79), is required for enzymatic activity.
Structure of a Trm112–MTase complex from H. volcanii
To gain insight into the molecular bases responsible for the interaction between HvoTrm112 and its interacting MTase partners, different HvoTrm112-MTase complexes were subjected to crystallization trials in the presence or in the absence of SAM. Crystals diffracting up to 1.35Å resolution were obtained for the Hvo_0019–Trm112 complex. The structure of this complex was solved at 2.5Å resolution by the Sulfur-SAD method (Supplementary Figure S8) by taking advantage of the high number of sulfur atoms in this complex (11, excluding the initial methionine from Hvo_0019, which is very likely to be excised in E. coli (80)). The structure was further refined at high resolution using a 1.35Å resolution native dataset to yield a final model with R and Rfree values of 18.5% and 20.7%, respectively. There are two virtually identical copies of the Hvo_0019–Trm112 complex in the asymmetric unit (Figure 4A; rmsd value of 0.27 Å over 211 Cα atoms). Each complex is bound to a SAH molecule, most probably co-purified with Hvo_0019–Trm112 complex.
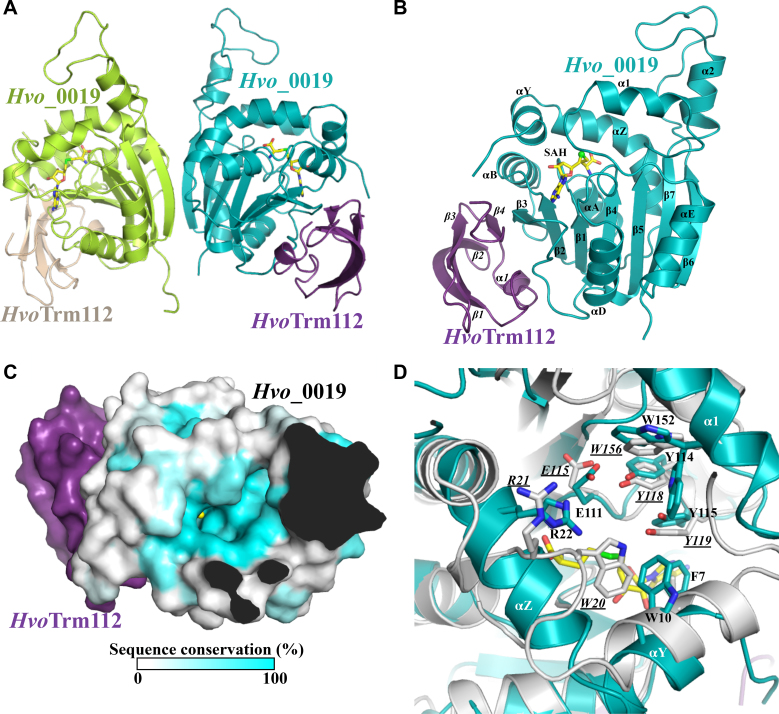
Figure 4.
Crystal structure of Hvo_0019–Trm112 complex. (A) Ribbon representation of the heterotetrameric Hvo_0019–Trm112 complex. The SAH molecule bound to each Hvo_0019 monomer is shown as yellow sticks. (B) Ribbon representation of the Hvo_0019–Trm112 heterodimer. (C) Sequence conservation mapped at the surface of Hvo_0019 protein structure. Only the heterodimer is shown for the sake of clarity. Conservation scores have been calculated from an alignment of 241 sequences using the Consurf server (91). The SAM methyl group modeled by superimposing the structure of S. cerevisiae Bud23 (33) onto the structure of Hvo_0019 is shown as a yellow sphere. Opaque regions correspond to the interior of the proteins. (D) Comparison of Hvo_0019 (blue) and NodS (grey) active sites with conserved residues shown as sticks. Labels for NodS residues are underlined and in italics. SAH is shown as yellow sticks.
