Abstract
Chronic low levels of survival motor neuron (SMN) protein cause spinal muscular atrophy (SMA). SMN is ubiquitously expressed, but the mechanisms underlying predominant neuron degeneration in SMA are poorly understood. We report that chronic low levels of SMN cause Senataxin (SETX)-deficiency, which results in increased RNA–DNA hybrids (R-loops) and DNA double-strand breaks (DSBs), and deficiency of DNA-activated protein kinase-catalytic subunit (DNA-PKcs), which impairs DSB repair. Consequently, DNA damage accumulates in patient cells, SMA mice neurons and patient spinal cord tissues. In dividing cells, DSBs are repaired by homologous recombination (HR) and non-homologous end joining (NHEJ) pathways, but neurons predominantly use NHEJ, which relies on DNA-PKcs activity. In SMA dividing cells, HR repairs DSBs and supports cellular proliferation. In SMA neurons, DNA-PKcs-deficiency causes defects in NHEJ-mediated repair leading to DNA damage accumulation and neurodegeneration. Restoration of SMN levels rescues SETX and DNA-PKcs deficiencies and DSB accumulation in SMA neurons and patient cells. Moreover, SETX overexpression in SMA neurons reduces R-loops and DNA damage, and rescues neurodegeneration. Our findings identify combined deficiency of SETX and DNA-PKcs stemming downstream of SMN as an underlying cause of DSBs accumulation, genomic instability and neurodegeneration in SMA and suggest SETX as a potential therapeutic target for SMA.
INTRODUCTION
Spinal muscular atrophy (SMA) is the leading neurodegenerative disease of the early childhood mortality and is caused by mutation or deletion of the survival of motor neuron 1 (SMN1) gene (1). In humans, another copy SMN2, similar to SMN1, undergoes alternative splicing due to a point mutation resulting in a majority of transcript lacking the last coding exon (exon 7) and encoding a truncated protein, SMNΔ7 (2). However, SMN2 also produces ∼10–20% full-length SMN protein. The low levels of SMN however are insufficient and cause manifestation of SMA in humans carrying the homozygous SMN1 mutation. Chronic low levels of SMN cause motor neuron degeneration, which leads to muscle atrophy, symmetric limb paralysis and death in SMA.
SMN is a ubiquitously expressed protein and is essential for cell viability. Since the discovery of the SMN1 gene over 20 years ago, it has remained enigmatic why deficiency of a ubiquitous protein causes a neurological disease (3,4). Molecular events stemming downstream of SMN-deficiency that lead to predominant neuron degeneration in SMA are largely unknown. However, some reports have provided insight into the degenerative and cell death pathways, including Rho/ROCK (5), Ubiquitin (6) and the JNK (7) signaling pathways, that are activated by SMN-deficiency and mediate neuronal cell death in SMA (8). However, these signaling pathways are known to operate at the inflection point between cell death and survival and are downstream of primary biochemical defects caused by the loss-of-function of SMN. One outstanding question in SMA pathogenesis is why spinal cord motor neurons predominantly degenerate due to deficiency of a ubiquitous protein, SMN. It has been shown that SMN plays a critical function in the assembly of spliceosomal small nuclear ribonucleo proteins (snRNPs), and SMN deficiency causes splicing defects (9). Studies have tried to identify neuron-specific molecular targets that might contribute to neurodegeneration but failed to identify any specific targets that could justify selective degeneration of neurons in SMA (10). However, downregulation of genes associated with synaptogenesis, AGRIN and ETV1, and of several genes associated with the activation of unfolded protein response pathway that mediates endoplasmic reticulum stress, including DNAJC10, is caused by defects in splicing in different tissues and cell types, including spinal cord motor neurons; these defects may contribute to neurodegeneration in SMA (11–13). Recent study identified the SMN-ZPR1-HoxA5 axis, in which ZPR1 transcriptionally regulates HoxA5 levels (14). This axis is downregulated in SMA and results in phrenic motor neuron degeneration, leading to respiratory failure in SMA mice (14). These studies provided important insight into possible mechanisms that may mediate neurodegeneration and SMN functions in diverse cellular processes, including critical role of SMN in splicing and mRNA biogenesis (15).
In the current study, we tested the hypothesis that the deficiency of a ubiquitous protein, SMN, may cause a common biochemical defect in all cell types, but which might specifically affect a neuron-specific process, leading to selective degeneration of motor neurons in SMA. We examined the molecular defects caused by (i) acute SMN-deficiency (knockdown) in dividing normal (non-SMA) cells and (ii) chronic SMN-deficiency (SMA) in SMA patient dividing cells (primary fibroblast) and terminally differentiated cells (spinal cord neurons) derived from SMA mice. In addition, we have examined spinal cord tissues from SMA mice and severe SMA type I patients. We show that SMN-deficiency causes DNA double-strand breaks (DSBs) and activation of the DNA damage response (DDR) pathways. We uncover that Senataxin (SETX), which is required for resolving RNA–DNA hybrids (R-loops), colocalize with SMN in sub-nuclear bodies and acute (knockdown) and chronic (SMA) SMN-deficiencies cause down-regulation of SETX and accumulation of R-loops. In normal (non-SMA) dividing cells, SMN-deficiency results in activation of DNA repair mechanisms, homologous recombination (HR) that is dependent on the activation of ataxia-telangiectasia mutated (ATM) and ataxia-telangiectasia and Rad3-related (ATR) kinases and breast cancer type 1 susceptibility protein (BRCA1) (16), and non-homologous end joining (NHEJ) that requires DNA-activated protein kinase catalytic subunit (DNA-PKcs) activity (17). We found that chronic low levels of SMN also cause deficiency of DNA-PKcs in dividing SMA patient cells and non-dividing SMA neurons. In SMA patient dividing cells, HR (ATM/BRCA1) is activated for DSB repair, but NHEJ is impaired due to the lack of DNA-PKcs activity. However, DSB repair by HR enables SMA patient cells to divide and proliferate. Analysis of spinal cord neurons and spinal cord tissues from SMA mice and patients show marked reduction in SETX and DNA-PKcs and activation of DNA damage pathway. Notably, defects identified in SMN-deficient cells, SETX-deficiency, DNA-PKcs-deficiency, R-loops and DNA damage accumulation were rescued by restoring SMN levels in SMA patient cells and spinal cord neurons. Furthermore, SETX overexpression reduces R-loops and DNA damage accumulation, and rescues degeneration of SMA mice spinal cord neurons. Because neurons predominantly use NHEJ for DSBs repair (18), our data support a model in which combined deficiency of SETX and DNA-PKcs-deficiency leads to elevated levels of R-loops and DSB formation, and defects in NHEJ-mediated DSBs repair result in DNA damage accumulation, which may contribute to genomic instability and predominant degeneration of motor neurons in SMA.
MATERIALS AND METHODS
Mice
The wild-type FVB/NJ and SMA carrier mice on FVB background [Smn−/+; SMN2+/+; SMNΔ7+/+] (19), purchased from the Jackson Laboratory, were bred to generate non-SMA (normal) [Smn+/+; SMN2+/+; SMNΔ7+/+] and SMA [Smn−/−; SMN2+/+; SMNΔ7+/+] littermates. Mice (7-day-old) were euthanized to collect spinal cord tissues for culturing motor neuron immunofluorescence (IF) analysis and immunoblot (IB) analysis with average postmortem interval <30 min. All experiments and procedures were approved and performed according to the guidelines and policies set by the Institutional Biosafety Committee. All animals were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Texas Tech University Health Sciences Center El Paso (TTUHSC EP). Animals were treated humanely and euthanasia was performed using methods approved by the American Veterinary Medical Association (AVMA).
SMA patient tissues
Human frozen spinal cord (lumbar region) tissues of (i) normal (non-SMA) control (GM083), age 69 days, male, postmortem interval 27 h (cause of death asphyxia), (ii) SMA type I (GM04583), age 77 days, male, postmortem interval 4 h (cause of death pneumonia) and (iii) SMA type I (GM04629), age 169 days, male, postmortem interval 3 h (cause of death cardiopulmonary arrest) were received from the NICHD Brain and Tissue Bank for Developmental Disorders, Baltimore, MD 21201, USA (7). The Institutional Review Board (IRB) of the TTUHSC EP approved the use of human tissues.
SMA patient primary fibroblast
Primary fibroblast (dividing cells) derived from normal human (non-SMA), fibroblasts WI-38 (SMN1+/+) and GM03814 (SMN1−/+), and derived from patients with Werdnig–Hoffman disease (SMA type I), fibroblasts GM03813 and GM09677 (SMN1−/−), were cultured in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal bovine serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin (20). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cell lysates were prepared from cultured cells using Triton lysis buffer (TLB) for IB analysis (21). Cells were cultured on glass coverslips (1 × 105 cells/well) in 6-well plates, transfected with pCMV/mycSMN plasmid (1 μg/well) using Lipofectamine®2000 transfection reagent according to the manufacturer’s protocol. Cells were infected with Ad-h-SETX (Vector Biolabs) at 200 MOI. Cells were harvested for IB analysis or fixed at 48 h post-transfection or infection and processed for IF analysis (20).
Knockdown of SMN in non-SMA cells by RNAi
Human HeLa and WI-38 (non SMA) cells were cultured in MEM/EBSS supplemented with 10% FBS, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 1 mM sodium pyruvate. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells (1 × 105 cells/well) were plated on to a 6-well plate with glass coverslips. Cells were then transfected with 100 nM SMART pool human SMN1 siRNA (M-011108-02-0050) or scramble siRNA (Dharmacon) using Lipofectamine®2000. To establish the specificity siRNA, we tested a single siRNA oligo against human SMN1 from the SMART pool, 5′-GAGCAAAAUCUGUCCGAUC-3′ (D-011108-23) (siSMN-D23). Cells were harvested 30 h post-transfection and examined by IB and IF analysis (21).
DNA damage analysis
Southern blot analysis
Genomic DNA from cells was extracted using the DNA easy kit according to the manufacturer's instruction and treated with RNase A. Extracted DNA (500 ng) was biotin labeled at the 3′-OH end using the Biotin 3′-end DNA Labeling Kit (Thermofisher Scientific). Biotin-labeled DNA was separated on 0.8% agarose gel and transferred overnight to Zeta-Probe GT membrane (Bio-Rad) by capillary action. DNA on the membrane was UV cross-linked using Spectrolinker XL-1500 UV crosslinker (Spectronics Corporation). Biotin-labeled DNA was detected using chemiluminescent nucleic acid detection kit (Thermofisher Scientific). Chemiluminescent signal was examined by LAS 4000 biomolecular imager and analyzed using ImageQuant™ software (GE Healthcare Life Sciences).
TUNEL assay
Cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.25% Triton-X100 for 20 min and labeled with EdUTP and terminal deoxynucleotidyl transferase (TdT) to detect DNA breaks using Click-iT TUNEL Alexa-594 kit (ThermoFisher Scientific) according to the manufacturer’s instruction. After EdUTP labeling, cells were stained for SMN using mouse anti-SMN antibody followed by Alexa488-conjugated goat anti-mouse IgG. Coverslips were processed for IF analysis.
Primary spinal cord motor neurons
Mouse spinal cord explants from 7-day-old normal and SMA mice were cultured in vitro for 12–14 days in 8-well chamber microscope slides, coated with poly-D-lysine/laminin using neurobasal medium supplemented with B-27, 700 mM Glucose, 2 mM L-glutamine, 2.5 mM KCl and 1× penicillin/streptomycin with some modifications as described previously (7,22). Half of the culture medium was replaced with freshly made neurobasal medium every 48 h. The identity and morphology of the spinal cord motor neurons and glial cells were established by staining with specific markers, including choline acetyl transferase (ChAT) and homeobox containing protein Hlxb9 (Hb9) (7,22). Neurons were fixed with 4% PFA and processed for examination by IF analysis. For rescue experiments, cultured neurons were infected with Ad-GFP and Ad-GFP-SMN1 (SignaGen laboratories) or Ad-h-SETX (Vector Biolabs) at 100 MOI in a volume of 200 μl/well of 8-well chamber and 400 μl of medium added after 4 h incubation and medium was replaced after 12 h. Neurons were either fixed (4% PFA) or harvested for protein extraction at 48 h post-infection for IF and IB analysis, respectively.
Immunoblot analysis
Protein extracts for conventional IB analysis were prepared from cells transfected with siSMN, mouse tissues and human SMA patient tissues using TLB (20 mM Tris–HCl, pH 7.4, 137 mM NaCl, 1% Triton-X100, 2 mM ethylenediaminetetraacetic acid, 10% glycerol, 25 mM β-glycerophosphate, pH 7.4, 2 mM sodium pyrophosphate) with following added freshly 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease inhibitor cocktail (Sigma-Aldrich) (23). Protein concentration was measured using Coomassie (Bradford) protein assay. Cell and tissue lysates from normal human and SMA patient dividing cells, normal (non-SMA) and SMA mouse tissues and normal (non-SMA) human and SMA patient spinal cord tissues were prepared and diluted to protein concentration of 0.2 mg/ml using sample preparation kit (Protein Simple) for automated capillary western blot system, WES System (Protein Simple), which utilizes capillary-based electrophoretic separation and detection of proteins. Mouse spinal cord, tissues were isolated from 7-day-old non-SMA and SMA mice. Cell or tissue extracts with equal protein concentration were mixed with 0.1× sample buffer and 5× fluorescent master mix. The protein samples and the biotinylated ladder were denatured by heating at 95°C for 5 min. Protein samples, biotinylated ladder, primary antibodies (diluted 1:100 with antibody diluent), horseradish peroxidase (HRP)-conjugated secondary antibodies, chemiluminescence substrate and wash buffer were dispensed into respective wells of the assay plate and placed in Wes equipment. Signal intensity (area) of the protein was normalized to the peak area of loading control β-actin or α-tubulin. Quantitative analysis was performed using Compass software (Protein Simple) and statistical analysis of data was performed as described in below in ‘Statistical Analysis’ section.
The following primary phospho and non-phospho antibodies were used for IB analysis, SMN (610647, BD Biosciences), γH2AX (pSer 139) (NB100-79967), H2AX (NBP2-27428) and CHK1 (NB100-1768) from Novus Biologicals, ATM and p-CHK1 (Ser345) (2348) from Cell Signaling, β-actin (A5316) and α-tubulin (T8203) from Sigma-Aldrich, CHK2 (AFFN-CHEK2-17D2) deposited to DHSB by EU Program Affinomics), p-CHK2 (Thr68) (bs-3721R, Bioss Antibodies), p-BRCA1 (Ser1423) (ab90528), p-DNAPKcs (Ser2056) (ab18192), total-DNAPKcs (ab53701) and p-ATM (Ser1981) (ab81292) from Abcam, p-ATR (Ser 428) (sc-109912), BRCA1 (sc-642) (SantaCruz Biotech), ATR (ab-428) (Genescript) and SETX (#ab220827, Abcam).
Real-time quantitative PCR analysis
Total RNA was isolated from cultured (i) HeLa cells that were untransfected (Control) or transfected with siRNA 100 nM) against SMN1 or scrambled siRNA sequence (Scramble) and incubated for 30 h and (ii) normal (non-SMA) dividing human cells, WI-38 and GM03814, and SMA patient cells, GM03813 and GM09677 (SMA) using RNase Mini Kit (Qiagen). Total RNA (100 ng) per sample was reverse-transcribed using SuperScript VILO Kit (Invitrogen). We used SYBR Green PCR Master Mix (Applied Biosystems) for real-time quantitative polymerase chain reaction (RT-qPCR) analysis. The relative mRNA expression levels were calculated using log2−ΔΔCT method (14). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a positive control. The primer sequences were as follows: GAPDH primers: forward, 5′- ATAGGCGAGATCCCTCCAA-3′; reverse, 5′- TGAAGACGCCAGTGGAC-3′, SETX primers, forward 5′-CTTCATCCTCGGACATTTGAG-3′; and reverse 5′-TTAATAATGGCACCACGCTTC-3′ (24) and DNA-PKcs primers, forward 5′-CATCCAGAGTAGCGAATACTTCC-3′, reverse 5′-TTGTTTCGCAACCAGTTCAC-3′ (25).
Immunofluorescence analysis
Cells cultured on glass coverslips were washed with phosphate-buffered saline (PBS) and fixed in pre-chilled methanol (−20°C) for 5 min followed by 2 min in pre-chilled acetone (−20°C) at −20°C (20). Cells were also fixed with 4% PFA as described in individual experimental sections. PFA fixed cells were washed with PBS, permeabilized by incubation with 0.1% Triton-X100 for 5 min and washed 3× with PBS. Cells were blocked with 3% BSA in PBS with 0.5% Tween 20 (PBS-T) for 30 min at room temperature (22). Dividing cells were double labeled by sequential incubation with primary antibody against SMN for 1 h, washed 3× with PBS-T for 5 min and incubated with secondary antibody Alexa488-conjugated anti-mouse IgG, washed 3× with PBS-T for 5 min and followed by staining with second primary antibody against either one of the markers for nuclear bodies, p80 Coilin (#NBP2-15939, Novus Biologicals), LSM11 (#NBP1-81938, Novus Biologicals) and PML (#PM001, MBL) or DDR markers, γH2AX (#ab26350, Abcam), 53BP1 (#4937, Cell Signaling), p-ATM, p-ATR, p-CHK1, p-CHK2, p-DNAPKcs, p-BRCA1 or RNA–DNA hybrid (Clone #S9.6) (#ENH001, Kerafast) or SETX (#ab220827) or RNA Pol II (#ab210527), RNA helicase A (RHA) (#ab26271) and p-RPA32/RPA2 (Thr21) (#ab109394) from Abcam, for 1 h, washed 3× with PBS-T for 5 min, incubated with secondary antibody Alexa594-conjugated anti-rabbit IgG, washed 3× with PBS-T for 5 min. For ribonuclease H (RNase H) enzyme treatment, control and siSMN transfected cells were extracted with CSK buffer containing 0.1% Triton X-100 for 5 min followed by washing with CSK buffer without Triton-X100. Cells were incubated in RNase H digestion buffer (20 mM HEPES, pH 8.0, 50 mM KCl, 4 mM MgCl2, 1 mM 1, 4-dithiothreitol (DTT), 50 μg/ml BSA, ∼10U RNase H/ml) for 20 min at 37°C (26). After RNAse H treatment cells were washed with PBS and fixed in 4% PFA and double stained for SMN and coilin. Processed coverslips were mounted on microscope slides with Vectashield mounting medium containing DAPI and edges sealed with a coat of clear nail polish.
PFA fixed primary spinal cord motor neurons were washed with PBS, permeabilized with 0.1% Triton-X100 for 5 min, washed 3× with PBS-T for 5 min, blocked with 3% BSA in PBS-T for 30 min and double labeled using sequential incubation with primary antibody against either one of the DDR markers, γH2AX, 53BP1, p-ATM, p-ATR, p-CHK1, p-DNAPKcs, p-BRCA1 or RNA–DNA hybrid (S9.6) or RHA or SETX for 1 h, washed 3× with PBS-T for 5 min, incubated with secondary antibody Alexa488-conjugated anti-rabbit IgG, washed 3× with PBS-T for 5 min and followed by staining with second primary antibody anti-β-tubulin class-III neuron-specific antibody (clone TUJ1, #MAB1195, R & D Systems), washed 3× with PBS-T for 5 min, incubated Alexa594-conjugated anti-mouse IgG secondary antibody and washed 3× with PBS-T for 5 min (7). Immunofluorescence of stained cells was examined using confocal laser scanning microscope (Leica-TCS-SP5 equipped with AOBS and UV (405 nm), IR (633 nm) and Visible range lasers). Quantification of IF of R-loop accumulation in the nucleus of dividing cells and motor neurons was performed using NIH ImageJ software.
Statistical analysis
The quantitative data is presented as mean ± s.e.m. Statistical analysis performed using either one-way analysis of variance (ANOVA) or Student’s t-test (unpaired) with GraphPad Prism (version 5.0d). The value P = 0.05 or less was considered significant. In all experiments with mice, ‘n’ represents the number of mice used per group and in experiments with tissues or cells ‘n’ represents the number of times experiment was performed. A minimum of n = 3 mice or number of times experiment performed was used in all the experiments, unless otherwise specified in an experiment.
RESULTS
SMN deficiency causes DSBs and DDR activation in dividing non-SMA cells
To address the main question of why neurons selectively degenerate in SMA, we examined the effects of SMN-deficiency in normal (non-SMA) dividing human cells, followed by analysis of dividing cells derived from SMA patients, spinal cord neurons (non-dividing cells) derived from SMA mice and spinal cord tissues from SMA mice and patients.
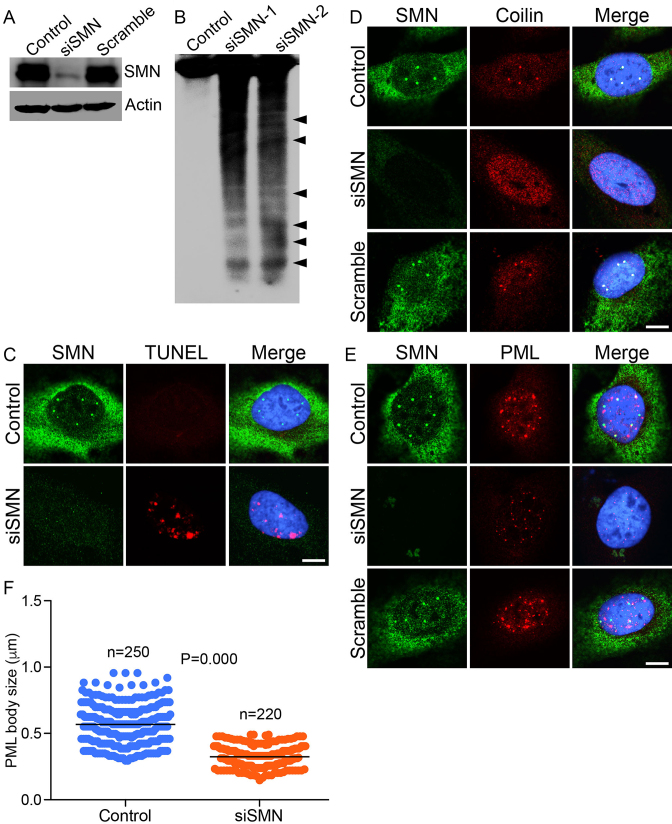
We began by examining the effects of acute (knockdown) and chronic (SMA) SMN-deficiencies on DNA damage and activation of DDR pathways. Using RNA interference (RNAi), we examined the effect of SMN knockdown on the integrity of genomic DNA. HeLa (non-SMA) cells treated with small interfering RNA against SMN (siSMN) show ∼80% reduction in SMN levels compared to control (mock-treated) and cells treated with scrambled siRNA (Scram) (Figure 1A). The specificity and efficiency of knockdown of SMN was established by examining the effect of a single oligonucleotide from the SMART siRNA pool. The single siRNA nucleotide shows knockdown of SMN similar to SMART pool (Supplementary Figure S1a). Analysis of genomic DNA isolated from cells treated with two concentrations of siRNA show marked DNA fragmentation suggesting that SMN-deficiency disrupts genomic DNA integrity (Figure 1B). Further, analysis of individual cells by the TUNEL assay, using in situ labeling of open DSB ends with EdUTP using terminal deoxynucleotidyl transferase, revealed accumulation of DSBs in SMN-deficient cells (Figure 1C). These data suggest that acute SMN-deficiency causes accumulation of DNA damage.
Figure 1.
Acute SMN deficiency (knockdown) causes DNA damage in non-SMA dividing cells. HeLa cells were untransfected (Control) or transfected with siRNA (100 nM) against SMN1 (siSMN) or scrambled siRNA sequence (Scramble) and incubated for 30 h. Cells were harvested to extract proteins and genomic DNA and fixed for staining with antibodies for IF analysis. (A) IB analysis of SMN and β-actin proteins in Control, siSMN and Scramble-treated cells. (B) Southern blot analysis of genomic DNA isolated from cells untransfected (Control) or transfected with siRNA, 100 nM (siSMN-1) and 200 nM (siSMN-2). Genomic DNA was isolated from Control and siRNA transfected cells (siSMN), 3′-end-labeled with biotin, separated by agarose gel electrophoresis, blotted to Zeta probe blotting membrane and detected using streptavidin–HRP-conjugated chemiluminescence kit. (C) Control and HeLa cells transfected with siRNA (siSMN) and scrambled siRNA (Scramble) were fixed and labeled with modified dUTP (EdU) nucleotide to detect strand breaks by TUNEL staining (red) and SMN is stained in green. Acute SMN deficiency causes disruption of sub-nuclear bodies known to contribute to genomic stability. (D) SMN (green) and coilin, marker of CB (red), (E) SMN (green) and PML, a marker of PML bodies (red). (F) Quantitation of number and size of PML bodies in control and siSMN-treated cells (10 cells/group). The average size of PML bodies in control is 0.55 ± 0.013 microns (n = 250) and in siSMN-treated cells is 0.32 ± 0.001 microns (n = 220). Immunofluorescence of stained cells was examined by confocal microscopy. Nuclei are stained with DAPI (blue). Scale bar is 10 μm.
In normal cells, SMN is present in the cytoplasm and in the nucleus where it accumulates in sub-nuclear bodies (NBs), including gems and Cajal bodies (CBs) (20). The function of sub-nuclear bodies, including CBs and promyelocytic leukemia (PML) bodies, has been associated with DNA damage and repair (27–29). To gain insight into the correlation between DNA damage and cellular organization of SMN+ nuclear bodies, we examined the effect of knockdown of SMN on CBs and PML bodies. SMN colocalizes with marker proteins p80 coilin (CBs) and PML protein (PML) in control (mock-treated) and scramble oligo treated cells (Figure 1D and E, Control)). Knockdown of SMN with RNAi (siSMN) in HeLa cells results in disruption of SMN-containing CBs (Figure 1D) that causes mislocalization and redistribution of coilin in the nucleoplasm. Moreover, SMN-deficiency causes reduction in the size of PML bodies, possibly due to disintegration of normal PML bodies into smaller sub-nuclear foci (Figure 1E). Quantitative analysis of PML bodies show a decrease in the average size (from 0.55 ± 0.013 to 0.32 ± 0.001 microns, P = 0.000) and number of sub-nuclear bodies in SMN-deficient cells (Figure 1F). These data suggest that the disruption of SMN+ NBs correlates with DNA damage accumulation in SMN-deficient cells.
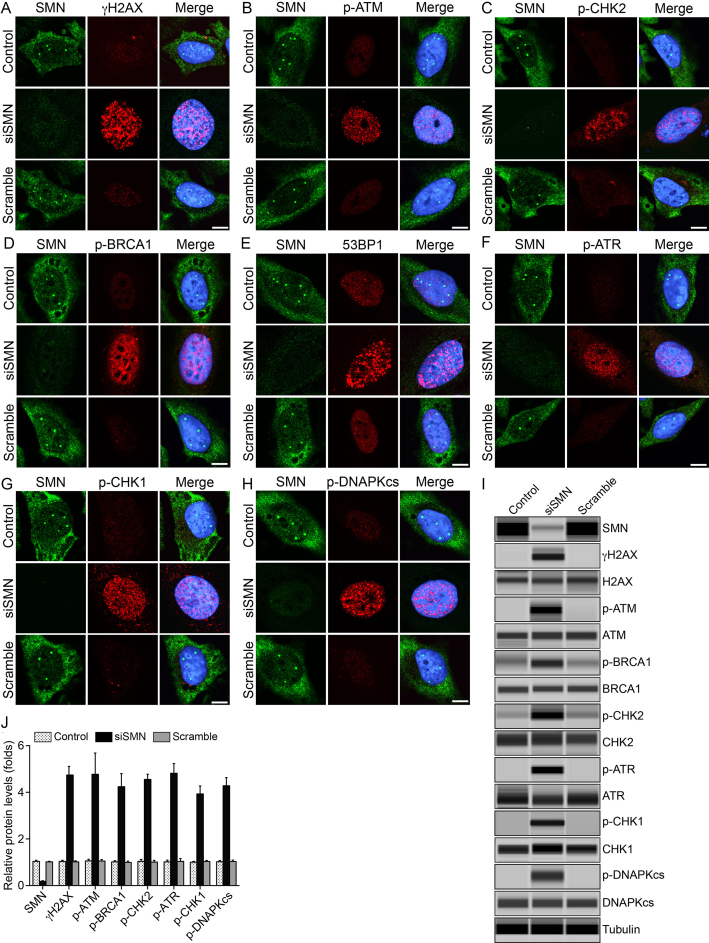
Analysis of DNA damage markers shows that SMN deficiency results in increased phosphorylation of the histone variant H2AX (γH2AX, phospho-H2AX, a marker for DSBs) (30) (Figure 2A and Supplementary Figure S2a, low magnification images), ATM kinase (p-ATM), a DNA damage sensor kinase that phosphorylates H2AX (Figure 2B) and DNA damage checkpoint kinase 2 (CHK2), required for cell cycle arrest upon DNA damage, in particular DSBs (Figure 2C). These data suggest that SMN deficiency causes DSB accumulation and activation of DNA damage checkpoints. Further, we examined the activation of markers of HR and NHEJ DNA repair pathways. In addition to increased phosphorylation of ATM (p-ATM), usually induced by DSB detection by the MRE11–RAD50–NBS1 complex (31), SMN-deficient cells show increased BRCA1 phosphorylation (p-BRCA1) (Figure 2D) and accumulation of 53BP1 in sub-nuclear foci (Figure 2E). These results indicate that the repair of 53BP1-marked DSBs occurs in part by a BRCA1-dependent HR pathway. For HR to initiate, DSBs are resected to generate single stranded (ss) DNA ends. Indeed, SMN-deficient cells showed increased phosphorylation also of the ATR kinase (Figure 2F), which responds to ssDNA and is activated following DSB processing (31) and of the ATR-downstream kinase, CHK1 (Figure 2G) (32). However, increase in the phosphorylation of DNA-PKcs in SMN-deficient cells (Figure 2H and Supplementary Figure S2b, low magnification images), suggests concomitant activation of DSB repair by NHEJ. Thus, both HR and NHEJ pathways are activated by the DSBs formed in response to SMN-deficiency.