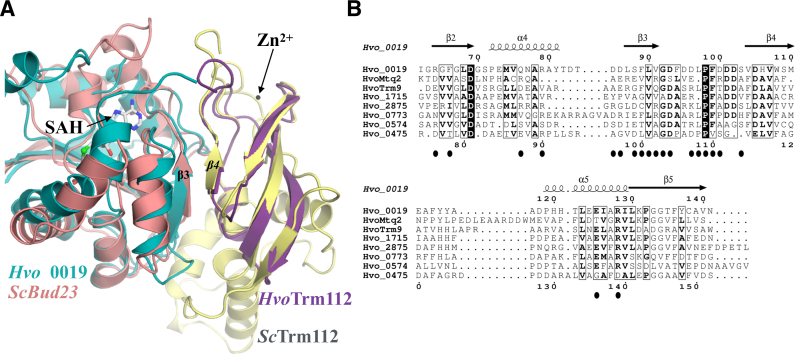
Structurally, Hvo_0019 adopts the typical class-I SAM-dependent MTase fold composed of a central seven stranded β-sheet surrounded by three α-helices (the C-terminal half of αZ, αA and αB) on one side and two on the other side (αD, αE; Figure 4B). On top of the central β-sheet, four α-helices (αY, the N-terminal half of αZ, α1 and α2) contribute to the formation of a cavity centered on the sulfur atom from the SAH molecule bound to the Hvo_0019 protein. SAH binds in a canonical manner compared to what was observed for other class I SAM-dependent MTases. HvoTrm112 structure consists of only the zinc knuckle domain previously observed in eukaryotic Trm112 proteins (rmsd values of 1.1–1.4 Å over around 55 Cα atoms; (25,27,33,47)), including a small N-terminal α-helix (α1) and a four-stranded anti-parallel β-sheet (Figure 4B). Contrary to eukaryotic Trm112 proteins and some structurally similar bacterial proteins (PDB code: 2KPI) of known structures, there is no zinc atom bound to HvoTrm112 in agreement with the absence of conservation of the cysteine residues involved in zinc coordination. The interface between HvoTrm112 and Hvo_0019 has an overall area of 975 Å2 and is slightly smaller than the previously described interfaces between eukaryotic Trm112 and its MTase partners (33,47). This most likely results from the absence of the helical domain, specific to eukaryotic Trm112 proteins, that was in part contributing to these interactions. Complex formation involves 20 and 21 residues from HvoTrm112 and Hvo_0019, respectively (Supplementary Figure S9). The core of this interface is formed by hydrophobic residues (M1, L5, I8, L9, P12, I50, P51, L53, L54, P55 and M58 from HvoTrm112 and F66, F89, L90, V91, L97, P98 and F99 from Hvo_0019; Supplementary Figure S9). Such a large hydrophobic interface rationalizes the strong solubilization effect that we observe for HvoTrm112 upon co-expression with Hvo_0019 in E. coli (Supplementary Figure S4A). This hydrophobic core is surrounded by polar residues (K2, D7, C10, K15, E29, N52 and R59 from HvoTrm112 and S2, R64, Q76, R79, D85, D86, S88, D93, D96, D100, S103, E125 and R128 from Hvo_0019). Six hydrogen bonds and three salt bridges are also observed at the interface (Supplementary Table S6). Two hydrogen bonds formed between main chain atoms of V91 from Hvo_0019 with P51 as well as L53 from HvoTrm112, are responsible for the formation of a β-zipper interaction between HvoTrm112 strand β4 and Hvo_0019 strand β3 (Figure 4B). Other hydrogen bonds are formed between P98, L97, R64 and D100 from Hvo_0019 and C10, K15, S4 and I8 from HvoTrm112, respectively. Finally, salt bridges are formed by K15 from HvoTrm112 with D96 and E125 from Hvo_0019, and by D7 from HvoTrm112 with R64 from Hvo_0019.
As stated above, there are two copies of Hvo_0019–Trm112 complex in the asymmetric unit and these are related by a two-fold symmetry axis (Figure 4A). These copies interact together through a large surface area of 840 Å2. As SEC-MALLS measurements on this complex have revealed that it forms heterotetramer in solution (Supplementary Figure S5; Table 2), this contact area most likely corresponds to the biological interface responsible for this oligomeric state. This homodimerization interface is exclusively formed by residues from Hvo_0019, more precisely from strands β6 and β7 and helix αZ. Hence, the MTase partner seems to govern the formation of oligomeric states, explaining why depending on the MTase, HvoTrm112-MTase complexes are either heterodimers, heterotetramers or heterohexamers in solution (Table 2).
BLAST searches for proteins sharing sequence similarity with Hvo_0019 identified orthologous proteins mostly from specific archaeal phyla such as halobacteriales and thaumarchaeota but also from some bacteria (Legionella shakespearei, Agrobacterium tumefaciens, rhodobacteriaceae, flavobacteriaceae, acidobacteria). Multiple sequence alignments revealed the presence of few highly conserved residues (Supplementary Figure S9B). Interestingly, in Hvo_0019 structure, these residues cluster around the expected position of the SAM methyl group and form a strongly conserved pocket, which is very likely to correspond to the enzyme active site (Figure 4C). Comparison of Hvo_0019 protein structure with previously described structures of MTases revealed a significant degree of conservation with the active site of NodS MTase from Bradyrhizobium japonicum (81). NodS is involved in the biosynthesis of the Nod factor, a modified chitosaccharide acting as a signal molecule in rhizobia. It is catalyzing the methylation of the NH2 group of Nod glucosamine moiety. Indeed, residues R22, E111, Y114, Y115 and W152 from Hvo_0019 structurally match with R21, E115, Y118, Y119 and W156 from NodS, respectively (Figure 4D). In addition, the side chains from Hvo_0019 W10 and NodS W20 are also in close vicinity. Altogether, this suggests that Hvo_0019 might modify a substrate with a hexose sugar ring. Further studies will be needed to characterize the enzymatic activity of the Hvo_0019–Trm112 complex.