Figure 2.
Acute SMN deficiency causes activation of DDR pathways in non-SMA dividing cells. HeLa cells were untransfected (Control) or transfected with siRNA (100 nM) against SMN1 (siSMN) or scrambled siRNA sequence (Scramble) and incubated for 30 h. Cells were fixed and stained with antibodies against marker proteins of DDR pathways. Representative images are presented for double-labeled immunostainings: (A) SMN (green) and phospho-H2AX (γH2AX) (red), (B) SMN (green) and phospho-ATM (p-ATM) (red), (C) SMN (green) and phospho-CHK2 (p-CHK2) (red), (D) SMN (green) and phospho-BRCA1 (p-BRCA1) (red), (E) SMN (green) and 53BP1 (red), (F) SMN (green) and phospho-ATR (p-ATR) (red), (G) SMN (green) and phospho-CHK1 (p-CHK1) (red) and (H) SMN (green) and phospho-DNAPKcs (red). Nuclei were stained with DAPI (blue). Immunofluorescence of stained cells was examined by confocal microscopy. Scale bar is 10 μm. Data from knockdown of SMN in normal human WI-38 (non-SMA) dividing cells is presented in Supplementary Figure S3. (I) IB analysis of protein extracts using automated capillary-based western blot system (ProteinSimple) to determine average levels of activation of DDR markers. Representative capillary-IB images of proteins, using antibodies against phospho-proteins and non-phospho-proteins are presented (full-length IBs are shown inSupplementary Figure S8). (J) Quantitative (mean ± s.e.m., n = 3) and statistical analysis (one-way analysis of variance, ANOVA) of relative levels of proteins normalized to α-tubulin show that the SMN knockdown with RNAi reduced SMN levels to (18.36 ± 2.68%, P = 0.000) and causes increase in phosphorylation (in folds) of DDR markers as follows: γH2AX (4.74 ± 0.37, P = 0.000), p-ATM (4.77 ± 0.91, P = 0.003), p-BRCA1 (4.24 ± 0.55, P = 0.000), p-CHK2 (4.55 ± 0.22, P = 0.000), p-ATR (4.81 ± 0.41, P = 0.000), p-CHK1 (3.93 ± 0.34, P = 0.000) and p-DNAPKcs (4.28 ± 0.34, P = 0.000) are presented as a bar graph. Analysis of levels of non-phospho-protein in control, siSMN and Scramble samples normalized to tubulin did not show any statistical differences between control and SMN-deficient cells (siSMN). Marked increase in levels of phospho-proteins that are components of DDR pathways show that the acute SMN-deficiency causes activation of DDR markers and DSB repair pathways, HR and NHEJ, in dividing non-SMA human cells.
Quantitative and statistical (one-way ANOVA) analysis of IBs (Figure 2I) shows that acute reduction of SMN levels to 18.36 ± 2.68% (P = 0.000) results in robust and significant increase (∼4- to 5-fold) in phosphorylation of DNA damage markers, γH2AX, p-ATM, p-CHK2 and DNA repair/DDR markers, p-ATR, p-CHK1, p-DNA-PKcs and p-BRCA1 (Figure 2J). Analysis of corresponding non-phospho-proteins in Control, siSMN and Scramble samples normalized to tubulin did not show any statistical differences between control and SMN-deficient cells (siSMN) suggesting that acute knockdown of SMN did not cause change in expression at protein levels and resulted in increase of phosphorylation (activation) of the DDR pathway markers (Figure 2I). In addition to human HeLa (non-SMA) cancer cell line, we also examined the effect of SMN deficiency in normal human (non-SMA) fibroblast cells (WI-38), obtaining similar results for all the above analyzed markers (Supplementary Figure S3). The consistent results on multiple DDR and DNA repair markers in the two analyzed normal cell lines suggest that acute SMN-deficiency causes DSB-related DNA damage that triggers activation of DDR and DSB repair pathways, HR and NHEJ, in dividing non-SMA cells.
SMN-deficiency causes downregulation of SETX and accumulation of R-loops
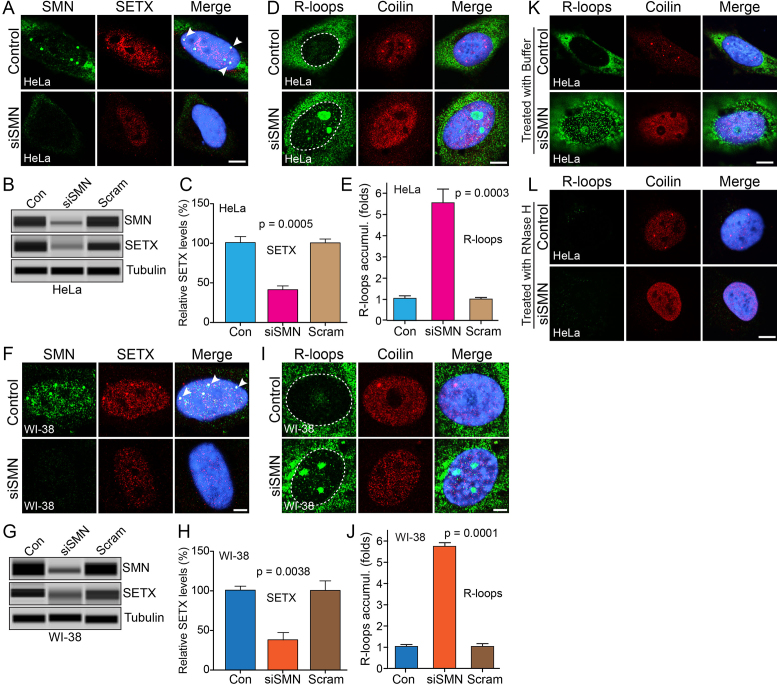
To further investigate the mechanism of DNA damage caused by SMN-deficiency, we examined the effect of acute SMN-deficiency on R-loop accumulation, one of the leading causes of genome instability associated with defects in transcription (33,34). We examined the cellular localization and levels of one of the key components, SETX, which is required to resolve RNA–DNA hybrids (R-loops) (24). Control cells show colocalization of SETX with SMN in sub-nuclear foci [Figure 3A (HeLa) and F (WI-38)]. Notably, knockdown of SMN (siSMN) shows loss of SETX containing nuclear bodies and reduced staining of SETX in the nucleus, indicating decrease in SETX levels [Figure 3A (HeLa) and F (WI-38)]. Quantitative and statistical (one-way ANOVA) analysis of IBs show SETX protein levels markedly decreased to (41.42 ± 4.79%, P = 0.0005) in SMN-deficient HeLa cells (Figure 3B and C) and (37.18 ± 9.33%, P = 0.0038) in SMN-deficient WI-38 cells (Figure 3G and H) compared to control and scramble (Scram) treated cells. Further, analysis of mRNA expression using real-time quantitative PCR method shows a decrease of ∼24% (P = 0.000) in SETX transcript levels in siSMN-treated cells compared to control and scrambled treated cells (Supplementary Figure S4a), suggesting that transcriptional downregulation contributes to reduced levels of SETX in SMN-deficient cells.
Figure 3.
Acute SMN deficiency causes downregulation of SETX and accumulation of RNA–DNA hybrids (R-loops) in non-SMA dividing human cells. HeLa and WI-38 cells were untransfected (Control) or transfected with siRNA (100 nM) against SMN1 (siSMN) or scrambled siRNA sequence (Scramble) and incubated for 30 h. (A) Immunofluorescence analysis show that SMN (green) and SETX (red) colocalize in sub-nuclear foci of (Control, arrowheads) HeLa cells and knockdown of SMN causes reduction in staining of SETX (red) and (B) IBs for SMN, SETX and Tubulin of cell lysates prepared from Control and siSMN and Scramble (Scram)-treated HeLa cells, (C) quantitative (mean ± s.e.m., n = 3) and statistical analysis (one-way ANOVA) of HeLa cells show knockdown of SMN levels to (22.90 ± 6.71%, P = 0.0003) decreased SETX levels to (41.42 ± 4.79%, P = 0.0005) compared to control and scramble (Scram). (D) Accumulation of RNA–DNA hybrids (R-loops) in SMN-deficient cells. R-loops were detected by monoclonal antibody (S9.6) against RNA–DNA hybrids (green). Coilin is a control stained in red. (E) Quantitative and statistical analysis (one-way ANOVA) of R-loop accumulation in cells using IF images and ImageJ software show cells treated with siRNA (siSMN) have increased R-loops 5.55 ± 0.68-fold (P = 0.0003) accumulation compared to Control and Scramble-treated cells. (F) Colocalization of SMN (green) and SETX (red) in nub-nuclear foci of WI-38 cells (Control, arrowheads) and knockdown of SMN causes reduction in staining of SETX (red). (H) IBs for SMN, SETX and Tubulin of cell lysates prepared from Control and siSMN and Scramble (Scram)-treated WI-38 cells, (I) quantitative (mean ± s.e.m., n = 3) and statistical analysis (one-way ANOVA) of WI-38 cells show knockdown of SMN levels to (31.87 ± 8.44%, P = 0.0008) decreased SETX levels to (37.18 ± 9.33%, P = 0.0038) compared to control and scramble (Scram). (G) Accumulation of R-loops (green) in SMN-deficient cells. (J) Quantitative and statistical analysis (one-way ANOVA) of R-loop accumulation in cells using IF images and ImageJ software show cells treated with siRNA (siSMN) have increased R-loops 5.74 ± 0.20-fold (P = 0.0001) accumulation compared to Control and Scramble-treated cells. Digestion of R-loops with the RNase H enzyme, but not buffer alone, reduces R-loop accumulation. R-loops nuclear levels were quantified from three cell culture replicates (30 cells each). Digestion of R-loops with the RNaseH enzyme, but not buffer alone, reduces R-loop accumulation. Cells transfected with siRNA (siSMN) were permeabilized and treated with (K) buffer only and (L) RNase H enzyme for 20 min, washed and fixed with 4% PFA. Cells were stained with antibodies and IF was examined by confocal microscopy; R-loops (green) and coilin (red). Nuclei were stained with DAPI (blue). Scale bar is 10 μm. (Full-length blots are included inSupplementary Figure S9).
To examine accumulation of R-loops in SMN-deficient cells, we stained HeLa and WI-38 cells with an antibody against RNA–DNA hybrids (S9.6). Control cells show presence of R-loops in the cytoplasm, which are known to be associated with mitochondrial replication and transcription (35), and low levels of R-loops in the nucleus [Figure 3D (HeLa) and I (WI-38)]. Knockdown of SMN (siSMN) causes accumulation of R-loops in small foci in the nucleus and large foci in the nucleolus, which may correspond to R-loops generated by highly transcribed ribosomal DNA [Figure 3D (HeLa), Supplementary Figure S2c (low mag images) and Figure 3I (WI-38)] (36). Quantitative and statistical (one-way ANOVA) analysis of nuclear IF show marked increase in R-loops accumulation (5.55 ± 0.68-fold, P = 0.0003) in SMN-deficient HeLa cells (Figure 3D and E) and (5.74 ± 0.20-fold, P = 0.0001) in SMN-deficient WI-38 cells (Figure 3I and J) compared to control cells. To test whether accumulated R-loops can be digested with recombinant RNase H, we treated control and transfected (siSMN) cells with permeabilization buffer (Figure 3K) or buffer containing RNase H (Figure 3L). Control and siSMN transfected cells show that R-loops in control and SMN-deficient (siSMN) cells can be resolved by digestion of RNA using exogenous RNase H (Figure 3L). Together, these findings suggest that acute SMN-deficiency causes down-regulation of SETX levels leading to accumulation of R-loops that may contribute to DNA damage and genome instability in SMN-deficient cells.
Chronic low levels of SMN cause SETX deficiency, R-loop accumulation and activation of DDR pathways in primary fibroblast (dividing cells) from severe SMA patients
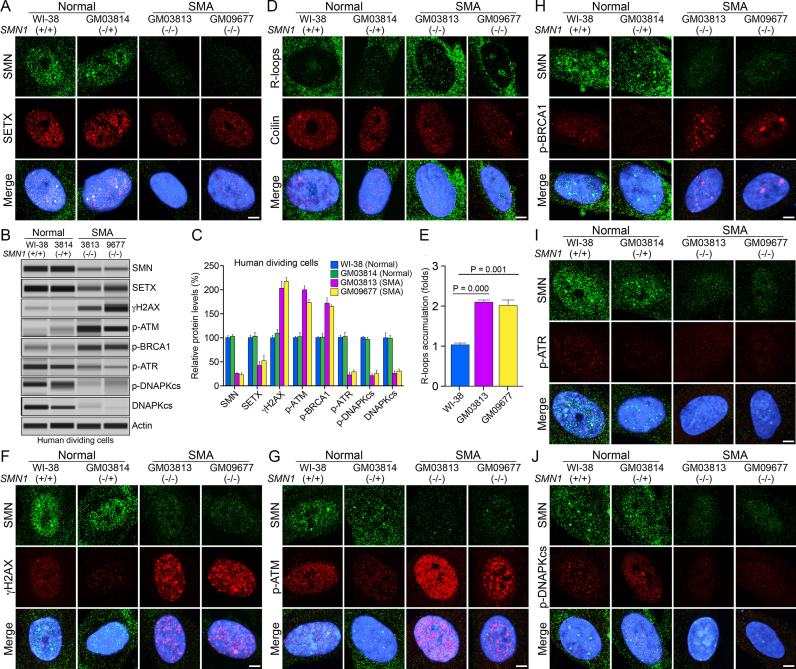
To test the relevance of defects caused by acute SMN-deficiency in the context of SMA pathogenesis, we examined SMA patient-derived primary fibroblast (dividing cells) that represent a pathophysiological state of chronic low levels of SMN. We used two control cell lines, WI-38 (SMN1+/+) and GM03814 (SMN1−/+) derived from one of the parents (carrier) heterozygous for SMN1−/+ (normal phenotype), and two patient cell lines GM03813 (SMN1−/−) and GM09677 (SMN1−/−) derived from severe SMA type I patients that have SMN1 deletion (20). To test whether chronic SMN-deficiency causes DNA damage, we examined DNA damage accumulation in dividing SMA patient cells, GM03813 and GM09677 using TUNEL assay. SMA patient cells show accumulation of DSBs compared to normal cells (WI-38) suggesting that chronic low levels of SMN cause DNA damage that is similar to the DNA damage caused by acute deficiency of SMN in non-SMA cells (Supplementary Figure S5 and Figure 1C). To find the mechanism and cause of DNA damage, we examined SETX levels, accumulation of R-loops and activation of DDR pathways in patient cells. Double staining of control human cells (Normal) with SMN and SETX show colocalization of SETX and SMN in nuclear bodies in WI-38 and GM3814 cells. No differences were detected between WI-38 (SMN1+/+) and GM03814 (SMN1−/+) cells suggesting that heterozygous SMN1−/+ genotype did not affect cellular phenotype, consistent with previous findings (20). Comparison of nuclear staining of SETX (Figure 4A) of SMA patient cells GM03813 and GM09677 with normal WI-38 and GM03814 shows mislocalization and downregulation of SETX in SMA patient cells. Comparison of levels of SMN, SETX, γH2AX, p-ATM, p-ATR, p-BRCA1, p-DNA-PKcs and total DNA-PKcs between normal (non-SMA) cells, WI-38 and GM03814, did not show any statistically significant differences (Figure 4B and C), which is consistent with IF analysis (Figure 4A, D, F–J). Average SETX protein levels in GM03813 and GM09677 cells showed decrease (53.34 ± 7.20%, P = 0.001) compared to normal WI-38 and GM03814 cells (Figure 4B and C). Analysis of mRNA expression also showed decrease of ∼25% (P = 0.001) in SETX transcripts in SMA patient cells GM03813 and GM09677 compared to normal WI-38 and GM03814 cells (Supplementary Figure S4b), suggesting that chronic SMN-deficiency causes transcriptional downregulation of SETX similar to acute SMN-deficiency (Supplementary Figure S4a) and contributes to reduced levels of SETX protein in SMA patient cells. Next, we examined R-loop accumulation using the monoclonal antibody (S9.6) against RNA–DNA hybrids, which revealed increase in R-loops foci in the nucleus of patient cells compared to normal cells (Figure 4D). Quantitative analysis of nuclear IF shows increased nuclear R-loops in GM03813 (2.09 ± 0.04-fold, P = 0.000) and GM9677 (2.01 ± 0.11-fold, P = 0.001) compared to normal cells (Figure 4E). These data suggest that chronic SMN-deficiency also causes mislocalization and >50% reduction in SETX levels and accumulation of R-loops, but to a smaller extent (∼2-fold) compared to acute SMN-deficiency (∼5-fold) with ∼60% loss of SETX.
Figure 4.
Chronic low levels of SMN cause deficiency of SETX and phospho-DNA-PKcs, and accumulation of R-loops and DNA damage in dividing cells derived from SMA patients. Dividing cells derived from human (WI-38, SMN1 +/+), non-SMA (Normal), GM03814 (carrier of disease, parent of GM03813, heterozygous for SMN1 ±, control) and SMA type I patients, GM03813 and GM09677 (SMA) were cultured, in petri dishes for IB analysis or on glass coverslips for IF analysis. Proteins were detected using antibodies against SMN and markers of DDR pathways. Representative images are presented for double-labeled immunostainings: (A) SMN (green) and SETX (red). (B) Protein extracts were prepared from normal (WI-38 and GM03814) and SMA patient cells, GM03813 and GM09677, and examined by IB analysis using automated capillary-based western blot system (ProteinSimple). Representative capillary-blot images of proteins and phospho-proteins are shown (full-length blots are included inSupplementary Figure S10). (C) Quantitative (mean ± s.e.m., n = 3) and statistical analysis (unpaired t-test) of relative levels of proteins normalized to β-actin in dividing normal and SMA patient cells are presented as a bar graph. Comparison of levels of different proteins between Normal, WI-38 and GM3814, cells did not show any statistically significant differences. Quantitative analysis show average of chronic SMN levels (GM03813 + GM09677) reduced to (24.45 ± 1.73%, P = 0.000) in SMA patient cells compared to Normal, WI-38 and GM03814 cells (100.6 ± 3.90%, P = 0.000), decrease in SETX levels to (47.21 ± 5.78%, P = 0.0018) and average increase in levels of DDR and HR repair phospho-proteins in SMA patient cells (GM03813 + GM09677), γH2AX (210.30 ± 10.25%, P = 0.000), p-ATM (187.00 ± 7.08%, P = 0.000), p-BRCA1 (168.80 ± 7.11%, P = 0.000) compared to WI-38 cells. p-ATR levels reduced to (26.09 ± 1.17%, P = 0.000). Levels of phospho-DNAPKcs (p-DNAPKcs) reduced to (23.48 ± 1.64%, P = 0.000) and total DNAPKcs (DNAPKcs) reduced to (28.38 ± 3.01%, P = 0.002) in SMA patient cells compared to non-SMA (Normal) cells. (D) Accumulation of R-loops (green) detected with S9.6 monoclonal antibody and coilin (red), used as control marker. (E) Quantitative (mean ± s.e.m., n = 3) and statistical (unpaired t-test) analysis of R-loop levels in cells using IF images and ImageJ software show increase in accumulation of R-loops in SMA patient cells, GM03813 (2.09 ± 0.04-fold, P = 0.000) and GM09677 and (2.01 ± 0.11-fold, P = 0.001) compared to WI-38 (Normal) cells. IF analysis of DDR markers shows that chronic low levels of SMN result in DNA damage accumulation and activation of HR repair pathway in SMA patient dividing cells: (F) SMN (green) and phospho-H2AX (γH2AX) (red), (G) SMN (green) and phospho-ATM (p-ATM) (red), (H) SMN (green) and phospho-BRCA1 (p-BRCA1) (red), (I) SMN (green) and phospho-ATR (p-ATR) (red). IB (B and C) and IF analysis show that chronic SMN deficiency causes decrease in levels of DNA-PKcs and its activation. (J) SMN (green) and phospho-DNA-PKcs (p-DNAPKcs) (red). Nuclei were stained with DAPI (blue). Scale bar is 5.0 μm.
Further, we examined activation of DDR markers in patient cells. Increased staining of γH2AX and its accumulation in nuclear foci suggests increased DSBs in SMA patient cells compared to normal cells (Figure 4F and Supplementary Figure S6a, low mag images). Examination of DDR and DSB repair proteins revealed increased phosphorylation (activation) of ATM (p-ATM) and BRCA1 (p-BRCA1) in SMA compared to normal cells (Figures 4G and H), which is similar to the activation caused by acute SMN-deficiency in non-SMA cells. However, in contrast to acute SMN-deficiency in non-SMA cells, chronic SMN-deficiency in SMA patient cells showed reduction in the levels of p-ATR compared to normal cells (Figure 4I). Quantification confirmed reduced SMN levels in SMA cells, GM03813 and GM09677, compared to normal cells, WI-38 and GM03814 cells (Figures 4B and C). Loss of ∼75% SMN causes (i) decrease in SETX levels in GM03813 (∼58%) and GM09677 (∼49%) compared to WI-38 cells and (ii) marked increase in levels of phosphorylated (activated) DDR markers, γH2AX, p-ATM and BRCA1 in GM03813 and GM09677 SMA cells compared to normal cells. Estimation of p-ATR shows its levels reduced (∼23%) in GM03813 and (∼29%) in GM09677 compared to WI38 cells suggesting that ATR, which responds to ssDNA, fails to be activated in SMA dividing cells. Thus, in SMA cells, endogenous SSBs may be converted into DSBs prior to ATR activation, and the DSBs may be less resected. Nevertheless, increased levels of γH2AX and p-ATM indicate more DSB-related DNA lesions, some of which are marked and potentially repaired by a BRCA1-dependent HR pathway. Together, these data suggest that chronic low levels of SMN cause average decrease of ∼54% in SETX levels and disruption of SETX-containing nuclear bodies, which may account for partial loss of its function to resolve R-loops. These defects result in accumulation of R-loops, which lead to DSB accumulation and activation of DNA damage repair pathways, including HR, in dividing SMA patient cells.
Chronic low levels of SMN cause deficiency of DNA-PKcs in SMA patient dividing cells
The above data show that the HR pathway is at least in part activated to repair the DNA damage arising in SMA patient cells. However, patient cells continue to show DSB accumulation and DDR activation. HR is responsible for the repair of a subset of lesions during replication and post-replication repair, whereas NHEJ plays important roles both in G1 and in G2/M. To gain insight into the status of the NHEJ pathway in SMA dividing cells, we examined the activation of DNA-PKcs, which is essential for NHEJ. Comparison of staining of phospho-DNA-PKcs (p-DNAPKcs) between normal and patient cells shows decrease in staining of p-DNA-PKcs in patient cells (Figure 4J and Supplementary Figure S6b, low magnification images), suggesting reduced DNA-PKcs activity in SMA patient cells. To find whether this is caused by a defect in phosphorylation (activation) of DNA-PKcs or due to deficiency of DNA-PKcs in patient cells, we examined the levels of p-DNA-PKcs and total DNA-PKcs proteins in normal and SMA patient cells. IBs showed marked reduction in levels of p-DNAPKcs in SMA, GM03813 and GM09677, compared to normal WI-38 and GM03814 cells (Figure 4B), consistent with IF data (Figure 4J). Moreover, examination of IBs of total DNA-PKcs revealed severe reduction in detectable amounts of total DNA-PKcs (DNAPKcs) in SMA patient cells (Figure 4B). Quantification of IBs showed marked reduction in phospho-DNA-PKcs and total DNA-PKcs protein levels. The levels of p-DNA-PKcs reduced to (21.36 ± 3.82%, P = 0.000) in GM03813 and (25.60 ± 7.06%, P = 0.000) in GM09677 compared to WI-38 cells suggesting severe loss of DNA-PKcs activity in SMA patient cells. Notably, levels of total DNA-PKcs were also markedly reduced; specifically, to (26.09 ± 4.82%, P = 0.002) and (30.66 ± 4.40%, P = 0.003) in SMA patient cells GM03813 and GM09677, respectively, compared to normal (WI-38) cells (Figure 4B and C). To determine whether SMN-deficiency has effect on the transcription levels of DNA-PKcs, we examined the effect of acute and chronic SMN-deficiencies on mRNA expression using RT-qPCR method. Acute SMN-deficiency causes a small decrease ∼13% (P = 0.050) in DNA-PKcs transcript levels in siSMN-treated cells compared to control and scrambled treated cells (Supplementary Figure S7a), suggesting that acute SMN deficiency reduces transcription of DNA-PKcs however small decrease in mRNA expression did not affect DNA-PKcs protein levels in HeLa (non-SMA) SMN-deficient cells (Figure 2I). Analysis of mRNA expression in SMA patient cells shows decrease of ∼35% (GM03813) and ∼31% (GM09677) in DNA-PKcs transcripts in SMA patient cells compared to normal WI-38 cells (Supplementary Figure S7b), suggesting that chronic SMN-deficiency causes transcriptional downregulation of DNA-PKcs ∼3-fold greater than acute SMN-deficiency and contributes to reduced levels of DNA-PKcs protein in SMA patient cells. Comparison of data from acute and chronic SMN deficiencies suggest that acute SMN deficiency results in activation of NHEJ pathway by activating DNA-PKcs, in contrast, chronic SMN-deficiency causes marked decrease in DNA-PKcs levels and its activation, resulting in inefficient NHEJ-mediated DNA repair in patient cells. However, DNA repair by HR may be sufficient to support the proliferation and growth of dividing SMA patient primary fibroblast cells. The potential for DNA repair in dividing SMA cells is supported by previous finding that showed cultured SMA mouse primary skeletal myocytes were competent of repairing DNA damage induced by γ-radiation and etoposide treatment (37).
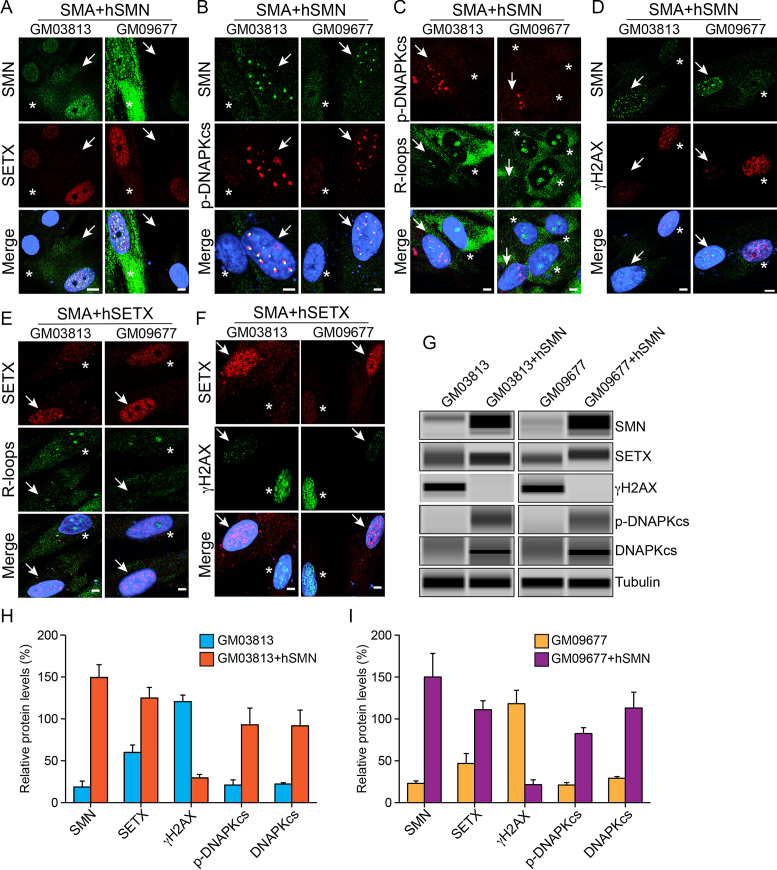
Rescue of SETX and DNA-PKcs deficiencies and DNA damage phenotype in SMA patient cells complemented with SMN or SETX
To test whether defects in SMA patient primary fibroblast (dividing cells) can be reversed by restoring SMN levels, we performed a rescue experiment and transfected patient cells with a plasmid containing recombinant human SMN (hSMN) for transient expression. Ectopic expression of SMN increases SMN levels (∼7-fold) and its accumulation in the nucleus of GM03813 and GM09677 cells as indicated by IF (Figure 5A, SMN) and IB and quantitative analysis (Figure 5G–I). Notably, restoration of SMN levels rescued SETX-deficiency by causing >2-fold increase in SETX levels (Figure 5H and I) and sub-cellular colocalization of SETX with SMN in nuclear bodies in transfected cells (arrows) compared to non-transfected (asterisks) cells (Figure 5A and G). Overexpression of SMN also up-regulated levels of p-DNA-PKcs (∼3-fold) and rescued deficiency of DNA-PKcs by increasing levels of total DNA-PKcs (∼3-fold) in patient cells (Figure 5B, G–I). To test whether elevation of SETX levels would reduce R-loop accumulation, we examined SMN overexpressing patient cells stained with antibodies against R-loops and p-DNA-PKcs that show decrease in R-loop accumulation in cells with restored levels of DNA-PKcs (or SMN) (arrows) compared to non-transfected (asterisks) cells, suggesting that restoration of SETX levels by SMN overexpression rescues R-loop accumulation phenotype in SMA patient cells (Figure 5C). Further, to test whether increase in SETX and DNA-PKcs and decrease in R-loop levels would rescue DNA damage accumulation in patient cells, we examined SMN overexpressing cells stained for γH2AX that show marked reduction in γH2AX foci in transfected cells (arrows) compared to non-transfected (asterisks) cells (Figure 5D) and the levels of γH2AX were reduced by ∼6-fold (Figure 5G–I). Together, the rescue of SETX and DNA-PKcs levels and DNA damage phenotype in patient cells suggest that chronic low levels of SMN are responsible for SETX-deficiency that results in accumulation of R-loops and DNA damage, and for DNA-PKcs-deficiency that may reduce NHEJ-mediated DNA repair and contribute to DSB accumulation in patient cells.