DISCUSSION
Protein translation is one of the most intricate processes in cell biology and requires numerous factors acting in a highly coordinated choreography. Most of these translational factors (tRNAs, rRNAs and proteins) are frequently subjected to post-transcriptional and post-translational modifications to perform correct functions. Methylation is so far known as the most prevalent modification of translation machinery. In yeast, effects of methylation on translation are perfectly illustrated by Trm112, an obligate activator for at least four methyltransferases contributing to ribosome biogenesis and function. The importance of this interacting network is also reflected by its conservation in human where these MTases are associated with diseases (28). Sequence analyses revealed that Trm112 orthologues are also found in archaea and bacteria. As most of the translational machinery components are very similar between eukaryotes and archaea, this raised the question of the existence of a similar Trm112 interacting network in archaea.
One protein, many MTase partners
In S. cerevisiae and human, Trm112/TRMT112 is known to directly interact with at least four class I SAM-dependent MTases, namely Bud23/BUD23, Trm9/ALKBH8, Trm11/TRMT11 and Mtq2/HEMK2 (28). Here, using co-immunoprecipitation of Flag-tagged Trm112 from H. volcanii under cross-linking conditions, we have observed a significant enrichment of MTases as potential Trm112 partners (Figure 1 and Table 1), i.e. about 36% of all the MTases predicted by bioinformatics analysis of H. volcanii genome. From this list, we selected nine class I SAM-dependent MTases for further validation and could demonstrate that at least eight interact directly with HvoTrm112 (Supplementary Figure S5). Hence, H. volcanii, and probably more generally archaeal, Trm112 interaction network is larger than anticipated from our current knowledge in eukaryotes, and we cannot exclude that additional MTase partners listed in Table 1 await for experimental validation. Our experimental approach proved very powerful as only three potential partners could have been identified by bioinformatics analyses based on their similarity with eukaryotic Mtq2, Trm9 and Trm11 proteins (no Bud23 ortholog could be found in H. volcanii and in archaea in general; (28,59)). These three MTases are found as potential HvoTrm112 partners in our co-IP experiments but we could validate their interaction with HvoTrm112 only for HvoTrm9 and HvoMtq2 as we were unable to over-express HvoTrm11 in a soluble form in E. coli. The presence of HvoTrm11 in the list of putative partners was somehow surprising as the orthologous proteins from two archaea (P. abyssii and T. kodakarensis) were previously shown to be sufficient to catalyze the mono- and di-methylation of G10 on some tRNAs in vitro (56,57). This could be explained by our recent analysis of the distribution of Trm112 orthologues within archaeal genomes. Indeed, Trm112 is absent in thermococcales and methanobacteriales, which include Pyrococcus abyssii and Thermococcus kodakarensis (28). Hence, we propose that when present in an archaeal organism, aTrm112 interacts directly with aTrm11 while when absent, aTrm11 has evolved to be active on its own. Studies aimed at clarifying this aspect are in progress.
The identification of at least six additional MTases able to directly interact with HvoTrm112, which is only composed of 61 amino acids, was completely unexpected. Bioinformatics analyses revealed that among those, three (Hvo_0773, Hvo_0475 and Hvo_0574) might be specific for halobacteriales phylum from euryarcheota, which includes H. volcanii. The three remaining MTases (Hvo_0019, Hvo_1715 and Hvo_2875) are found in some specific archaeal phyla such as halobacteriales (Hvo_0019, Hvo_1715 and Hvo_2875), thaumarchaeota (Hvo_0019) and thermococcales (Hvo_2875) but also in some bacteria. For instance, Hvo_0019 orthologues could be identified in Legionella shakespearei, Agrobacterium tumefaciens and Limnothrix rosea (Supplementary Figure S9B). Similarly, proteins sharing significant sequence identity with Hvo_1715 are present in Bacilli (such as Bacillus anthracis and Streptococcus pneumoniae) and in mycobacteria. This is also the case for Hvo_2875 present in nocardia. This suggests that in bacteria, Trm112 orthologues could also interact with MTases. This is further supported by several observations. First, the structures of bacterial Trm112 orthologues (PDB codes: 2KPI and 2JS4) are highly similar to the zinc-knuckle domains from H. volcanii and eukaryotic Trm112 proteins (rmsd of 1.4 to 1.8 Å). Second, superimposition of these bacterial structures onto those of Trm112-MTase complexes reveals that the Trm112 region involved in the interaction with MTases adopts the same structure in bacterial proteins and hence is compatible with MTase binding (28). Third, in some bacteria, genes encoding for Trm112 and MTases are fused and this is often an indication of physical interaction. Finally, a recent study aimed at identifying protein-protein interactions in Desulfovibrio vulgaris detected that Trm112 orthologue (DVU0656) interacts with three MTases (HemK, UbiE and Sun), some of which could also have a role in translation (82).