Figure 5.
Ectopic expression of either SMN or SETX reduce R-loop accumulation and rescue DNA damage phenotype in SMA patient cells. Dividing SMA patient dividing cells, GM03813 and GM09677 (SMA) were transfected with pCMV/mycSMN or infected with Ad-h-SETX virus and stained with antibodies against SMN, phospho-H2AX (γH2AX), phospho-DNA-PKcs (p-DNAPKcs) and SETX. (A–D) Transient expression of SMN restores levels of SETX and p-DNA-PKcs and reduces R-loop and DNA damage accumulation in SMA patient cells. (A) SMN (green) and SETX (red), (B) SMN (green) and p-DNAPKcs (red), (C) R-loops (green) and p-DNAPKcs (red) and (D) SMN (green) and γH2AX (red). (E and F) Transient expression of SETX also reduces R-loops and DNA damage accumulation, and rescues phenotype of SMA patient cells. (E) SETX (red) and R-loops (green) and (F) SETX (red) and γH2AX (green). Nuclei were stained with DAPI (blue). Scale bar is 5.0 μm. Arrows show transfected cells and asterisks indicate non-transfected cells. (G) Effect of SMN overexpression on changes in the levels of SETX, γH2AX, p-DNAPKcs, total DNAPKcs in SMA patient cells GM03813 and GM09677 examined by IB analysis (full-length blots are included inSupplementary Figure S11). Quantitation and comparison of SMN, γH2AX, SETX, p-DNA-PKcs and DNA-PKcs levels in (H) GM03813 and GM03813+hSMN cells show rescue of DNA damage in SMN complemented cells (GM03813+hSMN) by marked reduction in γH2AX levels to (29.55 ± 4.22%, P = 0.000) compared to control GM03813 cells (120.5 ± 7.85%) that is supported by increase in levels of SETX (64.87 ± 15.37%, P = 0.013), p-DNA-PKcs (71.97 ± 20.83%, P = 0.025) and total DNA-PKcs (69.50 ± 18.91%, P = 0.021) and (I) analysis of GM09677 and GM09677+hSMN cells show rescue of DNA damage in SMN complemented cells (GM09677+hSMN) by marked reduction in γH2AX levels to (21.63 ± 5.76%, P = 0.002) compared to control GM09677 cells (124.3 ± 13.99%) that is supported by increase in levels of SETX (64.12 ± 15.97%, P = 0.015), p-DNA-PKcs (61.52 ± 7.62%, P = 0.001) and total DNA-PKcs (83.77 ± 18.93%, P = 0.011).
Furthermore, to test whether SETX-deficiency found in patient cells is a critical cause of DNA damage phenotype in SMA, we performed another rescue experiment and infected patient cells with Ad-hSETX virus expressing recombinant human SETX (38). Analysis of patient cells double-stained with SETX and R-loop antibodies show that cells overexpressing SETX (arrows) display reduced staining for R-loops compared to non-infected (asterisks) cells (Figure 5E), suggesting decrease in R-loops accumulation by direct restoration of SETX levels. This result is consistent with the rescue of phenotype by indirect restoration of SETX levels using SMN overexpression in patient cells (Figure 5A and G). Further examination of cells double-stained with SETX and γH2AX show that cells overexpressing SETX (arrows) contain reduced numbers of γH2AX foci compared to non-infected (asterisk) cells (Figure 5F). Rescue of DNA damage phenotype by restoration of SETX levels in patient cells suggests that SETX-deficiency may be central to DNA damage accumulation caused by chronic low levels of SMN in SMA.
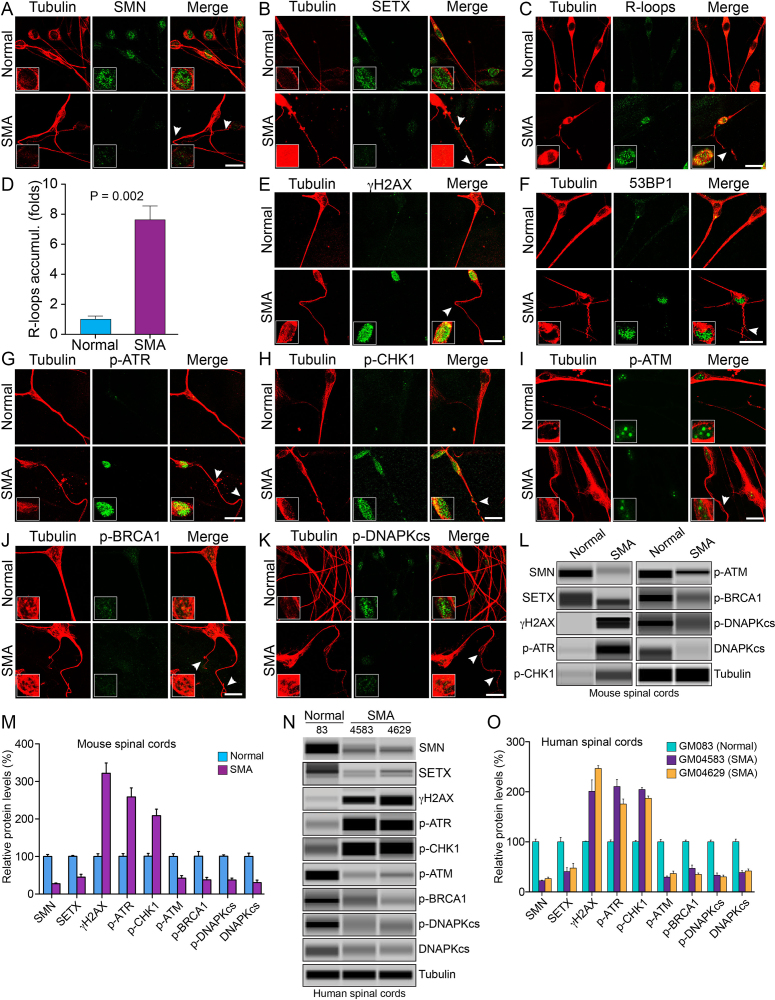
Chronic low levels of SMN cause down regulation of SETX and DNA-PKcs, accumulation of R-loops and activation of DDR pathways in spinal cord neurons from SMA mice
To gain insight into the mechanism of neurodegeneration, we tested if chronic low levels of SMN cause similar DNA damage accumulation in neurons (non-dividing cells). We examined cultured primary neurons isolated from the spinal cords of 7-day-old normal and SMA mice using a method that we previously developed (7). We have shown that cultured spinal cord neurons stain positive for known motor neuron markers, such as choline acetyltransferase (ChAT) and Hlxb9 (Hb9), suggesting that cultured neurons may have characteristics similar to motor neurons (7,22). In the control experiment, IF analysis of neurons double-stained with SMN and neuron-specific β-tubulin-III antibodies show reduced staining for SMN in SMA neurons compared to control neurons (Normal) from non-SMA mice (Figure 6A). Notably, chronic SMN-deficiency causes loosening, bending, retraction and degeneration of axons that are indicated by arrowheads, which are known to be associated with SMA pathogenesis (7), compared to neurons from non-SMA mice (Normal) (Figure 6). Next, we examined the status of SETX and accumulation of R-loops in SMN-deficient neurons derived from SMA mice. Examination of double-stained neurons with SETX and tubulin antibodies revealed marked reduction in the staining of SETX in neurons derived from SMA mice compared to normal mice (Figure 6B). Further, analysis of neurons stained with antibody against R-loops shows increased accumulation of R-loops in the nucleus of neurons derived from SMA mice compared to normal mice (Figure 6C). Quantification of nuclear IF shows (7.62 ± 0.97-fold, P = 0.002) higher accumulation of R-loops in SMA neurons compared to normal neurons and suggests that the SMN-deficiency causes accumulation of R-loops irrespective of a cell type (Figure 6D). Comparison of the increase in R-loop accumulation in dividing SMA patient cells (∼2-fold) and SMA motor neurons (∼7.6-fold) shows a larger accumulation of R-loops in SMA neurons. It is possible that greater accumulation of R-loops in neurons may result in increased genome instability in SMA neurons (non-dividing cells) compared to dividing SMA patient cells and may represent one of the factors that contribute to predominant degeneration of motor neurons in SMA.
Figure 6.
Chronic low levels of SMN cause SETX-deficiency, accumulation of R-loops and activation of DDR pathways in spinal cord neurons derived from SMA mice, and spinal cord tissues from SMA mice and SMA type I patients. Primary motor neurons were cultured using spinal cords isolated from 7-day-old wild-type (Normal) and SMAΔ7 mice (SMA). The spinal cord ex-plants were cultured in vitro for 12–14 days to allow differentiation and growth of motor neurons in laminin-coated 8-well chambers, fixed with 4% PFA and stained with antibodies and IF examined by confocal microscopy. (A) Low levels of SMN (green) in SMA neurons causes axonal defects as shown by staining with neuron-specific β-tubulin-III (clone TUJ1) (red). Axonal defects include bending, folding, retraction and ballooning of axons (arrowheads) that indicate degeneration of SMN-deficient neurons. Insets show a magnified view of a neuron cell body with the nucleus. (B) Staining for SETX (green) and β-tubulin-III (red) show reduced staining of SETX. (C) Staining of neurons with antibody (S9.6) against RNA–DNA hybrids (R-loops) (green) and β-tubulin-III (red) show increased accumulation of R-loops in SMA neurons compared to normal neurons. (D) Quantitative (mean ± s.e.m., n = 3) and statistical analysis (unpaired t-test) shows marked increase (7.62 ± 0.97, P = 0.002)-fold in accumulation of R-loops in SMA neurons compared to non-SMA (Normal) neurons. Staining for markers of DDR and DNA repair pathways: (E) β-tubulin-III (red) and phospho-H2AX (γH2AX) (green) and (F) β-tubulin-III (red) and 53BP1 (green) show increase in phosphorylation of H2AX and accumulation of 53BP1 foci in SMA neurons compared to normal neurons that suggest accumulation of DSB-related DNA damage in SMA motor neurons. (G) Staining of neurons with β-tubulin-III (red) and phospho-ATR (p-ATR) (green); (H) β-tubulin-III (red) and phospho-CHK1 (p-CHK1) (green), downstream target of ATR. (I) Neurons stained with β-tubulin-III (red) and phospho-ATM (p-ATM) (green); (J) β-tubulin-III (red) and phospho-BRCA1 (p-BRCA1) (green); (K) phospho-DNA-PKcs (p-DNAPKcs) (green) and β-tubulin-III (red). Scale bar is 20 μm. (L) Protein extracts were prepared from the spinal cords isolated from 7-day-old non-SMA (Normal) and SMA mice, and examined by IB analysis using automated capillary-based western blot system (ProteinSimple). Representative capillary-blot images of proteins and phospho-proteins are shown (full-length blots are included inSupplementary Figures S12 and 13). IB analysis of proteins in tissue lysates of the spinal cords isolated from 7-day-old non-SMA (Normal) and SMA pups shows increase in levels of phospho-H2AX (γH2AX) and phospho-ATR (p-ATR) in SMA mice compared to normal mice. (M) Quantitative (mean ± s.e.m., n = 3 mice/group) and statistical analysis (unpaired t-test) of IB data from mouse spinal cords show that SMN and SETX levels reduced to (27.67 ± 1.870%, P = 0.000) and (45.30 ± 7.66%, P = 0.002), respectively, in SMA mice (SMA) compared to non-SMA (Normal) mice. Analysis of DDR markers shows increase in levels of γH2AX (3.21 ± 0.27-fold, P = 0.001), p-ATR (2.58 ± 0.24-fold, P = 0.003) and p-CHK1 (2.08 ± 0.17-fold, P = 0.004). DSB repair markers of HR, p-ATM is reduced to (42.35 ± 6.94%, P = 0.004) and p-BRCA1 reduced to (37.85 ± 6.50%, P = 0.011) in SMA mice (SMA). DSB repair marker of NHEJ, phospho-DNA-PKcs (p-DNAPKcs) reduced to (37.47 ± 4.86%, P = 0.000) and total DNA-PKcs reduced to (30.54 ± 6.74%, P = 0.004) in SMA mice. (N) IB analysis of proteins in tissue lysates of human spinal cords (lumbar region) isolated from age matched non-SMA, GM083 (Normal) and severe SMA type I patients, GM04583 and GM04629 (SMA). (O) Quantitative and statistical analysis of IB data from human spinal cords show low levels of SMN in SMA patients, GM04583 (22.43 ± 1.07%, P = 0.000) and GM04629 (26.63 ± 2.91%, P = 0.000) compared to normal (non-SMA) human (100.4 ± 5.221%, P = 0.000) (GM083). SMA patient spinal cords show decrease in SETX levels GM04583 (59.53 ± 11.67%, P = 0.007) and GM04629 (52.52 ± 12.90%, P = 0.015) compared to normal (non-SMA) human (GM083). Analysis of DDR markers shows increase in levels of γH2AX in SMA patients (2.01 ± 0.22-fold, P = 0.011) in GM04583 and (2.46 ± 0.05-fold, P = 0.000) in GM04629, p-ATR (2.10 ± 0.14-fold, P = 0.001) in GM04583 and (1.75 ± 0.10-fold, P = 0.002) in GM04629, and p-CHK1 (2.04 ± 0.04-fold, P = 0.000) in GM04583 and (1.87 ± 0.04-fold, P = 0.000) in GM04629. Analysis of DDR and HR markers shows p-ATM reduced to (29.65 ± 2.18%, P = 0.000) (GM04583) and (36.63 ± 4.95, P = 0.000) (GM04629), and p-BRCA1 (47.09 ± 6.63, P = 0.001) (GM04583) and (35.17 ± 2.717, P = 0.000) (GM04629). Analysis of NHEJ pathway shows phospho-DNA-PKcs (p-DNAPKcs) reduced to (33.33 ± 4.512%, P = 0.000) (GM04583) and (30.06 ± 3.590, P = 0.000) (GM04629), and total DNA-PKcs (38.52 ± 3.910, P = 0.000) (GM04583) and (41.76 ±4.193, P = 0.001) (GM04629) suggesting that low levels of SMN cause deficiency and the loss of DNA-PKcs activity.