Structural studies of eukaryotic Trm112-MTase complexes have shown that all these MTases interact in a very similar way with Trm112 and hence directly compete (27,33,37,47). The crystal structure of the Hvo_0019–Trm112 complex solved here reveals striking similarities between archaea and eukaryotes. Indeed, Hvo_0019–Trm112 and eukaryotic Trm112-MTase complexes superimpose almost perfectly (Figure 5A; rmsd values ranging from 1.7 Å to 2.15 Å between the different heterodimers). A first hallmark common to all these complexes is the presence of a parallel β-zipper interaction formed between Trm112 strand β4 and MTase strand β3 (Figure 5A). The formation of this β-zipper relies on hydrogen bonds between amino acid main chain atoms from both partners and hence is less affected by substitutions of amino acid located in the MTase strand β4. This very likely explains that Trm112 proteins can have many partners adopting the same overall fold. A second conserved feature between eukaryotic and archaeal Trm112-MTase complexes is the presence of a hydrophobic core at the complex interface, which most likely rationalizes the solubilizing and stabilizing effects observed for Trm112 on several MTases (Supplementary Figure S4A; (25,30,42)). We propose that as in eukaryotes, all identified HvoMTases compete and interact in a very similar mode with HvoTrm112. This is further supported by the fact that all these archaeal MTases are SAM-dependent class I MTases and hence adopt the same overall three-dimensional structure. In addition, the comparison of Hvo_0019 residues directly contacting HvoTrm112 with the structurally equivalent residues in all other HvoTrm112 MTase partners identified here, highlights a significant degree of conservation (Figure 5B). Indeed, positions forming the hydrophobic core at the interface between Hvo_0019 and HvoTrm112 are largely occupied by hydrophobic residues in the other HvoTrm112 MTase partners.
Figure 5.
A conserved interaction mode between eukaryotic and archaeal Trm112-MTase complexes. (A) Superimposition of the crystal structures of Hvo_0019–Trm112 and S. cerevisiae Bud23–Trm112 complexes. Strands forming the β-zipper interaction between Trm112 and the MTases are labeled. The zinc atom bound to ScTrm112 is shown as a black sphere. The SAH molecule bound to Hvo_0019 MTase is shown as gray sticks. (B) Sequence alignment of all HvoMTases that have been experimentally confirmed to be direct partners of HvoTrm112. For the sake of clarity, only the region of the MTase domain contacting HvoTrm112 is shown. Secondary structure elements assigned from the Hvo_0019 crystal structure are indicated above the alignment. Black closed circles indicate Hvo_0019 positions involved in interaction with HvoTrm112. This figure was generated with the Espript server (92).
Then, similarly to what was described for eukaryotic Trm112 proteins, SAM-dependent class I MTases appear as privileged Trm112 partners in archaea. As some of these MTases are not strictly conserved within the archaeal domain of life, the number of MTase partners may vary between archaea. Finally, the similarities found for some of these archaeal proteins with bacterial MTases further suggest that Trm112 interacts with MTases in the three domains of life and that their interaction mode is conserved.
Functional characterization of translation-related complexes
Among the MTases validated as HvoTrm112 partners, two (HvoMtq2 and HvoTrm9) have convincing assigned functions based on their significant sequence similarity with eukaryotic proteins. We have experimentally demonstrated the biochemical functions of HvoMtq2–Trm112 and HvoTrm9–Trm112 complexes both in vitro and in vivo based on our current knowledge of biochemical functions of eukaryotic Mtq2-Trm112 and Trm9–Trm112 complexes.
First, we have shown that the HvoMtq2-Trm112 complex exhibits an in vitro MTase activity on wild-type (but not on the GGA mutant of the universally conserved GGQ motif) class I release factor aRF1 only in the presence of 3M KCl, a common trend of H. volcanii enzymes (Figure 2A). As observed for yeast and human Trm112-Mtq2 complexes, this activity is dependent on the presence of the GTP-bound form of class II translation termination factor aRF3 (25,26). This reflects the high similarity between eukaryotic and archaeal translation termination factors eRF1/aRF1 and eRF3/aRF3 (62). We also demonstrate that HvoaRF1 is methylated on the glutamine side chain of its GGQ motif in H. volcanii cells and that this modification requires HvoMtq2 protein but not HvoTrm112 (Figure 2B and C). As HvoMtq2 orthologues are found in all archaea (28), we propose that the glutamine side chain of the universally conserved GGQ motif from aRF1 is methylated in archaea in general. The conservation of this post-translational modification in the three domains of life strongly argues in favor of an important biological role. Indeed, despite radically different structures of bacterial RF1/RF2 on one side and eRF1/aRF1 on the other side, these organisms have independently evolved enzymatic machineries to add a methyl group on the NH2 group of the glutamine side chain of their universally conserved GGQ motif (24,62,83). In bacteria, the methylation of RF1/RF2 GGQ motif by PrmC has been shown to be important for normal growth, to increase the affinity of RF1/RF2 for ribosomes and to enhance translation termination (84,85). In eukaryotes and archaea, the role of this modification still remains to be clarified. However, the growth defect phenotype observed in S. cerevisiae and H. volcanii upon deletion of MTQ2 gene ((20), this study) together with the conservation of this modification in eukaryotes and archaea argue in favor of a strong functional importance for this post-translational modification.