Accumulations of R-loops in SMN-deficient neurons prompted us to further investigate its potential association with DNA damage and activation of DDR pathway that may contribute to neurodegeneration in SMA. Examination of DNA damage markers revealed increased γH2AX staining (Figure 6E) and accumulation of 53BP1 in foci (Figure 6F) in SMA neurons compared to normal neurons, suggesting accumulation of DSBs in SMN-deficient neurons. ATR and its downstream target CHK1 play essential roles in maintaining genome integrity and in the survival of neurons (32,39). SMA neurons show increase in phospho-ATR (p-ATR) and phospho-CHK1 (p-CHK1) staining compared to normal neurons (Figures 6G and H). These data suggest that the ATR/CHK1 pathway is activated to support genome integrity and survival of SMA neurons, but neurons continue to degenerate in SMA. To find the cause of neurodegeneration, we examined the activation of factors required for DSB repair by HR. Analysis of p-ATM and p-BRCA1 shows decreased intensity in SMA neurons compared to normal neurons indicating reduced or no activation of ATM/BRCA1 (Figures 6I and J). These data suggest that DDR markers associated with HR are not activated in motor neurons derived from SMA mice, a finding consistent with the fact that neurons are terminally differentiated cells that do not undergo DNA replication. Moreover, as the HR pathway is predominately activated to repair DNA damage during S and G2 phases of the cell cycle in dividing cells (40), it is not surprising that this pathway is not activated in spinal cord neurons regardless of the SMA disease status.
DSB repair in neurons (non-dividing, differentiated cells) is predominantly mediated by the NHEJ pathway, which relies on the activation of DNA-PKcs (17). Examination of SMA neurons shows marked reduction in staining for p-DNA-PKcs compared to control (Normal) neurons (Figure 6K). These results are consistent with the findings from SMA patient dividing cells and suggest that the loss of DNA-PKcs activity may impair NHEJ-mediated DSB repair and contribute to DNA damage accumulation in SMA neurons.
Combined deficiency of SETX and DNA-PKcs in SMA mice and patient spinal cords tissues causes DNA damage accumulation and activation of DDR pathways
Findings from in vitro cultured SMA patient cells and spinal cord neurons from SMA mice suggest that combined deficiency of SETX and DNA-PKcs deficiency may be a cause of gradual accumulation of unrepaired DNA damage in neurons leading to neurodegeneration in SMA. To test whether in vitro findings are consistent with in vivo SMA pathogenesis, we examined levels of SETX and DNA-PKcs and other critical components of DNA repair pathways, HR and NHEJ, in the spinal cord tissues from SMA mice and severe SMA type I patients. IB and quantitative analysis of spinal cords from normal and SMA mice show decrease in SETX (55.22 ± 7.99%, P = 0.002) levels in SMA compared to normal mice (Figures 6L and M). Comparison of SETX levels in human spinal cords from age matched non-SMA, GM083 (Normal) and SMA patients show decrease in SETX levels GM04583 (59.53 ± 11.67%, P = 0.007) and GM04629 (52.52 ± 12.90%, P = 0.015) in SMA patients compared to normal human (Figures 6N and O). Decrease in SETX levels in the spinal cord tissues from SMA mice and patients is consistent with cultured SMA patient cells that suggest a partial loss of SETX function due to mislocalization and reduced levels might be a cause of R-loop accumulation in SMA.
Analysis of activation of DNA damage marker H2AX shows robust increase in levels of γH2AX (3.21 ± 0.27-fold, P = 0.001) in SMA compared to normal mice (Figures 6L and M). Analysis of human spinal cords shows increase in levels of γH2AX in GM04583 (2.01 ± 0.22-fold, P = 0.011) and GM04629 (2.46 ± 0.05-fold, P = 0.000) in SMA patients compared to normal human (Figures 6N and O). Analysis of critical components of DDR, ATR and CHK1 show >2-fold increase in p-ATR and p-CHK1 levels in SMA mice spinal cord tissue and SMA patient spinal cord tissues (GM04583 and GM04629) suggesting activation of the ATR/CHK1 pathway due to increased SSB or ssDNA-associated DNA damage in SMA neurons. Analysis of DSB repair components, ATM and BRCA1, showed that p-ATM and p-BRCA1 levels were reduced to (42.35 ± 6.94%, P = 0.004) and (37.85 ± 6.50%, P = 0.011), respectively, in SMA mice compared to normal mice indicating that HR pathway is not activated in spinal cord tissues (Figures 6L and M). SMA patient spinal cord tissues also show that levels of p-ATM and p-BRCA1 are markedly reduced in SMA patients (GM04583 and GM04629) compared to GM083 (Normal). These data are consistent with data from SMA neurons and support previous findings that DSB repair by HR pathway may be restricted to actively dividing cells (40). At last, we examined the activation and levels of DNA-PKcs that is essential for NHEJ-mediated DNA repair in neurons. Levels of phospho-DNA-PKcs (p-DNA-PKcs) were reduced to (37.47 ± 4.86%, P = 0.000) and total DNA-PKcs levels were reduced to (30.54 ± 6.74%, P = 0.004) in the spinal cords of SMA mice compared to normal mice (Figures 6L and M). Analysis of DNA-PKcs activation in human spinal cord tissues also shows p-DNA-PKcs levels reduced to (33.33 ± 4.51%, P = 0.000) in GM04583 and (30.06 ± 3.59%, P = 0.000) in GM04629 compared to GM083 spinal cords (Figures 6N and O). Further, our analysis revealed marked reduction in levels of total DNA-PKcs (DNA-PKcs) protein. Levels of total DNA-PKcs were reduced to (38.52 ± 3.91%, P = 0.000) in GM04583 and (41.76 ± 4.19%, P = 0.001) in GM04629 compared to GM083 spinal cords (Figures 6N and O). Together, these data from SMA mice and patient spinal cord tissues are consistent with the data from SMA spinal cord neurons and suggest that the chronic low levels of SMN cause combined deficiency of SETX and DNA-PKcs that may be a major cause of R-loop accumulation, DSB formation and accumulation of DNA damage due to inefficient NHEJ-mediated DSB repair leading to neurodegeneration in SMA.
Rescue of DNA damage and degeneration of SMA neurons
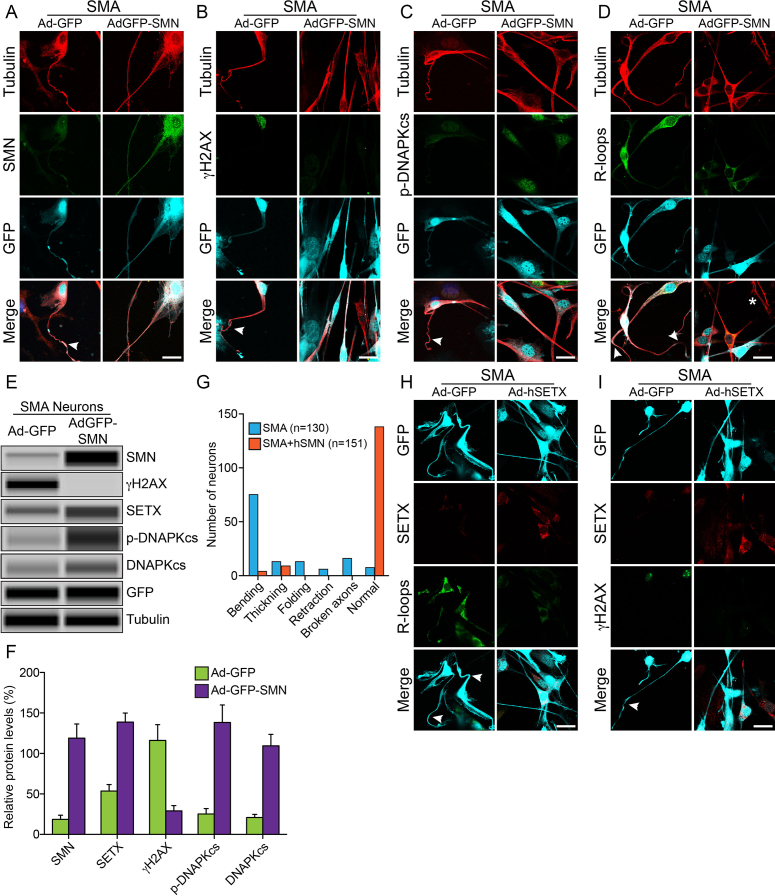
To test our emerging hypothesis that combined deficiency of SETX and DNA-PKcs is a major cause of DNA damage accumulation and neurodegeneration in SMA, we performed two rescue experiments using ectopic expression of either SMN or SETX in cultured spinal cord neurons from SMA mice. In the first rescue experiment, we infected neurons with adenovirus containing GFP (Ad-GFP) and recombinant human SMN (Ad-GFP-SMN1) for transient expression of SMN and examined phenotype of stained neurons and levels of SMN, SETX, DNA-PKcs, p-DNA-PKcs and γH2AX in neurons. Control SMA neurons expressing GFP show degeneration, including bending, retraction and folding (indicated by arrowheads) that is similar to degeneration found in experiments shown in Figure 6 and consistent with previous findings (7), suggesting that the expression of GFP did not change the phenotype of SMN-deficient spinal cord neurons from SMA mice (Figures 7A–D, Ad-GFP panels). Complementation of neurons with virus AdGFP-SMN increases SMN levels and rescues neurodegenerative phenotype by reducing degenerative features such as axonal bending, swelling, ballooning, retraction and folding in SMN expressing SMA neurons compared to GFP expressing SMA neurons (Figures 7A–D, AdGFP-SMN panels and Figure 7G). Notably, restoration of SMN levels by ∼6-fold increase (Figures 7A, E and F) reduced γH2AX levels by ∼4-fold (Figures 7B, E and F) and upregulated levels of SETX (∼2.5-fold), p-DNA-PKcs (∼5.4-fold) and total DNA-PKcs (∼5.2-fold) (Figures 7A–C, E and F), suggesting reduction in DSB accumulation and downregulation of DDR. Further, examination of neurons stained with antibodies against R-loops show reduction in staining of R-loops suggesting that restoration of SMN levels recuse SETX levels that reduce accumulation of R-loops (Figure 7D). These data show that the restoration of SMN levels in SMA neurons rescues SETX and DNA-PKcs levels, which reduce R-loop accumulation, DNA damage and rescues neuron degeneration phenotype (Figure 7G). Thus, these results suggest an important role for SMN in maintaining optimal levels of SETX, critical for resolving R-loops and of DNA-PKcs, essential for NHEJ-mediated DNA repair, to avert genomic instability and prevent neurodegeneration.
Figure 7.
Rescue of DNA damage and neuron degeneration of SMA spinal cord neurons by ectopic expression of SMN and SETX. Ectopic expression of recombinant SMN restores SETX and DNA-PKcs levels, reduces DDR activation and rescues SMA neuron degeneration. Cultured primary spinal cord neurons were infected adenovirus Ad-GFP and Ad-GFP-SMN1 or Ad-GFP and Ad-h-SETX at 100 MOI for 48 h, fixed with 4% PFA and stained with antibodies and IF examined by confocal microscopy. Low levels of SMN in SMA neurons causes axonal defects, bending, folding, retraction and ballooning (arrowheads), as shown by staining with neuron-specific β-tubulin-III, (clone TUJ1) (red) that lead to degeneration of SMA motor neurons. GFP positive neurons are pseudo-colored as cyan (GFP). Staining for markers of DDR and DNA repair pathways shows restoration of SMN levels, increased levels of DNA-PKcs, rescue of DNA damage and axonal degeneration in SMA neurons. (A) β-tubulin-III (red) and SMN (green), (B) β-tubulin-III (red) and phospho-H2AX (γH2AX) (green), (C) β-tubulin-III (red) and p-DNAPKcs (green) and (D) β-tubulin-III (red) and R-loops (green) show reduced staining for γH2AX and increased staining for p-DNA-PKcs, reduced staining for R-loops and rescue of axonal growth defects. (E) Neurons infected with adenovirus Ad-GFP and Ad-GFP-SMN1 were also harvested at 48 h post-infection to make total cell lysate for protein analysis using automated Wes System (ProteinSimple). Representative capillary-blot images of proteins and phospho-proteins are shown (full-length blots are included inSupplementary Figure S14). Tubulin and GFP blots are shown as loading controls. IB analysis shows restoration of SMN levels in SMA neurons (Ad-GFP-SMN), increased SETX, phospho-DNA-PKcs and total DNA-PKcs levels and reduction of phospho-H2AX (γH2AX) levels compared to control SMA neurons (Ad-GFP). (F) Quantitation and comparison of SMN, γH2AX, SETX, p-DNA-PKcs and DNA-PKcs levels in SMA+Ad-GFP and SMA+AdGFP-SMN neurons show rescue of DNA damage in SMN complemented neurons (SMA+ AdGFP-SMN) by marked reduction in γH2AX levels to (29.08 ± 6.44%, P = 0.013) compared to control SMA neurons (116.1 ± 19.53%) that is supported by increase in levels of SETX (85.19 ± 13.76%, P = 0.003), p-DNA-PKcs (113.0 ± 22.55%, P = 0.007) and total DNA-PKcs (88.55 ± 14.68%, P = 0.003) in (SMA+hSMN) neurons compared to Control (SMA+Ad-GFP) neurons. (G) Analysis of morphological features that represent neurodegeneration such as bending (that includes swelling and ballooning), thickening, folding and retraction of axons show reduction in neuron degeneration and improvement in axonal growth after rescue of DNA damage phenotype in neurons. Neurons stained for (H), SETX (red) and R-loops (green) and (I) SETX (red), phospho-H2AX (γH2AX) (green) show reduced staining for γH2AX and R-loops and rescue of axonal growth defects. Scale bar is 20 μm.