We have also reconstituted a tRNA MTase activity for HvoTrm9–Trm112 complex in vitro but only using tRNAs purified from a trm9Δ S. cerevisiae strain as substrate. As observed for HvoMtq2-Trm112, this enzymatic activity is salt-dependent (Figure 3A). The strict requirement of the cm5U modification at position 34 of tRNAs indicates that HvoTrm9–Trm112 complex catalyzes the formation of mcm5U34 similarly to eukaryotic Trm9–Trm112 complexes (42,44,47). This is further supported by the detection of the mcm5U nucleoside in tRNAs purified from WT but not from trm9Δ or trm112Δ strains (Figure 3B). To our knowledge, this is the first experimental evidence for the existence of the mcm5U modification in archaeal tRNAs. Surprisingly, tRNAs purified from a Δtrm9 H. volcanii strain were not substrates for the HvoTrm9–Trm112 complex. Indeed, we did not detect the cm5U modification, the known substrate of the Trm9–Trm112 complex, in tRNAs from both H. volcanii trm9Δ or trm112Δ strains (Figure 3C–D), indicating that this cm5U intermediate is unstable and rapidly converted into another modification, which could be ncm5U, similarly to what has been observed for some tRNAs in yeast (44). It is noteworthy that this modification, which biosynthetic pathway remains obscure, has been detected in tRNALeu(UAG) from T. acidophilum archaeon (86). Further studies will be needed to precisely characterize the chemical structures of the modifications found at position U34 from other H. volcanii tRNAs but also more generally in archaea. Due to the highly polar nature of modified uridines, which results in low ionization efficiency, this is very likely to be challenging. Hence, the similarity between archaeal HvoTrm112 and its eukaryotic orthologues is not solely restricted to its ability to interact with class I dependent MTases but also extends to the in vivo and in vitro biochemical functions of at least two MTase partners (HvoTrm9 and HvoMtq2) and most probably HvoTrm11, which all modify components contributing to mRNA translation. Together with the strong enrichment of translation factors or ribosome components in our co-IP experiments, this supports an important role of archaeal Trm112 in translation and strengthens the analogies between eukaryotic and archaeal translation apparatus.
CONCLUSION
Trm112 proteins, which are found in the three domains of life, have been characterized exclusively in eukaryotes so far, where these are partners and activators of several MTases modifying factors of the protein synthesis machinery. Here, we have characterized Trm112 network in archaea using Haloferax volcanii as model organism. In this organism, Trm112 interacts with at least eight different MTases. Some exhibit similar functions as their eukaryotic orthologues, further supporting the similarities between eukaryotic and archaeal translation machineries. Other MTases are orthologous to bacterial proteins, strongly suggesting that bacterial Trm112 proteins might also interact with MTases, in agreement with a recent study (82).
Members of Trm112 family are not unique in their ability to interact with several MTases. Indeed, it has been shown that Vea protein from Aspergillus nidulans acts as a hub protein interacting with at least four MTases, some of which being involved in development, secondary metabolism and fungal pathogenicity (87,88). Hence, precise understanding of molecular details ruling the association of a single protein with various proteins adopting the same fold but exhibiting low sequence identity is of importance to decipher the numerous interaction networks that contribute to most cellular pathways (89,90).
DATA AVAILABILITY
The coordinates and structure factors files are available from the Protein Data Bank (PDB) under accession code 6F5Z. The dataset for the mass spectrometry experiments has been deposited on the ProteomeXchange database under project accession number PXD008466.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J.-M. Strub for protein identification by peptide mass fingerprinting. We thank Dr A. Marchfelder for sharing reagents with us. We are grateful to Martin Savko for assistance and to the SOLEIL staff for smoothly running the facility. Experiments were performed on the Proxima-2 beamline at SOLEIL Synchrotron, France (proposal number 20150742).
Author Contributions: T.V.N., L.M., R.L.R., R.L., J.L. and N.U. performed the experiments. R.L.R., R.L., P.A.L., S.C. and M.G. designed research; T.V.N., R.L.R., P.A.L., V.d.C-L, S.C. and M.G. analyzed the data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National pour la Recherche Scientifique (CNRS) including a specific support by the ATIP-AVENIR program (to M.G.); Agence Nationale pour la Recherche (ANR) [ANR-14-CE09-0016-02]; Ecole Polytechnique; PhD fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche (MESR) (to T.V.N., L.M., J.L.); Ecole Polytechnique (to T.V.N.); CNRS and the University of Strasbourg (to S.C.); S.C. thanks the GIS IBiSA; French Proteomic Infrastructure (ProFI) [ANR-10-INBS-08-03]; Région Alsace for financial support; National Institutes of Health [R01 GM70641 to V.d.C.-L.]; Fondation de l’Ecole Polytechnique (to V.d.C.-L.); National Institutes of Health [R01 GM058843 to P.A.L.]. Funding for open access charge: Agence Nationale pour la Recherche (ANR) [ANR-14-CE09-0016-02].
Conflict of interest statement. None declared.
REFERENCES
- 1. McKenney K.M., Alfonzo J.D.. From prebiotics to Probiotics: The evolution and functions of tRNA modifications. Life. 2016; 6:E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 3. Sharma S., Lafontaine D.L.. ‘View From A Bridge’: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 4. Decatur W.A., Fournier M.J.. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 5. Natchiar S.K., Myasnikov A.G., Kratzat H., Hazemann I., Klaholz B.P.. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017; 551:472–477. [DOI] [PubMed] [Google Scholar]
- 6. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. . N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilbert W.V., Bell T.A., Schaening C.. Messenger RNA modifications: form, distribution, and function. Science. 2016; 352:1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Couttas T.A., Raftery M.J., Padula M.P., Herbert B.R., Wilkins M.R.. Methylation of translation-associated proteins in Saccharomyces cerevisiae: Identification of methylated lysines and their methyltransferases. Proteomics. 2012; 12:960–972. [DOI] [PubMed] [Google Scholar]
- 11. Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L., Lindahl L., Liljas A., Lipton J.M. et al. . A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014; 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lipson R.S., Webb K.J., Clarke S.G.. Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2010; 500:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davydova E., Ho A.Y., Malecki J., Moen A., Enserink J.M., Jakobsson M.E., Loenarz C., Falnes P.O.. Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2). J. Biol. Chem. 2014; 289:30499–30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dzialo M.C., Travaglini K.J., Shen S., Loo J.A., Clarke S.G.. A new type of protein lysine methyltransferase trimethylates Lys-79 of elongation factor 1A. Biochem. Biophys. Res. Commun. 2014; 455:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dzialo M.C., Travaglini K.J., Shen S., Roy K., Chanfreau G.F., Loo J.A., Clarke S.G.. Translational roles of elongation factor 2 protein lysine methylation. J. Biol. Chem. 2014; 289:30511–30524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jakobsson M.E., Davydova E., Malecki J., Moen A., Falnes P.O.. Saccharomyces cerevisiae eukaryotic elongation factor 1A (eEF1A) is methylated at Lys-390 by a METTL21-like methyltransferase. PLoS One. 2015; 10:e0131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamey J.J., Winter D.L., Yagoub D., Overall C.M., Hart-Smith G., Wilkins M.R.. Novel N-terminal and lysine methyltransferases that target translation elongation factor 1A in yeast and human. Mol. Cell. Proteomics: MCP. 2016; 15:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dincbas-Renqvist V., Engstrom A., Mora L., Heurgue-Hamard V., Buckingham R., Ehrenberg M.. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000; 19:6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heurgue-Hamard V., Champ S., Engstrom A., Ehrenberg M., Buckingham R.H.. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002; 21:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heurgue-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Kisselev L.L., Buckingham R.H.. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005; 280:2439–2445. [DOI] [PubMed] [Google Scholar]
- 21. Polevoda B., Span L., Sherman F.. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006; 281:2562–2571. [DOI] [PubMed] [Google Scholar]
- 22. Klaholz B.P. Molecular recognition and catalysis in translation termination complexes. Trends Biochem. Sci. 2011; 36:282–292. [DOI] [PubMed] [Google Scholar]
- 23. Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V.. Structural basis for stop codon recognition in eukaryotes. Nature. 2015; 524:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graille M., Heurgue-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., van Tilbeurgh H., Buckingham R.H.. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005; 20:917–927. [DOI] [PubMed] [Google Scholar]
- 25. Heurgue-Hamard V., Graille M., Scrima N., Ulryck N., Champ S., van Tilbeurgh H., Buckingham R.H.. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006; 281:36140–36148. [DOI] [PubMed] [Google Scholar]
- 26. Figaro S., Scrima N., Buckingham R.H., Heurgue-Hamard V.. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008; 582:2352–2356. [DOI] [PubMed] [Google Scholar]