Furthermore, to test whether SETX-deficiency found in SMA patients is critical for DNA damage accumulation and neurodegeneration in SMA, we performed a second rescue experiment and infected neurons with Ad-hSETX virus. Comparison of neurons double-stained with SETX and R-loop antibodies shows rescue of neuron degenerative phenotype and reduction in R-loop accumulation in neurons overexpressing SETX (Figure 7H), suggesting that direct restoration of SETX levels is sufficient to resolves R-loops and rescue neurons with chronic SMN-deficiency. This result is consistent with the rescue of phenotype by indirect restoration of SETX levels using SMN overexpression in SMA neurons (Figure 7E). Further analysis of neurons stained with SETX and γH2AX showed that SETX overexpression reduces γH2AX foci compared to GFP expressing neurons (Figure 7I). Together, these data show that the SETX-deficiency may be one of the major causes of DNA damage stemming downstream of chronic low levels of SMN in SMA and the restoration of SETX levels alone can rescue DNA damage accumulation and degenerative phenotype of SMN-deficient neurons suggest that SETX represents a potential therapeutic target to reduce or prevent neurodegeneration in SMA.
DISCUSSION
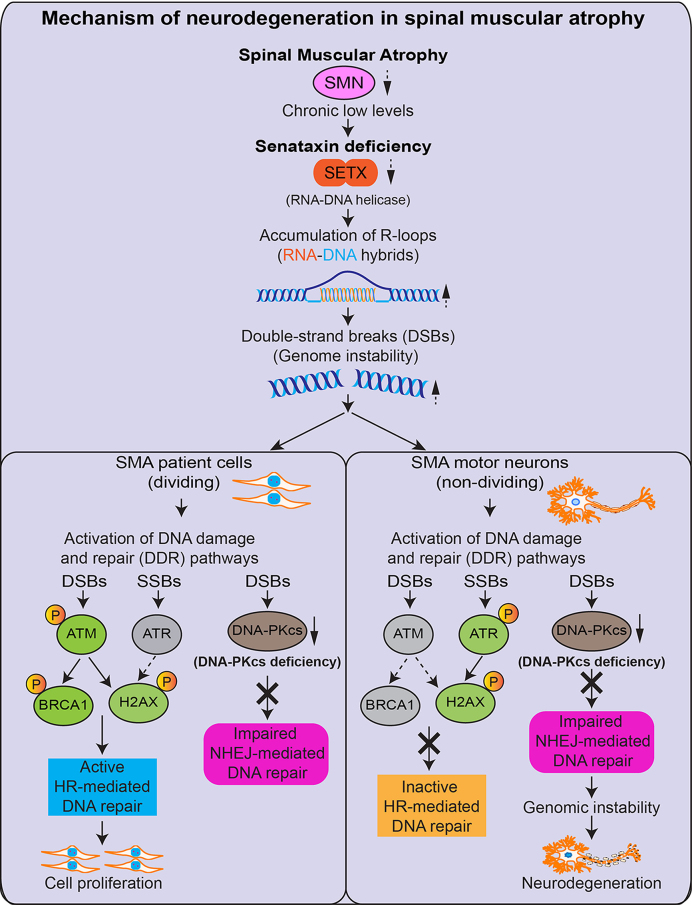
SMA is an autosomal recessive neurodegenerative disease caused by chronic low levels of SMN starting from the stage of conception of a child. Low levels of SMN are sufficient for embryonic development and survival but result in spinal motor neuron degeneration during embryonic and postnatal development, leading to muscle atrophy, respiratory failure and death in SMA patients. SMN is an ubiquitously expressed protein that plays critical roles in mRNA biogenesis, including snRNP assembly and splicing (15). However, one of the most intriguing and outstanding questions in SMA is why deficiency of a ubiquitous protein predominantly causes neuron degeneration and results in a neurological disease (4). Our study provides insight into the molecular mechanism of neurodegeneration associated with SMA pathogenesis by uncovering that chronic low levels of SMN cause combined deficiency of SETX and DNA-PKcs, which results in DNA damage accumulation leading to genome instability and predominant degeneration of motor neurons in SMA (Figure 8).
Figure 8.
Graphical summary of the molecular mechanism of neurodegeneration in SMA. Chronic low levels of SMN cause deficiency of SETX that results in accumulation of RNA–DNA hybrids (R-loops). Accumulation of R-loops causes DSBs and activation of DDR. In dividing cells, DSBs are repaired by HR and NHEJ, but neurons predominantly use NHEJ that requires DNA-activated protein kinase catalytic subunit (DNA-PKcs) activity. Chronic low levels of SMN cause deficiency of DNA-PKcs in neurons. In SMA dividing cells, HR pathway is active and repairs DNA damage that supports cellular proliferation. In SMA neurons, HR-mediated DNA repair is inactive and DNA-PKcs-deficiency causes defects in NHEJ-mediated DSB repair that leads to genome instability and predominant degeneration of motor neurons in SMA.
SETX deficiency causes R-loop accumulation and DNA damage in SMA
We show that the low levels of SMN cause downregulation of SETX levels in SMA patient cells and neurons. Our data show transcriptional downregulation of the SETX gene under acute and chronic low levels of SMN. SMN plays a critical role in the assembly of spliceosomal snRNPs and SMN deficiency causes defects in pre-mRNA splicing of genes with large number of introns in SMA (10). The human SETX gene is large (∼95 kb) and contains 33 exons (41), and therefore it is possible that splicing defects result in alternatively spliced transcripts with exon skipping, intron retention causing frameshift or nonsense coding, which fail to translate and may contribute to downregulation of SETX levels in SMA. Alternative splicing of the SETX gene is reported in AOA2 patients (42). In addition, disruption of SMN–SETX protein complex due to low levels of SMN may cause destabilization followed by degradation of SETX that may also contribute to overall downregulation of SETX. The SETX is an RNA/DNA helicase that is critical for resolution of RNA–DNA hybrids (R-loops) and SETX deficiency is known to causes R-loop and DSB accumulation leading to genome instability (33,43). Mutations in the human SETX gene are associated with neurodegenerative disorders, a recessive form of ataxia oculomotor apraxia type 2 (AOA2) characterized by the loss of Purkinje cells resulting in cerebellar atrophy and peripheral neuropathy (44,45) and the dominant juvenile form of amyotrophic lateral sclerosis type 4 (ALS4) characterized by progressive degeneration of motor neurons in the brain and spinal cord, leading to muscle weakness and atrophy (46). There are ∼120 mutations, including missense, nonsense, splice site, frameshift and insertion/deletion reported in the SETX gene that are associated with AOA2 (47–49). About 46 missense mutation, 20 in the NH2-terminal and 26 in the helicase domain of SETX that are reported in AOA2 are summarized in references (49–51). In AOA2 patients, the loss of function SETX mutations results in inefficient resolution of R-loops that are accumulated because of defects in transcription leading to activation of DDR pathways (52–54). SMA and ALS have similar characteristics such as degeneration of the spinal cord lower and upper motor neurons, respectively, and muscle weakness and atrophy. Several missense mutations, T3I, L389S, T1118I, C1554G, K2029E, R2136H and I2547T in the SETX gene have been reported to be associated with ALS4 (55). Two mutations, L389S and V891A (in trans), in the SETX gene are also reported in patients with autosomal dominant proximal SMA (ADSMA) (56).
Knockout of the Setx gene in mouse results in accumulation of R-loops and DSBs, which causes defects in disassembly of RAD51 filaments (43). The SMN interacting protein GEMIN2 binds to RAD51 and SMN-GEMIN2 fusion protein complex has been reported to enhance RAD51-mediated HR (57,58). SETX is required to maintain the integrity of replication fork during the collision with RNA polymerase II (RNAPII)-dependent transcription to prevent DNA damage, and it promotes BRCA1-mediated DNA repair (59,60). Interestingly, SETX knockout causes R-loop accumulation in mice, but SETX-deficient mice did not show neurodegenerative phenotype suggesting that deficiency of SETX alone is not sufficient to cause neuron degeneration (43). Our data show that SMA neurons have combined deficiency of SETX and DNA-PKcs; SETX deficiency causes accumulation of R-loops and DNA damage, and DNA-PKcs deficiency impairs NHEJ-mediated DNA repair leading to genomic instability and progressive degeneration of motor neurons in SMA. Together, these findings suggest that the loss of function mutations in SETX may be coupled with a deficiency or the loss of function of another cell-specific factor that may contribute to Purkinje cells and motor neurons degeneration in AOA2 and ALS4, respectively. Our study opens a new door for investigation of the molecular mechanisms of pathogenic neurodegeneration in human diseases associated with SETX mutations.
Transcription, R-loop resolution, splicing, DNA damage and DNA repair are tightly coupled nuclear processes that require precise temporal coordination. SETX interacts with many proteins, including RNAPII, SMN and BRCA1 that are involved in transcription, splicing and DNA repair, respectively (60–63). SMN is shown to interact with RNAPII and may play a role in transcription (64,65). Disruption of SMN–RNAPII interaction by mutation in RNAPII causes defects in transcription termination that result in R-loop accumulation (65). The precise function of SETX–SMN complex in R-loop biogenesis is unclear. However, it is possible that RNAPII, SMN and SETX proteins form complexes and collaborate to perform sequential functions required for RNA biogenesis, such as RNAPII–SMN complex may control transcription termination and the SETX–SMN complex may play a role in splicing where SETX resolves RNA–DNA hybrids, which is required prior to splicing, and the SMN–snRNPs complex initiates pre-mRNA splicing (24,61,65).
R-loops are naturally occurring RNA–DNA hybrids consisting of three nucleic acid strands, RNA–DNA hybridized strands and a complementary DNA strand, which are generated during transcription. R-loops may play roles in gene expression, DSB repair and contribute to genome instability (34,66). Defects in splicing result in transcriptional defects (67) that are associated with R-loop accumulation (68). Moreover, inactivation of the splicing factor ASF/SF2 results in genomic instability (10,69). SMN-deficiency causes defects in splicing and correlates with increase in R-loops but why R-loops are not resolved in SMN-deficient cells was unclear (70). Our data show SETX deficiency is the cause of accumulation of R-loops that results in DSBs and activation of DDR pathways in SMA. Notably, R-loop accumulation was ∼5.6-fold higher in SMA spinal cord neurons compared to dividing patient cells, suggesting that R-loop accumulation may gradually increase in terminally differentiated neurons compared to dividing cells where R-loops may be cleared/resolved during each mitotic cycle. Higher levels of R-loops in neurons compared to dividing cells suggests increased secondary DSBs that may contribute to predominant degeneration of motor neurons in SMA. Together, these findings support the idea that accumulation of R-loops in dividing and non-dividing SMA cells may be a cause of DSBs and genomic instability that may contribute to neurodegeneration associated with SMA pathogenesis.
Contribution of DNA-PKcs deficiency to neurodegeneration and SMA pathogenesis
Accumulation of R-loops and DSBs followed by activation of DDR pathways revealed that DSB repair by HR and NHEJ is triggered in normal dividing cells upon inducing SMN deficiency. Acute SMN-deficiency caused a small decease (∼13%) in DNA-PKcs transcript levels that did not affect the DNA-PKcs protein levels and the DNA-PKcs was phosphorylated, suggesting activation of the NHEJ pathway for DNA repair is triggered by SMN-deficiency. Interestingly, chronic low levels of SMN in patient dividing cells caused ∼35% decrease in DNA-PKcs transcripts and marked reduction (∼70%) in total DNA-PKcs protein levels resulting in the loss of activated phospho-DNA-PKcs and impairment of the NHEJ pathway in SMA. It is established that SMN-deficiency causes defects in the pre-mRNA splicing of genes with large number of introns (10), and the human PRKDC (DNA-PKcs) gene is very long ∼187 kb and the mouse Prkdc gene is ∼250 kb long and contains 86 exons (71). Alternative splicing of the DNA-PKcs gene is reported in cancer (72,73). Therefore, it is likely that the defects in pre-mRNA splicing of the DNA-PKcs gene might also contribute to DNA-PKcs protein deficiency in SMA cells. Rescue of DNA-PKcs levels by restoration of SMN levels in patient cells and SMA neurons suggests that SMN is critical for maintaining normal levels of DNA-PKcs. NHEJ-mediated DSB repair is dependent on DNA-PKcs activity. Neurons and other post-mitotic cells predominantly use NHEJ for DNA repair (18). DNA-PKcs deficiency and reduced DNA repair activity by NHEJ is reported in the brain tissue of Alzheimer’s disease patients (74). Because DNA-PKcs is required for NHEJ-mediated DNA repair in neurons and plays a critical role in maintaining genome integrity in neurons (75), our data suggest that DNA damage caused by SETX deficiency gradually increases due to inefficient NHEJ-mediated DNA repair in neurons, leading to genome instability that may contribute to predominant neuron degeneration in SMA.
In addition to neurodegeneration, DNA-PKcs deficiency may contribute to immune system defects in SMA. Mutations in DNA-PKcs that results in the loss of its activity or genetic inactivation of the Prkdc gene cause severe combined immune deficiency (76). Defects in immune system are reported in SMA, including abnormalities in spleen and pulmonary infection such as pneumonia, which are common in SMA patients (77). These findings call for further investigation of immune complications in SMA and provide basis for pre-emptive elimination of potential risk of infection in SMA patients, which would be extremely important for the success of ongoing and future clinical trials.