- 27. Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R.H., van Tilbeurgh H., Heurgue-Hamard V., Graille M.. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011; 39:6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bourgeois G., Letoquart J., van Tran N., Graille M.. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules. 2017; 7:E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White J., Li Z., Sardana R., Bujnicki J.M., Marcotte E.M., Johnson A.W.. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 2008; 28:3151–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Figaro S., Wacheul L., Schillewaert S., Graille M., Huvelle E., Mongeard R., Zorbas C., Lafontaine D.L., Heurgue-Hamard V.. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell. Biol. 2012; 32:2254–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ounap K., Kasper L., Kurg A., Kurg R.. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS One. 2013; 8:e75686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sardana R., White J.P., Johnson A.W.. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA. 2013; 19:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Letoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgue-Hamard V., Lafontaine D.L.. Structural and functional studies of Bud23–Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ounap K., Leetsi L., Matsoo M., Kurg R.. The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via Ubiquitin-Proteasome pathway. PLoS One. 2015; 10:e0133841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgue-Hamard V., Lafontaine D.L.. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015; 26:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Purushothaman S.K., Bujnicki J.M., Grosjean H., Lapeyre B.. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005; 25:4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bourgeois G., Marcoux J., Saliou J.M., Cianferani S., Graille M.. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 2017; 45:1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalhor H.R., Clarke S.. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003; 23:9283–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R.. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006; 59:677–688. [DOI] [PubMed] [Google Scholar]
- 40. Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., Begley T.J.. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007; 28:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu D., Brophy J.A., Chan C.T., Atmore K.A., Begley U., Paules R.S., Dedon P.C., Begley T.J., Samson L.D.. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010; 30:2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazauric M.H., Dirick L., Purushothaman S.K., Bjork G.R., Lapeyre B.. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010; 285:18505–18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Songe-Moller L., van den Born E., Leihne V., Vagbo C.B., Kristoffersen T., Krokan H.E., Kirpekar F., Falnes P.O., Klungland A.. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010; 30:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen C., Huang B., Anderson J.T., Bystrom A.S.. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One. 2011; 6:e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patil A., Chan C.T., Dyavaiah M., Rooney J.P., Dedon P.C., Begley T.J.. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012; 9:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deng W., Babu I.R., Su D., Yin S., Begley T.J., Dedon P.C.. Trm9-catalyzed tRNA modifications regulate Global Protein Expression by Codon-Biased Translation. PLos Genet. 2015; 11:e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Letoquart J., Tran N.V., Caroline V., Aleksandrov A., Lazar N., van Tilbeurgh H., Liger D., Graille M.. Insights into molecular plasticity in protein complexes from Trm9–Trm112 tRNA modifying enzyme crystal structure. Nucleic. Acids. Res. 2015; 43:10989–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shimada K., Nakamura M., Anai S., De Velasco M., Tanaka M., Tsujikawa K., Ouji Y., Konishi N.. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009; 69:3157–3164. [DOI] [PubMed] [Google Scholar]
- 49. Nakazawa Y., Arai H., Fujita N.. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res. 2011; 71:1146–1155. [DOI] [PubMed] [Google Scholar]
- 50. Tiedemann R.E., Zhu Y.X., Schmidt J., Shi C.X., Sereduk C., Yin H., Mousses S., Stewart A.K.. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012; 72:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stefanska B., Cheishvili D., Suderman M., Arakelian A., Huang J., Hallett M., Han Z.G., Al-Mahtab M., Akbar S.M., Khan W.A. et al. . Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin. Cancer Res. 2014; 20:3118–3132. [DOI] [PubMed] [Google Scholar]
- 52. Jangani M., Poolman T.M., Matthews L., Yang N., Farrow S.N., Berry A., Hanley N., Williamson A.J., Whetton A.D., Donn R. et al. . The Methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J. Biol. Chem. 2014; 289:8931–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivera M.C., Jain R., Moore J.E., Lake J.A.. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:6239–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yutin N., Makarova K.S., Mekhedov S.L., Wolf Y.I., Koonin E.V.. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 2008; 25:1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lyu Z., Whitman W.B.. Evolution of the archaeal and mammalian information processing systems: towards an archaeal model for human disease. Cell. Mol. Life Sci. 2017; 74:183–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Armengaud J., Urbonavicius J., Fernandez B., Chaussinand G., Bujnicki J.M., Grosjean H.. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004; 279:37142–37152. [DOI] [PubMed] [Google Scholar]
- 57. Hirata A., Nishiyama S., Tamura T., Yamauchi A., Hori H.. Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 2016; 44:6377–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grosjean H., Gaspin C., Marck C., Decatur W.A., de Crecy-Lagard V.. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics. 2008; 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Phillips G., de Crécy-Lagard V.. Biosynthesis and function of tRNA modifications in Archaea. Curr. Opin. Microbiol. 2011; 14:335–341. [DOI] [PubMed] [Google Scholar]
- 60. Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984; 259:9461–9471. [PubMed] [Google Scholar]
- 61. Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D.. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000; 100:311–321. [DOI] [PubMed] [Google Scholar]
- 62. Kobayashi K., Saito K., Ishitani R., Ito K., Nureki O.. Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex. Nucleic Acids Res. 2012; 40:9319–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bitan-Banin G., Ortenberg R., Mevarech M.. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 2003; 185:772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lestini R., Duan Z., Allers T.. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair (Amst.). 2010; 9:994–1002. [DOI] [PubMed] [Google Scholar]
- 65. Fischer S., Benz J., Spath B., Maier L.K., Straub J., Granzow M., Raabe M., Urlaub H., Hoffmann J., Brutschy B. et al. . The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010; 285:34429–34438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993; 26:795–800. [Google Scholar]
- 67. Schneider T.R., Sheldrick G.M.. Substructure solution with SHELXD. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1772–1779. [DOI] [PubMed] [Google Scholar]
- 68. Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 2004; 11:49–52. [DOI] [PubMed] [Google Scholar]
- 69. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D.. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C. et al. . 2016; Cambridge: Global Phasing Ltd. [Google Scholar]
- 74. Florens L., Carozza M.J., Swanson S.K., Fournier M., Coleman M.K., Workman J.L., Washburn M.P.. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006; 40:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schubert H.L., Blumenthal R.M., Cheng X.. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003; 28:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meury J., Kohiyama M.. ATP is required for K+ active transport in the archaebacterium Haloferax volcanii. Arch. Microbiol. 1989; 151:530–536. [Google Scholar]
- 77. Gupta R. Transfer RNAs of Halobacterium volcanii: sequences of five leucine and three serine tRNAs. System. Appl. Microbiol. 1986; 7:102–105. [Google Scholar]
- 78. Glatt S., Letoquart J., Faux C., Taylor N.M., Seraphin B., Muller C.W.. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012; 19:314–320. [DOI] [PubMed] [Google Scholar]
- 79. Glatt S., Zabel R., Kolaj-Robin O., Onuma O.F., Baudin F., Graziadei A., Taverniti V., Lin T.-Y., Baymann F., Seraphin B. et al. . Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016; 23:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hirel P.H., Schmitter M.J., Dessen P., Fayat G., Blanquet S.. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cakici O., Sikorski M., Stepkowski T., Bujacz G., Jaskolski M.. Crystal structures of NodS N-methyltransferase from Bradyrhizobium japonicum in ligand-free form and as SAH complex. J. Mol. Biol. 2010; 404:874–889. [DOI] [PubMed] [Google Scholar]
- 82. Shatsky M., Allen S., Gold B.L., Liu N.L., Juba T.R., Reveco S.A., Elias D.A., Prathapam R., He J., Yang W. et al. . Bacterial Interactomes: Interacting protein partners share similar function and are validated in independent assays more frequently than previously reported. Mol. Cell. Proteomics: MCP. 2016; 15:1539–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Graille M., Figaro S., Kervestin S., Buckingham R.H., Liger D., Heurgue-Hamard V.. Methylation of class I translation termination factors: structural and functional aspects. Biochimie. 2012; 94:1533–1543. [DOI] [PubMed] [Google Scholar]
- 84. Pavlov M.Y., Freistroffer D.V., Dincbas V., MacDougall J., Buckingham R.H., Ehrenberg M.. A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol. 1998; 284:579–590. [DOI] [PubMed] [Google Scholar]
- 85. Mora L., Heurgue-Hamard V., de Zamaroczy M., Kervestin S., Buckingham R.H.. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007; 282:35638–35645. [DOI] [PubMed] [Google Scholar]
- 86. Tomikawa C., Ohira T., Inoue Y., Kawamura T., Yamagishi A., Suzuki T., Hori H.. Distinct tRNA modifications in the thermo-acidophilic archaeon, Thermoplasma acidophilum. FEBS Lett. 2013; 587:3575–3580. [DOI] [PubMed] [Google Scholar]
- 87. Sarikaya-Bayram O., Bayram O., Feussner K., Kim J.H., Kim H.S., Kaever A., Feussner I., Chae K.S., Han D.M., Han K.H. et al. . Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev. Cell. 2014; 29:406–420. [DOI] [PubMed] [Google Scholar]
- 88. Sarikaya-Bayram O., Palmer J.M., Keller N., Braus G.H., Bayram O.. One juliet and four Romeos: VeA and its methyltransferases. Front. Microbiol. 2015; 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O. et al. . A protein complex network of Drosophila melanogaster. Cell. 2011; 147:690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K. et al. . The BioPlex network: a Systematic exploration of the human interactome. Cell. 2015; 162:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N.. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010; 38:W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Robert X., Gouet P.. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014; 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors files are available from the Protein Data Bank (PDB) under accession code 6F5Z. The dataset for the mass spectrometry experiments has been deposited on the ProteomeXchange database under project accession number PXD008466.