Mechanism of neurodegeneration in SMA
In this study, we have uncovered that the low levels of SMN disrupt colocalization of SETX with SMN in sub-nuclear bodies and causes downregulation of SETX levels in patient cells, SMA mice neurons and spinal cord tissues from SMA mice and patients. SETX-deficiency leads to generation of excessive amounts of unresolved R-loops. Accumulation of R-loops is one of the major causes of genomic instability (34), which results in DNA damage, particularly DSBs and activation of DDR pathways. In dividing cells, HR and NHEJ mechanisms repair DSBs, but neurons predominantly use NHEJ that is dependent on DNA-PKcs activity (17,78). Further, we uncovered that chronic low levels of SMN is also a cause of DNA-PKcs deficiency in SMA mice, patient cells and spinal cords tissues suggesting that DSB repair may be insufficient due to impaired NHEJ-mediated DNA repair in patient cells and SMA neurons. However, activation of ATM/BRCA1 in patient cells, suggests that HR may repair the majority of DSBs in dividing cells. In SMA neurons, deregulation of p-ATM and p-BRCA1 levels indicate non-active HR, which is consistent with the experimental evidences that DSB repair by HR pathway may exclusively occur in dividing cells (40), suggesting HR may not contribute to DSB repair in neurons. Therefore, neurons may primarily rely on DSB repair by NHEJ. Thus, deficiency of DNA-PKcs may cause defects in NHEJ-mediated DSB repair and result in gradual increase in DNA damage accumulation in neurons leading to genomic instability and neurodegeneration in SMA. The mechanism of neurodegeneration caused by chronic low levels of SMN in SMA is presented as a graphical summary in Figure 8. Rescue of SETX and DNA-PKcs deficiency, DNA damage and neurodegeneration by restoring SMN levels suggests that normal levels of SMN are required for optimal levels of SETX and DNA-PKcs. Furthermore, overexpression of only SETX rescues DNA damage by reducing R-loop accumulation and degeneration of SMA neurons, supporting the idea that SETX deficiency stemming downstream of the low levels of SMN may be central to causing DNA damage phenotype in SMA. Together, these findings identify SETX as a new potential therapeutic target for the treatment of SMA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nancy Jiang and Kayla Waits for technical assistance. We thank Dr Saif Ahmad for assistance with confocal microscopy. We thank summer accelerated biomedical research (SABR) student Roshan Tom and graduate student Leann Rodriguez for assistance with quantitative analysis of sub-nuclear bodies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health, Research Grant [R01 NS064224 to L.G.]; Muscular Dystrophy Association [MDA 480210 to L.G.]; Italian Association for Cancer Research (AIRC), Research Grant [IG 18976 to D.B.]. Funding for open access charge: Institutional Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. . Identification and characterization of a spinal muscular atrophy- determining gene. Cell. 1995; 80:155–165. [DOI] [PubMed] [Google Scholar]
- 2. Lorson C.L., Hahnen E., Androphy E.J., Wirth B.. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burghes A.H., Beattie C.E.. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick. Nat. Rev. Neurosci. 2009; 10:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005; 48:885–896. [DOI] [PubMed] [Google Scholar]
- 5. Bowerman M., Shafey D., Kothary R.. Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J. Mol. Neurosci. 2007; 32:120–131. [DOI] [PubMed] [Google Scholar]
- 6. Wishart T.M., Mutsaers C.A., Riessland M., Reimer M.M., Hunter G., Hannam M.L., Eaton S.L., Fuller H.R., Roche S.L., Somers E. et al. . Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J. Clin. Invest. 2014; 124:1821–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Genabai N.K., Ahmad S., Zhang Z., Jiang X., Gabaldon C.A., Gangwani L.. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum. Mol. Genet. 2015; 24:6986–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmad S., Bhatia K., Kannan A., Gangwani L.. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J. Exp. Neurosci. 2016; 10:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gubitz A.K., Feng W., Dreyfuss G.. The SMN complex. Exp. Cell Res. 2004; 296:51–56. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G.. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008; 133:585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arnold A.S., Gueye M., Guettier-Sigrist S., Courdier-Fruh I., Coupin G., Poindron P., Gies J.P.. Reduced expression of nicotinic AChRs in myotubes from spinal muscular atrophy I patients. Lab. Invest. 2004; 84:1271–1278. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z., Dreyfuss G.. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19348–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng S.Y., Soh B.S., Rodriguez-Muela N., Hendrickson D.G., Price F., Rinn J.L., Rubin L.L.. Genome-wide RNA-seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell. 2015; 17:569–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Genabai N.K., Kannan A., Ahmad S., Jiang X., Bhatia K., Gangwani L.. Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci. Rep. 2017; 7:8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singh R.N., Howell M.D., Ottesen E.W., Singh N.N.. Diverse role of survival motor neuron protein. Biochim. Biophys. Acta. 2017; 1860:299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cortez D., Wang Y., Qin J., Elledge S.J.. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999; 286:1162–1166. [DOI] [PubMed] [Google Scholar]
- 17. Davis A.J., Chen B.P., Chen D.J.. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst). 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rass U., Ahel I., West S.C.. Defective DNA repair and neurodegenerative disease. Cell. 2007; 130:991–1004. [DOI] [PubMed] [Google Scholar]
- 19. Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H.. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005; 14:845–857. [DOI] [PubMed] [Google Scholar]
- 20. Gangwani L., Mikrut M., Theroux S., Sharma M., Davis R.J.. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat. Cell Biol. 2001; 3:376–383. [DOI] [PubMed] [Google Scholar]
- 21. Gangwani L. Deficiency of the zinc finger protein ZPR1 causes defects in transcription and cell cycle progression. J. Biol. Chem. 2006; 281:40330–40340. [DOI] [PubMed] [Google Scholar]
- 22. Ahmad S., Wang Y., Shaik G.M., Burghes A.H., Gangwani L.. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum. Mol. Genet. 2012; 21:2745–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gangwani L., Mikrut M., Galcheva-Gargova Z., Davis R.J.. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J. Cell Biol. 1998; 143:1471–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nishida Y., Mizutani N., Inoue M., Omori Y., Tamiya-Koizumi K., Takagi A., Kojima T., Suzuki M., Nozawa Y., Minami Y. et al. . Phosphorylated Sp1 is the regulator of DNA-PKcs and DNA ligase IV transcription of daunorubicin-resistant leukemia cell lines. Biochim. Biophys. Acta. 2014; 1839:265–274. [DOI] [PubMed] [Google Scholar]
- 26. Nichols N.M., Yue D.. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Enzymatic manipulation of DNA and RNA: ribonucleases. Current Protocols in Molecular Biology. 2008; 84:USA: John Wiley & Sons, Inc; 3.13.1–3.13.8. [Google Scholar]
- 27. Wang Q., Sawyer I.A., Sung M.H., Sturgill D., Shevtsov S.P., Pegoraro G., Hakim O., Baek S., Hager G.L., Dundr M.. Cajal bodies are linked to genome conformation. Nat. Commun. 2016; 7:10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr. Opin. Cell Biol. 2012; 24:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeung P.L., Denissova N.G., Nasello C., Hakhverdyan Z., Chen J.D., Brenneman M.A.. Promyelocytic leukemia nuclear bodies support a late step in DNA double-strand break repair by homologous recombination. J. Cell Biochem. 2012; 113:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mah L.J., El-Osta A., Karagiannis T.C.. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010; 24:679–686. [DOI] [PubMed] [Google Scholar]
- 31. Branzei D., Foiani M.. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008; 9:297–308. [DOI] [PubMed] [Google Scholar]
- 32. Cimprich K.A., Cortez D.. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamperl S., Cimprich K.A.. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst). 2014; 19:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 35. Brown T.A., Tkachuk A.N., Clayton D.A.. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008; 283:36743–36751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Rubio M.L., Perez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A.. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fayzullina S., Martin L.J.. DNA damage response and DNA repair in skeletal myocytes from a mouse model of spinal muscular atrophy. J. Neuropathol. Exp. Neurol. 2016; 75:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walker C., Herranz-Martin S., Karyka E., Liao C., Lewis K., Elsayed W., Lukashchuk V., Chiang S.C., Ray S., Mulcahy P.J. et al. . C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat. Neurosci. 2017; 20:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye W., Blain S.W.. Chk1 has an essential role in the survival of differentiated cortical neurons in the absence of DNA damage. Apoptosis. 2011; 16:449–459. [DOI] [PubMed] [Google Scholar]
- 40. Iyama T., Wilson D.M. 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst). 2013; 12:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Humphray S.J., Oliver K., Hunt A.R., Plumb R.W., Loveland J.E., Howe K.L., Andrews T.D., Searle S., Hunt S.E., Scott C.E. et al. . DNA sequence and analysis of human chromosome 9. Nature. 2004; 429:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fogel B.L., Lee J.Y., Perlman S.. Aberrant splicing of the senataxin gene in a patient with ataxia with oculomotor apraxia type 2. Cerebellum. 2009; 8:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Becherel O.J., Yeo A.J., Stellati A., Heng E.Y., Luff J., Suraweera A.M., Woods R., Fleming J., Carrie D., McKinney K. et al. . Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013; 9:e1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moreira M.C., Klur S., Watanabe M., Nemeth A.H., Le Ber I., Moniz J.C., Tranchant C., Aubourg P., Tazir M., Schols L. et al. . Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004; 36:225–227. [DOI] [PubMed] [Google Scholar]
- 45. Lavin M.F., Yeo A.J., Becherel O.J.. Senataxin protects the genome: Implications for neurodegeneration and other abnormalities. Rare Dis. 2013; 1:e25230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y.Z., Bennett C.L., Huynh H.M., Blair I.P., Puls I., Irobi J., Dierick I., Abel A., Kennerson M.L., Rabin B.A. et al. . DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 2004; 74:1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Groh M., Albulescu L.O., Cristini A., Gromak N.. Senataxin: genome guardian at the interface of transcription and neurodegeneration. J. Mol. Biol. 2017; 429:3181–3195. [DOI] [PubMed] [Google Scholar]
- 48. Fogel B.L., Perlman S.. Novel mutations in the senataxin DNA/RNA helicase domain in ataxia with oculomotor apraxia 2. Neurology. 2006; 67:2083–2084. [DOI] [PubMed] [Google Scholar]
- 49. Anheim M., Monga B., Fleury M., Charles P., Barbot C., Salih M., Delaunoy J.P., Fritsch M., Arning L., Synofzik M. et al. . Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain. 2009; 132:2688–2698. [DOI] [PubMed] [Google Scholar]
- 50. Chen X., Muller U., Sundling K.E., Brow D.A.. Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human Senataxin that elicit cerebellar ataxia. Genetics. 2014; 198:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Y.Z., Hashemi S.H., Anderson S.K., Huang Y., Moreira M.C., Lynch D.R., Glass I.A., Chance P.F., Bennett C.L.. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol. Dis. 2006; 23:97–108. [DOI] [PubMed] [Google Scholar]
- 52. Fogel B.L., Cho E., Wahnich A., Gao F., Becherel O.J., Wang X., Fike F., Chen L., Criscuolo C., De Michele G. et al. . Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum. Mol. Genet. 2014; 23:4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuce O., West S.C.. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol. 2013; 33:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. De Amicis A., Piane M., Ferrari F., Fanciulli M., Delia D., Chessa L.. Role of senataxin in DNA damage and telomeric stability. DNA Repair (Amst). 2011; 10:199–209. [DOI] [PubMed] [Google Scholar]
- 55. Arning L., Epplen J.T., Rahikkala E., Hendrich C., Ludolph A.C., Sperfeld A.D.. The SETX missense variation spectrum as evaluated in patients with ALS4-like motor neuron diseases. Neurogenetics. 2013; 14:53–61. [DOI] [PubMed] [Google Scholar]
- 56. Rudnik-Schoneborn S., Arning L., Epplen J.T., Zerres K.. SETX gene mutation in a family diagnosed autosomal dominant proximal spinal muscular atrophy. Neuromuscul. Disord. 2012; 22:258–262. [DOI] [PubMed] [Google Scholar]
- 57. Takaku M., Tsujita T., Horikoshi N., Takizawa Y., Qing Y., Hirota K., Ikura M., Ikura T., Takeda S., Kurumizaka H.. Purification of the human SMN-GEMIN2 complex and assessment of its stimulation of RAD51-mediated DNA recombination reactions. Biochemistry. 2011; 50:6797–6805. [DOI] [PubMed] [Google Scholar]
- 58. Takizawa Y., Qing Y., Takaku M., Ishida T., Morozumi Y., Tsujita T., Kogame T., Hirota K., Takahashi M., Shibata T. et al. . GEMIN2 promotes accumulation of RAD51 at double-strand breaks in homologous recombination. Nucleic Acids Res. 2010; 38:5059–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brambati A., Colosio A., Zardoni L., Galanti L., Liberi G.. Replication and transcription on a collision course: eukaryotic regulation mechanisms and implications for DNA stability. Front. Genet. 2015; 6:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L. et al. . BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015; 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suraweera A., Lim Y., Woods R., Birrell G.W., Nasim T., Becherel O.J., Lavin M.F.. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 2009; 18:3384–3396. [DOI] [PubMed] [Google Scholar]
- 62. Ursic D., Chinchilla K., Finkel J.S., Culbertson M.R.. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004; 32:2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pellizzoni L., Kataoka N., Charroux B., Dreyfuss G.. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998; 95:615–624. [DOI] [PubMed] [Google Scholar]
- 64. Pellizzoni L., Charroux B., Rappsilber J., Mann M., Dreyfuss G.. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 2001; 152:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao D.Y., Gish G., Braunschweig U., Li Y., Ni Z., Schmitges F.W., Zhong G., Liu K., Li W., Moffat J. et al. . SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016; 529:48–53. [DOI] [PubMed] [Google Scholar]
- 66. Ohle C., Tesorero R., Schermann G., Dobrev N., Sinning I., Fischer T.. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016; 167:1001–1013. [DOI] [PubMed] [Google Scholar]
- 67. Chabot B., Shkreta L.. Defective control of pre-messenger RNA splicing in human disease. J. Cell Biol. 2016; 212:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 69. Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 70. Jangi M., Fleet C., Cullen P., Gupta S.V., Mekhoubad S., Chiao E., Allaire N., Bennett C.F., Rigo F., Krainer A.R. et al. . SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2347–E2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fujimori A., Araki R., Fukumura R., Saito T., Mori M., Mita K., Tatsumi K., Abe M.. The murine DNA-PKcs gene consists of 86 exons dispersed in more than 250 kb. Genomics. 1997; 45:194–199. [DOI] [PubMed] [Google Scholar]
- 72. Connelly M.A., Zhang H., Kieleczawa J., Anderson C.W.. Alternate splice-site utilization in the gene for the catalytic subunit of the DNA-activated protein kinase, DNA-PKcs. Gene. 1996; 175:271–273. [DOI] [PubMed] [Google Scholar]
- 73. Abbaszadeh F., Clingen P.H., Arlett C.F., Plowman P.N., Bourton E.C., Themis M., Makarov E.M., Newbold R.F., Green M.H., Parris C.N.. A novel splice variant of the DNA-PKcs gene is associated with clinical and cellular radiosensitivity in a patient with xeroderma pigmentosum. J. Med. Genet. 2010; 47:176–181. [DOI] [PubMed] [Google Scholar]
- 74. Kanungo J. DNA-PK deficiency in Alzheimer's disease. J. Neurol. Neuromed. 2016; 1:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Enriquez-Rios V., Dumitrache L.C., Downing S.M., Li Y., Brown E.J., Russell H.R., McKinnon P.J.. DNA-PKcs, ATM, and ATR interplay maintains genome integrity during neurogenesis. J. Neurosci. 2017; 37:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jhappan C., Morse H.C. 3rd, Fleischmann R.D., Gottesman M.M., Merlino G.. DNA-PKcs: a T-cell tumour suppressor encoded at the mouse scid locus. Nat. Genet. 1997; 17:483–486. [DOI] [PubMed] [Google Scholar]
- 77. Deguise M.O., Kothary R.. New insights into SMA pathogenesis: immune dysfunction and neuroinflammation. Ann. Clin. Transl. Neurol. 2017; 4:522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McKinnon P.J. Maintaining genome stability in the nervous system. Nat. Neurosci. 2013; 16:1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.